Abstract
The penC resistance gene was previously characterized in a FA19 penA mtrR penB gonococcal strain (PR100) as a spontaneous mutation that increased resistance to penicillin and tetracycline. We show here that antibiotic resistance mediated by penC is the result of a Glu-666 to Lys missense mutation in the pilQ gene that interferes with the formation of the SDS-resistant high-molecular-mass PilQ secretin complex, disrupts piliation, and decreases transformation frequency by 50-fold. Deletion of pilQ in PR100 confers the same level of antibiotic resistance as the penC mutation, but increased resistance was observed only in strains containing the mtrR and penB resistance determinants. Site-saturation mutagenesis of Glu-666 revealed that only acidic or amidated amino acids at this position preserved PilQ function. Consistent with early studies suggesting the importance of cysteine residues on stability of the PilQ multimer, mutation of either of the two cysteine residues in FA19 PilQ led to a similar phenotype as penC: increased antibiotic resistance, loss of piliation, intermediate levels of transformation competence, and absence of SDS-resistant PilQ oligomers. These data show that a functional secretin complex can enhance the entry of antibiotics into the cell and suggest that the PilQ oligomer forms a pore in the outer membrane through which antibiotics diffuse into the periplasm.
INTRODUCTION
For nearly 30 years, sexually transmitted infections caused by Neisseria gonorrhoeae were routinely treated with penicillin. During that time, however, clinical isolates gradually became more resistant until treatment failure occurred and penicillin was discontinued as a therapeutic agent. Resistance to penicillin in the gonococci occurs by two routes: the plasmid-encoded production of a TEM-1-like ß-lactamase, and multiple chromosomally mediated mutations that increase resistance in a stepwise manner (Cannon and Sparling, 1984). As shown in an elegant series of studies by Sparling and colleagues, chromosomally mediated resistance genes can be transferred from a high-level resistant strain (e.g. FA6140) to a susceptible strain (e.g. FA19) by transformation and homologous recombination (Maness and Sparling, 1973; Sarubbi et al., 1974).
The mechanisms underlying chromosomally mediated penicillin resistance in the gonococci are complex and not fully understood. Multiple gene mutations are required for high-level resistance, but none of these mutations increases resistance dramatically. Only when these mutations are combined do they increase resistance to levels above that achieved clinically. There are five gene mutations known to contribute to high-level penicillin resistance (Minimum Inhibitory Concentration (MIC) ≥ 2 μg/ml) in chromosomally mediated resistant N. gonorrhoeae (CMRNG) strains (Ropp et al., 2002), and these genes are transferred from a resistant strain to a susceptible strain in a specific order. The first resistance determinant transferred is penA, which encodes altered forms of penicillin-binding protein 2 (PBP 2) with decreased rates of acylation by penicillin (Brannigan et al., 1990; Dowson et al., 1989). The second resistance locus transferred is mtrR, which carries a mutation in the promoter region of the MtrR repressor that abolishes transcription of the mtrR gene and results in overexpression of the MtrC-MtrD-MtrE efflux pump (Hagman et al., 1995; Pan and Spratt, 1994). Overexpression of the MtrC-MtrD-MtrE efflux pump increases penicillin resistance only slightly, but is required for further increases in resistance mediated by subsequent resistance genes (Hagman et al., 1997; Veal et al., 2002). The third resistance determinant transferred, designated penB, corresponds to porin alleles carrying mutations on loop 3 that decrease the concentration of antibiotics in the periplasmic space (Gill et al., 1998; Olesky et al., 2002).
Transformation of a third-level resistant strain (FA19 penA mtrR penB) to a higher level of resistance is very difficult to achieve in the laboratory (Dougherty, 1986; Faruki and Sparling, 1986). We previously investigated the role of the ponA gene encoding PBP 1, the other essential PBP in N. gonorrhoeae, in high-level penicillin resistance (Ropp and Nicholas, 1997; Ropp et al., 2002). The ponA gene from high-level penicillin-resistant strains contains a single base mutation that decreases the rate of acylation of PBP 1 by 3- to 4-fold. Reversion of the mutant ponA allele (ponA1) back to the wild type sequence in the CMRNG strain FA6140 decreased the MIC for penicillin by 2-fold, conclusively demonstrating that ponA1 is involved in high level resistance. However, additional determinants are required to enable ponA1 to increase resistance, since no increase in resistance was observed when ponA1 was transferred to PR100 (FA19 penA mtrR penB), a third-level transformant strain.
