Abstract
EngA, a unique GTPase containing a KH-domain preceded by two consecutive G-domains, displays distinct nucleotide binding and hydrolysis activities. So far, Escherichia coli EngA is reported to bind the 50S ribosomal subunit in the guanosine-5′-trihosphate (GTP) bound state. Here, for the first time, using mutations that allow isolating the activities of the two G-domains, GD1 and GD2, we show that apart from 50S, EngA also binds the 30S and 70S subunits. We identify that the key requirement for any EngA–ribosome association is GTP binding to GD2. In this state, EngA displays a weak 50S association, which is further stabilized when GD1 too binds GTP. Exchanging bound GTP with guanosine-5′-diphosphate (GDP), at GD1, results in interactions with 50S, 30S and 70S. Therefore, it appears that GD1 employs GTP hydrolysis as a means to regulate the differential specificity of EngA to either 50S alone or to 50S, 30S and 70S subunits. Furthermore, using constructs lacking either GD1 or both GD1 and GD2, we infer that GD1, when bound to GTP and GDP, adopts distinct conformations to mask or unmask the 30S binding site on EngA. Our results suggest a model where distinct nucleotide-bound states of the two G-domains regulate formation of specific EngA–ribosome complexes.
INTRODUCTION
Guanosine-5′-trihosphate (GTP) binding proteins or GTPases make use of conformational changes associated with GTP binding and hydrolysis and thereby switch between three distinct nucleotide-bound states, i.e. an ‘empty’ nucleotide-free state, a GTP-bound ‘ON’ state and a guanosine-5′-diphosphate (GDP)-bound ‘OFF’ state. In doing so, several well characterized GTPases act as molecular switches and impart a tight control over important biological processes ranging from signal transduction, translation, intracellular transport and so on (1–3). On the other hand, little is known about the function of 11 universally conserved bacterial GTPases (4). Although initially thought to be present in prokaryotes, some of these are well conserved in eukaryotes too. Several recent reports suggest that these GTPases bind ribosomal subunits and possibly play key roles in their biogenesis or assembly (4–14). Of these, Escherichia coli Obg is implicated in the process of ribosome assembly where it interacts with both the 30S and 50S subunits (7). HflX was recently shown to bind the 50S (8). Of the circularly permuted GTPases, YjeQ and YloQ bind the 30S ribosomal subunit (9,10), RbgA or YlqF participates in the late step of 50S subunit assembly in Bacillus subtilis (11) and YqeH binds 30S in the GTP-bound form (B. Anand et al. unpublished results) to participate in its assembly (12). Era is known to bind the 30S ribosomal subunit in a nucleotide-free state (6). EngA has been shown to bind the 50S subunit (13–15) and it has been reported that YphC (EngA homologue in B. subtilis) and YsxC function together in 50S assembly (14).
Of the aforementioned proteins, EngA is unique as it is the only GTPase known to possess two contiguous G-domains, GD1 and GD2. A KH-domain that follows the G-domains is usually known to be involved in RNA binding or participate in protein–protein interactions (16,17). Based on the crystal structure of Der, an EngA homologue in Thermotoga maritima, it was postulated that the KH-domain participates in protein–protein interactions (16). A recently determined structure of YphC from B. subtilis, suggests its role in binding the ribosomal RNA (17). As several prokaryotic GTPases bind ribosomal subunits, we began our investigations to find out if EngA too would bind the ribosome. Reports by Hwang and Inouye (15), Bharat et al. (13) and Schaefer et al. (14) concur with our observation that EngA binds the 50S ribosomal subunit specifically when bound to a non-hydrolysable GTP analogue, guanosine-5-[β,γ-imido]triphosphate (GMPPNP). Bharat et al. (13) further suggest that both G-domains are important for ribosome binding. The crystal structure of YphC too, suggests the involvement of GD1 in ribosome binding, wherein a large conformational change (∼60 Å) is observed in GD1 between its GTP and GDP-bound forms. By comparing structures of Der (16) and YphC, Muench et al. (17) hypothesize that the GTP-bound state of GD1 exposes a positively charged RNA-binding region on the surface of KH-domain, which is masked due to the large movement of GD1. Schaefer et al. (14) indicate that the ribosome exits YphC in its GDP-bound state. On the whole, these studies indicate an importance for the two G-domains of EngA in binding the ribosomal subunits.
Hitherto, EngA was shown to bind the 50S ribosomal subunit. Here, for the first time, we show that E. coli EngA not only binds the 50S but also 30S and 70S. We identify two distinct ribosome-bound states of EngA—one where it binds 50S alone and another where it binds 50S, 30S and 70S. We associate this differential binding to the different nucleotide occupancy states of GD1 and GD2, based on co-fractionation experiments with ribosomal subunits using EngA mutants, which either disable nucleotide binding or change specificity from guanine to xanthine nucleotides (18). Surprisingly in a very recent report BipA, a highly conserved prokaryotic GTPase, was also shown to bind 70S and 30S subunits, although the latter is achieved in presence of ppGpp, an alarmone synthesized during stress. The 70S binding to BipA is realized in presence of GMPPNP (19). In contrast, here we show that EngA achieves the two distinct ribosome-bound states by varying the GTP or GDP-bound states of the two G-domains.
MATERIALS AND METHODS
Plasmids
The gene coding for EngA (503 amino acids) was amplified using forward (CTAGCTAGCGTGCGTTGTCTGATGAT) and reverse (CCGCTCGAGTTATTTATTTTTCTTGATGTG) primers from E. coli k12 genomic DNA, using Pfu DNA polymerase (Fermentas). Amplicon was digested with NheI/XhoI and cloned into corresponding sites in modified pGEX expression vector. Two truncated constructs of EngA, ΔGD1–EngA (213–503 amino acids) and ΔGD1–ΔGD2EngA (386–503 amino acids), were generated in a similar way using forward primers (CGCGCTAGCAGTCTGCCGATCAAACTGC) and (CGCGCTAGCAGCTCCACCCGTCGTGTGGG), respectively. His-YphC construct was prepared similarly by amplifying the gene coding for YphC using forward (CCGCATATGATGGGTAAACCTGTCGTA) and reverse primers (CGCGCGGCCGCTTATTTTCTAGCTCTC) from B. subtilis genomic DNA, using Pfu DNA polymerase (Fermentas). Amplicon was digested with NdeI/NotI and cloned into corresponding sites in pQE2 expression vector. The clones were confirmed by DNA sequencing.
Site-directed mutagenesis
All the mutants of EngA i.e. K28A, K228A, D134N and D337N and YphC-Y134A/D297N used in this study were generated by overlapping PCR technique using EngA full length as a template. Primers were designed with the desired change in the codon sequence. Single mutants K28A, K228A, D134N and D337N were generated using primers (GGGCGCCCTAACGTAGGAGCATCCACGTTA), (CCGAACGTAGGTGCGTCTACACTCACT), (GTGGCAAACAAAACTAACGGTCTGGATCCC) and (GGTGAATAAGTGGAATGGCTTAAGTCAGGAAGTG), respectively. His-YphC double mutant Y134A/D297N was also generated similarly using primers (GCGAATATTTATGATTTTGCATCGCTAGGCTTTGGC, GTAAACAAATGGAATGCTGTTGACAAAGAT) and His-YphC full length as a template. All mutations were confirmed by DNA sequencing.
