Summary
Drosophila HNF4 (dHNF4) is the single ancestral ortholog of a highly conserved subfamily of nuclear receptors that includes two mammalian receptors, HNFα and HNFγ, and 269 members in C. elegans. We show here that dHNF4 null mutant larvae are sensitive to starvation. Starved mutant larvae consume glycogen normally, but retain lipids in their midgut and fat body, and have increased levels of long chain fatty acids, suggesting that they are unable to efficiently mobilize stored fat for energy. Microarray studies support this model, indicating reduced expression of genes that control lipid catabolism and β-oxidation. A GAL4-dHNF4;UAS-lacZ ligand sensor can be activated by starvation or exogenous long chain fatty acids, suggesting that dHNF4 is responsive to dietary signals. Taken together, our results support a feed-forward model for dHNF4, in which fatty acids released from triglycerides activate the receptor, inducing enzymes that drive fatty acid oxidation for energy production.
Keywords: lipid metabolism, transcription, mitochondria, gene regulation, nuclear receptor signaling
Introduction
Nuclear receptors are ligand-regulated transcription factors that act at the interface between chemical signaling and transcriptional control. They are defined by a conserved zinc finger DNA binding domain (DBD) and a C-terminal ligand binding domain (LBD) that can impart multiple functions, including hormone binding, receptor dimerization, and transactivation. Nuclear receptors are regulated by a wide range of lipophilic compounds that include dietary signals and metabolic intermediates, such as sterols, fatty acids, and bile acids. Binding of these compounds to the LBD triggers a conformational change that switches the regulatory function of the receptor, directing coordinate changes in downstream target gene expression. The resulting transcriptional programs often drive feedback or feed-forward pathways that act upon the specific class of compounds bound by the receptor (Chawla et al., 2001). Functional studies of several nuclear receptor subclasses, including ERR, LXR, and PPAR, have defined roles for these factors in regulating mitochondrial function, sterol homeostasis, and lipid metabolism – pathways that are central to human health, as manifested by the alarming rise in diabetes and obesity among human populations worldwide.
In this study, we focus on Hepatic Nuclear Factor 4 (HNF4), a nuclear receptor subclass that is represented by two paralogs in mammals, HNF4α and HNF4γ. Mutations in human HNF4α are associated with MODY1, an autosomal dominant genetic disorder that is characterized by early onset type 2 diabetes (Yamagata et al., 1996). MODY1 patients are hyperglycemic and hypoinsulinemic, have reduced levels of circulating lipids, and display defects in the expression of genes involved in glucose and lipid metabolism (Stoffel and Duncan, 1997; Shih et al., 2000). Interestingly, similar phenotypes have been observed in HNF4α mutant mice. Targeted disruption of HNF4α leads to early embryonic lethality with defects in the expression of visceral endoderm secretory proteins that are required for maintaining gastrulation (Chen et al., 1994; Duncan et al., 1997). Tissue-specific loss of HNF4α in the mouse liver results in increased lipid deposits in hepatocytes, hepatomegaly, and early lethality (Hayhurst et al., 2001). Circulating cholesterol, phospholipid, and triglyceride levels are reduced in these animals, consistent with the decreased expression of hepatic genes involved in lipid metabolism and transport. An HNF4γ null mutant mouse strain has been reported as viable with slightly increased body weight, decreased food intake, and decreased nighttime activity, leaving it unclear what contribution this paralog might have to overall HNF4 function (Gerdin et al., 2006).
Remarkably, the HNF4 family underwent a significant expansion during the evolution of nematodes, resulting in 269 family members in C. elegans (Robinson-Rechavi et al., 2005). One of these, NHR-49, has been characterized in detail (Van Gilst et al., 2005a). Like liver-specific mouse HNF4α mutants, nhr-49 mutants accumulate fat and display early lethality. By examining the expression of 65 genes involved in energy metabolism, significant effects were observed in mitochondrial β-oxidation, the pathway by which fatty acids are utilized for energy production, and fatty acid desaturation, increasing the ratio of stearic acid to oleic acid in mutant worms. Overexpression of a mitochondrial acyl-CoA synthetase that is down-regulated in nhr-49 mutants was sufficient to suppress the high fat phenotype, suggesting that this defect arises from reduced levels of β-oxidation.
Biochemical studies have provided insights into the molecular mechanisms of HNF4 function. The basal transcriptional activity of HNF4 can be enhanced by fatty acyl-CoA thioesters, suggesting that HNF4 may be modulated by a fatty acid-derived ligand (Hertz et al., 1998). Structural analysis of the HNF4α and HNF4γ LBD revealed C14–C18 long chain fatty acids (LCFAs) almost filling a well-defined hydrophobic pocket, suggesting that they are natural ligands for the receptor (Dhe-Paganon et al., 2002; Wisely et al., 2002). These fatty acids, however, are tightly bound and could not be dissociated from the receptor under non-denaturing conditions (Wisely et al., 2002). Mutating conserved amino acids within the LBD that disrupt fatty acid binding significantly decreases HNF4 transcriptional activity, suggesting that this interaction is essential for receptor function (Wisely et al., 2002; Aggelidou et al., 2004). Both active and inactive LBD conformations of HNF4α have bound LCFAs, implying that another mechanism is required for HNF4 transcriptional activation (Dhe-Paganon et al., 2002). Taken together, these data support a model whereby HNF4, with a tightly bound LCFA, acts as a constitutive transcriptional activator, although regulation by posttranslational modifications, cofactor interactions, or ligand exchange in vivo, remains a possibility.
The presence of multiple HNF4 paralogs in mice and C. elegans has complicated our understanding of the physiological functions of this nuclear receptor subclass. In contrast, only a single HNF4 ortholog is encoded in the Drosophila genome, with 89% identity to HNF4α in its DBD and 61% identity in its LBD. Moreover, most of the basic metabolic pathways and regulatory interactions that maintain homeostasis are conserved in the fly, providing an ideal context for genetic and biological studies of HNF4 function (Baker and Thummel, 2007; Leopold and Perrimon, 2007). Here we show that dHNF4 null mutant larvae are sensitive to starvation. This starvation sensitivity is associated with a reduced ability to generate energy from stored lipid, as manifested by increased levels of triglycerides and LCFAs in starved animals, along with significant reduction in the transcription of genes involved in lipolysis and β-oxidation. In addition, we show that the HNF4 LBD changes it activation status during development, and can be activated by starvation or exogenous LCFAs. Taken together, our data defines the ancestral function of HNF4 as a key regulator of lipid mobilization and β-oxidation in response to nutrient deprivation.
Results
dHNF4 is expressed in the major tissues that control metabolism
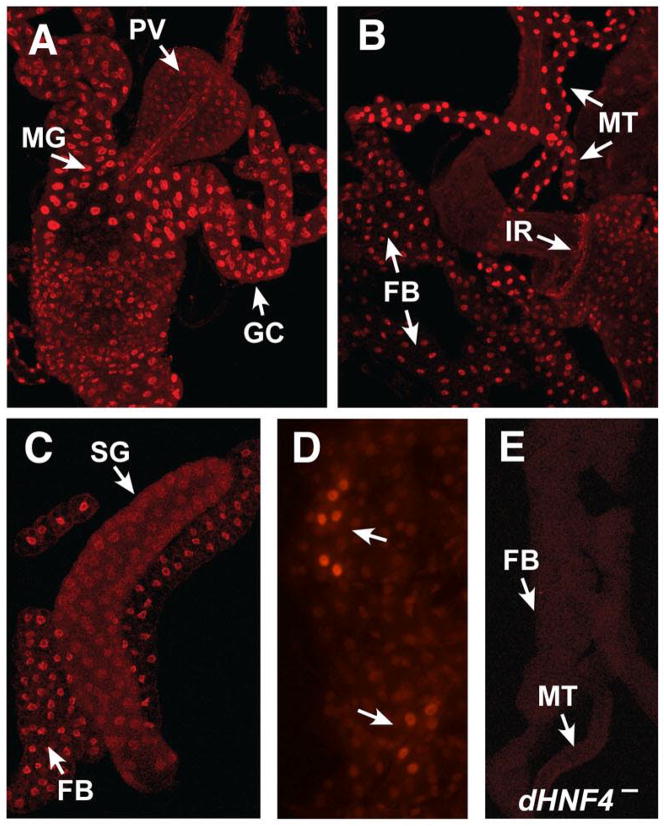
As a first step toward dHNF4 functional characterization, we used affinity purified antibodies to determine its spatial pattern of expression in organs dissected from third instar larvae. dHNF4 protein is restricted to the nucleus, consistent with its role as a transcription factor (Figure 1). It is most highly expressed in four tissues: the midgut and attached gastric caeca (Figure 1A), the fat body (Figure 1B,C), the Malpighian tubules (Figure 1B), and the oenocytes (Figure 1D). The gastric caeca are four anterior projections off the midgut that provide a primary site for digestive enzyme secretion and absorption of nutrients into the circulatory system. Nutrients are metabolized and stored, primarily as glycogen and triglycerides (TAG), in the fat body, the insect equivalent of the mammalian liver and white adipose tissue. Lipid mobilization is facilitated by the oenocytes, which are small clusters of cells that are embedded within the epidermis (Gutierrez et al., 2007). Waste products are transferred back into the circulatory system and absorbed by the Malpighian tubules, the principal osmoregulatory and excretory organ of the insect. Low levels of dHNF4 expression were detected in the proventriculus (Figure 1A), salivary glands (Figure 1C), epidermis (Figure 1D), brain, and ring gland (Supplemental Figure 1). dHNF4 was not detected in imaginal discs, although it is expressed in the ring of imaginal cells that will develop into the adult hindgut during metamorphosis (Figure 1B). dHNF4 is also not detected in the median neurosecretory cells that produce insulin-like peptides (IPCs, Supplemental Figure 1). A similar overall spatial distribution of dHNF4 expression is seen in embryos by in situ hybridization (Zhong et al., 1993), suggesting that this represents a stable pattern of expression. Northern blot analysis indicates that dHNF4 is expressed at a constant level throughout development (Sullivan and Thummel, 2003).
