Abstract
In T cells, the stochasticity of protein expression could contribute to the useful diversification of biological functions within a clonal population or interfere with accurate antigen discrimination. Combining computer modeling and single-cell measurements, we examined how endogenous variation in the expression levels of signaling proteins might affect antigen responsiveness during T cell activation. We found that the CD8 coreceptor fine-tunes activation thresholds, while SHP-1 phosphatase digitally regulates cell responsiveness. Stochastic variation in the expression of these proteins generates substantial diversity of activation within a clonal population of T cells, but co-regulation of CD8 and SHP-1 levels ultimately limits this very diversity. These findings reveal how eukaryotic cells can draw on regulated variation in gene expression to achieve phenotypic variability in a controlled manner.
In biological systems, stochasticity of protein expression can contribute to useful phenotypic variability within a clonal population (1), yet it also may compromise the reliability of a cellular response. How cells achieve robust physiological performance while experiencing substantial variation in the expression of molecules that control their functional activities has yet to be fully explained (2). Theoretical and experimental studies at the single-cell level have described the nature and origin of fluctuations in gene and protein expression (3–7), and have identified key molecular components controlling phenotypic variability. Most of these have been conducted on single-cell organisms, focusing on bacterial chemotaxis and competence (8–10).
Understanding the consequences of variation in protein expression is especially relevant for T lymphocyte activation in the immune system. Confrontation of these cells with foreign and self ligands that bind their receptors initiates a signaling-based decision between productive activation and tolerance (11). Reliability of this process is critical for the proper functioning of the immune system and avoidance of autoimmunity (12). Nevertheless, ligand discrimination must also be flexible during T cell development (13), and diverse to allow differentiation between short-lived effector and long-lived memory cells during antigen-induced signaling responses (14, 15). Here we examined how differences in protein expression within a clonal population might affect these dual features of robust and yet adaptable T cell responses to antigenic ligands.
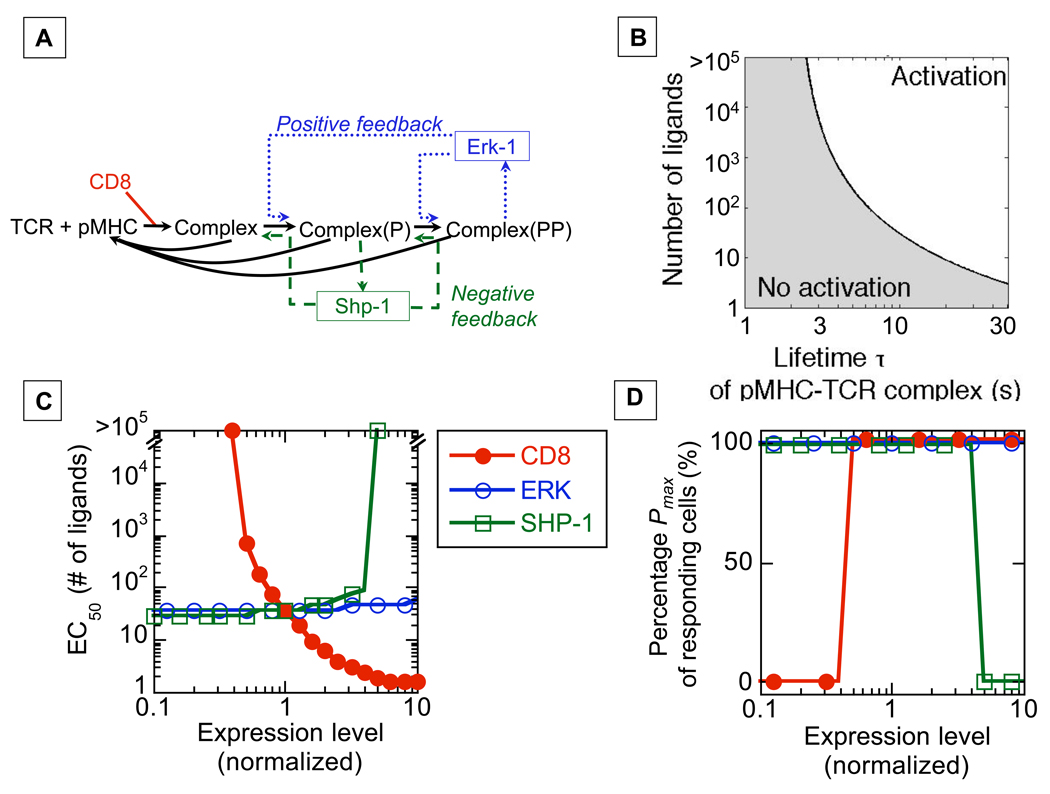
We first theoretically tested how the natural variability in signaling components expression within T cells might affect their response. To this end, a previously established computer model of T cell activation (16–19) was employed wherein kinetic proofreading, enforced by two competing signaling feedback loops, accounts for sharp ligand discrimination (Fig. 1A)‥ Signal transduction by the T cell receptor (TCR) of cytotoxic T cells is initiated when the coreceptor CD8 co-associates with TCRs engaged with their ligands (peptide-bound MHC class I molecules or pMHC). Negative feedback associated with the phosphatase SHP-1, limits spurious stimulation by ligands that have weak affinity to the TCR (20). Positive feedback, involving the activation of the kinase ERK-1, maintains strong signaling if the cell is confronted with more strongly binding ligands (20). Because ERK-1 activation is digital (‘all-or-none’) in T cells (19), we defined the half-maximal effective concentration EC50 as the minimal number of ligands able to trigger phosphorylation of ERK-1 into ppERK in 50% of the cells. Our model predicts that EC50 is sharply dependent on the lifetime of the TCR-pMHC complex (Fig. 1B).