Protein expression and purification
All constructs and mutants of EngA were expressed in E. coli BL21 (DE3). Cells were grown up to OD600 0.6–0.8 at 37°C in presence of 100 µg/ml of ampicillin in Luria Bertani media (Himedia) and were induced with 50 µM isopropyl-β-D-1-thiogalacto-pyranoside (IPTG) (Sigma) at 18°C for 10 h. Cells were harvested by centrifugation at 4000×g for 5 min at 4°C and washed with Buffer A (50 mM sodium-phosphate pH 7.4 and 100 mM NaCl). Similarly His-YphC and its double mutant were expressed in E. coli DH5α cells. Cells were grown similarly, but induced with 250 µM IPTG (Sigma) at 30°C for 5 h. Cells were harvested by centrifugation at 4000×g for 5 min at 4°C and washed with Buffer B (50 mM Tris–HCl pH 8.0 and 250 mM NaCl). To purify YphC protein, cells were lysed in Buffer B containing 5 mM MgCl2, 350 mM NaCl, 0.01 mM AEBSF [4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride] and 1 mg/ml lysozyme by repeated freeze-thaw cycles. Lysates, treated with DNAse and RNAse, were centrifuged at 30 000×g for 1 h at 4°C. Supernatant was loaded on a 5 ml His-trap affinity column (Amersham), pre-equilibrated with Buffer B. Unbound protein was washed (with Buffer B) and protein was eluted in the same buffer using 0–500 mM gradient of imidazole. Fractions containing the protein were analysed by size exclusion chromatography on sephacryl 200 column (Amersham). GST–EngA was purified as described below (‘RNA association with EngA’), except the use of RNAase and 450 mM NaCl during cell lysis.
Fluorescent nucleotide binding assays
Fluorescent nucleotide binding studies were carried out using LS 55 Fluorescence Spectrometer (PerkinElmer Life Sciences). The N-methyl-3′-O-anthranoyl (mant) group attached to the nucleotides was monitored with an excitation wavelength of 355 nm (slit width of 5 nm) and emission wavelength of 400–600 nm (slit width of 10 nm). Emission profiles of mant-nucleotides were generated from 0.4 µM free mant-nucleotides in Buffer F containing 50 mM Tris–HCl (pH 8.0), 400 mM KCl, 150 mM NaCl and 2 mM MgCl2. Protein mant-nucleotide complexes were generated by incubating 8.0 µM of the respective proteins with 0.4 µM mant-nucleotides in Buffer F at room temperature. The binding of D134N, D337N and Y134A/D297N mutants for xanthine nucleotides was studied by measuring mant-XDP binding and the specificity was assessed in presence of a 100 fold excess of GDP (40 µM) over mant-XDP, resulting in a GDP:mant-XDP ratio of 100:1.
Malachite green assay
GTP hydrolysis by GST–EngA and EngA was measured in a reaction buffer. Equimolar (500 nM) amount of protein was incubated with 500 µM of GTP in Buffer F for 25 min at room temperature. Released phosphate was measured by using a colorimetric malachite green assay as described in (20). A separate reaction was setup without any protein to estimate the amount of GTP hydrolysed during the time of the reaction. This was negated from the aforesaid measurements to estimate the amount of inorganic phosphate released due to the GTP hydrolysis by the proteins. All the reactions were setup in triplicates.
RNA association with EngA
GST–EngA was purified using affinity chromatography. The complete experiment was carried out in an RNAse-free environment. Cells were lysed in Buffer A containing 1% Tween-20, 5 mM MgCl2, 50 mM NaCl, 0.01 mM AEBSF and 1 mg/ml lysozyme by repeated freeze-thaw cycles. Lysates, treated with DNAse, were centrifuged at 30 000×g for 1 h at 4°C. Supernatant was loaded on a 5 ml GSTrap affinity column (Amersham), pre-equilibrated with Buffer A. Unbound protein was washed (with Buffer A) and protein was eluted in the same buffer containing 10 mM glutathione. Fractions containing protein were analysed by size exclusion chromatography on HiLoad 16/60 Superdex 200 column (Amersham). Co-eluted rRNA was recovered from the protein by phenol–chloroform extraction and was analysed on 1% agarose–formaldehyde gel. The identity of the rRNA was further confirmed by RT-PCR using AMV reverse transcriptase enzyme (Genei) using specific primers for 16S (forward—AAATTGAAGAGTTTGATCATGGCTCAGATT and reverse—TAAGGAGGTGATCCAACCGCAGGTTCCCC) and 23S rRNA (forward—GGTTAAGCGACTAAGCGTACACGGGTGGATG and reverse—AAGGTTAAGCCTCACGGTTCATTAGTACCG).
Ribosome profiling
Wild type (wt) and mutants of EngA were expressed in E. coli BL21 (DE3) as described above with the following modification. Before harvesting the cells, chloramphenicol was added to a final concentration of 100 µg/ml on ice for 10 min to trap the polysomes. Cells, harvested and washed with Buffer C (50 mM Tris–HCl pH 7.5, 10 mM MgCl2 and 50 mM NH4Cl), were lysed in an RNAse-free environment, in the same buffer with appropriate nucleotides (xanthine-5-[β,γ-imido]triphosphate (XMPPNP) (Jena Bioscience), GDP or/and GMPPNP (Sigma)) at 1 mM concentrations and with 50–350 mM NH4Cl, as the case may be. The lysate was centrifuged at 18 000×g for 30 min. The ribosomal subunits (A254 10 units) were resolved on a 17 ml continuous sucrose gradient (15–43%) in Buffer D (20 mM Tris–HCl pH 7.5, 10 mM MgCl2 and 50–350 mM NH4Cl). Gradient was subjected to ultra centrifugation at 28 000 RPM (Sorvall Sure-Spin 630 rotor) for 10 h. Thirty equal fractions (seven drops each) were collected from top to bottom using ISCO Gradient Maker, while monitoring A254. Fractions 5–30 (corresponding to lanes 1–26 in all Figures 1, 2 and 3) were resolved on SDS–PAGE followed by immuno-blotting using anti-GST antibodies (Santa Cruz Biotechnology) as all constructs of EngA described here contain an N-terminal GST fusion tag.
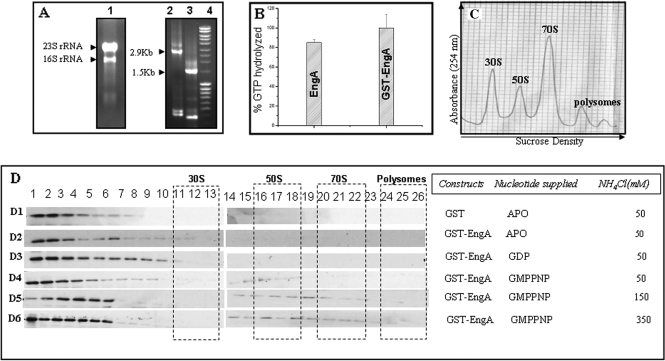
Figure 1.
