Abstract
No genes for any of the known uracil DNA glycosylases of the UDG superfamily are present in the genome of Methanothermobacter thermautotrophicus ΔH, making it difficult to imagine how DNA-U repair might be initiated in this organism. Recently, Mth212, the ExoIII homologue of M. thermautotrophicus ΔH has been characterized as a DNA uridine endonuclease, which suggested the possibility of a novel endonucleolytic entry mechanism for DNA uracil repair. With no system of genetic experimentation available, the problem was approached biochemically. Assays of DNA uracil repair in vitro, promoted by crude cellular extracts, provide unequivocal confirmation that this mechanism does indeed operate in M. thermautotrophicus ΔH.
INTRODUCTION
Water is the universal solvent for all biochemistry; DNA, on the other hand, is vulnerable to hydrolysis. Spontaneous hydrolytic deamination of cytosine residues, in particular, is a constant mutagenic threat to every living cell (1). This reaction results in DNA uracil residues and, therefore, in U/G mismatches which, left unrepaired, lead to C→T transition mutations in half of the progeny. Initiation of DNA uracil repair has so far been considered to be exclusively mediated by uracil DNA glycosylases (UDGs) throughout all domains of life (2–5). These enzymes catalyze hydrolysis of the glycosidic bond in a uracil-specific fashion, leaving behind a base-free (‘AP’) site which is processed further by other enzymes; the entire pathway being referred to as base excison repair (6).
For the typical growth temperatures of thermophilic and hyperthermophilic bacteria and archaea, fundamental principles of physics demand a dramatically increased rate of C-deamination and it is therefore expected that such microorganisms possess particularly efficient biochemical mechanisms that help them cope with the problem.
It is therefore surprising that archaea, in general, seem to be devoid of the otherwise ubiquitous family 1 UDGs (7). Several completely sequenced archaeal genomes, including that of Methanothermobacter thermautotrophicus ΔH (8), do not harbor any gene belonging to one of the five known UDG families and no corresponding activity could be detected in cellular extracts of M. thermautotrophicus ΔH (9).
A possible solution to this puzzle was offered by our recent discovery that Mth212, an ExoIII homologue of M. thermautotrophicus ΔH (10), is unusually equipped with an endonucleolytic activity directed against DNA uridine residues. This prompted us to hypothesize that in M. thermautotrophicus ΔH it is Mth212 that initiates DNA-U repair by endonucleolytic incision on the 5′-side of the 2′-d-uridine residue (9). Here we demonstrate biochemically that this novel mechanism is indeed employed by M. thermautotrophicus ΔH and that initiation of DNA-U repair is completely and exclusively dependent on the DNA uridine endonuclease activity of Mth212.
MATERIALS AND METHODS
Strains
Escherichia coli DH5α (Invitrogen, Carlsbad, CA): F−, ø80dlacZΔM15, Δ(lacZYA-argF)U169, deoR, recA1, endA1, hsdR17 (rk−, mk+), phoA, supE44, thi-1, λ-, gyrA96 (Nalr), relA1, (lacZYA-argF) U169.
Escherichia coli BL21_UX [(9), based on BL21-CodonPlus(DE3)-RIL, Stratagene, La Jolla, CA]. E. coli B, F−, ompT, hsdS (rB−mB−), dcm+, TetR, gal λ(DE3), endA, Hte [argU, ileY, leuW, CmR], Δung::kan.
Escherichia coli BL21_UXX (based on BL21_UX in which the kan gene was eliminated using plasmid pCP20 (11) as described (12). Escherichia coli B, F−, ompT, hsdS (rB−mB−), dcm+, TetR, gal λ(DE3), endA, Hte [argU, ileY, leuW, CmR], Δung.
Methanothermobacter thermautotrophicus ΔH (DSM 1053) was cultured as described earlier (13) and harvested in an early stationary phase.
Plasmids
pCR-Blunt II-TOPO vector for cloning of blunt-ended PCR-products was purchased from Invitrogen (Carlsbad, CA) as part of the ‘Zero Blunt® TOPO’ kit. pET-28a was from Novagen (San Diego, CA).
Enzymes and chemicals
Restriction endonucleases HindIII, NdeI, BamHI and T4 DNA ligase were from Fermentas (St Leon-Rot, Germany), chemicals were from either Roth (Karlsruhe, Germany) or Merck (Darmstadt, Germany).
Antibodies
Custom rabbit antisera to heterologously produced and highly purified Mth212 (9) and Mth.PolB (14), respectively, were raised by BioGenes (Berlin, Germany). For the analyses described here, purified whole IgG fractions prepared by this company were used. Anti-rabbit IgG conjugated with alkaline phosphatase was purchased from Sigma-Aldrich (Steinheim, Germany).
Substrates
All oligonucleotides were purchased from PURIMEX (Grebenstein, Germany) in preparative polyacrylamide gel electrophoresis purified grade. F: fluorescein moiety, U: 2′-d-uridine residue, P: phosphate residue.