Figure 1. dHNF4 is expressed in tissues that control metabolic homeostasis.
Affinity-purified antibodies directed against dHNF4 were used to determine its spatial pattern of expression in organs dissected from third instar larvae. (A) dHNF4 protein is localized to the nucleus and is present at relatively high levels in the gastric caeca (GC) and anterior midgut (MG) of the digestive system, with lower levels in the proventriculus (PV). (B,C) Relatively high levels of expression are also seen in the fat body (FB), Malpighian tubules (MT), and hindgut imaginal ring (IR), with lower levels of expression in the salivary gland (SG). (D) dHNF4 is highly expressed in the larval oenocytes (arrows), with low level expression in the surrounding epidermis. (E) No expression is detectable in tissues dissected from dHNF4 mutant tissues, indicating that the antibody is specific for dHNF4 protein.
dHNF4 mutants are sensitive to starvation
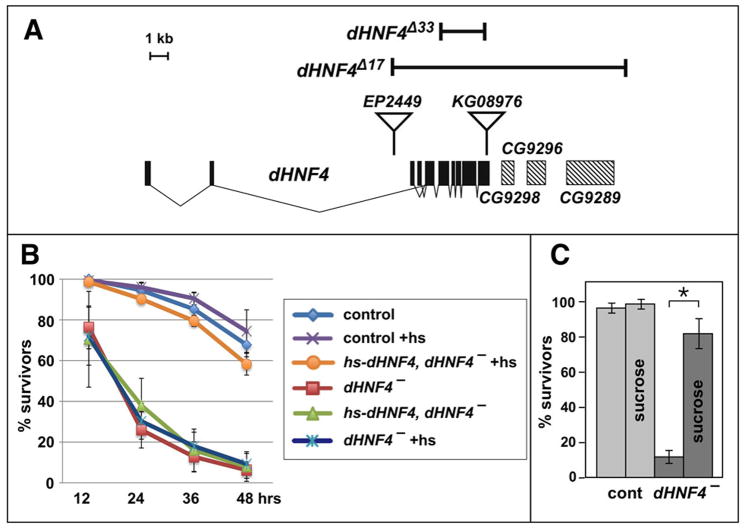
Imprecise excision of two P-element insertions in dHNF4 was used to create deletions for the locus (Figure 2A). A 8193 bp deletion of EP2449, designated dHNF4Δ17, removes most of the dHNF4 coding region along with three adjacent genes. Imprecise excision of KG08976 resulted in the isolation of dHNF4Δ33, a 1939 bp deletion specific to dHNF4 that removes sequences encoding most of the DBD and the entire LBD. A transheterozygous combination of these two alleles, dHNF4Δ33/dHNF4Δ17, was used for all functional studies of dHNF4. These mutants have no detectable dHNF4 protein, as expected for a specific dHNF4 null allele. (Figure 1E). Larvae that were transheterozygous for precise excisions of the EP2449 and KG08976 P-elements were used as genotypically matched controls.
Figure 2. dHNF4 null mutants are sensitive to starvation.
(A) A schematic representation of the dHNF4 locus is depicted, with the gene structure shown below along with three 3′-flanking genes, CG9298, CG9296, and CG9289. The locations of the two P-element insertions in dHNF4, EP2449 and KG08976 are shown, along with the dHNF4Δ17 and dHNF4Δ33 deletions. (B) A time course of the viability of starved control and dHNF4 mutant larvae, with and without a wild type hs-dHNF4 transgene. Whereas most larvae that are transheterozygous for precise excisions of the EP2449 and KG08976 P-elements (control) can survive two days of starvation, most dHNF4Δ33/dHNF4Δ17 mutants (dHNF4−) die. Heat treatment (+hs) has no effect on these animals. Heat treatment of dHNF4Δ33/dHNF4Δ17 mutants that carry a hs-dHNF4 rescue construct (hs-dHNF4, dHNF4− +hs), however, regain their starvation resistance. (C) Control (cont) or dHNF4 mutant (dHNF4−) late second instar larvae were either maintained on moist filter paper or on a 20% sucrose diet (sucrose) for two days at 25°C, after which the percent of surviving animals was scored and plotted. Whereas dHNF4 mutant larvae are sensitive to starvation, they survive normally on a sucrose diet. A representative of four independent experiments is presented, with N=10–40 animals in each experiment. Error bars are +/− S.E. * p = 1.3×10−3.
dHNF4 mutants maintained under normal culture conditions progress through development until adult eclosion, when many of the animals die with their head protruding from the pupal case. The remaining dHNF4 mutant adults die within a day of eclosion. Maintaining dHNF4 mutants at low density under ideal culture conditions, however, results in many animals surviving this lethal period and developing into morphologically normal adults. This dependence of dHNF4 mutant lethality on culture conditions suggests that this phenotype arises, at least in part, from metabolic defects due to the nutritional status of the animal.
As a first step toward assessing possible metabolic functions of dHNF4, we examined the ability of mutants to survive a period of starvation. Whereas most control larvae survive two days in the absence of nutrients, almost all dHNF4 mutants succumb during this period (Figure 2B). This sensitivity can be rescued by a heat-inducible wild type dHNF4 transgene (hs-dHNF4), indicating that it is specific to dHNF4 (Figure 2B). Significant rescue can also be achieved with expression of wild-type dHNF4 in the midgut or fat body, but not the ring gland, oenocytes, or IPCs (Supplemental Figure 2). Importantly, dHNF4 mutants maintain the ability to survive on a sugar diet (Figure 2C). This suggests that these mutant larvae develop normally and that their basic metabolic activities (glycolysis and downstream mitochondrial energy producing pathways) remain functional.
dHNF4 mutants display defects in lipid metabolism
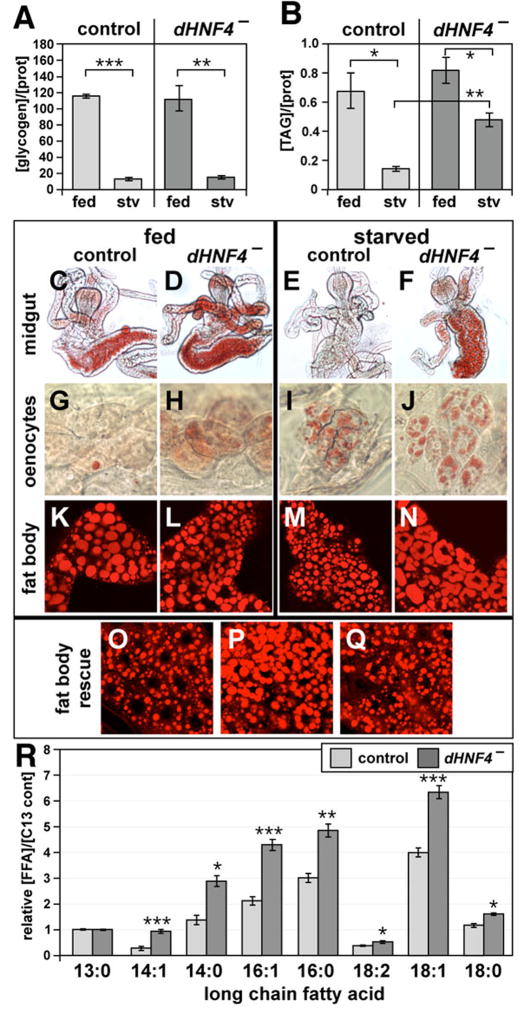
In order to identify possible causes for the starvation sensitivity of dHNF4 mutants, glycogen and TAG levels were measured in control and dHNF4 mutants that were either fed or starved for one day (Figure 3A,B). Levels of both metabolites did not change significantly in fed control and mutant larvae, consistent with the apparently normal developmental progression of these animals. In addition, glycogen appears to be consumed normally in dHNF4 mutants upon starvation (Figure 3A). Starved dHNF4 mutants, however, have significantly higher levels of TAG than starved control larvae (Figure 3B). This observation suggests that dHNF4 mutants are unable to properly mobilize their lipid stores upon nutrient deprivation. Starvation-induced autophagy, however, is induced normally in dHNF4 mutant larvae (Supplemental Figure 3) (Britton et al., 2002; Rusten et al., 2004).