EngA interacts with rRNA and ribosomal subunits. (A) 16S rRNA and 23S rRNA co-elute with EngA. The E. coli BL21 (DE3) cells overexpressing EngA were lysed in RNAse-free environment and EngA was purified as described in Materials and methods section. Lane 1 shows the rRNA following phenol–chloroform extraction of a fraction containing EngA. The identity of the rRNA was confirmed by RT-PCR using specific primers for 16S and 23S rRNA. Lanes 2 and 3 show the DNA fragments amplified from the co-eluted rRNA, using 23S rRNA and 16S rRNA primers, respectively. DNA ladder (Fermentas, Mass ruler SM0403) loaded in lane 4, suggest that the sizes of the amplified DNA fragments correspond to these rRNAs. (B) The ability of GST–EngA and EngA to hydrolyse GTP was analysed by using malachite green assay (see Materials and methods section). Bar plots indicate that the amount (%) of GTP hydrolysed by GST-tagged and untagged EngA are similar. (C) EngA–ribosome co-fractionation experiments were conducted using the same cells. Cells lysed either in absence of nucleotides or in presence of GDP or GMPPNP, were sedimented through a 15–43% sucrose gradient at 28 000 RPM (Sorvall Sure-spin 630 rotor) and fractions were collected while monitoring A254. A profile indicating the different ribosomal subunits is shown. (D) The fractions were analysed on a 12% SDS–PAGE and followed by immuno-blotting with anti-GST antibody. EngA does not associate with ribosomes (D2) in absence of nucleotides (Apo) or (D3) in presence of GDP. However, it associates with 50S ribosomal subunit in the presence of GMPPNP at (D4) 50 mM, (D5) 150 mM and also at (D6) 350 mM NH4Cl. The influence of the GST-tag on these interactions is negated in D1, where GST (lacking EngA) was used as a negative control in these experiments.
Figure 2.
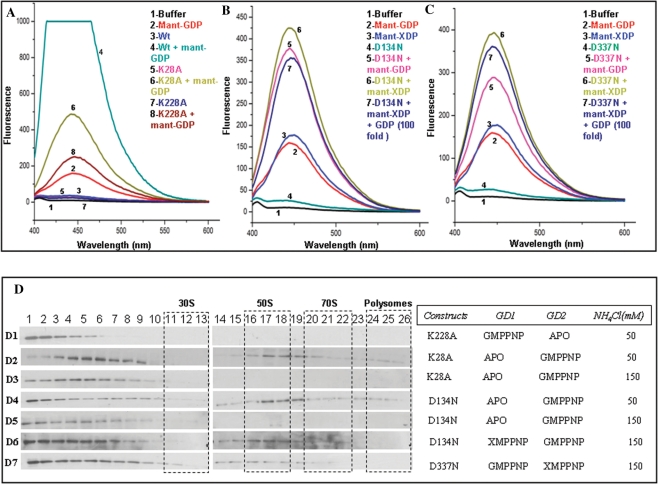
The importance of GD1 and GD2 in binding 50S. Nucleotide binding was assayed by recording emission spectra (λex 355 nm) (A–C) upon incubating 8 µM of protein with 0.4 µM fluorescent mant-nucleotides (mant-GDP and/or mant-XDP). Spectra corresponding to various nucleotide-bound forms of the protein are indicated in different colors. The protein–nucleotide combinations used are indicated in the insets in corresponding colors and numbers. (A) GDP binding to K28A and K228A mutants is reduced, when compared with the binding to wt-EngA, due to impaired nucleotide binding in the mutated G-domains. (B and C) Proteins carrying D134N and D337N mutations, preferentially bind mant-XDP even in presence of a 100-fold excess of GDP (over mant-XDP). The mutant proteins also bind mant-GDP since the mutation to preferentially bind xanthine nucleotides is created only in one of the G-domains, allowing the other to bind mant-GDP. (D) These mutants were subjected to ribosome co-fractionation experiments as in Figure 1D, with varying salt concentrations and nucleotides, as indicated. (D1) K228A mutant in presence of GMPPNP, which results in EngA[GTP:Apo] state, shows no interaction with ribosomal subunits at 50 mM NH4Cl. Whereas EngA[Apo:GTP] state, generated using K28A, shows an association with 50S (D2) at 50 mM NH4Cl which is abolished (D3) at 150 mM NH4Cl. EngA[Apo:GTP] state was also generated using the mutant D134N. An association with 50S seen (D4) at 50 mM NH4Cl, is abolished (D5) at 150 mM NH4Cl. EngA–50S interaction can be restored (D6) if XMPPNP, too, is provided to the D134N mutant to achieve EngA [GTP:GTP] state. (D7) This effect is verified using D337N mutant, which under similar conditions binds the 50S.
Figure 3.
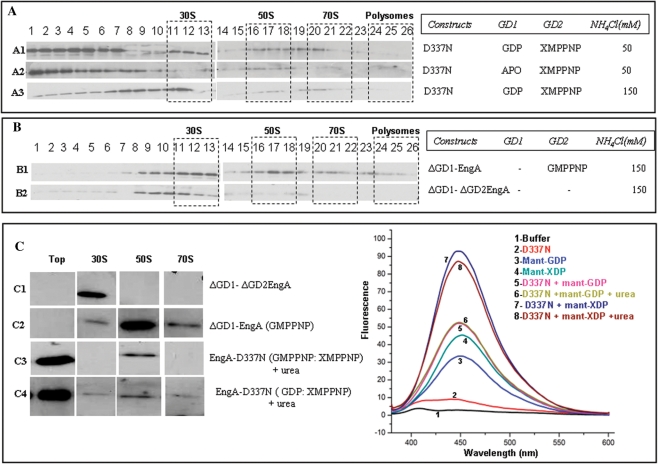
GD1 plays a role in EngA binding the 30S subunit. (A) The D337N mutant and constructs ΔGD1–EngA, ΔGD1–ΔGD2EngA were used in ribosome co-fractionation experiments as in Figure 1D. An EngA[GDP:GTP] state was achieved using D337N mutant, with GDP and XMPPNP. It associates with 50S, 30S and 70S subunits (A1) at 50 mM NH4Cl. In this mutant, when GD1 was not supplied with GDP, resulting in EngA[Apo:GTP] state, it showed no interaction (A2) with 30S. Stable interactions with 30S can be restored (A3) even at 150 mM NH4Cl, if GDP is supplied to GD2, i.e. EngA[GDP:GTP] state. (B) ΔGD1–EngA interacts with 30S, 50S and 70S in presence of GMPPNP at 150 mM NH4Cl (B1), while ΔGD1–ΔGD2EngA binds only the 30S (B2). (C) In the in vitro ribosome interaction assays, proteins were incubated with crude ribosome (five A254) and appropriate nucleotides as described in Materials and methods section. The reaction mixtures were resolved on a manually prepared 17 ml step gradient of sucrose and 30 fractions were collected from top to bottom while monitoring A254. RNA was isolated from each of the fractions and analysed by formaldehyde–agarose gel for the presence of 16S and 23S rRNA. Based on this, top fraction devoid of ribosomes, peak fractions corresponding to 30S, 50S and 70S subunits were analysed on a SDS–PAGE. Immuno-blots, using anti-GST antibodies, for the ‘top’, 30S, 50S and 70S fractions are shown. The ΔGD1–ΔGD2EngA associates with the 30S (C1) and ΔGD1–EngA with 30S, 50S and 70S (C2). Urea treated D337N mutant incubated with GMPPNP, XMPPNP, restored 50S interactions (C3) and also the interactions with 30S, 50S and 70S when incubated with GDP and XMPPNP (C4). Fluorescent nucleotide binding assays were conducted (as in Figure 2) with the D337N mutant in presence of various nucleotides and urea (1.5 M), as indicated. Emission spectra show that urea does not alter nucleotide binding to the protein.