Fig. 1.
Computational model of T cell activation links heterogeneity in protein expression with variability in cell response. A) Model of proximal T cell signaling involved in ligand discrimination. B) Predictions for digital ERK activation by different numbers of pMHC ligands, with varied lifetimes t of interaction with TCR. We define activation as 100% of ERK becoming doubly phosphorylated and inactivation as no doubly phosphorylated ERK. The dependence of EC50 on τ is delineated by the black curve. C-D) Predictions for the effect of varied expression of proteins on EC50 for an agonist ligand (τ =10s) and on the percentage Pmax of cells responding with EC50<105 (see Fig. S8 for other ligands). Expression levels of proteins are normalized to the average wild-type level.
This model was then probed to examine how variability in expression of signaling components might impact the T cell response to antigen (21), revealing that signaling components would fall into three categories: non-critical components, defined as signaling proteins whose variation in expression within the observed range does not measurably affect ligand response; analog regulators, which are signaling proteins whose level of expression fine-tunes EC50; and digital regulators whose level of expression switches a cell between responsive and unresponsive states (among responsive cells, the level of digital regulator does not affect the EC50).
Despite its key role in regulating ligand discrimination, our model predicted ERK-1 to be a non-critical regulator because its average expression is in excess, such that variation within physiological range would not affect the T cell response (Fig. 1C). The CD8 co-receptor was predicted by the model to be an analog regulator (Fig. 1C): higher levels of expression of CD8 considerably decrease the minimum number of ligands required for effective activation, without affecting the percentage of cells that can respond to the maximum dose of ligands (Pmax). In contrast, the model predicts that SHP-1 would be a digital regulator. Thus, below a critical level (four times above the average level of SHP-1) the TCR signaling network responds consistently to strong agonists, with little variation in activation threshold EC50 (Fig. 1C). However, in cases where SHP-1 exceeds the critical level, the EC50 for the signaling network becomes infinite, and Pmax becomes null (Fig. 1D). This means that, as SHP-1 levels increase, an increasing fraction of T cells becomes unresponsive to any physiologically relevant amount of ligands, while the other cells remain as sensitive as cells expressing low levels of SHP-1.
Simple mechanisms can explain how CD8 and SHP-1 differently regulate TCR signaling. Because Lck, a kinase associated with the CD8 coreceptor, directly phosphorylates antigen-engaged TCRs, lower levels of CD8 may be compensated by increased ligand availability, leading to analog mode regulation. In contrast, at a low level of SHP-1, ligands fail to activate enough of that phosphatase to prevent an early digital ERK response that, in turn, protects TCRs from dephosphorylation by SHP-1. This makes the precise level of the SHP-1 irrelevant under these conditions. At higher levels of SHP-1, negative feedback associated with phosphatase activity rapidly affects enough TCRs to prevent ERK activation and suppress effective signaling: the cells become unresponsive to any level of ligand. In combination, these latter effects lead to digital regulation within the T cell population.