Figure 3. Starved dHNF4 mutants have excess stored lipid and increased free fatty acids.
Late second instar larvae that were transheterozygous for precise excisions of the EP2449 and KG08976 P-elements (control) or dHNF4Δ33/dHNF4Δ17 mutant larvae (dHNF4−) were either fed or starved for 24 hrs, after which homogenates were subjected to (A) glycogen or (B) TAG assays. Amounts of glycogen (in μg) and TAG (in mg) were normalized to the amount of protein (in mg), shown on the y axis. Glycogen amounts drop to similar levels in starved control and dHNF4 mutant larvae, while TAG levels remain relatively high in starved dHNF4 mutants. N=25 animals for each sample, with each experiment using six samples. A representative of four (glycogen) or six (TAG) experiments is depicted. Error bars are +/− S.E. *p<0.01, **p<1×10−4, ***p<1×10−8 (C–F) Oil Red O staining of midguts and (G–J) oenocytes dissected from fed or 24 hour starved, control or dHNF4 mutant larvae. Whereas lipid levels drop significantly in the midguts of control larvae upon starvation (C,E), lipid is inefficiently depleted from the midguts of starved dHNF4 mutants (D,F). An increase in the lipid content of oenocytes occurs normally in starved dHNF4 mutants relative to starved controls (I,J). Fed dHNF4 mutants, however, display a slight increase in lipids in both the midgut and oenocytes relative to fed controls (C,D,G,H). (K–Q) Nile Red staining of larval fat bodies. Whereas the large lipid droplets in the fat bodies of fed controls become smaller upon starvation (K,M), lipid droplets appear to aggregate in starved dHNF4 mutant fat bodies (L,N). (O–Q) Tissue-specific rescue of lipid droplet morphology as assayed by Nile Red stains of fat bodies from starved animals. Starved UAS-dHNF4 dHNF4−/+ control animals (O) display wild-type lipid droplet morphology (compare to M). Starved dHNF4Δ33/fb-GAL4 dHNF4Δ17 mutants display lipid droplet aggregation (P), and this phenotype is largely rescued in starved UAS-dHNF4 dHNF4Δ33/fb-GAL4 dHNF4Δ17 larvae (Q). (R) GC/MS analyses of lipid extracts reveal that dHNF4 mutant early third instar larvae contain relatively higher levels of most free long chain fatty acids upon starvation than control larvae. N=20 larvae for each sample, with eight samples tested. Similar results were obtained from an independent experiment. The Y axis represents the concentration of each free fatty acid (FFA) relative to a 13C-labeled tridecanoate internal standard. Error bars are +/− S.E. *p<1×10−3, **p<1×10−4, ***p<1×10−5.
If dHNF4 mutants retain TAG upon starvation, then this defect should be evident using a cellular based assay to detect stored lipids. Accordingly, tissues were dissected from fed or starved control or dHNF4 mutant larvae and stained with lipophilic dyes to detect neutral lipids (Figure 3C–N). After one day of starvation, lipids are effectively cleared from the midgut of control animals (Figure 3C,E) and accumulate in the oenocytes (Figure 3G,I), similar to published results (Gutierrez et al., 2007). Higher levels of lipid, however, are evident in the midguts of starved dHNF4 mutants compared to starved controls (Figure 3E,F). In addition, slightly elevated lipid levels are seen in the midguts and oenocytes of fed dHNF4 mutants relative to fed controls (Figure 3C,D,G,H), suggesting that metabolic dysfunction is also present in the fed state. Many small lipid droplets are evident in the fat body cells of starved control larvae relative to fed controls, consistent with the mobilization of stored lipid for energy production (Figure 3K,M). Moreover, fed dHNF4 mutants have essentially normal lipid droplet morphology (Figure 3L). These droplets, however, appear to aggregate abnormally in starved mutants (Figure 3N). This observation, together with the elevated levels of lipid in the midguts of starved dHNF4 mutants, are consistent with the retention of TAG as measured by enzymatic assay (Figure 3B).
Tissue-specific rescue was performed to further characterize the lipid metabolic defect seen in the fat body of starved dHNF4 mutant larvae. As expected, small lipid droplets are present in the fat body of a starved control animal that carries only one mutant copy of dHNF4 (Figure 3O), similar to that seen in wild type animals (Figure 3M). Similarly, lipid droplet aggregation is seen in the fat body of a starved dHNF4 mutant that carries only the fat body-specific fb-GAL4 driver (Figure 3P). Essentially normal lipid droplet morphology, however, is recovered when wild-type dHNF4 is expressed in the fat bodies of starved dHNF4 mutants (Figure 3Q). This result suggests that the lipid mobilization defects seen in the fat body of starved dHNF4 mutants is tissue-autonomous.
Finally, gas chromatography and mass spectrometry (GC/MS) were used to analyze the lipid composition of fed and starved control and dHNF4 mutant larvae. Although no significant changes were evident under fed conditions (data not shown), starved dHNF4 mutants display reproducibly increased levels of free LCFAs compared to controls (Figure 3R). Thus, both TAG and LCFAs accumulate upon starvation in dHNF4 mutant larvae.
Lipolysis and β-oxidation genes are down-regulated in dHNF4 mutants
Microarray studies were performed to investigate the molecular mechanisms by which dHNF4 contributes to the starvation response. RNA extracted from fed or starved, control or dHNF4 mutant larvae was labeled and hybridized to Affymetrix Drosophila 2.0 microarrays. All experiments were conducted in triplicate to facilitate statistical analysis. The raw expression data files were imported into dChip (Li and Hung Wong, 2001), the background was normalized, and gene expression changes were determined by using SAM, with < 5% estimated false positives in each comparison (Tusher et al., 2001).
This study revealed that 3138 transcripts are significantly affected by starvation in wild type larvae (≥1.5-fold), with 1809 genes up-regulated and 1329 genes down-regulated (Supplemental Table 1). Comparison of this dataset with the 2185 transcripts identified by Zinke et al (2002) in their analysis of the 12 hour starvation response in larvae revealed that 977 (45%) transcripts are shared in common. Of these, 941 transcripts (96%) reported as either up-regulated or down-regulated by Zinke et al. (2002) show the same response in our study. Similarly, 145 of the 398 transcripts (36%) identified in starved adult Drosophila are present in our list of affected genes (Gronke et al., 2005), with 115 (79%) of these common transcripts regulated in the same manner (up or down). As expected, genes involved in gluconeogenesis and lipid mobilization, transport, and β-oxidation are up-regulated in our list of starvation response genes, while genes involved in glycolysis, lipid synthesis, and mitochondrial protein biosynthesis are down-regulated (Supplemental Table 2).
Comparison of the transcriptional profiles in fed control and dHNF4 mutant larvae revealed that only 85 transcripts displayed a significant change in expression level of ≥1.3-fold (Supplemental Table 3). This number increased to 330 affected transcripts in starved dHNF4 mutants compared to starved controls, with 45 transcripts shared in common between fed and starved conditions (Supplemental Table 5). The majority of affected mRNAs are down-regulated in dHNF4 mutants, consistent with the role of HNF4 family members as transcriptional activators (Hertz et al., 1998; Van Gilst et al., 2005b).
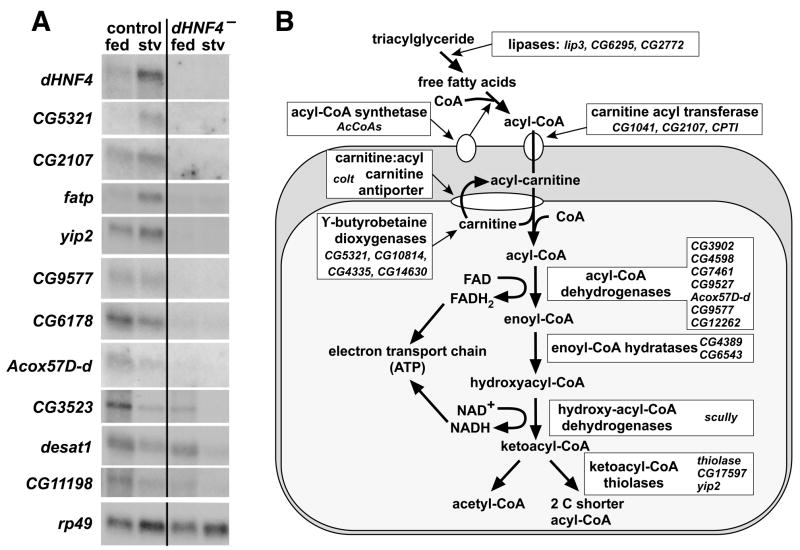
GOstat was used to identify predicted gene functions corresponding to transcripts that are mis-regulated in either fed or starved dHNF4 mutants relative to controls (Beissbarth and Speed, 2004). Interestingly, the top gene ontology categories, under both fed and starved conditions, all relate to different aspects of mitochondrial β-oxidation – the pathway by which fatty acids are converted into high energy electron donors for energy production (Table 1). These include oxidoreductase enzymes in the β-oxidation pathway, transferases that could facilitate acyl-CoA import into mitochondria, and γ-butyrobetaine dioxygenases that synthesize the carnitine intermediate required for fatty acid import into mitochondria (Table 1, Figure 4B). Similarly, comparison of a list of genes in Drosophila metabolic pathways with our dHNF4 mutant microarray lists demonstrated major effects on β-oxidation in both fed and starved animals (Supplemental Tables 4,6). These effects on gene expression are consistent with the results of metabolic assays described above, in that reduced flux through the β-oxidation pathway would result in an accumulation of fatty acids and TAG under starvation conditions.