Ribosome binding experiments using purified components
For Figure 3C, 100 pmol protein(s) were incubated with five A254 crude ribosomes in presence of appropriate nucleotides (1 mM) in Buffer C. The reaction mixtures were then resolved on a manually prepared 17 ml step gradient of sucrose in Buffer D. The gradient was subjected to ultra centrifugation at 28 000 RPM (Sorvall Sure-Spin 630 rotor) for 10 h. Thirty equal fractions (seven drops each) were collected from top to bottom using ISCO Gradient Maker, while monitoring A254. RNA was isolated from the fractions and was analysed by formaldehyde–agarose gel (1.5% agarose) for the presence of 16S and 23S rRNA. Based on this, peak fractions containing 30S, 50S and 70S subunits were analysed on a SDS–PAGE followed by immuno-blotting using anti-GST antibodies.
For Figure 4, ribosomes were purified from B. subtilis strain using the protocol employed for ribosome profiling. Fractions were analysed for the presence of various ribosomal subunits, as above. To generate a pure sample of the ribosomal subunits, only the peak fractions corresponding to the various ribosomal subunits were pooled, concentrated and were snap frozen in liquid nitrogen and stored at −80°C for further use.
Figure 4.
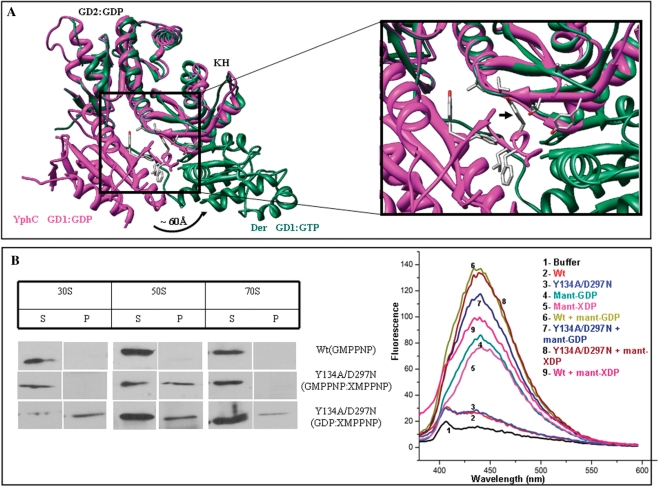
GD1–KH interactions are important for ribosome binding. (A) A structural superposition of EngA homologues, Der (PDB:1MKY) and YphC (PDB:2HJG), showing a 60 Å movement of GD1 between the GTP- and GDP-bound forms. Tyr134 of YphC, a key residue stabilizing the GD1–KH interface is shown by an arrow. (B) In vitro interactions with the purified 30S, 50S and 70S ribosomal subunits, as described in Materials and methods section were carried out using wt-His-YphC and the double mutant (Y134A, D297N). Immuno-blots using anti-His antibodies, for the supernatant and pellet fractions are shown. The wt-His-YphC did not display any interactions in presence of GMPPNP whereas the YphC double mutant restored interactions with the 50S in presence of GMPPNP and XMPPNP. Interaction of the double mutant with 30S, 50S and 70S was also found in presence of GDP and XMPPNP. Fluorescent nucleotide binding assays were carried out with wt-His-YphC and the double mutant (Y134A, D297N) in presence of mant-XDP and mant-GDP. Emission spectra confirms binding of mant-GDP to wt-His-YphC and both mant-GDP and mant-XDP to the double mutant.
The 100 pmol protein were incubated with five A254 of the purified ribosomal subunits in presence of appropriate nucleotides (1 mM) in Buffer C. The reaction mixtures were loaded on a 4 ml cushion of 25% sucrose in Buffer D and were then subjected to ultra centrifugation at 40 000 RPM (TH 660 rotor) for 2 h. The supernatant and pellet fractions were analysed for the presence of the protein by immuno-blotting using anti-His antibodies.
Structural superposition
Structures of B. subtilis YphC (PDB:2HJG) and T. maritima Der (PDB:1MKY) were superimposed using Coot (21). Figures 4A and 5C were prepared using CHIMERA (22).
Figure 5.
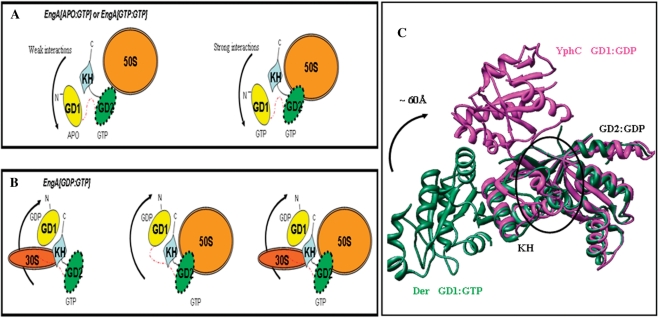
Two distinct ribosome-bound states of EngA. (A) The first state that only binds the 50S, is generated by EngA[Apo:GTP] or EngA[GTP:GTP]. (B) The second state that binds 50S, 30S, and 70S is generated by EngA[GDP:GTP]. (C) A structural superposition of EngA homologues, Der (PDB:1MKY) and YphC (PDB:2HJG), showing a 60 Å movement of GD1 between the GTP- and GDP-bound forms. Here, GD2 is bound to GDP. The RNA binding site proposed by Muench et al. (17) is indicated by a circle.
RESULTS
EngA binds the 50S subunit in the GTP-bound state
The E. coli RrmJ, a heat shock controlled rRNA methyltransferase modifies the 23S rRNA in intact 50S ribosomal subunits. The rrmJ null mutants lead to significant accumulation of ribosomal subunits at the expense of functional 70S particles, which are restored to normal levels upon overexpressing EngA (23). This suggests a functional relationship between EngA and 23S rRNA or the 50S subunit. We first wanted to examine if EngA binds 23S rRNA, as RrmJ does.
EngA, cloned in pQE2 and pET28A vectors to produce His6-tagged protein, led to highly insoluble aggregates, upon overexpression. The GST fused EngA proteins produced in pGEX vector, increased protein solubility significantly (see Materials and methods section). Hence, GST–EngA was used for further experimentation. The GST–EngA overexpressing (E. coli) cells were lysed in an RNase-free environment and the protein (GST–EngA) was purified by GSTrap affinity purification. When this was subjected to size-exclusion chromatography experiments, it eluted in the void volume fractions (data not shown). A phenol–chloroform extraction of the fractions containing GST–EngA suggests that both 23S rRNA and 16S rRNA co-elute with the protein (Figure 1A, lane 1). The identity of the rRNAs was verified using RT-PCR employing primers specific for 16S rRNA and 23S rRNA and the co-eluted rRNA as a template. In Figure 1A, lane 2 shows the DNA amplified using 23S rRNA primers and lane 3 shows the DNA amplified using 16S rRNA primers. Size of the amplified DNA fragments was found to be 2.9 and 1.5 kb, respectively, in accordance with the sizes of 23S and 16S rRNAs. Figure 1A thus reveals that not only 23S rRNA co-elutes with the protein, as we had anticipated, but also the 16S rRNA. In agreement with this observation, Hwang and Innoye (15) also found that der-depleted cells accumulate 16S and 23S rRNA precursors.
EngA belongs to the family of universally conserved bacterial GTPases, of which some like Era have been shown to bind the 16S rRNA, but not 23S rRNA (24). Most members of this family are now known to interact with ribosomal subunits in a specific nucleotide-bound state of the protein (4–15). These observations, together with the co-elution of 16S and 23S rRNA, suggested that EngA would bind the 30S and 50S ribosomal subunits. To verify ribosome binding, co-fractionation experiments were designed using cells overexpressing GST–EngA. Prior to this, to rule out any additional effect the bulky GST-tag may have on the biochemical behaviour of the protein, we analysed the enzymatic activity, i.e. the ability to hydrolyse GTP, of GST–EngA and EngA using the malachite green assay (20). Figure 1B shows that GTP hydrolysis by both GST-tagged and untagged EngA are similar.