HINDIII_U_T1 (30-mer) 5′ F-CCTGCCGAGTGCACCTGCGAAGUTTGATGT 3′
HINDIII_C_T1 (30-mer) 5′ F-CCTGCCGAGTGCACCTGCGAAGCTTGATGT 3′
HINDIII_T_T1 (30-mer) 5′ F-CCTGCCGAGTGCACCTGCGAAGTTTGATGT 3′
HINDIII_T2 (30-mer) 5′ P-ACATGCAGGGTCGCACGCTGTTACTGATAA 3′
HINDIII_LIG_F (20-mer) 5′ F-CCTGCCGAGTGCACCTGCGA 3′
HINDIII_LIG_P (40-mer) 5′ P-AGCTTGATGTACATGCAGGGTCGCACGCTGTTCATGATAA 3′
HINDIII_55-G (55-mer) 5′ CAGCGTGCGACCCTGCATGTACATCAAGCTTCGCAGGTGCACTCGGCAGGTCTAG 3′
HINDIII_23-Flap (23-mer) 5′ GGCTAGCCTCCGCTGCTGAGCTC 3′
HINDIII_55-G-Flap (55-mer) 5′ CAGCGTGCGACCCTGCATGTACATCAAGAGCTCAGCAGCGGAGGCTAGCCTCTAG 3′
HINDIII_M_212 (22-mer) 5′ F-CCTGCCGAGTGCACCTGCGAAG 3′
HINDIII_C_50_F (50-mer) 5′ F-CCTGCCGAGTGCACCTGCGAAGCTTGATGTACATGCAGGGTCGCACGCTG 3′
Preparation of 60-mer oligonucleotides containing a U, C or T residue at position 23 via ligation
Since 60-mer oligonucleotides were difficult to purify to a state, where they did not contain chains shorter by just one or few nucleotides, these oligonucleotides necessary for substrate hybridization were obtained by ligation. To this end, 20 pmol of HINDIII_U_T1, HINDIII_C_T1 or HINDIII_T_T1, respectively, 100 pmol of HINDIII_T2, and 50 pmol of HINDIII_55_G as a splint oligonucleotide were annealed in 50 µl SSC buffer (150 mM NaCl, 15 mM trisodiumcitrate) according to the procedure described (9). The hybridization mixtures were incubated with 4 u T4 DNA Ligase in a total of 60 µl (40 mM Tris–HCl pH 7.8, 10 mM MgCl2, 10 mM DTT, 0.5 mM ATP, 125 mM NaCl, 12.5 mM trisodiumcitrate) for 3 h at 37°C. The reactions were stopped by adding 20 µl of loading dye (98% formamide, 10 mM EDTA pH 8.0, 0.025% xylencyanole FF, 0.025% bromphenol blue). Forty microlitres of the mixtures were applied to an 11% denaturing polyacrylamide gel (20 cm length, acrylamide:bisacrylamide = 30:1, gel buffer: 106.8 mM Tris–base, 106.8 mM boric acid, 3 mM EDTA, 7 M urea) and electrophoresed at 1.5 V/cm2 for ca. 50 min with 89 mM Tris–base, 89 mM boric acid, 2.5 mM EDTA as running buffer. Fluorescein-labeled oligonucleotides were visualized by UV-light (312 nm). The bands corresponding to the 60-mer oligonucleotides were excised and the DNA was eluted overnight at 65°C into 500 µl H2O by diffusion followed by ethanol precipitation. Concentrations of the purified oligonucleotide solutions were determined by quantitative comparison of the fluorescence of aliquots with different aliquots of HIND_III_C_50_F as a standard after electrophoresis in an ALF DNA sequencer.
Preparation of substrates
All hybridizations for preparation of substrates were carried out as decribed (9). Repair substrates: 10 pmol of the ligated U, C or T containing 60-mer oligonucleotides (see above), 15 pmol of HINDIII_55_G, in 50 µl SSC. Polymerase substrate: 50 pmol of HINDIII_LIG_F, 75 pmol of HINDIII_55_G, in 100 µl SSC. After hybridization, the mixture was diluted to 250 µl with water. Flap endonuclease substrate: 10 pmol of the ligated 60mer oligonucleotide containing C at postion 23 (see above), 30 pmol of HINDIII_23-Flap, 15 pmol of HINDIII_55_G, in 50 µl SSC. Ligase substrate: 50 pmol of HINDIII_LIG_F, 150 pmol of HINDIII_LIG_P, 75 pmol of HINDIII_55_G in 100 µl SSC. After hybridization, the mixture was diluted to 250 µl with water.
Preparation of cell extract
Five grams of wet-cell mass of M. thermautotrophicus ΔH were resuspended in 20 ml PBS (10 mM Na2HPO4, 1.8 mM KH2PO4 pH 7.3, 140 mM NaCl, 2.7 mM KCl) containing in addition 10 mM EDTA and 5 mM DTT. Cells were sonicated twice for 3 min on ice with a Branson Sonifier 250 (Danbury, CT, output level 5, duty cycle 50%, microtip) and then passed through a Constant System Ltd (Daventry, England) cell disrupter at 180 MPa. The lysate was centrifuged with 15 000 rpm for 60 min at 4°C in an SS-34 rotor (Sorvall, Thermo Electron Corp., Waltham, MA). The supernatant was slowly mixed with four volumes of saturated (4.1 M) ammonium sulfate solution (final concentration: 80% saturation) and the precipitate was collected by centrifugation at 15 000 rpm. for 10 min at 4°C in an SS-34 rotor. The pellet was dissolved in 10 ml PBS and dialyzed overnight against 5 l PBS at 4°C. Protein concentration was determined by a Bradford assay (15) using bovine serum albumin (BSA) as the standard.
Immunodepletion of Mth212 from cell extracts
An amount of 100 µl Dynabeads® Protein G (Invitrogen, Carlsbad, CA) were washed three times with 500 µl buffer A (0.1 M Na-Citrate/Citric acid pH 5.0). (Beads were always collected by placing the tube in a magnetic stand.) The washed beads were resuspended in 100 µl anti-Mth212 total IgG-fraction (∼1 mg IgGs) and incubated for 40 min at room temperature with gentle shaking. The supernatant was discarded and the beads were washed three times with 500 µl buffer A and three times with 1 ml 0.2 M triethanolamine pH 8.2. Bound IgGs were crosslinked to protein G by incubation of the beads for 30 min at room temperature with gentle shaking in 1 ml freshly prepared 20 mM dimethylpimelimidate in 0.2 M triethanolamine pH 8.2. Beads were then incubated for 15 min at room temperature in 1 ml 50 mM Tris–HCl pH 7.5. Finally, beads were washed three times with 1 ml PBS (10 mM Na2HPO4, 1.8 mM KH2PO4 pH 7.3, 140 mM NaCl, 2.7 mM KCl).
The thus prepared IgG-loaded protein G beads were incubated with gentle shaking in 50 µl cell extract (∼500 µg) from M. thermautotrophicus ΔH (see above) for 1 h at room temperature. For regeneration, beads were incubated three times with 50 µl 3 M NaCl in PBS for 15 min to elute bound material and washed once with 1 ml PBS. Regenerated beads were stored in PBS containing 0.05% TWEEN® 20 at 4°C. In total, extracts were subjected to three consecutive rounds of immunodepletion using either freshly prepared or regenerated IgG protein G beads.