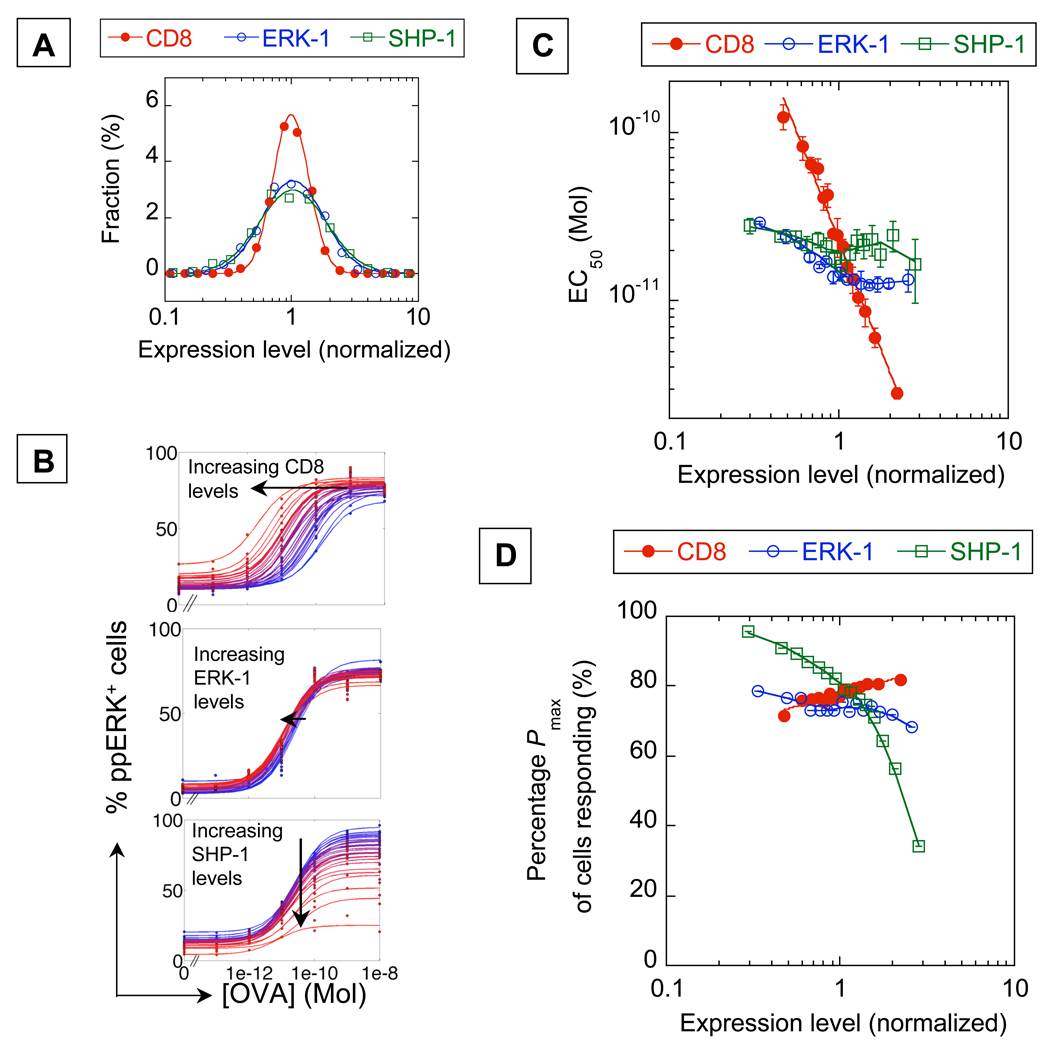
To test these predictions, we used a quantitative assessment of protein expression in individual cells, based on antibody staining and flow cytometry (Fig. S1–Fig. S4). We studied OT-1 TCR-transgenic Rag1 knock-out CD8+ T cell clones i.e. cells that express only a single TCR specific for Kb/OVA pMHC (13). For these cells, the distribution of expression for CD8, ERK-1, and SHP-1 were lognormal with coefficients of variations equal to 0.32, 0.56, and 0.62 respectively (Fig. 2A). Thus, 99% of cells express CD8 within a 3-fold range of one another, and ERK-1 and SHP-1 within a 6-fold range. The large range in expression levels of these key signaling components, in conjunction with our computer predictions of response variability (Fig. 1C-D), raised the possibility that a clonal population of T cells would not respond homogeneously to antigen stimulation.