The co-fractionation experiments designed to verify interactions of EngA with ribosomal subunits, were carried out with cells overexpressing the GST vector alone (as a negative control) and the cells overexpressing GST–EngA. The cells were lysed in the presence of appropriate nucleotides, whenever indicated. The immuno-blots in Figure 1D, using an anti-GST antibody, identify the protein present in the various fractions obtained following a sucrose density gradient centrifugation (see Materials and methods section). The negative control, i.e. cells expressing GST alone, shows no interaction with the ribosomal subunits (Figure 1D, D1) and thus negates the possibility of the GST-tag generating interactions with the ribosome. In addition, the pGEX vector employed here was modified to have a TEV protease cleavage site following the GST and thrombin cleavage site, resulting in a large linker region (∼15 residues) between GST and the protein. This allows sufficient domain movements between the two. Therefore, GST–EngA was employed for all further experiments. EngA, from here on, implies GST–EngA.
The experiment with cells overexpressing EngA reveals that it co-fractionates with the 50S subunit only in presence of non-hydrolysable GTP analogue, GMPPNP (Figure 1D, D4), but not GDP (Figure 1D, D3) or in absence of nucleotides (Figure 1D, D2). In concurrence with existing reports (13,15) EngA seems to be a GTP-dependent 50S binding protein. As the strength of ribosomal interactions can be assessed by increasing the amount of salt used in the co-fractionation experiments (9,25), we employed the same to assess EngA–50S interaction in presence of GMPPNP. Figure 1D (D5 and D6) show that the interactions are stable even at 150 and 350 mM NH4Cl.
While 50S binding accounts for 23S rRNA co-elution (Figure 1A), 16S rRNA co-elution remains unexplained. Hence, further experiments were designed to explore conditions that promote EngA–30S interactions.
GTP binding to GD2 is a primary requirement for ribosome binding
The two G-domains of EngA have contrasting nucleotide binding affinities and GTP hydrolysis rates. Robinson et al. (16) suggest that GD2 binds nucleotides with higher affinity and has slow GTP hydrolysis rates in comparison to GD1, which has lower nucleotide binding affinity and faster GTP hydrolysis rates. Hence, it could be that the two G-domains of EngA render distinct affects towards ribosome binding. To investigate the same, ribosome binding by EngA was tested while varying the nucleotide occupied states of the two G-domains. From hereon, for enhanced clarity, the GTP or GDP-bound states of GD1 and GD2 are denoted as GD1:GTP, GD1:GDP and GD2:GTP, GD2:GDP, respectively. Similarly, the nucleotide state of EngA, which is a consequence of the nucleotide-bound states of its two G-domains, is denoted as follows: for example, EngA[Apo:GTP] indicates that GD1 is in a nucleotide-free state and GD2 is in a GTP-bound state, while EngA[GDP:GTP] implies GD1 is in a GDP-bound state and GD2 is in a GTP-bound state, and so on.
In order to assess the significance of the two G-domains towards 50S binding, we generated single mutants K28A and K228A. These residues are present in the P-loops of GD1 and GD2, respectively. Typically, the lysine residues in the P-loops (GxxxxGKS/T) of GTPases coordinate the phosphates of GTP and GDP and their mutation is known to disrupt nucleotide binding (26,27). In the crystal structure of YphC (EngA homologue in B. subtilis) too, these lysines show similar interactions with the nucleotide (17). Hence, GD1 or GD2 can be arrested in a nucleotide-free state, by single mutants K28A or K228A, respectively, while simultaneously controlling the nucleotide-bound state of the other G-domain (without a mutation) by supplying appropriate nucleotides during cell lysis.
However, to confirm the nucleotide-free states of GD1 and GD2 in K28A and K228A mutants, respectively, fluorescence nucleotide binding experiments were carried out using mant-nucleotides. The mant-nucleotides are sensitive fluorescent probes which show an increase in fluorescence when present in a hydrophobic environment (28) i.e. when the mant-nucleotide binds to the protein in our experiments. Figure 2A shows a decrease in mant-GDP binding by mutants K28A and K228A in comparison to wt-EngA that has two intact G-domains to bind the nucleotide. Interestingly, mutant K28A, that disrupts nucleotide binding at GD1, shows a higher binding in comparison with K228A, in which binding is disrupted at GD2. This is in agreement with the reported higher nucleotide binding affinity of GD2 (16). These experiments confirm the nucleotide-free states generated by K28A and K228A.
Co-fractionation experiments were carried out with cells overexpressing mutants K28A or K228A, and lysed in presence of GMPPNP to ensure either the EngA[Apo:GTP] or EngA[GTP:Apo] state. Compared with the fluorescent nucleotide binding experiments where the protein is in excess (protein:nucleotide ratio 20:1), in all the co-fractionation experiments, the nucleotides (∼1 mM) were provided in excess (over the protein ∼µM) to ensure that a majority of protein molecules attain the desired nucleotide-bound states. Figure 2D (D1), corresponding to K228A mutant, representing EngA[GTP:Apo] state, in presence of 50 mM NH4Cl, shows no interactions with the ribosome. This suggests an importance of GD2:GTP in ribosome binding. In contrast, the K28A mutant (Figure 2D, D2) representing the converse state, i.e. EngA[Apo:GTP], shows an interaction with 50S at 50 mM NH4Cl. It is surprising that while the wt-protein retains 50S interaction even at 350 mM salt (Figure 1D, D6), K28A mutant representing EngA[Apo:GTP] state, shows no interaction at 150 mM NH4Cl (Figure 2D, D3), suggesting a weak interaction. Hence, these experiments suggest that it is GTP binding to GD2, but not to GD1, which is a primary requirement to observe any 50S interaction.
GTP binding to GD1 stabilizes 50S binding
The above data suggests that EngA–50S interaction requires a GD2:GTP state. However, the strong interaction seen for the wt-protein (Figure 1D, D5 and D6) suggests that the weak interactions found in EngA[Apo:GTP] state (Figure 2D, D2) may be further stabilized in the EngA[GTP:GTP] state, i.e. when GD1 also binds GTP. To substantiate this, we designed the following mutants that allow simultaneous variation of the nucleotide-bound states of GD1 and GD2, as the lysine mutants K28A or K228A only lead to a nucleotide-free (Apo) state.
Five conserved signature motifs (G1–G5) in the G-domain participate in stabilizing the bound nucleotide and its hydrolysis. Of these, a conserved Asp in G4 motifs (N/TKxD) renders specificity to bind guanine nucleotides by making bifurcated hydrogen bonds to the guanine base of GTP/GDP. In the structure of YphC too, the corresponding residues D134, D337 in G4 motifs make similar interactions with the guanine ring (17). Hence, D134N and D337N mutants of GD1 and GD2, respectively, were generated, as they would change specificity from guanine to xanthine nucleotides [(18) and references therein]. Like with the lysine mutants, D134N and D337N mutants were employed in fluorescent nucleotide binding experiments using mant-XDP and mant-GDP to verify nucleotide binding and change in specificity towards xanthine nucleotides. Figure 2B indicates D134N to bind mant-XDP at GD1, where the mutation was created and mant-GDP at GD2, which is not mutated. Similarly, Figure 2C indicates binding of mant-GDP at GD1 and mant-XDP at GD2, in D337N mutant. The selective binding to xanthine nucleotides by these mutants was further confirmed by measuring mant-XDP binding in presence of excess GDP (GDP:mant-XDP = 100:1). This does not affect mant-XDP binding significantly, as seen in Figure 2B and C. This verifies that the single mutants D134N and D337N allow for isolating nucleotide binding events at GD1 and GD2 and provide the ability to control the nucleotide states of both G-domains simultaneously, whereas wt-EngA preferentially binds mant-GDP (see Figure S1, Supplementary Material). In these, the domain carrying the mutation would bind xanthine nucleotides (when supplied) and allow guanine nucleotides to selectively bind the other.