Western blot analysis
Western blot analysis was carried out using standard procedures. Briefly, 10 µl of immunodepleted or untreated cell extract were separated on a 15% polyacrylamide gel and transferred to a nitrocellulose membrane (Protran BA79, Schleicher & Schuell BioScience, Dassel, Germany) by semi-dry blot. Membranes were blocked with nonfat dry milk and incubated overnight with either anti-Mth212 or anti-Mth.PolB antibodies (each diluted 1:2000). Following washing, incubation with anti-rabbit IgG conjugated with alkaline phosphatase (working dilution 1:10 000) as secondary antibody and final washing signals were detected with chromogenic substrates (0.15 mg/ml 5-bromo-4-chloro-3-indolyl phosphate and 0.3 mg/ml nitro blue trazolium [BCIP/NBT, Applichem, Darmstadt, Germany] in 100 mM Tris–HCl pH 9.5, 100 mM NaCl, 50 mM MgCl2).
Cloning, production and purification of 5′-flap endonuclease (Mth1633)
Using standard procedures gene mth1633 was amplified by PCR from M. thermautotrophicus ΔH genomic DNA with primer pair 5′ CATATGGGAGTTAAACTCAGGGATGTTG 3′ and 5′ GGATCCTCAGAACCAGTCCTCCA GGCT 3′ (NdeI and BamHI sites are underlined). PCR product was cloned into pCR-Blunt II-TOPO as described by the supplier. NdeI/BamHI fragment was recovered from recombinant TOPO plasmid and inserted into pET-28a using standard procedures. The resulting construct was confirmed by DNA sequence analysis.
The protein carries an N-terminal His-tag; it was produced and purified as described earlier for Mth212 (9), exceptions being the use of BL21_UXX as production strain and MonoQ anion exchange chromatography instead of heparin affinity chromatography. For MonoQ column (5/50 GL, GE Healthcare, Uppsala, Sweden) 20 mM Tris–HCl pH 7.8 was used as running buffer with a gradient from 0 to 1 M NaCl. Protein eluted at ca. 300 mM NaCl.
Preparation of TTUDGA
TTUDGA was prepared as described (16), except that BL21_UX was used as production strain and HS cation exchange chromatography was replaced by heparin affinity chromatography. The concentration of purified TTUDGA was determined by a Bradford assay using BSA as standard.
Activity assays
Assays with cell extract
A total amount of 0.25 pmol oligonucleotide substrates were preincubated at 65°C for 10 min in a total volume of 45 μl 44 mM Tris–HCl pH 7.8, 5.5 mM MgCl2, 11% PEG 6000, 110 mM NH4Cl (assay buffer) with or without 1 nmol dNTPs (each) and 100 nmol ATP as indicated in the figures. Total 50 µg (5 µl) of cell extract were added and the reaction incubated for 30 min at 65°C (final volume: 50 µl). The reaction was stopped by extraction with one volume of TE-saturated phenol followed by a phenol/chloroform (1:1, v:v) extraction. DNA was precipitated with ethanol and dissolved in 20 µl H2O. For detection of single activities, 10 µl loading dye (95% formamide, 20 mM EDTA pH 8.0, 50 mg/ml Dextran Blue) were added to the mixture and the reaction products subjected to gel electrophoretic analysis as described below. To detect complete repair reactions, 10 µl of the reaction products dissolved in water (see above) were incubated with 10 u HindIII in a total volume of 15 µl reaction buffer (10 mM Tris–HCl pH 8.5, 10 mM MgCl2, 100 mM KCl, 0.1 mg/ml BSA) for 60 min at 37°C prior to gel electrophoresis. The reaction was stopped with 7.5 µl of loading dye and the products analyzed by gel electrophoresis as described below.
Flap endonuclease assay of Mth1633
Assay was performed as specified above but instead of cell extract 2.5 pmol of purified Mth1633 were used.
Glycosylase assay of TTUDGA
Total 0.25 pmol of U/G-, C/G- or T/G-substrates were preincubated in a total volume of 50 µl assay buffer (see above) for 10 min at 65°C. After addition of 2.5 pmol TTUDGA the mixture was incubated for 30 min. Reaction was stopped by adding NaOH to a final concentration of 100 mM following incubation at 95°C for 10 min. The solution was mixed with 25 µl loading dye and analyzed as described below.
Gel electrophoretic analysis
After incubation at 95°C for 5 min 7 µl of the samples were loaded onto an 11% polyacrylamide gel (30 cm length, composition and running buffer see section ‘Substrates’). Gels were run at 52°C in an ALF DNA sequencer (GE Healthcare, Uppsala, Sweden) at a constant power of 40 W and 2 mW laser power. The fluorescence of the 60-mer oligonucleotide was detected after 180 min under these conditions. In case of low signal intensity as a result of loss of fluorescent material during sample preparation, gel electrophoresis was repeated with a maximum sample volume of 15 µl. Peak areas were quantitated with the software Fragment Manager (GE Healthcare, Uppsala, Sweden).
RESULTS
Demonstration of all core activities of the repair pathway under a common set of conditions
Mth212 recognizes DNA uracil residues within double-stranded DNA and cleaves the phosphodiester bond at the 5′-side of the damaged nucleotide leaving behind a 3′-hydroxy and a 5′-phosphate residue. The initial nick may be further processed to a gap by the 3′→5′ exonuclease activity of the enzyme (9). Starting with either the nick or the gap, additional biochemical functions are needed to complete repair: (i) a DNA polymerase has to extend the primer. If the relevant DNA polymerase does not possess 5′→3′ exonuclease activity, primer extension is accompanied by strand displacement with the DNA uridine residue at the 5′-terminus of the flap formed. This requires (ii) the action of a 5′-flap endonuclease before (iii) DNA ligase seals the previously damaged DNA strand. If, alternatively, the DNA polymerase involved possesses 5′→3′ exonuclease activity, nick translation occurs and 5′-flap endonuclease is dispensable.