Table 1.
Top Affected Gene Functions in Fed or Starved dHNF4 Mutants Reveal a Central Role in Fatty Acid β-Oxidation
| Fed Animals | ||
|---|---|---|
| GO category | # genes (total) | p-value |
| oxidoreductase activity | 17 (541) | 4.8e-07 |
| carnitine o-acyl transferase activity | 4 (6) | 2.3e-06 |
| mitochondrion | 15 (521) | 6.2e-06 |
| transferase activity, other than amino-acyl groups | 7 (75) | 1.0e-05 |
| acyltransferase activity | 7 (75) | 1.0e-05 |
| cytoplasmic part | 22 (1537) | 1.8e-05 |
| lipid particle | 8 (129) | 1.9e-05 |
| transferase activity, transferring acyl groups | 7 (88) | 1.9e-05 |
| fatty acid metabolic process | 5 (35) | 4.9e-05 |
| fatty acid β-oxidation | 3 (5) | 6.3e-05 |
| fatty acid oxidation | 3 (6) | 1.1e-04 |
| o-acyltransferase activity | 4 (21) | 1.4e-04 |
| Starved Animals | ||
| GO category | # genes (total) | p-value |
| oxidoreductase activity | 29 (541) | 3.9e-14 |
| fatty acid metabolic process | 10 (35) | 3.4e-09 |
| cellular lipid metabolic process | 15 (113) | 3.4e-09 |
| lipid metabolic process | 15 (125) | 1.1e-08 |
| fatty acid β-oxidation | 5 (5) | 4.9e-08 |
| monocarboxylic acid metabolic process | 10 (53) | 1.4e-07 |
| fatty acid oxidation | 5 (6) | 2.1e-07 |
| transferase activity, other than amino-acyl groups | 11 (75) | 2.3e-07 |
| acyltransferase activity | 11 (75) | 2.3e-07 |
| hydrolase activity | 44 (1550) | 9.5e-07 |
| transferase activity, transferring acyl groups | 11 (88) | 1.1e-06 |
| γ-butyrobetaine dioxygenase activity | 4 (4) | 1.6e-06 |
GOstat analysis of either the 74 genes that are down-regulated in fed dHNF4 mutants compared to fed control larvae (Supplemental Table 3) or the 197 genes that are down-regulated in starved dHNF4 mutants compared to starved control larvae (Supplemental Table 5). The top twelve GO categories are listed, from top to bottom, along with the number of affected genes in that category, the total number of genes in that category in parentheses, and the p-value of the match (http://gostat.wehi.edu.au/cgi-bin/goStat.pl).
Figure 4. dHNF4 regulates all levels of the β-oxidation pathway.
(A) Second instar larvae that were transheterozygous for precise excisions of the EP2449 and KG08976 P-elements (control), or dHNF4Δ33/dHNF4Δ17 mutants (dHNF4−), were fed or starved for 24 hours, after which RNA was isolated and analyzed by northern blot hybridization. dHNF4 mRNA is induced upon starvation, with no transcript seen in mutant animals. CG5321, CG2107, fatp, yip2, CG9577, CG6178, and Acox57D-d, which function in β-oxidation, are significantly down-regulated in the dHNF4 mutant, consistent with the microarray results. CG3523, which encodes a predicted fatty acid synthetase, desat1, which encodes a predicted stearoyl-CoA desaturase, and CG11198, which encodes a predicted acetyl-CoA carboxylase, are all down-regulated upon starvation in both control larvae and dHNF4 mutants, and their overall level of expression is reduced in the mutant. (B) A schematic representation of the mitochondrial β-oxidation pathway is depicted. At the top, stored lipid in the form of triglycerides are hydrolyzed into free fatty acids by lipases. Acyl-CoA synthetases, which reside in the outer mitochondrial membrane, convert fatty acids into acyl-CoA for entry into the β-oxidation pathway. The acyl group is transported through the outer and inner mitochondrial membranes via a carnitine intermediate, and then processed through four enzymatic steps, as depicted. Each cycle generates one FADH2 and one NADH, which donate their high energy electrons to the electron transport chain for ATP production. Each cycle of β-oxidation results in an acyl-CoA that is shortened by two carbons. This acyl-CoA can be processed through successive cycles to produce more FADH2 and NADH. The names of genes that are down-regulated in dHNF4 mutants are listed next to their predicted enzymatic or transport functions.
Northern blot hybridizations were conducted to test the response of selected dHNF4-regulated genes in both fed and starved animals (Figure 4A). This study showed that dHNF4 is significantly induced upon starvation, consistent with its role in mediating lipid mobilization, and is not detectable in the mutant, as expected. CG5321, CG2107, fatp, yip2, CG9577, CG6178, and Acox57D-d, many of which are predicted to act in β-oxidation (Figure 4B), are up-regulated in starved controls, and down-regulated in both fed and starved dHNF4 mutants (Figure 4A). Three genes were also tested that could contribute to lipid metabolism: CG3523 (predicted to encode a fatty acid synthase), desat1 (predicted to encode a stearoyl-CoA desaturase), and CG11198 (predicted to encode a acetyl-CoA carboxylase). These genes are down-regulated in starved control animals, and their expression is reduced in dHNF4 mutants. Taken together, these results validate the dHNF4 targets identified by microarray and demonstrate that the receptor acts at all levels of the β-oxidation pathway.
The dHNF4 LBD can be activated by starvation and exogenous fatty acids
If dHNF4 plays a central role in maintaining lipid homeostasis in Drosophila, then it may mediate this effect by changing its transcriptional activity in response to developmental cues or dietary conditions. To test this possibility, we used the GAL4-dHNF4 ligand sensor to follow dHNF4 LBD activation during larval development, as the animal terminates feeding and growing in preparation for entry into metamorphosis. This system uses a heat-inducible transgene that encodes the yeast GAL4 DNA binding domain fused to the dHNF4 LBD (hsp70-GAL4-dHNF4), in combination with a UAS-nlacZ reporter gene that directs the synthesis of nuclear-localized β-galactosidase. The temporal and spatial patterns of β-galactosidase expression in ligand sensor transgenic animals accurately reflect nuclear receptor LBD activation at different stages of development and in response to exogenous ligands (Palanker et al., 2006).
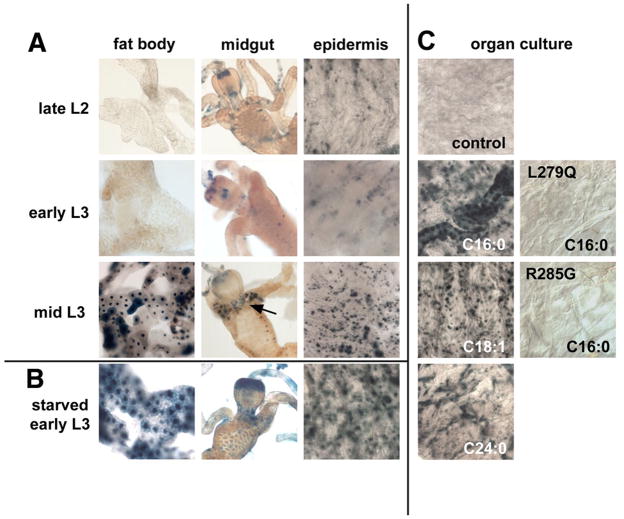
GAL4-dHNF4 shows low levels of LBD activation in late second instar or early third instar larvae (Figure 5A). A distinct change, however, is seen in mid-third instar larvae, with high levels of LBD activation in the fat body, at the base of the gastric caeca, and in the epidermis. Animals undergo a widespread change in physiology at this time, including down-regulation of PI3K activity and initiation of autophagy, as they prepare for the cessation of feeding, the onset of wandering behavior, and metamorphosis (Britton et al., 2002; Rusten et al., 2004). Accordingly, we asked if the dHNF4 ligand sensor could be activated by starvation, which is known to induce lipolysis and autophagy in third instar larvae (Britton et al., 2002; Rusten et al., 2004; Scott et al., 2004). Interestingly, whereas little activation is seen in early third instar larvae maintained on a regular diet or on yeast paste, high activation is seen in the fat body and epidermis of starved animals (Figure 5A,B and data not shown). Larvae fed a sugar diet, however, show no increase in activation, indicating that this response requires complete nutrient deprivation (data not shown).