In a search for additional mutations that would increase resistance above that conferred by the first three determinants, we identified a mutation, termed penC, that increased resistance to both penicillin and tetracycline (Ropp et al., 2002). Resistance due to penC is the result of a spontaneous mutation and is readily transferable between strains. Surprisingly, introduction of the ponA1 allele into PR100 increased resistance two-fold, indicating that the penC allele conferred the ability to increase resistance to ponA1 strains. However, the absence of penC in CMRNG strains suggests that clinical isolates use another as yet undetermined mechanism to further increase resistance levels.
In this report, we show that the increased resistance due to penC is the result of a point mutation in the pilQ gene (pilQ2). PilQ (previously known as Outer Membrane Protein-Macromolecular Complex or OMP-MC), a member of the secretin family of proteins, is a major component of the gonococcal outer membrane (Newhall et al., 1980). PilQ is required for formation of the Type IV pilus, which is involved in twitching motility, cell attachment and invasion of epithelia, and DNA transformation (Tonjum and Koomey, 1997). PilQ forms a ring-shaped multimeric complex, most likely a dodecamer (Collins et al., 2001), with the pilus formed by a repeating polymer of pilin subunits protruding from the central cavity (Collins et al., 2003; Collins et al., 2004). The pilQ2 mutation disrupts the formation of an SDS-resistant multimeric complex, causes a loss of piliation, and decreases transformation competence by 50-fold. Increased resistance to a wide range of antibiotics due to acquisition of pilQ2 is observed only in strains containing the mtrR and penB resistance determinants. The increase in MICs of a variety of antibiotics in these strains highlights the importance of a functional PilQ multimer in the entry of antibiotics into the cell.
RESULTS
Antibiotic resistance due to penC is the result of a point mutation in pilQ
We previously reported the isolation of spontaneously arising colonies of PR100 (FA19 penA mtrR penB; see Table 1) having 2-fold increases in the MICs for both penicillin and tetracycline (Ropp et al., 2002). This mutation, termed penC, was not due to additional mutations in the penA, mtrR, or penB genes. Because genomic DNA from PR100 penC transformed PR100 to increased penicillin resistance with high frequency, we cloned the penC gene by generating a size-fractionated ClaI plasmid library of genomic DNA from one of the PR100 penC colonies and identifying plasmids capable of transforming PR100 to increased penicillin resistance (see Materials and Methods for more details). Three individual clones were identified that harbored a plasmid with an identical 10.6 kb insert. Transformation experiments with PCR fragments amplified from the DNA insert localized the penC mutation to the pilQ gene, and sequencing of the pilQ gene revealed that the penC phenotype was the result of a single G to A mutation at nucleotide 1996 in the pilQ gene that changes Glu-666 to Lys (numbering from the initiating methionine). We refer to the penC allele hereafter as pilQ2.
Table 1. Strains and plasmids used in this study.
| Strain or plasmid | Descriptiona | Reference |
|---|---|---|
| pUC18-pilQ2-kan | Plasmid containing bp 1333-2196 of pilQ2, a kan resistance cassette, and an additional 300 bp of downstream sequence |
This study |
| pUC18-pilQFA19-kan | Same as above without the mutation at Glu-666 | This study |
| pUNCH290 | Plasmid containing pilQ disrupted by the Ω fragment | (Chen et al., 2004) |
| pMO/porIB-G120K-erm | Plasmid containing the mature coding sequence of PIB with a G120K mutation, an erm resistance cassette, and additional 300 bp of downstream sequence |
(Olesky et al., 2002) |
| pSY6 | Plasmid containing gonococcal DNA fragment encoding nalidixic acid resistance |
(Stein et al., 1991) |
| FA19 | Penicillin-susceptible laboratory strain | (Maness and Sparling, 1973) |
| FA6140 | Penicillin-resistant clinical isolate | (Danielsson et al., 1986) |
| FA19 penA4 | FA19 × FA6140 DNA (0.03 μg/ml penb) | (Ropp et al., 2002) |
| FA19 penA4 pilQ2 | FA19 penA4 X pUC18-pilQ2-kan (50 μg/ml kan) | This study |