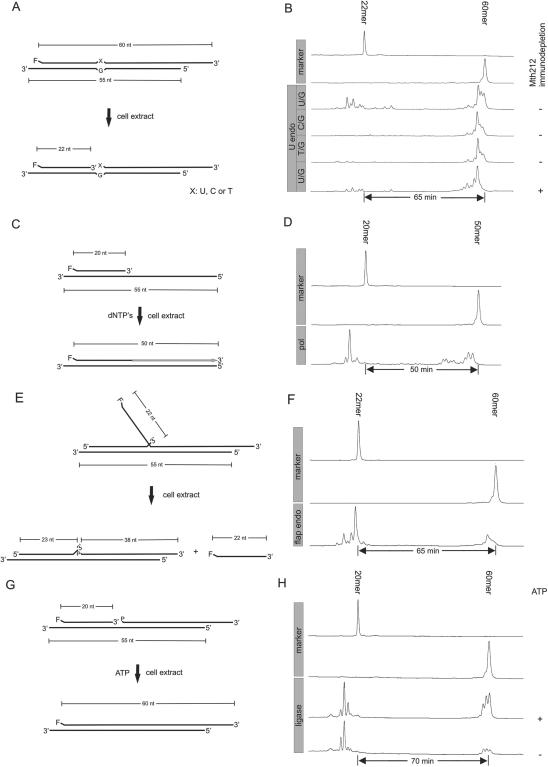
In the first round of experiments, common reaction conditions were established under which strand incision, primer elongation, 5′-flap removal and ligation could individually be documented. All assays were based on electrophoretic mobility of fluorescently labeled oligonucleotides, essentially as described earlier (9,17). Figure 1 gives schematic illustrations of substrates and documents results achieved with them.
Figure 1.
Demonstration of enzymatic activities in cell extracts inferred to be required for uridine excision repair. (A), (C), (E), (G): Schematic drawings of substrates and expected reaction products (F: fluorescein moiety, nt: nucleotides). (B), (D), (F), (H): Trackings of fluorescence readouts as recorded by an ALF DNA sequencer. In lanes labeled ‘marker’, 5′-fluorescein conjugated oligonucleotides of known length were run; numbers of residues are indicated above respective peaks. Numbers of minutes refer to run time differences between marker oligonucleotides. (A), (B) DNA uridine endonuclease assay. (A) Three substrates differing with respect to the identity of residue ‘X’ (U, C or T), but otherwise being identical were tested. The left 3′-overhang is 5 nt in length, the right one 10 nt. (B) Reaction products of different substrates as indicated. α-Mth212 antibodies: ‘−’ or ‘+’ denote no treatment or pretreatment, respectively, of cell extract with α-Mth212 antibodies prior to reaction. (C), (D) DNA polymerase assay. C: Right 3′-overhang is as in (A). dNTPs: reaction was performed in presence of dNTP′s (20 µM each) in assay buffer. (E), (F) Flap endonuclease assay. (E) The flap branch point can move by one nucleotide yielding either a 22-nt or a 23-nt flap. Only the former structure is shown. 3′-overhangs are as in (A). ‘P’ denotes a 5′-phosphate residue. (G), (H): DNA ligase assay. G: 3′-overhangs are as in (A). ‘P’ denotes a 5′-phosphate residue. Ligase assay was carried out with or without ATP (5 mM) added to the assay buffer, as indicated in the right margin of (H). Conditions for all reactions were identical as described in detail in ‘Materials and Methods’ section.
DNA-U endonuclease activity (Figure 1A and B)
Two different reactions are taking place. (i) The 60-mer is partially shortened by a few residues. This reaction, apparently brought about by an exonuclease, is independent of the nature of the X in X/G. Since Mth212 does not act upon 3′-protruding DNA ends (this laboratory, unpublished), at least one additional 3′→5′ exonuclease seems to be present in the extract. This notion is corroborated by the fact that the reaction persists after Mth212 is immunochemically removed from the extract (bottom lane). (ii) A set of oligonucleotides with chain lengths of less than 22 residues is produced in a substrate-specific manner, i.e. with a U/G opposition present in the starting DNA duplex. The products are most easily explained as resulting from U-specific incision, followed by exonucleolytic processing of the reaction intermediate. Incomplete incision observed with the U/G substrate is due to the assay conditions which were tuned to optimal performance of the extract in complete repair (compare below). After immunodepletion of the extract for Mth212, no material in the shorter range of chain lengths is observed (bottom lane) consistent with the notion of ExoIII homologue Mth212 being the carrier of the U endo activity (9). This result has further implications for assays of complete DNA repair (compare Figure 4).
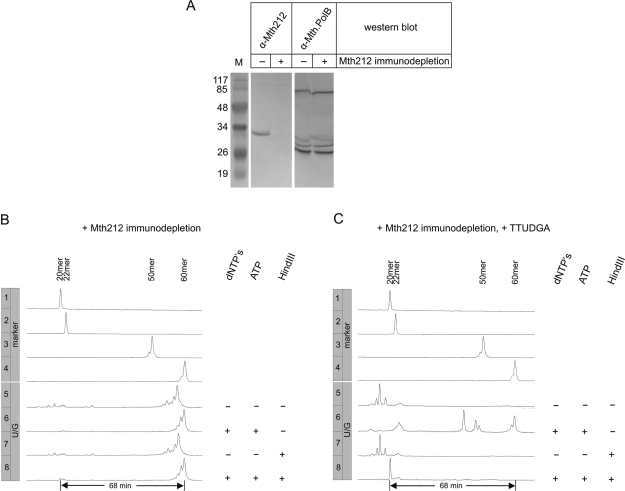
Figure 4.
DNA-U repair assays with extracts from M. thermautotrophicus ΔH immunochemically depleted for Mth212. (A) Western blot analysis of Mth212 immunodepletion. Untreated (‘−’) or Mth212 immunodepleted (‘+’) M. thermautotrophicus ΔH cell extracts were analyzed for the presence of Mth212 or Mth.PolB (control) by western blot as described in ‘Materials and methods’ section. M: Marker proteins (Fermentas) with corresponding molecular masses (×10−3). Calculated relative molecular weight for Mth212 is 30 350 and for the two subunits of Mth.PolB (14) 67 950 and 25 500. (B) Gel electrophoretic analysis of products after incubation of repair substrate (compare Figure 3) with cell extract immunochemically depleted for Mth212. dNTPs: presence (‘+’) or absence (‘−’) of dNTP's (20 µM each) in assay buffer. ATP: presence (‘+’) or absence (‘−’) of ATP (2 mM) in assay buffer. HindIII: no incubation (‘−’) or incubation (‘+’) of reaction products with HindIII. Lengths of 5′-fluorescein-labeled oligonucleotides are indicated above the respective peaks in lanes labeled ‘marker’. (C) Gel electrophoretic analysis of products after incubation of repair substrate with M. thermautotrophicus ΔH cell extract supplemented with 25 pmol of purified TTUDGA and then immunochemically depleted for Mth212. Other labels as in (B).