Figure 5. GAL4-dHNF4 can be activated by starvation and exogenous long chain fatty acids.
(A) Fed larvae carrying both the GAL4-dHNF4 and UAS-nlacZ transgenes were heat-treated, allowed to recover for ~6 hours, and dissected organs were stained with X-gal for β-galactosidase activity. Low levels of ligand sensor activity are seen in the fat body, midgut, and epidermis of late second instar (late L2) or early third instar larvae (early L3), while high levels of activity are detected in mid-third instar larvae (mid L3). GAL4-dHNF4 activity in the mid-third instar midgut is restricted to the anterior region, adjacent to the proventriculus (arrow). No ligand sensor activity is detected in the oenocytes, although negative results with the ligand sensor system are difficult to interpret (Palanker et al., 2006). (B) Late second instar GAL4-dHNF4; UAS-nlacZ larvae were fasted or fed for 3 hrs, heat-treated, and fasted or fed for another 3– 4 hrs, after which dissected organs were stained with X-gal for β-galactosidase activity. Starvation leads to strong activation of GAL4-dHNF4 in the fat body and epidermis of early third instar larvae compared to fed controls (A, early L3). (C) Late second instar GAL4-dHNF4; UAS-nlacZ larvae were heat-treated to induce GAL4-dHNF4 expression, allowed to recover, and then bisected, inverted, and cultured overnight in either control medium or medium supplemented with fatty acids as shown. Palmitic acid (C16:0), oleic acid (C18:1), and the very long chain fatty acid, lignoceric acid (C24:0), activate the ligand sensor in epidermis. This activation, however, is not seen in animals that express either the L279Q or R285G GAL4-dHNF4 mutant ligand sensors. Activation is also seen in the trachea, as shown in the C16:0-treated control tissue.
Fasting normally triggers rapid breakdown of TAG into LCFAs as a first step toward β-oxidation and energy production. Given that LCFAs can bind to the HNF4 LBD, are required for its activity (Dhe-Paganon et al., 2002; Wisely et al., 2002; Aggelidou et al., 2004), and the dHNF4 LBD copurifies with palmitic acid (Yang et al., 2006) (H. Krause, personal communication), we wanted to test if the dHNF4 ligand sensor could be activated by fatty acids. Consistent with this possibility, increased lipolysis driven by ectopic expression of the Brummer lipase results in increased ligand sensor activity in larval fat bodies (Supplemental Figure 4) (Gronke et al., 2005). To extend this observation, organ culture studies were conducted using exogenous purified fatty acids. Addition of palmitic acid (C16:0) or oleic acid (C18:1) led to efficient GAL4-dHNF4 activation (Figure 5C). Activation was also seen upon addition of a very long chain fatty acid, lignoceric acid (C24:0; Figure 5C,) or an isobranched fatty acid, 14-methyl palmitic acid (Supplemental Table 7). Although these compounds are not predicted to activate HNF4 based on structural studies (Dhe-Paganon et al., 2002; Wisely et al., 2002), it is possible that metabolites derived from these compounds could bind to and directly activate the dHNF4 LBD. Other LCFAs are capable of activating GAL4-dHNF4, but not cAMP, the PPAR agonist bezafibrate, vitamin D, chenodeoxycholic acid, 9-cis retinoic acid, cholesterol, or 20-hydroxyecdysone (Supplemental Table 7). Although relatively high concentrations of fatty acids were added to the culture medium to see an effect (0.1–1 mM) it is likely that the resulting intracellular concentration is much lower.
If fatty acids activate dHNF4 through its LBD, then specific LBD mutations that disrupt fatty acid binding should reduce its transactivation function. Accordingly, transgenic lines were established that carry GAL4-dHNF4 constructs with point mutations corresponding to specific critical amino acids in the LBD conserved from Drosophila to mammals. One of these, R285 in dHNF4 (R226 in HNF4α), orients the fatty acid within the ligand binding pocket via an ion pair between the carboxylic acid headgroup of the fatty acid and the guanidinium group of arginine (Dhe-Paganon et al., 2002). Mutation of this amino acid to glycine in HNF4α has no apparent effect on receptor homodimerization, DNA binding, or coactivator recruitment, but eliminates its transactivation function (Aggelidou et al., 2004; Iordanidou et al., 2005). The second LBD mutation tested, L279Q in dHNF4 (L220Q in HNF4α), changes a hydrophobic amino acid that is predicted to interact with the fatty acid ligand (Dhe-Paganon et al., 2002). This mutation in HNF4α also has no apparent effect on receptor homodimerization or DNA binding, but eliminates the receptor transactivation function and is unresponsive to coactivators (Aggelidou et al., 2004; Iordanidou et al., 2005). Heat-induced expression of either mutated construct resulted in approximately equal levels of GAL4-dHNF4 protein in transgenic animals (Supplemental Figure 5), but eliminated the activation seen in mid-third instar larvae (Supplemental Figure 6) and in starved third instar larvae (Supplemental Figure 7). Similarly, both mutant GAL4-dHNF4 ligand sensors show no detectable response to exogenous palmitic acid, indicating that key amino acids required for fatty acid binding are needed for GAL4-dHNF4 transcriptional activity and its responsiveness to exogenous LCFAs (Figure 5C).
Discussion
The presence of multiple HNF4 family members in mice and C. elegans complicates our understanding of the physiological functions of this nuclear receptor subclass. Here we characterize the single ancestral HNF4 in Drosophila, demonstrating its critical role in regulating the adaptive response to starvation. Our results support a feed-forward model for dHNF4 function in which fatty acids freed from triglycerides activate the receptor, inducing the expression of enzymes that drive lipid mobilization and β-oxidation for energy production.
dHNF4 plays an essential role in lipid catabolism
Remarkably, the expression pattern of HNF4 has been conserved through evolution, from flies to mammals, with abundant dHNF4 expression in the midgut, fat body, and Malpighian tubules –tissues that are the functional equivalents of the intestine, liver, and kidney, respectively, where mammalian HNF4 is expressed (Duncan et al., 1994) (Figure 1). The only exception is a lack of dHNF4 expression in the IPCs, the functional equivalent of mammalian pancreatic β cells (Rulifson et al., 2002). dHNF4 is also expressed in the oenocytes, which have hepatocyte-like properties and contribute to lipid mobilization (Gutierrez et al., 2007). The starvation sensitivity of dHNF4 mutants, however, cannot be rescued by expression of wild-type dHNF4 in the oenocytes, and starvation-induced lipid accumulation occurs normally in the oenocytes of dHNF4 mutants, leaving it unclear what role, if any, dHNF4 plays in these cells.
Although dHNF4 mutants can survive to adulthood under ideal culture conditions, significant defects become evident upon food deprivation, including an inability to efficiently mobilize stored lipid in the midgut and fat body, increased levels of LCFAs and TAG, and premature death. This retention of lipid is similar to the accumulation of lipids seen in the guts of nhr-49 mutant worms and the steatosis seen in liver-specific HNF4α mutant mice (Hayhurst et al., 2001; Van Gilst et al., 2005a). The ability of dHNF4 mutant larvae to survive on a sugar diet indicates that glycolysis and oxidative phosphorylation are intact in these animals. Rather, starved dHNF4 mutant larvae appear to die from an inability to breakdown TAG and use the resulting free fatty acids for energy production via β-oxidation. Similar phenotypes are associated with defects in β-oxidation in mice and humans, which result in sensitivity to starvation, accumulation of lipid in the liver, and increased levels of free fatty acids (Rinaldo et al., 2002). β-oxidation takes place in the mitochondria or peroxisomes of most organisms, with very long chain fatty acids (VLCFAs) as the substrate for the peroxisomal pathway. Although peroxisomes are present in the gut and Malpighian tubules of Drosophila adults (Beard and Holtzman, 1987), and VLCFAs accumulate in a VLCFA acyl-CoA synthase mutant (Min and Benzer, 1999), the existence of peroxisomes in Drosophila larvae remains unclear.
dHNF4 regulates genes that act in lipid β-oxidation
Like mammalian HNF4α, dHNF4 mRNA is significantly up-regulated in response to starvation (Figure 4A; Yoon et al., 2001). In addition, many genes in the β-oxidation pathway are up-regulated upon starvation, and display significantly reduced expression in dHNF4 mutant larvae (Figure 4A). These include acetyl-CoA synthetase (AcCoAs), γ–butyrobetaine dioxygenases, and carnitine acyl transferases, the rate-limiting step in acyl import into mitochondria. In addition, genes that encode the four enzymatic steps of β-oxidation are all dependent on dHNF4 for their proper level of expression (Figure 4). Importantly, this effect is also seen in fed dHNF4 mutant larvae, where only 86 transcripts change ≥1.3-fold compared to wild type, many of which encode components of the β-oxidation pathway (Supplemental Tables 3,4). These include yip2, Acox57D-d, thiolase, scully, and CPTI (carnitine palmitoyltransferase) among the most highly down-regulated genes in fed dHNF4 mutants (1.8 to 4-fold as determined by dChip). Thus dHNF4 is required for both basal and starvation-induced β-oxidation gene transcription. Moreover, many of the genes in this pathway have at least one canonical binding site for an HNF4 homodimer within 1 kb of their predicted 5′ end, suggesting that dHNF4 directly regulates their transcription (Supplemental Table 8).