Fig. 2.
Experimental single-cell analysis classifies CD8, ERK-1, and SHP-1 respectively as analog, non-critical, and digital regulators of T cell response. A) Distribution of endogenous levels of these signaling proteins in OT-1 T lymphocytes. B) Single-cell analysis of OT-1 T cell responses to varying concentrations of OVA peptide, for different levels of expressions of CD8, ERK-1, and SHP-1 (blue: low levels, red: high levels). C) EC50 and (D) Pmax for different levels of signaling proteins, obtained by fitting the curves in B. The full lines are eye guides. Error bars are standard errors for triplicate samples.
To examine this, multiparameter flow cytometry was used to simultaneously analyze individual cells for both endogenous protein expression and the TCR signaling response to antigen. Ligand response was measured by intracellular antibody staining for ppERK (22) five minutes after stimulation by peptide-pulsed antigen presenting cells (4–7,21). Consistent with our model predictions, variations in the levels of CD8, ERK-1, and SHP-1 were found to have substantially different effects on T cell responsiveness (Fig. 2B and S6). CD8 functioned as a positive analog regulator, such that subpopulations with varied CD8 levels displayed different EC50, but had nearly equal Pmax (Fig. 2B). A sixfold difference in membrane CD8 expression was associated with a 100-fold change in EC50 (Fig. 2C and S5). Our model predicted that very low CD8 levels would reduce Pmax, although this would occur only beyond the normal physiological range of CD8 expression (Fig. 1C and Fig. 2A). SHP-1 acted as a negative digital regulator, again consistently with the predictions of the model: subpopulations with varied levels of SHP-1 expression had different Pmax, while the responsive fractions had the same EC50 (Fig. 2C). A 2-fold increase in SHP-1 levels was associated with a 2.5-fold decrease in Pmax (Fig. 2D). In contrast to CD8 and SHP-1, and also in agreement with the simulations, variation in ERK-1 expression only marginally affected the T cell response in terms of either EC50 or Pmax (Fig. 2C-D).
These observations demonstrate that a clonal population of T cells can display substantial phenotypic variability based on stochastic variation in the expression of signaling proteins (here CD8 and SHP-1). In contrast, experimental measurements of IFNγ (23), calcium (24), or ppERK (19) responses suggest that the T cell antigen response can in some circumstances be highly uniform (robust) within a clonal population. We then identified two different mechanisms, associated with the different categories of signaling components, which enforce such robustness despite stochasticity in protein expression. Digital regulators like SHP-1 maintain constant EC50 but at the expense of a fraction of cells made unresponsive (Fig. 2D), a parameter not often examined in published studies. Analog regulators modulate the cell’s responsiveness in a sensitive manner, so their variability in expression levels must be constrained to limit variability in response (6,7), and this is seen for CD8 (Fig. 2A & C).