DNA polymerase (Figure 1C and D)
The primer (20-mer) is completely used up in the reaction (Figure 1D, bottom lane). There are two groups of product peaks, one corresponding to chains significantly shorter than 20 residues and one migrating only slightly faster than the 50-mer marker. There appears to be competition between 3′→5′ exonuclease and DNA polymerase activities acting upon the substrate. A fraction of primer seems to be degraded to material too short to still stably hybridize to the template strand. Using the OligoCalc algorithm (18), we estimate the minimal length of duplex required for stable hybridization under assay conditions to be 17 residues. In agreement with this, the major peak in the left group seems to correspond to 16-mer. Clearly, primer elongation occurs, although no full-length run-off product is discernible. The group of peaks extending from about 40–50 nt could either be due to secondary 3′→5′ exonuclease activity acting on the full-length run-off product or reflect incomplete primer extension. Both explanations are compatible with proper function of the polymerase activity in DNA repair.
5′-Flap endonuclease activity (Figure 1E and F)
The substrate was designed to allow the branch point to alternate between two positions one nucleotide apart. This measure proved necessary, as already described in the literature (19–22). For clarity, Figure 1E depicts only the alternative with the branch point moved left and the 3′-teminus of the upstream (unlabeled) oligonucleotide unpaired. (In the alternative form, the non-fluorescent upper strand oligonucleotide is completely base-paired to the lower strand and the free flap extended from 22 to 23 nt; for details of substrate design refer to Supplementary Data.)
The small amount of starting material left after incubation (Figure 1F, bottom lane) is partially trimmed by exonuclease as observed before (Figure 1B). The bulk of the material corresponds to 5′-flap endonuclease product, with a predominant length of 22 nt. By analogy to earlier results obtained by other laboratories (19–21), the 23-mer would have been expected as the major product of 5′-flap endonuclease reaction. This difference is significant since removal of a 23-mer leaves behind a nicked duplex which is a substrate for ligation without further nucleolytic trimming. The occurrence of oligonucleotides shorter than 22 residues again hints at the presence of an exonuclease acting on single-stranded DNA (compare above). This left open the possibility of the 22-mer already being a processing product of the 23-mer as a necessary intermediate. To clarify this point, the 5′-flap endonuclease of M. thermautotrophicus ΔH, product of orf1633 (21), was produced and purified using standard techniques. When the reaction was repeated with pure enzyme, the 22-mer was observed as the sole product (Supplementary Figure 1). The substrate requirements of the 5′-flap endonuclease seem to be more complex than previously believed. The presence of 5′-flap endonuclease activity in the extract leaves unknown the nature of the relevant DNA polymerase (with/without 5′→3′ exonuclease activity, compare above).
DNA ligase activity (Figure 1G and H)
A large proportion of the substrate is converted to ligation product (Figure 1H, third lane from top), again partially trimmed at its 3′-end (compare above); whereas a second set of products is shorter than the starting 20-mer. Note that in the absence of DNA polymerase reaction (no dNTPs were added to the assay mixture) exonucleolytic removal of a single nucleotide from the starting fluorescent strand precludes ligation and irreversibly directs the oligonucleotide to degradation (final length about 16-mer, compare above: discussion of DNA polymerase reaction). Some ligation occurs without ATP being added (Figure 1H, bottom lane). We interpret this result as an indication of covalent AMP/ligase adduct (a mandatory intermediate of the ligation reaction) still being present in the extract and being able to promote a single turnover.
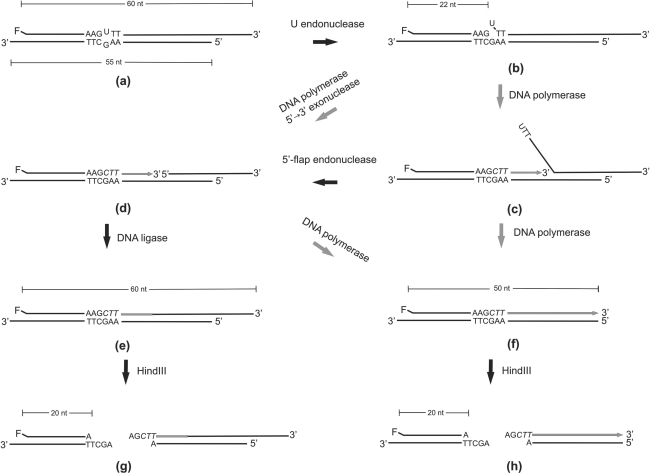
Design of repair assay
To demonstrate full repair of U/G sites by the crude extract, an assay was needed which demonstrates replacement of the mismatched uridine by a cytidine residue while distinguishing between run-off polymerase reaction and repair completed by DNA ligation. The assay was based on the same DNA substrate as was used for monitoring DNA-U endonuclease activity (Figure 1A); its logic is outlined in Figure 2. The core element is restoration of a restriction site by repair of a base/base mismatch (23). In particular, repair of the U/G mismatch in structure (a) to C/G restores susceptibility of the hexanucleotide sequence to cleavage by HindIII (24). Once repair is initiated by DNA-U endonuclease (Figure 2, structure b) and DNA polymerase has taken over the primer end (Figure 2, structure c), the fluorescently labeled strand can have one of two alternative fates: Run-off synthesis leads to a labeled 50-mer (Figure 2, structure f), whereas full repair results in conservation of the 3′-protrusion of 10 nt and thus in a labeled 60-mer (Figure 2, structure e). In both cases, and in contrast to the starting material, incubation with HindIII liberates a labeled 20-mer (Figure 2, structures g and h). Thus, repair can be analyzed by measuring both length and HindIII-sensitivity of the fluorescently labeled DNA strand (5).
Figure 2.
Outline of DNA-U repair assay. DNA polymerase reactions are represented by grey arrows, newly synthesized DNA is indicated by grey lines and letters in italics. The rationale underlying the assay is described in detail in the text.