The effects of dHNF4 are not, however, restricted to lipid oxidation. Three predicted lipase transcripts, lip3, CG6295, and CG2772, are significantly reduced in starved dHNF4 mutants compared to starved controls (Supplemental Table 5), suggesting a direct role for the receptor in the release of LCFAs from TAG. Similarly, the CGI-58 homolog encoded by CG1882, which activates lipolysis in mammals, is up-regulated in starved dHNF4 mutants, while adipokinetic hormone receptor (AKHR) is down-regulated, consistent with its normal role in mobilizing stored lipids analogous to mammalian β-adrenergic signaling (Grönke et al., 2007). No effect, however, is seen on expression of brummer, which encodes adipose triglyceride lipase, suggesting that dHNF4 does not directly impact brummer-mediated lipolysis (Gronke et al., 2005). A VLCFA-CoA ligase gene (bubblegum; Min and Benzer, 1999) and LCFA-CoA ligase gene (CG6178; Oba et al., 2004) are also affected in starved dHNF4 mutants. These enzymes activate free fatty acids for either catabolic or anabolic processes. We also see effects on lipid synthesis, with down-regulation of a predicted fatty acid synthase (CG3523), a predicted acetyl-CoA carboxylase (CG11198), and two predicted glycerol-3-phosphate acyltransferases (GPATs) (CG3209 and CG17608) in starved dHNF4 mutants. Two predicted stearoyl-CoA desaturase genes, CG15531 and desat1, which catalyze the production of monounsaturated fatty acids, are also down-regulated in the mutant. A similar result has been reported for NHR-49, where a stearoyl-CoA desaturase gene is a key functional target of the receptor (Van Gilst et al., 2005a). The glyoxylate pathway is also affected in nhr-49 mutants, under both fed and starved conditions (Van Gilst et al., 2005b). This pathway is analogous to mammalian ketogenesis, in which fatty acid β-oxidation products are used for energy production. Genes for the central enzymes in this pathway, malate synthase and isocitrate lyase, have not yet been identified in D. melanogaster. However, CG12208, which is predicted to contribute to glyoxylate catabolism, is down-regulated in both fed and starved dHNF4 mutants. Taken together, these effects on transcription define a central role for dHNF4 in balancing lipid anabolic and catabolic pathways in response to dietary conditions.
Although genome-wide ChIP suggests that HNF4α is a promiscuous regulator of many actively transcribed genes in pancreatic islets and hepatocytes, relatively few targets have been identified in functional studies (Odom et al., 2004). These include multiple apolipoprotein genes, NTCP, and L-FABP, indicating roles in very low density lipoprotein secretion, high density lipoprotein synthesis, and bile acid homeostasis (Duncan et al., 1997; Hayhurst et al., 2001). Only a few β-oxidation genes have been studied in HNF4α mutant mice, with contradictory results. HNF4α can bind to the promoters of liver CPT-I and MCAD (which encodes an acyl-CoA dehydrogenase), and CPT-I mRNA levels are reduced in HNF4α mutant livers (Carter et al., 1993; Louet et al., 2002). In contrast, other studies show that CPT-I, CPT-II and MCAD transcripts are elevated in HNF4α mutant livers (Hayhurst et al., 2001; Rhee et al., 2003). Given our results, it would be interesting to resolve these contradictory observations and more broadly examine other steps in the β-oxidation pathway in HNF4α mutant mice.
A feed-forward model for dHNF4 in driving fatty acid β-oxidation for energy production
Although dHNF4 is widely expressed in the larval midgut (Figure 1A), its activity appears to be spatially restricted, primarily to a band of cells at the anterior end of the midgut that lie at the base of the gastric caeca (Figure 5A, arrow) (Palanker et al., 2006). This is the primary site for nutrient absorption into the circulatory system, suggesting that dHNF4 is responsive to dietary signals (Chapman, 1998). In the fat body, the dHNF4 ligand sensor is inactive during most of larval development, when fat storage is favored over fat breakdown, but becomes activated at the end of larval growth when lipolysis and autophagy are initiated in the fat body (Britton et al., 2002; Rusten et al., 2004) (Figure 5A). Similarly, a significant increase in GAL4-dHNF4 activation is seen in the fat body of starved larvae (Figure 5B), concurrent with the increase in fatty acid β-oxidation that is required to survive these conditions. Consistent with this model, the dHNF4 LBD can be activated by ectopic bmm expression or exogenous LCFAs in cultured larval organs. The most effective LCFAs, palmitic acid (C16:0) and oleic acid (C18:1) are relatively abundant in Drosophila larvae (Figure 3R), and are the primary constituents of triglycerides that are hydrolyzed upon starvation, making them logical signaling intermediates (Chapman, 1998). In addition, the dHNF4 LBD copurifies with palmitic acid, indicating that fatty acid binding has been conserved through evolution (Yang et al., 2006) (H. Krause, personal communication). Two different point mutations that change conserved amino acids, each of which is predicted to disrupt dHNF4 fatty acid binding, block the ability of the LBD to respond to starvation or exogenous fatty acids, suggesting that direct fatty acid binding is essential for dHNF4 transcriptional activity (Figure 5C).
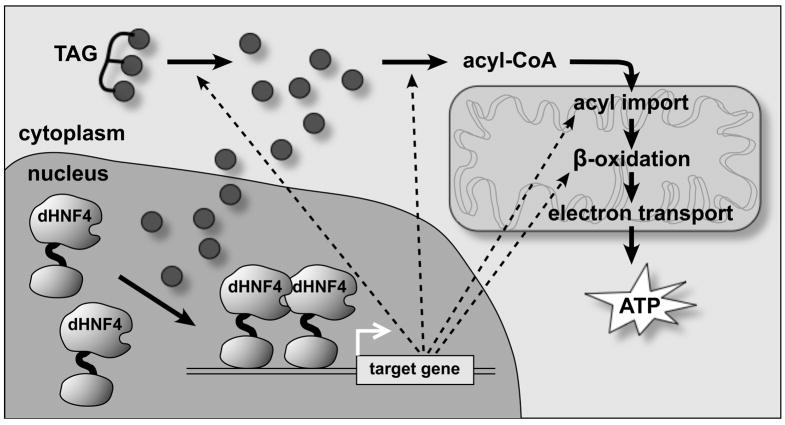
Taken together, our studies support a feed-forward model in which dHNF4 functions as a sensor for free fatty acid levels in the larva, driving their catabolism through β-oxidation (Figure 6). Upon nutrient deprivation, TAG is hydrolyzed into free LCFAs. As has been shown in mammalian cells, it is likely that these cytosolic LCFAs can translocate into the nucleus, activating dHNF4 (Wolfrum et al., 2001). This, in turn, leads to the transcriptional induction of key genes involved in TAG breakdown, acyl-CoA production, acyl import into mitochondria, and β-oxidation (Figure 6). The net result of this response is consumption of the dHNF4 activating signal (LCFAs), maintaining energy homeostasis through ATP production. This feed-forward model is consistent with the phenotypes of dHNF4 mutants, which are starvation sensitive and accumulate TAG and LCFAs.
Figure 6. A feed-forward model for dHNF4 in fatty acid β-oxidation.
In the absence of dietary nutrients, triglycerides (TAGs) are cleaved by lipases to yield long chain fatty acids (black circles). We propose that these fatty acids can directly or indirectly activate dHNF4 in the nucleus, inducing target gene transcription. The dHNF4 target genes encode lipases as well as enzymes and transporters that use fatty acids as a substrate for β-oxidation in the mitochondria, removing the activating signal for dHNF4 and maintaining energy homeostasis. See text for more details.