When D134N mutant was tested for 50S binding (Figure 2D, D4) in a co-fractionation experiment carried out at 50 mM NH4Cl and in presence of GMPPNP but not XMPPNP (i.e. EngA[Apo:GTP] state), it shows interactions with 50S, like the K28A mutant in Figure 2D (D2). As anticipated, upon increasing NH4Cl concentration to 150 mM, the interaction (Figure 2D, D5) was completely abolished like in Figure 2D (D3). Interestingly, the 50S interaction can be restored, as in the wt-protein, if XMPPNP is also provided to achieve EngA[GTP:GTP] state (Figure 2D, D6). This restoration was also verified using D337N mutant in lieu of D134N in a similar experiment. Here too, 50S binding is restored (Figure 2D, D7) as above. These experiments lend support to the observations made from K28A, K228A mutants (Figure 2D, D1–D3), that GD2:GTP initiates EngA–50S interactions and GD1:GTP further stabilizes it.
The role of GD1 in binding specific ribosomal subunits
The above experiments show that both GD1:GTP and GD2:GTP are needed for efficient 50S binding. Since the D134N and D337N mutants additionally facilitate varying nucleotides occupied by the two G-domains, we questioned if doing so would alter ribosome binding. The EngA[GDP:GTP] analogous state was achieved using D337N mutant, in co-fractionation experiments containing GDP and XMPPNP. At 50 mM NH4Cl, this state, surprisingly, shows interactions not only with the 50S, but also with 30S and 70S subunits (Figure 3A, A1). To determine if the 30S and 70S interactions are specific to GD1:GDP, EngA[Apo:GTP] state was generated with the D337N mutant. Here, when GD1 is devoid of GDP, no interaction is observed with 30S and 70S (Figure 3A, A2), suggesting the importance of EngA[GDP:GTP] state for these interactions. These interactions could be restored upon supplying GDP at 150 mM NH4Cl too (Figure 3A, A3), indicating stable binding.
So far it appears that EngA, by employing the GD1:GDP state (Figure 3A) generates additional interactions for binding 30S. To further understand the role of GD1 in regulating EngA–ribosome interactions, we generated a construct ΔGD1–EngA, in which GD1 was truncated. When ΔGD1–EngA was employed in co-fractionation experiments, the protein bound 30S, 50S and 70S, even at 150 mM NH4Cl (Figure 3B, B1), like EngA[GDP:GTP)]. Interestingly, another construct, ΔGD1–ΔGD2EngA, where both GD1 and GD2 were truncated (i.e. only the KH-domain was present), largely bound the 30S (Figure 3B, B2). Possibly, this experiment suggests that KH-domain possesses sites for 30S interaction, and taken together with Figure 3A (A–A3), it appears that the 30S site is accessible only in the GD1:GDP state.
For ease of presentation, we define the EngA–ribosome interactions (discussed thus far) examined from the cells overexpressing EngA, as in vivo experiments. On the other hand, interactions described from now on, carried out using purified EngA and ribosomal subunits, would be referred as in vitro experiments.
Unfastening inter-domain interactions facilitates EngA–ribosome interactions
The in vitro EngA–ribosome interaction assays using purified EngA and ribosomal subunits, were carried out in presence of appropriate nucleotides. Surprisingly, the full length EngA constructs i.e. wt or the D to N mutants, failed to show any interaction in the EngA[GTP:GTP] or EngA[GDP:GTP] states. This led us to suspect the presence of an unknown factor in the cell lysate enabling these interactions in the in-vivo studies. To examine this, the whole cell lysate devoid of ribosomes, was added to the reaction mixtures. However, this too did not result in any EngA–ribosome association (data not shown).
Surprisingly, ΔGD1–ΔGD2EngA, where both GD1 and GD2 were truncated (i.e. only the KH-domain was present), or a construct ΔGD1–EngA, where GD1 was truncated, when incubated with the purified ribosomal subunits, interacted with specific ribosomal subunits, as in the in vivo assays (Figure 3B). In these studies, ΔGD1–ΔGD2EngA associated with the 30S and ΔGD1–EngA with 30S, 50S and 70S (Figure 3C, C1–C2). These experiments suggest that EngA needs to be devoid of GD1 to restore interactions with ribosomal subunits in the in vitro experiments. We reasoned that, in the full length protein, the inter-domain interactions provided by GD1 may lock the enzyme in a conformational state inappropriate for binding the ribosome. Often, mild concentrations of urea, guanidium hydrochloride or SDS loosen inter-domain interactions, which may be facilitated, in vivo, by a factor present in the cell lysate. To examine this, the full length D337N mutant was incubated on ice for 20 min with increasing concentrations of urea (from 500 mM to 2 M). From this, 100 pmol of protein was added to the crude ribosome in presence of appropriate nucleotides. The final concentration of urea in reaction mixtures, following this dilution, was ∼15–45 mM. Surprisingly, incubating D337N mutant in 1.5–2 M of urea, restored interactions with 50S in the EngA[GTP:GTP] state (Figure 3C, C3) and with 30S, 50S and 70S in the EngA[GDP:GTP] state (Figure 3C, C4). However, it appears from Figure 3C that only a small fraction of the protein molecules associate with the ribosomal subunits, as most of the protein remains in the top fractions. Perhaps this could be associated to the mild urea concentrations (∼15–45 mM) employed here. To clarify if urea interferes with nucleotide binding, fluorescent nucleotide binding assays were conducted in the presence and absence of 1.5 M urea. Emission spectra show that guanine and xanthine nucleotide binding to the protein (D337N mutant) remains unaltered even at 1.5 M urea (Figure 3C).
Role of GD1–KH interactions in ribosome association
In vitro experiments with urea (Figure 3C) made us examine interactions at the domain interfaces of EngA, which may be disrupted by urea to achieve an appropriate conformational state of EngA that binds the ribosome. In the crystal structure of YphC (EngA homologue in B. subtilis), GD1, bound to GDP, provides hydrophobic interactions for the GD1–KH interface (Figure 4A). GTP binding to GD1 disrupts these interactions as evident from the large conformational change (∼60 Å) in the position of GD1 (Figure 4A). This hydrophobic nature of GD1–KH interface is conserved in several homologs of EngA. We, therefore, reasoned that in the aforementioned in vitro experiments (Figure 3C), urea may disrupt these interactions at GD1–KH interface and allow ribosome binding. To examine this, Tyr134, a key residue contributed by GD1 to this interface (Figure 4A), was mutated (see Figure S2, Supplementary Material). A His-YphC double mutant (Y134A, D297N) and the wt-His-YphC were employed in the in vitro interactions. In YphC, D297 is identical to D337 of E. coli EngA and hence its mutation to N would alter the specificity of GD2 from guanine to xanthine. When wt-HisYphC was incubated with purified ribosomal subunits, it did not display any interactions in EngA[GTP:GTP] state as noted for GST–EngA in the in vitro experiments. As anticipated, the YphC double mutant restored interactions with the 50S in EngA[GTP:GTP] state and also with 30S, 50S and 70S in the EngA[GDP:GTP] state (Figure 4B). These results are in accordance with the in vivo interactions displayed by GST–EngA (Figures 1D and 3A). The mant-XDP and mant-GDP binding to wt-HisYphC and the double mutant were verified, as shown in Figure 4B.