Demonstration of repair
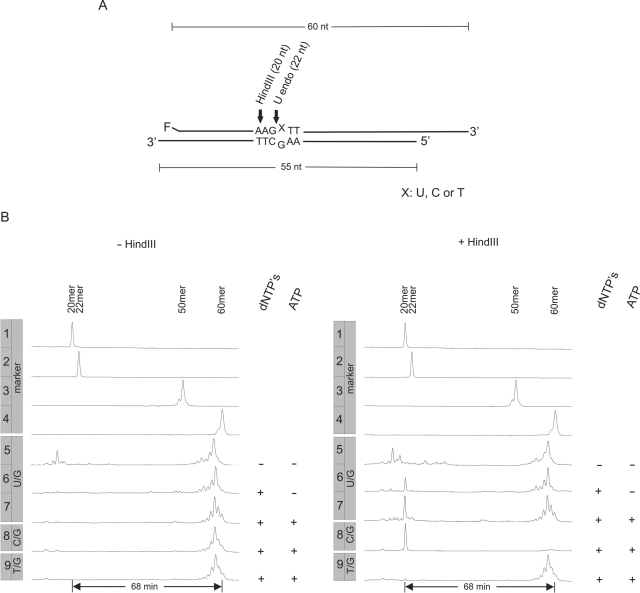
Results of DNA-U repair assays are documented in Figure 3B. Lanes carrying the same numbers illustrate products of reactions carried out under identical conditions, with the one exception that mixtures represented in the right hand row were incubated with HindIII, whereas those in the left hand row were not. Lane 5 of this row (no dNTPs, no ATP added) corresponds to the DNA-U endonuclease assay of Figure 1A and B and, consequently, it shows a very similar result (incomplete incision, followed by exonucleolytic degradation of the reaction intermediate and some exonucleolytic resection of the protruding 3′-end of the labeled substrate strand). When DNA polymerase and DNA ligase reactions are allowed to occur by addition of dNTPs and ATP (lanes 7, left and right), no short chain products are visible in the reaction mixture that did not undergo HindIII treatment (lane 7, left), whereas a pronounced peak at chain length 20 residues appears upon incubation of that mixture with HindIII (lane 7, right). Clearly, U/G was replaced by C/G in a sizable proportion of total substrate. In addition, there is very little run-on/run-off product visible in lane 7, left; therefore the 20-mer formed by HindIII treatment must largely be derived from full-length oligonucleotide, i.e. from products of complete repair. To test this interpretation, an experiment was carried out in which the material represented by the 60-mer group of peaks was eluted from the gel and subjected to HindIII treatment. The results were essentially indistinguishable from those displayed in lane 7, right (data not shown). The 20-mer visible in lane 7, right, disappears upon omission of dNTPs and ATP from the assay mixture (lane 5, right). This lends further support to the notion that formation of this product is repair-dependent.
Figure 3.
DNA-U repair in cell extracts from M. thermautotrophicus ΔH. (A) Schematic representation of repair substrates (also compare figure 2). Arrows indicate cleavage points of U endonuclease (U endo, X = U) and HindIII (X = C, the C residue being provided with the substrate or introduced in the course of repair). (B) Gel electrophoretic analysis of reaction products after incubation of repair substrate with 50 µg cell extract from M. thermautotrophicus ΔH. dNTPs: presence (‘+’) or absence (‘−’) of dNTPs (20 µM each) in assay buffer. ATP: presence (‘+’) or absence (‘−’) of ATP (2 mM) in assay buffer. Sizes in nucleotides of 5′-fluorescein labeled oligonucleotides are given above the respective peaks in the lanes labeled ‘marker’. Left row of lanes (‘−HindIII’): without addition of HindIII. Right row of lanes (‘+ HindIII’): same reactions as in left panel, but after additional incubation with HindIII.
Remarkably, even in the absence of ATP (required for DNA ligase reaction) little run-off product is detected (compare lanes 6 and 7, left). In addition, HindIII liberates significant amounts of 20-mer under ‘no ATP’ assay conditions. A gel elution/HindIII cleavage performed on the 60-mer group of products unequivocally indicated most of the 20-mer to be derived from full-length products (data not shown). These observations are in accord with the finding that there is significant ligation without ATP added (compare above: Figure 1H).
As expected, the control assays with X = C and X = T yield indistinguishable products before HindIII treatment, the only visible reaction being exonucleolytic attack on the 3′-ends (lanes 8 and 9, left). Upon incubation with Hind III, C/G yields the 20-mer as the virtually exclusive product (lane 8, right), whereas T/G (lane 9, right) shows none of this.
Dependence of repair on Mth212
To test whether Mth212 is the sole or predominant initiator of DNA-U repair in crude extracts of M. thermautotrophicus ΔH, repair was measured with extracts that had been immunochemically depleted for Mth212. As shown in Figure 4A, no band corresponding to Mth212 (Mr: 30 350) can be detected in western blots after immunodepletion, whereas the amount of Mth.PolB (two bands expected at Mr 67 950 and 25 500) is apparently not affected by this procedure. Hence, Mth212 immunodepletion was efficient and specific on this account. The depletion of Mth212 was accompanied by loss of repair activity: treatment of the extract abolishes 20-mer formation almost completely (Figure 4B, bottom lane; compare Figure 3B, lane 7, right).
In principle, the observed lack of repair could be due to some downstream function being inhibited by the polyclonal antibodies. This was tested by adding to the extract thermostable uracil DNA glycosylase TTUDGA of Thermus thermophilus HB27 (16) prior to the immunochemical treatment; results are documented in Figure 4C. With TTUDGA present, the starting material is almost completely converted to material yielding 20-mer upon HindIII treatment (Figure 4C, bottom lane). Apparently, the extract contains enough AP endonuclease to be fully competent in base excision repair. The genome of M. thermautotrophicus ΔH contains a gene for an EndoIV-type AP endonuclease (orf mth1010). This is a plausible candidate for contributing the activity to Mth212-depleted extracts but some residual Mth212 cannot be excluded (it should be kept in mind that the AP endo activity of Mth212 is much more pronounced than its U endo activity). Not surprisingly (compare above), enough 3′→5′ exonuclease activity also remains to convert the intermediate 22-mer to short DNA species in the absence of dNTPs and ATP (Figure 4C, lane 5). The mixture of repair products before HindIII treatment (Figure 4C, lane 6) contains a conspicuously higher proportion of run-on and run-off products than was seen with the untreated extract (Figure 3B, left row, lane 7). In the latter case, however, a large proportion of the 60-mer group of peaks is derived from unprocessed substrate and is not repair products (compare above). In the present experiment, enough TTUDGA could be added to force strand cleavage to completion. In contrast, maximizing Mth212 activity in untreated extract, lowers the overall repair efficiency (compare above). The production and characterization of the particular batch of TTUDGA used here is documented in online Supplementary Figure 2.