| FA19 penA4 pilQ::Ω | FA19 penA4 X pUNCH290 (100 μg/ml spt) | This study |
| FA19 penA4 mtrR | FA19 penA4 X FA6140 DNA (1 mg/ml Triton X-100) | (Ropp et al., 2002) |
| FA19 penA mtrRpilQ2 | FA19 penA4 mtrR X pUC18-pilQ2-kan (50 μg/ml kan) | This study |
| FA19 penA mtrR pilQ::Ω | FA19 penA4 mtrR X pUNCH290 (100 μg/ml spt) | This study |
| FA19 penB | FA19 X pMO/porIB-G120K-erm (6 μg/ml erm) | This study |
| FA19 penB pilQ2 | FA19 penB X pUC18-pilQ2-kan (50 μg/ml kan) | This study |
| FA19 mtrR penB | FA19 mtrR X porIB-G120K-erm (6 μg/ml erm) | This study |
| FA19 mtrR penB pilQ2 | FA19 mtrR penB X pUC18-pilQ2-kan (50 μg/ml kan) | This study |
| PR100 | FA19 penA4 mtrR penB5 | (Ropp et al., 2002) |
| PR100 pilQ2 | PR100 X pUC18-penC-kan (50 μg/ml kan) | This study |
| PR100 pilQ::Ω | PR100 X pUNCH290 (100 μg/ml spt) | This study |
| FA6140 pilQ2 | FA6140 X pUC18-pilQ2-kan (50 μg/ml kan) | This study |
| FA6140 pilQ::Ω | FA6140 X pUNCH290 (100 μg/ml spt) | This study |
| RM11.2 recA6 | FA1090 with IPTG-regulatable recA allele | (Long et al., 1998) |
| RM11.2 recA6 pilQFA19 | RM11.2 recA6 X pUC18-pilQFA19-kan (50 μg/ml kan) | This study |
| RM11.2 recA6 pilQ2 | RM11.2 recA6 X pUC18-pilQ2-kan (50 μg/ml kan) | This study |
| SZ3 | RM11.2 recA6 penA4 mtrR penB5 | This study |
| SZ3 ΔpilE | pilE deletion strain of SZ3 | This study |
| SZ3 pilT::erm | SZ3 X RM11.2 recA6 pilT::erm (6 μg/ml erm) | This study |
| SZ3 ΔpilP | SZ3 X pUC18-ΔpilP (0.5 μg/ml pen) | This study |
| SZ3 pilQ2 | SZ3 X pUC18-pilQ2-kan (50 μg/ml kan) | This study |
| SZ3 pilQ::Ω | SZ3 X pUNCH290 (100 μg/ml spt) | This study |
Strains were constructed by homologous recombination as described in Materials and Methods and are shown as parental strain X donor DNA (selection agent). All relevant genotypes were confirmed by DNA sequencing.
The abbreviations are: pen, penicillin; kan, kanamycin; spt, spectinomycin; erm, erythromycin.
Effect of pilQ2 on pilus formation and transformation efficiency
Transformation of PR100 with pUC18-pilQ2 plasmid DNA to increased penicillin resistance resulted in a nonpiliated colony morphology, but how pilQ2 influences pilus formation was unclear. Thus, we generated isogenic strains of RM11.2 recA6, a defined pilin variant of strain FA1090 (Long et al., 2001) with a regulatable recA gene that allows for IPTG control of pilin antigenic variation (Seifert, 1997), containing either the FA19 pilQ allele (pilQFA19; FA19 and FA1090 PilQ differ by two amino acids in the C-terminal domain) or the pilQ2 allele. We examined the piliation state of these strains by immunogold transmission electron microscopy with an anti-pilin peptide antibody (Long et al., 2003). All of the RM11.2 recA6 derivatives were confirmed to have identical pilE sequences, which assured that differences in piliation were not due to pilin variation. Approximately 50% of RM11.2 recA6 or RM11.2 recA6 pilQFA19 cells had pili and the majority of pili were in bundles (Fig. 1A, left and middle panels). In contrast, two independently derived RM11.2 recA6 pilQ2 mutants had no observable distinct pili on 955 cells examined (Fig. 1A, right panel). However, in a small number of RM11.2 recA6 pilQ2 cells (∼0.6%), we observed short, membrane-attached projections that interacted specifically with the anti-pilin antibody (Fig. 1B). These projections did not have the same morphology as pili (Fig. 1A) and therefore represent an alternative expression of the pilin protein on the cell surface of pilQ2 strains.
Figure 1. Electron micrographs of immunogold-labeled pili from strain RM11.2 recA6 with different pilQ alleles.
(A) RM11.2 recA6 (left panel), RM11.2 recA6 pilQFA19 (middle panel), and RM11.2 recA6 pilQ2 (right panel) were constructed as described in Materials and Methods. Strains were grown on GCB plates and colonies were lifted directly onto Formavar-coated grids. Pili were detected with an anti-pilin antibody and detected with a gold-conjugated secondary antibody as described previously (Long et al., 2003). Whereas most of the RM11.2 recA6 pilQ2 cells were as shown in the right panel of (A), a small number of cells had small projections that were labeled with the anti-pilin antibody (B).