DISCUSSION
EngA is essential for bacterial cell growth. Cells depleted with engA show slower growth patterns (29). Current reports show that it interacts with the ribosome (13,15) and participates in its biogenesis (14). EngA is particularly interesting and distinctive due to the two tandem G-domains, GD1 and GD2, it possesses, apart from the KH-domain. The results provided here demonstrate that by the use of distinct nucleotide-bound states of the two G-domains, EngA regulates ribosome binding.
A previous report has shown that a null mutant of rrmJ, an rRNA methylase, displays an increase in the level of unassembled 30S and 50S subunits and a decrease in 70S. This effect was restored upon overexpressing EngA or Obg (23), indicating their role in ribosome assembly (formation of 70S). Apart from this, Obg and EngA are known to be involved in ribosome biogenesis (7,14) and Obg is also known to bind both 30S and 50S subunits (7). Similarly, we found EngA to co-elute with both 23S and 16S rRNA (Figure 1A), although RrmJ only binds and modifies the 23S rRNA. However, like other investigators (13,15), we too found EngA to interact only with the 50S subunit in a GTP dependent manner, but not with the 30S (Figure 1D, D4–D6). 50S interaction accounts for the co-elution of 23S rRNA, while 16S rRNA co-elution remains elusive. Driven by these facts, we reasoned that EngA would bind both 50S and 30S ribosomal subunits if appropriate conditions were provided and that the two G-domains have a role in generating these.
The importance of the G-domains is underscored by their contrasting GTP binding and hydrolyzing activities (16). Also, Brown and co-workers (13) suggest that both G-domains play critical roles for the cellular function of EngA. By using single mutants K28A or K228A that arrest GD1 or GD2, respectively, in a nucleotide-free state (Figure 2A), we deduce that GTP binding to GD2 is a primary requirement for any EngA–50S interaction (Figure 2D, D1–D2).
The significance of GD1 is revealed in experiments where it is held in a GTP-bound state. Using salt-sensitivity of an interaction as an estimated indicator of the binding strength (9,25), we infer that a weak interaction with 50S in EngA[Apo:GTP] state, is further strengthened in EngA[GTP:GTP] state (Figure 2D). It appears that 50S interactions are initiated by the GD2:GTP conformation, but both GD1:GTP and GD2:GTP result in an efficient and sustained EngA–50S interaction.
The importance of nucleotide occupancies of the two G-domains is further revealed by mutants D134N and D337N, which allow specific binding to xanthine nucleotides (Figure 2B and C) (18). By segregating nucleotide binding events at the two G-domains using guanine and/or xanthine nucleotides with these mutants, we could identify conditions where EngA not only binds the 50S subunit but also the 30S (Figure 3A, A1–A3). The 30S interaction thus rationalizes 16S rRNA co-elution found in Figure 1A. In addition, it also accounts for the recent observation that EngA interacts with the structural protein S7 of the 30S subunit (30). However, the most significant finding based on these experiments is that EngA exists in two distinct ‘ribosome-bound states’ as depicted in Figure 5. The first state (Figure 5A) provided by EngA[GTP:GTP)] or EngA[Apo:GTP] binds 50S, (Figure 2D) and the second (Figure 5B) provided by EngA[GDP:GTP] binds 30S, 50S and 70S (Figure 3A). These two ribosome-bound states of EngA are distinguished by the GTP and GDP-bound forms of GD1 indicating an important regulatory role for it.
A recently determined crystal structure of YphC reveals a large conformational change (∼60 Å) of GD1 between its GTP and GDP-bound forms (Figure 5C) (17). Based on structural similarity to the C-terminal domain of S3 ribosomal protein, the authors define an ON state for EngA, where it binds rRNA via a region contributed by KH-domain and GD2 (indicated by a circle, Figure 5C). They speculate that EngA switches to an OFF state in GD1:GDP and results in a large conformational change (of GD1), which makes this region inaccessible to RNA. The proposed ON state is EngA[GTP:GDP] and the OFF state is EngA[GDP:GDP]. In contrast, our work shows that when GD2 is devoid of GTP, EngA does not bind any of the ribosomal subunits. Given this, the implications of the structural similarity and the proposed rRNA binding site observed by Muench et al. (17) are intriguing. Interestingly, we find that when GD1 is truncated, the construct ΔGD1–EngA binds 30S, 50S and 70S subunits (Figure 3C, C1), essentially behaving like the EngA[GDP:GTP] state. Hence, this construct seems to provide analogous interactions and accomplish a state similar to the ‘second ribosome-bound state’ of EngA. However, ΔGD1–ΔGD2EngA, where both GD1 and GD2 are truncated, binds the 30S, implying that KH-domain alone is sufficient to provide the 30S binding interactions (Figure 3C, C2). As ΔGD1–EngA and EngA[GDP:GTP] bind 30S, while EngA[GTP:GTP] does not, it is possible that GD1, upon GTP binding undergoes a conformational change (see arrows in Figure 5A) and masks the 30S site on the KH-domain.
In addition to this, in vitro experiments, carried out using E. coli EngA and B. subtilis YphC, indicate a need for the disruption of GD1–KH interface to promote ribosome binding, together with GTP hydrolysis at GD1. The wt-proteins, GST–EngA and His-YphC, fail to interact with ribosomal subunits, unless supplied with a mild concentration of urea (Figure 3C) or till the inter-domain interactions are compromised by mutating Tyr134 at the GD1–KH interface (Figure 4). Perhaps, this indicates the need for an additional factor in the cellular environment to facilitate the disruption of GD–KH interface and appropriately position GD1 to promote ribosome binding.
Together with the inferences drawn based on this study, and the available structural data (16,17), the rRNA binding proposed by Muench et al. may be further clarified as follows. Assuming there would be no large conformational change between GD2:GTP and GD2:GDP, the site proposed by Muench et al. is likely responsible for 50S binding, as it is proposed to be at the interface of GD2 and KH (17). Furthermore, GD1 makes use of GTP binding to mask the 30S binding site provided by the KH-domain and also stabilizes 50S interaction. The GTP hydrolysis at GD1 would then result in unmasking the 30S binding site (see arrows in Figure 5B). On the other hand, Muench et al. do not find any other exposed and conserved region to bind rRNA (17) that can account for a 30S site, necessitating further crystal structure analysis.
Our work, for the first time, presents evidence that EngA binds both 50S and 30S, and most importantly, depicts two distinct conformational states of EngA with varied specificity for ribosomal subunits. Taken together with the large conformational change in GD1 (17) and the role of EngA in ribosome biogenesis (14), it is tempting to speculate that EngA, having completed the 50S assembly in the EngA[GTP:GTP] state, promotes 50S–30S interaction in the EngA[GDP:GTP] state to assemble the complete 70S particles.