DISCUSSION
To date, base excision repair acting on DNA uracil residues has been the sole accepted mechanistic paradigm for the avoidance of mutations resulting from hydrolytic deamination of DNA cytosine residues. The results presented here establish an alternative mechanism for general DNA-U repair, i.e. the direct endonucleolytic cleavage of the phosphodiester bond on the 5′-side of the U residue with creation of an extendable primer end immediately upstream of the damaged nucleotide.
Besides direct damage reversal, this constitutes the simplest DNA repair pathway characterized so far. Simplicity intuitively suggests that the mechanism should be widely spread phylogenetically. There are, indeed, a number (albeit limited) of known precedents falling into that category, such as the ubiquitous AP endonucleases (25), Vsr endonuclease of E. coli (26) and UV endonuclease of Neurospora crassa (27) and Schizosaccharomyces pombe (28). In addition, AP endonucleases of the ExoIII and the EndoIV families have been implied in similar reactions on benzetheno-derivatives of adenine and cytosine residues (29,30).
These examples encompass several different types of DNA damage and also several different structural classes of enzymes. The latter fact clearly indicates that this mechanism has been found along several independent trajectories of molecular evolution.
Vsr endonuclease cleaves next to the T residue of T/G mismatches that are embedded in the substrate site of Dcm DNA cytosine methyltransferase (26), which means it protects E. coli from the genetic hazard associated with hydrolytic deamination of 5-meC residues. Vsr endonuclease accepts U instead of T residues and thus comes close to the properties and function of Mth212. Vsr endonuclease, however, accepts DNA-U residues only in the proper sequence context and when mismatched to G (31). The DNA-U endonuclease activity of Vsr endonuclease therefore is a by-product owed to lack of discrimination between T and U and the enzyme does not qualify as an initiator of general DNA-U repair.
AP endonucleases are normally considered auxiliary enzymes in the context of base excision repair. Generation of AP-sites by uncatalyzed hydrolytic loss of undamaged bases from DNA is, however, a kinetically significant reaction. Indeed, AP sites occur much more frequently as primary pre-mutagenic lesions than as mere intermediates in the course of repairing the product of C-deamination (1), which implies that DNA-U repair should be the evolutionary latecomer. We envisage a history with DNA-U repair being grafted upon pre-existing AP repair in at least two alternative ways: (i) by evolution of U-specific DNA glycosylase activity in some other protein, resulting in the foundation of the UDG superfamily and (ii) by expanding the substrate spectrum of an ExoIII-type AP endonuclease itself such that it accepts, in addition, DNA-U residues as substrates, as is the case with Mth212. Note that these options are not mutually exclusive. It seems possible that in any one phylogenetic lineage the two events occurred sequentially. The resulting redundancy with respect to DNA-U repair capacity could later have been set back by loss of one or the other function (and may even still exist in some organisms not studied yet).
This present work has expanded our knowledge about the nature of enzymes with initiating function in DNA-U repair and their distribution across the archaeal domain, but this knowledge is still incomplete. Methanococcus maripaludis S2, for example, has no UDG gene (32) and its Mth212 (ExoIII) homologue is devoid of U endonuclease activity (33). There are still other archaea such as Methanopyrus kandleri AV19 (34) and Methanosphaera stadtmanae (35) which have neither UDG nor ExoIII genes. For the time being, the nature of DNA-U repair in such organisms remains unclear.
A second open question concerns the identity of the DNA polymerase involved in the newly discovered pathway in M. thermautotrophicus ΔH. Annotation of the M. thermautotrophicus ΔH genome (8) points to the presence of three different DNA polymerases (PolB, PolII and PolX). By the criterion of structural families to which they belong, none of the three polymerases would be expected to have 5′→3′ exonuclease activity. For Mth.PolB, this has been confirmed experimentally (14). No experimental data are available for Mth.PolII; its homologue from Methanocaldococcus jannaschii, however, has been characterized as a replicative DNA polymerase (36). On the other hand, a DNA polymerase activity of unknown identity, associated with 5′→3′ exonuclease activity has been reported for Methanothermobacter marburgensis (37); the enzyme is described as a single-chain polypeptide having a relative molecular mass of 72 000, which does not correlate to any of the three relevant proteins of M. thermautotrophicus ΔH. Clearly, more experiments are required to answer these salient questions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Georg-August-Universität Göttingen.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Elena Ciirdaeva for western blot analysis and Christiane Preiß for expert technical assistance.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Dianov G, Lindahl T. Reconstitution of the DNA base excision-repair pathway. Curr. Biol. 1994;4:1069–1076. doi: 10.1016/s0960-9822(00)00245-1. [DOI] [PubMed] [Google Scholar]
- 3.Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 4.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sartori AA, Jiricny J. Enzymology of base excision repair in the hyperthermophilic archaeon Pyrobaculum aerophilum. J. Biol. Chem. 2003;278:24563–24576. doi: 10.1074/jbc.M302397200. [DOI] [PubMed] [Google Scholar]
- 6.Seeberg E, Eide L, Bjoras M. The base excision repair pathway. Trends Biochem. Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 7.Pearl LH. Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res. 2000;460:165–181. doi: 10.1016/s0921-8777(00)00025-2. [DOI] [PubMed] [Google Scholar]