We also examined the effect of the pilQ2 mutation on transformation competence. The transformation frequency of a gyrB allele encoding resistance to nalidixic acid (Stein et al., 1991) was determined with SZ3 (RM11.2 recA6penA mtrR penB; see Table 1) alone and SZ3 harboring either the pilQ2, ΔpilE, pilT::erm, or pilQ::Ω alleles. As shown in Fig. 2, the pilQ2 allele resulted in a 50-fold reduction in transformation frequency, whereas the pilT, pilE, and pilQ::Ω alleles reduced the transformation frequency to ≤1 × 10−6. Thus, although the pilQ2 mutation appears to disrupt piliation based on electron microscopy, the mutation has a much less dramatic affect on transformation frequency. Taken together, these data suggest that the pilQ2 mutation results in PilQ complexes that are not functional for pilus assembly, but retain the partial ability to facilitate DNA transformation. It is possible that the pilin-expressing membraneous projections provide a means for transforming DNA to enter the bacterial cell.
Figure 2. Transformation competence of SZ3 (RM11.2 recA6 penA mtrR penB) and SZ3 with pilQ2, pilQ::Ω, pilE, or pilT::erm alleles.
The indicated strains were prepared as described in Materials and Methods and incubated with 100 ng of pSY6, which confers resistance to nalidixic acid (NALr), for 5 hrs before plating. The transformation efficiency is expressed as the number of NALr transformants per CFU. The dotted line represents the lowest transformation efficiency that was measurable. The average ± S.D. for three independent experiments is shown.
Effect of the pilQ2 mutation on PilQ oligomerization
To understand the effects of the pilQ2 mutation on PilQ, we analyzed cell lysates from PR100 and PR100 pilQ2 by SDS-PAGE and Western blotting with a PilQ antibody. As shown in Fig. 3A, PilQ from PR100 migrated at two positions: at the top of the gel, which corresponds to an SDS-resistant high-molecular-mass (HMM) multimer (Drake et al., 1997) (most likely a dodecamer (Collins et al., 2001; Collins et al., 2004)), and at 75 kDa, which corresponds to a PilQ monomer. In contrast, PilQ from PR100 pilQ2 showed no evidence of the SDS-resistant multimer, but instead migrated as a band at 75 kDa with increased amounts of PilQ breakdown products. PR100 with a disrupted pilQ gene (pilQ::Ω), in which the Ω fragment encoding spectinomycin resistance (Prentki and Krisch, 1984) replaced codons 266 to 367 of the pilQ gene (Chen et al., 2004), showed a complete loss of PilQ protein (Fig. 3A). We also tested the effects of temperature on the capacity of the mutant PilQ to resist denaturation by SDS. However, no HMM PilQ multimer was observed regardless of the temperature at which the samples in SDS-PAGE loading buffer were incubated before electrophoresis (Fig. 3B).
Figure 3. Western blots of PilQ from strains with wild type and pilQ2 alleles.
Western blots of whole cell lysates (A, B) or outer membranes (C) probed with a polyclonal antibody against PilQ. The large arrow indicates the SDS-resistant high-molecular-mass PilQ multimer, whereas the small arrow indicates the PilQ monomer. In panels (A) and (C), samples in SDS-loading buffer were incubated at 100°C for 5 min before loading, while in panel (B), samples were treated for 5 min at the indicated temperature before loading. The outer membranes in (C) were prepared as described by Heckels (1977). In panel (D), proteins from FA19 (left) or FA19 pilQ2 (right) membranes extracted into SB-10 were submitted to gel filtration on Sephacryal S-500. The elution of PilQ oligomer (left graph) and total PilQ (right graph) was assessed by quantitation of Western blots as described in Materials and Methods. The arrows at the top denote the positions of protein standards (MWs are listed above the arrows). In panel (E), Western blots of whole cell lysates were probed with a monoclonal antibody directed against the pilin protein. Percentage of total pilin protein (RM11.2 recA6 taken as 100%) and S-pilin (reported as the % of total pilin in that lane) were determined by densitometry and are listed below each lane.
Next, we investigated whether the pilQ2 mutation interfered with transport of PilQ subunits to the outer membrane, where it is assembled into SDS-resistant HMM oligomers, or whether it interfered with oligomer assembly directly. Outer membranes were isolated (Heckels, 1977) and subjected to SDS-PAGE and Western blotting. As shown in Fig. 3C, a banding pattern for PilQ essentially identical to whole cell lysates was observed in each strain, suggesting that the PilQ mutant protein is transported to the outer membrane but is not present as an SDS-resistant HMM oligomer.