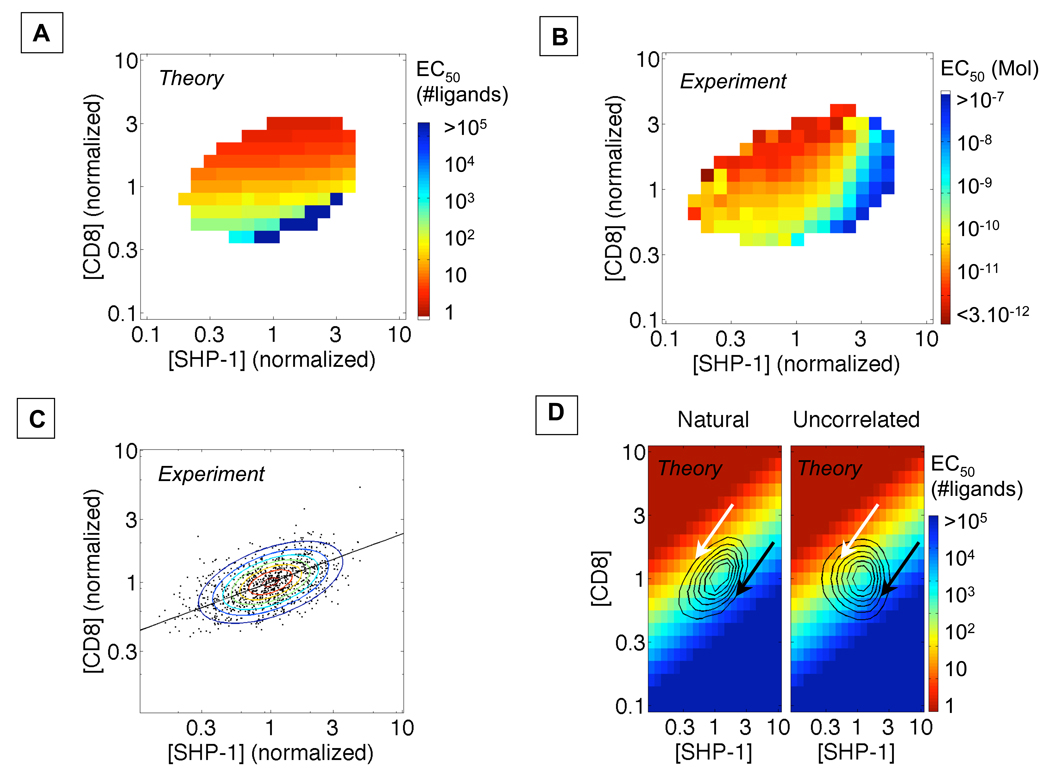
We also considered a third, less obvious, constraint mechanism, based on recent evidence for co-variation in gene expression among components in functional networks (7, 9). The contradictory effects of positive and negative modulators might cancel out if their expression levels were co-regulated, reducing response variation in the population. To examine this hypothesis, we examined computer-modeled responses for the effects of varying levels of both CD8 and SHP-1 (Fig. 3A). These analyses revealed that, for any increase of SHP-1 level, one can find a corresponding increased CD8 level that maintains a constant EC50 (Fig. 3A), implying that co-regulation of SHP-1 and CD8 expression would limit the diversity of the T cell activation.
Fig. 3.
Covariation of CD8 and SHP-1 levels limits the frequencies of hypo- and hyperresponsive cells. Computer model (A) and single-cell experimental measurements (B) for T cell activation demonstrate how the EC50 for ppERK responsiveness is determined by both CD8 and SHP-1 levels. C) Single-cell analysis of OT-1 T cell blasts shows a correlation between expression of SHP-1 and CD8. The line is the fit log([CD8])=0.4*log([SHP-1]). D) Overlay of a fitted colormap of EC50 with the contour plot of the distribution of the levels of CD8 and SHP-1 (left panel: for the endogenous/co-regulated population; right panel: for a theoretical population with uncorrelated levels of CD8 and SHP-1 keeping their individual measured distributions).CD8/SHP-1 co-regulation limits the number of hypo- and hyper-responsive cells (indicated by black and white arrows respectively).
To explore this prediction experimentally, we analyzed the ppERK dose-response for different CD8 and SHP-1 levels in individual OT-1 T cell blasts and found a good match with our computer simulations (Fig. 3B). Moreover, our analyses revealed that T cell blasts co-regulate their CD8 and SHP-1 levels (Fig. 3C). To quantify how this correlation might limit response functional diversity, we estimated the distribution of EC50 by integrating over the distribution of CD8 and SHP-1 levels (Fig. 3B-C) (21). For the natural distribution of signaling proteins (Fig. 3D – left panel), we found that the resulting distribution of EC50 was lognormal with a coefficient of variation of 5.0 (Fig. S9). For a hypothetical distribution of CD8 and SHP-1, with the same individual distributions but without co-regulation (Fig. 3D – right panel), the distribution of EC50 would also be lognormal but with a larger coefficient of variation of 18 (Fig. S9). In other words, the actual fraction of hyper-responsive OT-1 T cell blasts whose EC50 is below 0.1pM is 0.03% but this fraction would be predicted to be ten-fold higher (0.3%) without co-regulation of CD8 and SHP-1 levels (see arrows in Fig. 3D). Thus, co-regulation decreases the number of hyper-responsive cells and potentially limits the risk of selfresponsiveness and autoimmune activation among T cells. Several non-exclusive mechanisms may account for this co-regulated expression, include gene locus colocalization (4), control of transcription by shared factors, and targeting of transcripts by the same miRNA, and will require further study to elucidate.