Crystal structure studies of mammalian HNF4α have shown that the fatty acid-bound form of the LBD can assume both open and closed positions, with helix 12 either extended or held against the body of the LBD, while the HNF4γ LBD appears to be locked in the open configuration (Dhe-Paganon et al., 2002; Wisely et al., 2002). Our data, however, indicates that the Drosophila HNF4 LBD is responsive to the nutritional status of the animal and can be activated by exogenous LCFAs. Although it is possible that fatty acids act as exchangeable ligands for dHNF4 in vivo, our model is also consistent with a role for fatty acid binding in constitutive dHNF4 transcriptional activation. dHNF4 mRNA is significantly induced upon starvation (Figure 4A). The resultant newly synthesized protein could act as a sensor for free fatty acids, binding those molecules and thereby forming active dHNF4 transcription complexes. In contrast, dHNF4 protein synthesized during stages with less metabolic demand would have access to lower free fatty acid levels and thus remain less active. This model is consistent with our ligand sensor studies, where newly-synthesized GAL4-dHNF4 LBD is inactive in the presence of abundant nutrients (late L2 and early L3, Figure 5A), and is highly active upon food deprivation (Figure 5B). Our work sets the stage for biochemical studies that address the mechanisms of dHNF4 activation in vivo, and whether protein turnover, posttranslational modification, and/or differential cofactor binding modulate the activation of dHNF4 by LCFAs. It also raises the interesting possibility that vertebrate HNF4 may function as a fatty acid sensor.
dHNF4 functions like PPARα in vertebrates
Our feed-forward model for dHNF4 function is reminiscent of that ascribed to a mammalian nuclear receptor, PPARα. PPARα binds LCFAs and directly regulates genes that are orthologous to dHNF4 targets, including many genes in the β-oxidation pathway (Michalik et al., 2006). PPARα mutant mice are defective in the adaptive response to starvation and display increased plasma free fatty acids, and fatty liver with enlarged hepatocyte lipid droplets (Kersten et al., 1999; Leone et al., 1999; Hashimoto et al., 2000) – phenotypes shared with dHNF4 mutants. Van Gilst et al. (2005a) reach a similar conclusion in their characterization of C. elegans nhr-49 mutants. There are no orthologs of the PPAR nuclear receptor subclass in lower organisms, including Drosophila and worms. This is in spite of the fact that PPARα functions – the ability to sense nutrient deprivation and mobilize stored forms of energy to maintain homeostasis – are essential for animal survival. Indeed, even unicellular fungi have an analogous pathway, where the Oaf1/Pip2 transcription factor heterodimer binds the fatty acid oleate and regulates peroxisomal β-oxidation (Karpichev et al., 1997; Phelps et al., 2006). We propose that the ancestral function of HNF4 has been adopted by PPARα during the course of evolution. Our studies also indicate additional functions for dHNF4 beyond lipid mobilization that provide possible directions for future research. In addition, phenotypic characterization of dHNF4 mutants under different environmental and dietary conditions may provide new insights into dHNF4 activities, and raise new implications for the regulation and function of its vertebrate counterparts.
Experimental Procedures
Fly stocks
Transposable P-element insertions EP2449 (Bloomington #17249) and KG08976 (Bloomington #16471) were used to generate dHNF4 mutants by imprecise excision, scoring w− progeny for lethality in combination with Df(2L)Te30Cb-1. The extent of the deletion in each dHNF4 mutant was determined by PCR and DNA sequencing. The hs-dHNF4 transgenic line was made by PCR amplifying the dHNF4 coding sequence (isoform A; FlyBase) using primers: 5′-GCGGCCGCATCGGAGAGCCACATAATGC-3′ and 5′-TCTAGAACTGCATATTGCACCGTTCG-3′. The resulting 2050 bp fragment was inserted into the pCaSPeR-hs-act P-element vector using NotI and XbaI, and transgenic lines were established using standard methods. Flies were maintained on Standard Bloomington Stock Center medium (with malt) at 25°C, with added dry yeast, unless otherwise specified.
Metabolic assays
For TAG assays, 25 larvae were homogenized in 100 μl PBS, 0.1% Tween 20 and immediately incubated at 70°C for 5 minutes. Heat-treated homogenate (20 μl) was incubated with either 20 μl PBS or Triglyceride Reagent (Sigma) for 30 minutes at 37°C, after which the samples were centrifuged at maximum speed for 3 minutes. Thirty μl was transferred to a 96 well plate and incubated with 100 μl of Free Glycerol Reagent (Sigma) for 5 minutes at 37°C. Samples were assayed using a BioTek Synergy HT microplate spectrophotometer at 540 nm. TAG amounts were determined by subtracting the amount of free glycerol in the PBS-treated sample from the total glycerol present in the sample treated with Triglyceride reagent. TAG levels were normalized to protein amounts in each homogenate, using a Bradford assay (Bio-Rad), and data was analyzed using a Student’s t-test.
For glycogen assays, animals were prepared in an identical manner as described for the TAG assays, except that larvae were homogenized in PBS. Heat-treated homogenate was diluted 1:10 in PBS and centrifuged at maximum speed for 3 minutes, after which 30 μl supernatant was transferred to each of three wells of a 96 well plate. One well was treated with 100 μl PBS, the second was treated with 100 μl of glucose reagent (Sigma), and the third was treated with 100 μl of glucose reagent and 0.3 U of amyloglucosidase (Sigma). The plate was incubated at 37°C for ~30 min, after which color intensity was measured using a BioTek Synergy HT microplate reader at 540 nm. Glucose and glucose plus glycogen amounts were determined using a standard curve, and the amount of glycogen was determined by subtracting the glucose from the glucose plus glycogen. Total glycogen was normalized to protein amount as described above.
For Nile Red stains, larvae were dissected on a glass slide in 0.00002% Nile Red (Sigma), 75% glycerol and incubated for five minutes before mounting with a coverslip. Slides were imaged within one hour of dissection using a Leica TCS SP2 confocal microscope at 40X magnification with an excitation wavelength of 543 nm and an emission of ~626 nm. For Oil Red O stains, larvae were dissected and fixed for 30 minutes at room temperature in 4% formaldehyde in PBS. Larval organs were washed twice with PBS and twice in 100% propylene glycol, after which they were incubated with 0.5% Oil Red O in propylene glycol at 60°C. They were then washed twice in 85% propylene glycol and twice in PBS, both at room temperature, and mounted in 75% glycerol. Slides were imaged on a Zeiss Axioskop 2 plus microscope using a CoolSnap Pro camera and Image Pro Plus software.
Microarrays
Staged late second instar control larvae that were transheterozygous for precise excisions of the EP2449 and KG08976 P-elements, and dHNF4Δ33/dHNF4Δ17 mutant larvae, were either fed or starved for 24 hrs. RNA was extracted from the apparently healthy animals that molted to the third instar using Trizol (Gibco) and purified on RNAeasy columns (Qiagen). All samples were prepared in triplicate to facilitate subsequent statistical analysis. Probe labeling, hybridization to Affymetrix GeneChip® Drosophila Genome 2.0 Arrays, and scanning, were performed by the University of Maryland Biotechnology Institute Microarray Core Facility. Raw data was normalized using dChip (Li and Hung Wong, 2001) and analyzed using SAM 2.0 (Tusher et al., 2001), with a 5% false positive rate. Comparison between microarray datasets was performed using Microsoft Access.
Organ culture of GAL4-dHNF4, UAS-nlacZ larvae
Late second instar hs-GAL4-dHNF4; UAS-nlacZ larvae (Palanker et al., 2006) were heat treated at 37°C for 30 min and allowed to recover for 1–2 hours at 25°C. These animals were bisected and the anterior half was rinsed in PBS, everted, and placed in oxygenated Grace’s Insect Medium (Invitrogen) containing 0.01% NP-40. Compounds were administered at 0.1–1 mM in freshly oxygenated Grace’s Medium plus 0.01% NP-40. The addition of 0.01% Nonidet P-40 to the culture medium is required to reduce the background level of GAL4-dHNF4 activation. Animals were cultured, fixed and stained with X-gal as described (Palanker et al., 2006).
Statistical Analyses
Statistical significance was calculated using an unpaired two-tailed Student’s t-test with unequal variance. All quantitative data are reported as the mean ± SEM.
Supplementary Material
Acknowledgments
We thank A-F. Ruaud and M. Sieber for helpful discussions on metabolism and associated protocols, J. Evans for help with collecting staged animals, R. Beckstead and K. Baker for help with antibody affinity purification, R. Beckstead and M. Horner for assistance with microarray analysis, A. Godinez for performing the Affymetrix microarray hybridizations and data collection, J. Cox for help with GC/MS, H. Krause for suggesting the use of NP-40 to reduce ligand sensor background activity, and lab members for critical comments on the manuscript. L.P. was supported by an NIH Predoctoral Training Grant in Genetics, T32 GM007464. This research was supported by NIH grant 1R01 DK075607.