Evidently, rigorous experimentation will be needed to examine the biological significance of GD1 in enabling EngA to switch between the two ‘ribosome-bound states’. Determining the crystal structures of EngA in the various nucleotide states of its G-domains could relate the large conformational changes to the function of the protein, i.e. ribosome biogenesis and/or assembly. Our attempts so far have led to poorly diffracting crystals and efforts are underway to improve their quality to elucidate the structural mechanisms prevalent in EngA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wellcome Trust, UK (in the form of International Senior Research Fellowship to B.P., grant number 73616); Department of Biotechnology, Government of India; CARE funds, IIT Kanpur. Funding for open access charge: Wellcome Trust,UK.
Conflict of interest statement. None declared
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof S. Sarkar and Mr M. Saxena for sharing facilities to carry out Fluoroscence nucleotide binding studies. We thank Neeraj Kumar, Deepak Joshi and Navdeep Yadav for excellent technical and secretarial assistance. We also thank S. K. Verma, S. Mathew, A. Baskaran, D. Dey, Drs A. Bandopadhaya, S. Ramaswamy and V. K. Nandicoori for constructive criticism and suggestions. S.K.T. acknowledges Ministry of Human Resource Development (MHRD) India and N.D. acknowledges Council of Scientific and Industrial Research (CSIR) for financial assistance.
REFERENCES
- 1.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 2.Sprang SR. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 3.Mohr D, Wintermeyer W, Rodnina MV. GTPase activation of elongation factors Tu and G on the ribosome. Biochemistry. 2002;41:12520–12528. doi: 10.1021/bi026301y. [DOI] [PubMed] [Google Scholar]
- 4.Caldon CE, March PE. Function of the universally conserved bacterial GTPases. Curr. Opin. Microbiol. 2003;6:135–139. doi: 10.1016/s1369-5274(03)00037-7. [DOI] [PubMed] [Google Scholar]
- 5.Anand B, Verma SK, Prakash B. Structural stabilization of GTP-binding domains in circularly permuted GTPases: implications for RNA binding. Nucleic Acids Res. 2006;34:2196–2205. doi: 10.1093/nar/gkl178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sayed A, Matsuyama S, Inouye M. Era, an essential Escherichia coli small G-protein, binds to the 30S ribosomal subunit. Biochem. Biophys. Res. Commun. 1999;264:51–54. doi: 10.1006/bbrc.1999.1471. [DOI] [PubMed] [Google Scholar]
- 7.Sato A, Kobayashi G, Hayashi H, Yoshida H, Wada A, Maeda M, Hiraga S, Takeyasu K, Wada C. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells. 2005;10:393–408. doi: 10.1111/j.1365-2443.2005.00851.x. [DOI] [PubMed] [Google Scholar]
- 8.Jain N, Dhimole N, Khan AR, De D, Tomar SK, Sajish M, Dutta D, Parrack P, Prakash B. E. coli HflX interacts with 50S ribosomal subunits in presence of nucleotides. Biochem. Biophys. Res. Commun. 2008;379:201–205. doi: 10.1016/j.bbrc.2008.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daigle DM, Brown ED. Studies of the interaction of Escherichia coli YjeQ with the ribosome in vitro. J. Bacteriol. 2004;186:1381–1387. doi: 10.1128/JB.186.5.1381-1387.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell TL, Daigle DM, Brown ED. Characterization of the Bacillus subtilis GTPase YloQ and its role in ribosome function. Biochem. J. 2005;389:843–852. doi: 10.1042/BJ20041873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uicker WC, Schaefer L, Britton RA. The essential GTPase RbgA (YlqF) is required for 50S ribosome assembly in Bacillus subtilis. Mol. Microbiol. 2006;59:528–540. doi: 10.1111/j.1365-2958.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 12.Uicker WC, Schaefer L, Koenigsknecht M, Britton RA. The essential GTPase YqeH is required for proper ribosome assembly in Bacillus subtilis. J. Bacteriol. 2007;189:2926–2929. doi: 10.1128/JB.01654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bharat A, Jiang M, Sullivan SM, Maddock JR, Brown ED. Cooperative and critical roles for both G domains in the GTPase activity and cellular function of ribosome-associated Escherichia coli EngA. J. Bacteriol. 2006;188:7992–7996. doi: 10.1128/JB.00959-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaefer L, Uicker WC, Wicker-Planquart C, Foucher AE, Jault JM, Britton RA. Multiple GTPases participate in the assembly of the large ribosomal subunit in Bacillus subtilis. J. Bacteriol. 2006;188:8252–8258. doi: 10.1128/JB.01213-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang J, Inouye M. The tandem GTPase, Der, is essential for the biogenesis of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 2006;61:1660–1672. doi: 10.1111/j.1365-2958.2006.05348.x. [DOI] [PubMed] [Google Scholar]
- 16.Robinson VL, Hwang J, Fox E, Inouye M, Stock AM. Domain arrangement of Der, a switch protein containing two GTPase domains. Structure. 2002;10:1649–1658. doi: 10.1016/s0969-2126(02)00905-x. [DOI] [PubMed] [Google Scholar]
- 17.Muench SP, Xu L, Sedelnikova SE, Rice DW. The essential GTPase YphC displays a major domain rearrangement associated with nucleotide binding. Proc. Natl Acad. Sci. USA. 2006;103:12359–12364. doi: 10.1073/pnas.0602585103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shan SO, Walter P. Induced nucleotide specificity in a GTPase. Proc. Natl Acad. Sci. USA. 2003;100:4480–4485. doi: 10.1073/pnas.0737693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLivron MA, Robinson VL. Salmonella enterica serovar Typhimurium BipA exhibits two distinct ribosome binding modes. J. Bacteriol. 2008;190:5944–5952. doi: 10.1128/JB.00763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baykov AA, Evtushenko OA, Avaeva SM. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal. Biochem. 1988;171:266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallog. Sect. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 23.Tan J, Jakob U, Bardwell JC. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 2002;184:2692–2698. doi: 10.1128/JB.184.10.2692-2698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hang JQ, Meier TI, Zhao G. Analysis of the interaction of 16S rRNA and cytoplasmic membrane with the C-terminal part of the Streptococcus pneumoniae Era GTPase. Eur. J. Biochem. 2001;268:5570–5577. doi: 10.1046/j.1432-1033.2001.02493.x. [DOI] [PubMed] [Google Scholar]
- 25.Hesterkamp T, Hauser S, Lutcke H, Bukau B. Escherichia coli trigger factor is a prolyl isomerase that associates with nascent polypeptide chains. Proc. Natl Acad. Sci. USA. 1996;93:4437–4441. doi: 10.1073/pnas.93.9.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 27.Van der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J. Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remmers AE, Posner R, Neubig RR. Fluorescent guanine nucleotide analogs and G protein activation. J. Biol. Chem. 1994;269:13771–13778. [PubMed] [Google Scholar]
- 29.Hwang J, Inouye M. An essential GTPase, Der, containing double GTP-binding domains from Escherichia coli and Thermotoga maritime. J. Biol. Chem. 2001;276:31415–31421. doi: 10.1074/jbc.M104455200. [DOI] [PubMed] [Google Scholar]
- 30.Lamb HK, Thompson P, Elliott C, Charles IG, Richards J, Lockyer M, Watkins N, Nichols C, Stammers DK, Bagshaw CR, et al. Functional analysis of the GTPases EngA and YhbZ encoded by Salmonella typhimurium. Protein Sci. 2007;16:2391–402. doi: 10.1110/ps.072900907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.