- 8.Smith DR, Doucette-Stamm LA, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J. Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georg J, Schomacher L, Chong JP, Majernik AI, Raabe M, Urlaub H, Muller S, Ciirdaeva E, Kramer W, Fritz HJ. The Methanothermobacter thermautotrophicus ExoIII homologue Mth212 is a DNA uridine endonuclease. Nucleic Acids Res. 2006;34:5325–5336. doi: 10.1093/nar/gkl604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfeifer S, Greiner-Stoffele T. A recombinant exonuclease III homologue of the thermophilic archaeon Methanothermobacter thermautotrophicus. DNA Repair. 2005;4:433–444. doi: 10.1016/j.dnarep.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 11.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guy CP, Majernik AI, Chong JP, Bolt EL. A novel nuclease-ATPase (Nar71) from archaea is part of a proposed thermophilic DNA repair system. Nucleic Acids Res. 2004;32:6176–6186. doi: 10.1093/nar/gkh960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelman Z, Pietrokovski S, Hurwitz J. Isolation and characterization of a split B-type DNA polymerase from the archaeon Methanobacterium thermoautotrophicum deltaH. J. Biol. Chem. 1999;274:28751–28761. doi: 10.1074/jbc.274.40.28751. [DOI] [PubMed] [Google Scholar]
- 15.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 16.Starkuviene V, Fritz HJ. A novel type of uracil-DNA glycosylase mediating repair of hydrolytic DNA damage in the extremely thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 2002;30:2097–2102. doi: 10.1093/nar/30.10.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gläsner W, Merkl R, Schmidt S, Cech D, Fritz HJ. Fast quantitative assay of sequence-specific endonuclease activity based on DNA sequencer technology. Biol. Chem. Hoppe. Seyler. 1992;373:1223–1225. doi: 10.1515/bchm3.1992.373.2.1223. [DOI] [PubMed] [Google Scholar]
- 18.Kibbe WA. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35:W43–W46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyamichev V, Mast AL, Hall JG, Prudent JR, Kaiser MW, Takova T, Kwiatkowski RW, Sander TJ, de Arruda M, Arco DA, et al. Polymorphism identification and quantitative detection of genomic DNA by invasive cleavage of oligonucleotide probes. Nat. Biotechnol. 1999;17:292–296. doi: 10.1038/7044. [DOI] [PubMed] [Google Scholar]
- 20.Lyamichev V, Brow MA, Varvel VE, Dahlberg JE. Comparison of the 5′ nuclease activities of Taq DNA polymerase and its isolated nuclease domain. Proc. Natl Acad. Sci. USA. 1999;96:6143–6148. doi: 10.1073/pnas.96.11.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser MW, Lyamicheva N, Ma W, Miller C, Neri B, Fors L, Lyamichev VI. A comparison of eubacterial and archaeal structure-specific 5′-exonucleases. J. Biol. Chem. 1999;274:21387–21394. doi: 10.1074/jbc.274.30.21387. [DOI] [PubMed] [Google Scholar]
- 22.Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 23.Lu AL, Clark S, Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc. Natl Acad. Sci. USA. 1983;80:4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shenoy S, Daigle K, Ehrlich KC, Gehrke CW, Ehrlich M. Hydrolysis by restriction endonucleases at their DNA recognition sequences substituted with mismatched base pairs. Nucleic Acids Res. 1986;14:4407–4420. [PMC free article] [PubMed] [Google Scholar]
- 25.Mol CD, Hosfield DJ, Tainer JA. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3′ ends justify the means. Mutat. Res. 2000;460:211–229. doi: 10.1016/s0921-8777(00)00028-8. [DOI] [PubMed] [Google Scholar]
- 26.Hennecke F, Kolmar H, Brundl K, Fritz HJ. The vsr gene product of E. coli K-12 is a strand- and sequence-specific DNA mismatch endonuclease. Nature. 1991;353:776–778. doi: 10.1038/353776a0. [DOI] [PubMed] [Google Scholar]
- 27.Yajima H, Takao M, Yasuhira S, Zhao JH, Ishii C, Inoue H, Yasui A. A eukaryotic gene encoding an endonuclease that specifically repairs DNA damaged by ultraviolet light. EMBO J. 1995;14:2393–2399. doi: 10.1002/j.1460-2075.1995.tb07234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takao M, Yonemasu R, Yamamoto K, Yasui A. Characterization of a UV endonuclease gene from the fission yeast Schizosaccharomyces pombe and its bacterial homolog. Nucleic Acids Res. 1996;24:1267–1271. doi: 10.1093/nar/24.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hang B, Chenna A, Fraenkel-Conrat H, Singer B. An unusual mechanism for the major human apurinic/apyrimidinic (AP) endonuclease involving 5′ cleavage of DNA containing a benzene-derived exocyclic adduct in the absence of an AP site. Proc. Natl Acad. Sci. USA. 1996;93:13737–13741. doi: 10.1073/pnas.93.24.13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hang B, Chenna A, Sagi J, Singer B. Differential cleavage of oligonucleotides containing the benzene-derived adduct, 1,N6-benzetheno-dA, by the major human AP endonuclease HAP1 and Escherichia coli exonuclease III and endonuclease IV. Carcinogenesis. 1998;19:1339–1343. doi: 10.1093/carcin/19.8.1339. [DOI] [PubMed] [Google Scholar]
- 31.Gläsner W, Merkl R, Schellenberger V, Fritz HJ. Substrate preferences of Vsr DNA mismatch endonuclease and their consequences for the evolution of the Escherichia coli K-12 genome. J. Mol. Biol. 1995;245:1–7. doi: 10.1016/s0022-2836(95)80033-6. [DOI] [PubMed] [Google Scholar]
- 32.Hendrickson EL, Kaul R, Zhou Y, Bovee D, Chapman P, Chung J, Conway de Macario E, Dodsworth JA, Gillett W, Graham DE, et al. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 2004;186:6956–6969. doi: 10.1128/JB.186.20.6956-6969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schomacher L. University of Göttingen. 2007. Ein neu entdeckter Weg der Reparatur hydrolytisch geschädigter DNA-Cytosinreste, etabliert im thermophilen Archaeon Methanothermobacter thermautotrophicus ΔH. Doctoral thesis. [Google Scholar]
- 34.Slesarev AI, Mezhevaya KV, Makarova KS, Polushin NN, Shcherbinina OV, Shakhova VV, Belova GI, Aravind L, Natale DA, Rogozin IB, et al. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl Acad. Sci. USA. 2002;99:4644–4649. doi: 10.1073/pnas.032671499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fricke WF, Seedorf H, Henne A, Kruer M, Liesegang H, Hedderich R, Gottschalk G, Thauer RK. The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J. Bacteriol. 2006;188:642–658. doi: 10.1128/JB.188.2.642-658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishino Y, Komori K, Cann IK, Koga Y. A novel DNA polymerase family found in Archaea. J. Bacteriol. 1998;180:2232–2236. doi: 10.1128/jb.180.8.2232-2236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klimczak LJ, Grummt F, Burger KJ. Purification and characterization of DNA polymerase from the archaebacterium Methanobacterium thermoautotrophicum. Biochemistry. 1986;25:4850–4855. doi: 10.1021/bi00365a019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.