To determine in more detail the effects of the pilQ2 mutation on the oligomeric structure of pilQ in the bacterial envelope, we extracted total membranes from FA19 and FA19 pilQ2 with the zwitterionic detergent SB-10 and fractionated the extracts by gel filtration. The levels of PilQ oligomer (FA19) and monomer (FA19 pilQ2) from individual fractions were assessed by Western blotting and densitometry (Fig. 3D). In the wild type cells, the SDS-resistant PilQ oligomer eluted as a broad peak at a position consistent with its predicted molecular mass (∼1 MDa). Interestingly, PilQ protein from FA19 pilQ2 was found in two distinct peaks: the first peak eluted at the same position as the PilQ oligomer, whilst the second more prominent peak eluted with an approximate molecular mass of ∼200 kDa. Although larger than the mass of a PilQ monomer (75 kDa), this latter peak eluted at the same position as the SDS-sensitive PilQ monomer present in wild type cells (data not shown). This peak potentially represents the trimer structure of meningococcal PilQ reported by Collins et al. (Collins et al., 2004). These results strongly suggest that a fraction of the Glu-666 to Lys mutant of PilQ exists in the membrane in an oligomeric form, but the mutation prevents its maturation into an SDS-resistant form.
Finally, because we did not observe any pilin fibers in the pilQ2 or pilQ::Ω strains, we examined the level of pilin proteins in whole cells by Western blot analysis (Fig. 3E). The total amounts of pilin in RM11.2 recA6, RM11.2 recA6 pilQFA19, RM11.2 recA6 pilQ2, and RM11.2 recA6 pilQ::cat were very similar. The biggest difference between the individual strains was observed in the band corresponding to the proteolytic breakdown product, S-pilin. Compared to RM11.2 recA6, the small portion of pilin migrating as S-pilin was ∼2-fold higher in RM11.2 recA6 FA19 pilQ, whereas the amount of S-pilin in RM11.2 recA6 penC and RM11.2 recA6 pilQ::cat was ∼2-fold lower (Fig 3E). Thus, the pilQ2 mutation had no significant effects on the total levels of pilin, but appeared to cause a modest decrease in the levels of the minority S-pilin form.
Effect of pilQ2 on antibiotic resistance
To further investigate the effect of pilQ2 on gonococcal antibiotic resistance, we generated strains with various combinations of resistance genes in concert with either the pilQ2 or pilQ::Ω allele. When either pilQ2 or pilQ::Ω was introduced into FA19, FA19 penA, or FA19 penA mtrR, no increase in the MIC of penicillin or tetracycline was observed (Fig. 4, left panels). In contrast, introduction of either of the two pilQ alleles into PR100 (FA19 penA mtrR penB) or FA6140, a high level penicillin-resistant CMRNG isolate (Danielsson et al., 1986), conferred a 2- to 3-fold increase in the MICs for penicillin and tetracycline (Fig. 4, middle panels). The levels of resistance conferred by pilQ2 were essentially identical to those conferred by pilQ::Ω. Acquisition of pilQ2 by strains with only one of the two resistance determinants mtrR or penB was not sufficient to confer resistance to penicillin, consistent with previous data demonstrating that penB is unable to increase resistance to penicillin in the absence of mtrR (Olesky et al., submitted; (Sparling et al., 1975; Veal et al., 2002). In contrast, acquisition of pilQ2 by a strain harboring both mtrR and penB resulted in increases in both penicillin and tetracycline resistance (Fig. 4, right panels).
Figure 4. Penicillin and tetracycline resistance of strains containing wild type, pilQ2, and pilQ::Ω alleles.
FA19 harboring various combinations of the penA (mutations in PBP 2), mtrR (increased expression of the MtrC-MtrD-MtrE efflux pump) and penB (mutations in porin; see Table 1 for more information) resistance determinants was transformed with plasmids encoding mutant (pilQ2) and inactivated pilQ (pilQ::Ω) alleles and the MICs of penicillin G and tetracycline for the resulting strains were determined as described in Materials and Methods. pilQ2 and pilQ::Ω were also introduced into FA6140, a chromosomally mediated penicillin-resistant clinical isolate (Danielsson et al., 1986). *P<0.01 relative to the parental strain.