In conclusion, this study predicted theoretically and confirmed experimentally that signaling molecules within a network can have qualitatively distinct effects on cell responsiveness. It also revealed that intraclonal differences in protein expression generate dispersion in responsiveness among individual cells, and demonstrated how co-regulation of expression among these proteins limits this phenotypic variability. For T lymphocytes, these insights help explain how ligand discrimination can be tuned through regulation of signaling components (13, 24) and diversified (14) such that pathogen reactivity is preserved without encouraging autoimmunity. More generally, these observations suggest how cells may leverage regulated stochasticity of intracellular processes to generate controlled variability in physiological performance.
Supplementary Material
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. Their manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and Notes
- 1.Kaern M, Elston TC, Blake WJ, Collins JJ. Nat Rev Genet. 2005;6:451. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 2.Barkai N, Leibler S. Nature. 1997;387:913. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 3.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Science. 2002;297:1183. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 4.Becskei A, Kaufmann BB, van Oudenaarden A. Nat Genet. 2005;37:937. doi: 10.1038/ng1616. [DOI] [PubMed] [Google Scholar]
- 5.Newman JR, et al. Nature. 2006;441:840. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 6.Bar-Even A, et al. Nat Genet. 2006;38:636. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 7.Sigal A, et al. Nature. 2006;444:643. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- 8.Korobkova E, Emonet T, Vilar JM, Shimizu TS, Cluzel P. Nature. 2004;428:574. doi: 10.1038/nature02404. [DOI] [PubMed] [Google Scholar]
- 9.Kollmann M, Lovdok L, Bartholome K, Timmer J, Sourjik V. Nature. 2005;438:504. doi: 10.1038/nature04228. [DOI] [PubMed] [Google Scholar]
- 10.Suel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Science. 2007;315:1716. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- 11.Davis MM, et al. Annu Rev Immunol. 2007;25:681. doi: 10.1146/annurev.immunol.24.021605.090600. [DOI] [PubMed] [Google Scholar]
- 12.Germain RN. Science. 2001;293:240. doi: 10.1126/science.1062946. [DOI] [PubMed] [Google Scholar]
- 13.Hogquist KA, et al. Cell. 1994;76:17. [Google Scholar]
- 14.Chang JT, et al. Science. 2007;315:1687. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 15.Stemberger C, et al. Immunity. 2007;27:985. doi: 10.1016/j.immuni.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 16.Germain RN, Stefanova I. Annu Rev Immunol. 1999;17:467. doi: 10.1146/annurev.immunol.17.1.467. [DOI] [PubMed] [Google Scholar]
- 17.Qi SY, Groves JT, Chakraborty AK. Proc Natl Acad Sci USA. 2001;98:6548. doi: 10.1073/pnas.111536798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stefanova I, et al. Nat Immunol. 2003;4:248. doi: 10.1038/ni895. [DOI] [PubMed] [Google Scholar]
- 19.Altan-Bonnet G, Germain RN. PLoS Biol. 2005;3:e356. doi: 10.1371/journal.pbio.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feinerman O, Germain RN, Altan-Bonnet G. Molecular Immunology. 2008;45:619. doi: 10.1016/j.molimm.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.See supporting materials on Science Online.
- 22.Irish JM, et al. Cell. 2004;118:217. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 23.Viola A, Lanzavecchia A. Science. 1996;273:104. doi: 10.1126/science.273.5271.104. [DOI] [PubMed] [Google Scholar]
- 24.Li QJ, et al. Cell. 2007;129:147. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 25.We thank M. Van der Brink for the generous use of the flow cytometer; and J. Allison, N. Altan-Bonnet, P. Cluzel, L. Denzin, M. Huse, E. O'Shea, N. Socchi and D. Sant′Angelo for comments and discussion
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.