Footnotes
Supplemental Data
Supplemental data include Supplemental Experimental Procedures, eight Supplemental Tables, and seven Supplemental Figures, and can be found with this article online at http://
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggelidou E, Iordanidou P, Tsantili P, Papadopoulos G, Hadzopoulou-Cladaras M. Critical role of residues defining the ligand binding pocket in hepatocyte nuclear factor-4alpha. J Biol Chem. 2004;279:30680–30688. doi: 10.1074/jbc.M401120200. [DOI] [PubMed] [Google Scholar]
- Baker KD, Thummel CS. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab. 2007;6:257–266. doi: 10.1016/j.cmet.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard ME, Holtzman E. Peroxisomes in wild-type and rosy mutant Drosophila melanogaster. Proc Natl Acad Sci U S A. 1987;84:7433–7437. doi: 10.1073/pnas.84.21.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Drosophila’s insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell. 2002;2:239–249. doi: 10.1016/s1534-5807(02)00117-x. [DOI] [PubMed] [Google Scholar]
- Carter ME, Gulick T, Raisher BD, Caira T, Ladias JA, Moore DD, Kelly DP. Hepatocyte nuclear factor-4 activates medium chain acyl-CoA dehydrogenase gene transcription by interacting with a complex regulatory element. J Biol Chem. 1993;268:13805–13810. [PubMed] [Google Scholar]
- Chapman R. The Insects: Structure and Function. Cambridge UK: Cambridge University Press; 1998. [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, Darnell JE., Jr Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos . Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- Dhe-Paganon S, Duda K, Iwamoto M, Chi YI, Shoelson SE. Crystal structure of the HNF4 alpha ligand binding domain in complex with endogenous fatty acid ligand. J Biol Chem. 2002;277:37973–37976. doi: 10.1074/jbc.C200420200. [DOI] [PubMed] [Google Scholar]
- Duncan SA, Manova K, Chen WS, Hoodless P, Weinstein DC, Bachvarova RF, Darnell JE., Jr Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc Natl Acad Sci U S A. 1994;91:7598–7602. doi: 10.1073/pnas.91.16.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SA, Nagy A, Chan W. Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: tetraploid rescue of Hnf-4(−/−) embryos. Development. 1997;124:279–287. doi: 10.1242/dev.124.2.279. [DOI] [PubMed] [Google Scholar]
- Gerdin AK, Surve VV, Jonsson M, Bjursell M, Bjorkman M, Edenro A, Schuelke M, Saad A, Bjurstrom S, Lundgren EJ, et al. Phenotypic screening of hepatocyte nuclear factor (HNF) 4-gamma receptor knockout mice. Biochem Biophys Res Commun. 2006;349:825–832. doi: 10.1016/j.bbrc.2006.08.103. [DOI] [PubMed] [Google Scholar]
- Grönke S, Hirsch J, Fellert S, Andreou A, Haase T, Jäckle H, Kühnlein RP. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 2007;5:e137. doi: 10.1371/journal.pbio.0050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, Jackle H, Kuhnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Gutierrez E, Wiggins D, Fielding B, Gould AP. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2007;445:275–280. doi: 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Cook WS, Qi C, Yeldandi AV, Reddy JK, Rao MS. Defect in peroxisome proliferator-activated receptor alpha-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J Biol Chem. 2000;275:28918–28928. doi: 10.1074/jbc.M910350199. [DOI] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz R, Magenheim J, Berman I, Bar-Tana J. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4alpha. Nature. 1998;392:512–516. doi: 10.1038/33185. [DOI] [PubMed] [Google Scholar]
- Iordanidou P, Aggelidou E, Demetriades C, Hadzopoulou-Cladaras M. Distinct amino acid residues may be involved in coactivator and ligand interactions in hepatocyte nuclear factor-4alpha. J Biol Chem. 2005;280:21810–21819. doi: 10.1074/jbc.M501221200. [DOI] [PubMed] [Google Scholar]
- Karpichev IV, Luo Y, Marians RC, Small GM. A complex containing two transcription factors regulates peroxisome proliferation and the coordinate induction of beta-oxidation enzymes in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:69–80. doi: 10.1128/mcb.17.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci U S A. 1999;96:7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold P, Perrimon N. Drosophila and the genetics of the internal milieu. Nature. 2007;450:186–188. doi: 10.1038/nature06286. [DOI] [PubMed] [Google Scholar]
- Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol . 2001;2:RESEARCH0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louet JF, Hayhurst G, Gonzalez FJ, Girard J, Decaux JF. The coactivator PGC-1 is involved in the regulation of the liver carnitine palmitoyltransferase I gene expression by cAMP in combination with HNF4 alpha and cAMP-response element-binding protein (CREB) J Biol Chem. 2002;277:37991–38000. doi: 10.1074/jbc.M205087200. [DOI] [PubMed] [Google Scholar]
- Michalik L, Auwerx J, Berger JP, Chatterjee VK, Glass CK, Gonzalez FJ, Grimaldi PA, Kadowaki T, Lazar MA, O’Rahilly S, et al. International Union of Pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacol Rev. 2006;58:726–741. doi: 10.1124/pr.58.4.5. [DOI] [PubMed] [Google Scholar]
- Min KT, Benzer S. Preventing neurodegeneration in the Drosophila mutant bubblegum. Science. 1999;284:1985–1988. doi: 10.1126/science.284.5422.1985. [DOI] [PubMed] [Google Scholar]
- Oba Y, Ojika M, Inouye S. Characterization of CG6178 gene product with high sequence similarity to firefly luciferase in Drosophila melanogaster. Gene. 2004;329:137–145. doi: 10.1016/j.gene.2003.12.026. [DOI] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanker L, Necakov AS, Sampson HM, Ni R, Hu C, Thummel CS, Krause HM. Dynamic regulation of Drosophila nuclear receptor activity in vivo. Development. 2006;133:3549–3562. doi: 10.1242/dev.02512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps C, Gburcik V, Suslova E, Dudek P, Forafonov F, Bot N, MacLean M, Fagan RJ, Picard D. Fungi and animals may share a common ancestor to nuclear receptors. Proc Natl Acad Sci U S A. 2006;103:7077–7081. doi: 10.1073/pnas.0510080103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee J, Inoue Y, Yoon JC, Puigserver P, Fan M, Gonzalez FJ, Spiegelman BM. Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proc Natl Acad Sci U S A. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo P, Matern D, Bennett MJ. Fatty acid oxidation disorders. Annu Rev Physiol. 2002;64:477–502. doi: 10.1146/annurev.physiol.64.082201.154705. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Maina CV, Gissendanner CR, Laudet V, Sluder A. Explosive lineage-specific expansion of the orphan nuclear receptor HNF4 in nematodes. J Mol Evol. 2005;60:577–586. doi: 10.1007/s00239-004-0175-8. [DOI] [PubMed] [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Rusten TE, Lindmo K, Juhász G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Shih DQ, Dansky HM, Fleisher M, Assmann G, Fajans SS, Stoffel M. Genotype/phenotype relationships in HNF-4alpha/MODY1: haploinsufficiency is associated with reduced apolipoprotein (AII), apolipoprotein (CIII), lipoprotein(a), and triglyceride levels. Diabetes. 2000;49:832–837. doi: 10.2337/diabetes.49.5.832. [DOI] [PubMed] [Google Scholar]
- Stoffel M, Duncan SA. The maturity-onset diabetes of the young (MODY1) transcription factor HNF4alpha regulates expression of genes required for glucose transport and metabolism. Proc Natl Acad Sci USA. 1997;94:13209–13214. doi: 10.1073/pnas.94.24.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan AA, Thummel CS. Temporal profiles of nuclear receptor gene expression reveal coordinate transcriptional responses during Drosophila development. Mol Endocrinol. 2003;17:2125–2137. doi: 10.1210/me.2002-0430. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 2005a;3:e53. doi: 10.1371/journal.pbio.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gilst MR, Hadjivassiliou H, Yamamoto KR. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc Natl Acad Sci U S A. 2005b;102:13496–13501. doi: 10.1073/pnas.0506234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisely GB, Miller AB, Davis RG, Thornquest AD, Jr, Johnson R, Spitzer T, Sefler A, Shearer B, Moore JT, Miller AB, et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure. 2002;10:1225–1234. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- Wolfrum C, Borrmann CM, Borchers T, Spener F. Fatty acids and hypolipidemic drugs regulate peroxisome proliferator-activated receptors alpha - and gamma-mediated gene expression via liver fatty acid binding protein: a signaling path to the nucleus. Proc Natl Acad Sci U S A. 2001;98:2323–2328. doi: 10.1073/pnas.051619898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, Fajans SS, Signorini S, Stoffel M, Bell GI. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- Yang P, Sampson HM, Krause HM. A modified tandem affinity purification strategy identifies cofactors of the Drosophila nuclear receptor dHNF4. Proteomics. 2006;6:927–935. doi: 10.1002/pmic.200500230. [DOI] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- Zhong W, Sladek FM, Darnell JE., Jr The expression pattern of a Drosophila homolog to the mouse transcription factor HNF-4 suggests a determinative role in gut formation. Embo J. 1993;12:537–544. doi: 10.1002/j.1460-2075.1993.tb05685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinke I, Schutz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. Embo J. 2002;21:6162–6173. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.