To examine the specificity of the effect of PilQ on the influx of antibiotics, we determined the MICs of a variety of antibiotics for PR100 and PR100 pilQ2 (data not shown). The MICs of ampicillin, ceftriaxone, and ciprofloxacin each increased ∼1.5- to 2-fold in PR100 pilQ2 compared to PR100, suggesting that these antibiotics readily enter the cell in a PilQ-dependent manner. In contrast, rifampin, chloramphenicol, and erythromycin showed much more modest increases (∼1.2-fold) in their MICs for PR100 pilQ2 compared to PR100. Thus, PilQ promotes the entry of a range of antibiotics into the bacterial cell, although there is some preference for hydrophilic antibiotics.
The role of Type IV Pilus accessory subunits in conferring penicillin resistance
Since the only known role for PilQ in the gonococci is for pilus assembly, we investigated the role of several accessory proteins involved in type IV pilus biogenesis on antibiotic sensitivity. We introduced deletions of pilT, pilE, and pilP into SZ3 (RM11.2 penA mtrR penB), which has a normal pilQ allele, and determined the MIC of penicillin in each of these strains (Fig. 5). Pilin proteins form the pilus fiber that requires PilQ for expression outside of the bacterial cell (Wolfgang et al., 2000), and we examined whether the presence of pilin fibers might influence the entry of penicillin into the periplasm. A ΔpilE mutant showed only a small increase in penicillin resistance, indicating that pilin fibers neither interfere nor assist with antibiotic diffusion into the periplasm. We also examined whether the pilus retraction-associated ATPase, PilT (Maier et al., 2002; Merz et al., 2000), was involved in helping penicillin enter the cell. When the pilT::erm allele was introduced into SZ3, no change in resistance was observed, demonstrating that pilus retraction is not involved in the entry of penicillin. Finally, we examined the role of PilP, which has been reported to influence PilQ multimerization (Drake et al., 1997), in altering antibiotic resistance. Deletion of the pilP gene in SZ3 resulted in decreased levels of multimeric PilQ in whole-cell lysates by Western blotting, while the levels of PilQ monomer were similar to those of the parent strain (data not shown). The MIC of penicillin for SZ3 ΔpilP increased, but the level of resistance was lower than that observed for SZ3 pilQ2 or SZ3 pilQ::Ω (Fig. 5). These data confirm that deletion of PilP decreases the amount of the SDS-resistant PilQ complex in the outer membrane, and, similar to the pilQ2 point mutation, results in a decrease in penicillin entry and a subsequent increase in resistance.
Figure 5. MICs of penicillin for SZ3 with deletions in Type IV pilus-associated genes.
SZ3 (RM11.2 recA6 penA4 mtrR penB5; see Table 1) was transformed with plasmids or genomic DNA containing inactivated or deleted alleles of pilT, pilE and pilP, and the MICs of penicillin for multiple colonies from each transformation and for SZ3 pilQ2 and SZ3 pilQ::Ω were determined as described in Materials and Methods. *P<0.01 relative to the parental strain.
Random mutagenesis of Glu-666
To investigate in more detail the importance of Glu-666 in formation of an SDS-resistant PilQ oligomer, we randomized the codon for Glu-666 in the wild type pilQ gene and transformed SZ3 to increased penicillin resistance. The pilQ gene was amplified from resistant colonies, and those genes with a silent NsiI site, which was present only in the randomized clones, were chosen for further analysis (Table 2). Sequencing of the pilQ genes from these colonies and subsequent Western blotting (data not shown) showed that substitution of Glu-666 with almost any uncharged polar or nonpolar amino acid resulted in the same phenotype as pilQ2: i.e. increased penicillin resistance (to the same level as pilQ2; Table 2) and inhibition of formation of SDS-resistant PilQ oligomers (data not shown). To our surprise, we did not identify the original Glu-666 to Lys mutation of pilQ2 in our random screening, nor did we identify any acidic, amide, or other basic residues. To investigate whether these three classes of amino acids were capable of substituting for Glu-666, we employed site-directed mutagenesis to change Glu-666 to Asp, Arg, or Gln. Transformation of SZ3 with the individual mutant plasmids revealed that only the Glu-666 to Arg mutation gave rise to penicillin-resistant colonies with frequencies above controls. The Glu-666 to Arg mutation also interfered with formation of the HMM PilQ multimer as determined by Western blotting (data not shown). These data demonstrate that only acidic or amidated amino acids are capable of replacing Glu-666 and allowing formation of the SDS-resistant PilQ multimer. Moreover, alignment of N. gonorrhoeae PilQ near the region of Glu-666 with several of the most homologous PilQ secretins from other bacterial species revealed that the amino acid in the same position as Glu-666 was either Glu, Asp, or Asn (Fig. S1), suggesting that this residue may play an important role in other secretins as well.
Table 2. Site-saturation mutagenesis of codon-666 of PilQ.
The amino acids at position-666 of PilQ that increased penicillin resistance and disrupted formation of the SDS-resistant multimer.
| Amino Acid at Position-666 | No. Clones Isolated | MICpen (μg/ml) |
|---|---|---|
| Glutamate | pilQ Wild Type | 0.5 |
| Lysine | pilQ2 | 1.2 |
| Alanine | 2 | 1.2 |
| Cysteine | 6 | 1.2 |
| Glycine | 7 | 1.2 |
| Isoleucine | 4 | 1.2 |
| Leucine | 3 | 1.2 |
| Phenylalanine | 1 | 1.2 |
| Serine | 1 | 1.2 |
| Threonine | 3 | 1.2 |
| Tryptophan | 1 | 1.2 |
| Tyrosine | 1 | 1.2 |
| Valine | 4 | 1.2 |
| Argininea | Site-directed | 1.2 |
| Aspartic Acida | Site-directed | (0.5)b |
| Glutaminea | Site-directed | (0.5)b |
Constructed by site-directed mutagenesis.
Not determined; transformation of SZ3 with pUC18-pilQ E666D or pUC18-pilQ E666Q did not give rise to resistant colonies above negative controls.
Importance of Cys-644 and Cys-653 in PilQ oligomerization
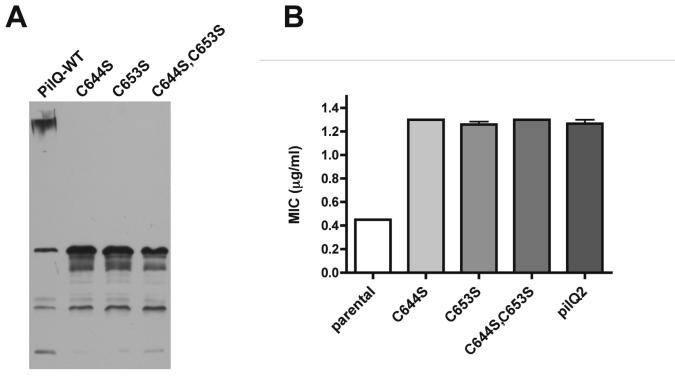
Early studies on PilQ (then named OMC or OMP-MC) revealed that the high molecular mass complex of PilQ obtained on SDS-PAGE could be converted quantitatively to a single 75 kDa subunit on SDS-PAGE following reduction and alkylation (Hansen and Wilde, 1984; Newhall et al., 1980). There are only two cysteine residues in FA1090 and FA19 PilQ, Cys-644 and Cys-653, both of which are relatively close to the site of the pilQ2 mutation described herein. To determine whether mutation of these two cysteine residues would also alter PilQ properties, we mutated the codons for the two cysteine residues of PilQ to serine either individually or together and transformed the mutant genes into strain SZ3. Strikingly, strains containing pilQ mutations of either cysteine (or both) resulted in a very similar phenotype as SZ3 pilQ2. Western blots of PilQ showed the complete absence of the high molecular mass dodecamer, increased levels of the monomer subunit (Fig. 6A), and identical MICs of penicillin compared to SZ3 pilQ2 (Fig. 6B). Transformation competence of SZ3 containing the pilQ cysteine mutants also was decreased to a similar extent as SZ3 pilQ2 (data not shown). As expected, strains harboring the mutant cysteine pilQ alleles displayed a non-piliated colony morphology. These data indicate that cysteine mutations prevent formation of an SDS-resistant PilQ complex and affect antibiotic resistance to the same extent as pilQ2.
Figure 6. Effects of Cys to Ala mutations in PilQ when expressed in SZ3.
Cys-643 and Cys-654 were changed individually or together to Ser and the mutant alleles were transformed into SZ3. (A) Western blots of PilQ in whole cell lysates. (B) MICs of penicillin for SZ3 with wild type (parental) and the indicated mutant pilQ alleles.
DISCUSSION
In the search for genes involved in high-level penicillin resistance (MIC ≥ 2 μg/ml), we identified in PR100 a spontaneously arising resistance determinant, termed penC, that conferred an increase in both penicillin and tetracycline resistance, suggesting that penC alters the general permeability of antibiotics (Ropp et al., 2002). We show here that penC results from a point mutation in the pilQ gene (pilQ2), which changes Glu-666 to Lys and prevents formation of an SDS-resistant high-molecular-mass multimer of PilQ without altering the levels of PilQ in the cell. These observations suggest that the PilQ secretin can facilitate the entry of small molecules into the bacterial cell in addition to its more well-defined role in pilin biogenesis and DNA transformation.
Two major models can account for our results. The most straightforward model is that antibiotics enter the periplasm through the pore made by the PilQ multimer and that the pilQ2 mutation blocks entry by destabilizing the pore. This model is consistent with reports that the pIV and PulD secretins function as gated pores when reconstituted into planar lipid bilayers (Marciano et al., 1999; Nouwen et al., 1999). In addition, the structure of meningococcal PilQ obtained by cryo-transmission electron microscopy (Collins et al., 2003) reveals a central cavity 65 Å in diameter at the largest end that tapers to a closed point at the other end, which needs to open to allow the pilus to transverse the outer membrane. This structure is consistent with the idea that PilQ, much like pIV and PulD, can function as a pore through which antibiotics diffuse into the periplasm.
The second major model is that the presence of a nonfunctional PilQ in the outer membrane alters the function of porin or the MtrC-MtrD-MtrE efflux pump, leading to a decrease in antibiotic entry or an increase in antibiotic efflux respectively. However, we believe this second model is unlikely for several reasons. First, both the pilQ2 and pilQ::Ω alleles had identical effects on penicillin resistance when introduced into PR100 or SZ3, even though in a pilQ2 strain the protein is still present in the outer membrane while in a pilQ::Ω strain it is absent. Secondly, neither pilQ2 nor pilQ::Ω had any effect on penicillin or tetracycline resistance when introduced into strains containing the mtrR or penB gene alone (Fig. 4). Finally, the pilQ2 mutation appears to increase resistance to hydrophilic antibiotics more than to hydrophobic antibiotics, which is opposite of what would be expected if the pilQ2 mutation increased the activity of the MtrC-MtrD-MtrE efflux pump (the pump shows preferences for hydrophobic agents and antibiotics; (Hagman et al., 1995)). Taken together, our data are most consistent with the idea that the PilQ secretin forms a pore across the outer membrane that allows the diffusion of antibiotics into the periplasm.
The model of PilQ as a pore-forming protein is also consistent with a previous study investigating heme import into N. gonorrhoeae (Chen et al., 2004). In that study, the authors identified several classes of spontaneously arising colonies of FA1090 harboring a deletion in the hemoglobin receptor protein HpuA that were capable of utilizing hemoglobin as the sole source of iron and porphyrins. One class of such mutants contained a F595L mutation in the pilQ gene (pilQ1), which appeared to alter the pore formed by the PilQ multimer to allow the diffusion of free heme released from hemoglobin into the cell. The pilQ1 mutation also increased antibiotic sensitivities to several antibiotics and Triton X-100, reinforcing the idea that a measurable influx of antibiotics can occur through the PilQ secretin complex. Interestingly, the pilQ1 mutation required the pilus-associated ATPase, PilT, for growth on hemoglobin, while we have shown that increase of penicillin resistance conferred by the pilQ2 mutation is PilT-independent (Fig. 6). This difference may reflect different processes, however, as the heme must enter into the cytoplasm to support growth, whereas penicillin need only diffuse into the periplasm. Indeed, Chen et al. (2004) showed that increased sensitivity to ampicillin in the pilQ1 strain was not dependent on PilT.
Other studies have documented the effects of mutations and/or insertions in secretins on outer membrane permeability. For example, Wall et al. (Wall et al., 1999) characterized a mutant allele of Myxococcus xanthus pilQ, which is involved in pilin biogenesis and social motility in this organism. The mutant allele contained two missense mutations (G741S and N762G) in its C-terminus that conferred hypersensitivity to vancomycin. Similarly, Russel (Russel, 1994) identified mutants in the pIV secretin involved in export of the filamentous phage f1 that conferred hypersensitivity to both vancomycin and deoxycholate. These pilQ mutants therefore resemble the F595L mutation characterized by Chen et al. (Chen et al., 2004) in that the alterations increased the susceptibility of the organism to antibiotics and deoxycholate, presumably by increasing the size of the PilQ channel or by changing the conformation of a region that constricts the channel. Interestingly, alignment of the PilQ secretins from M. xanthus and N. gonorrhoeae shows that one of the two mutations in the mutant M. xanthus pilQ allele (N762G) maps to within seven residues of Phe-595 of gonococcal pilQ1 (Chen et al., 2004), whereas the other mutation is ∼25 amino acids distant (data not shown). The similar location of these mutants in PilQ homologues suggests that this area is involved in defining the size of the PilQ channel.
In contrast to these other studies, in which mutations increased the sensitivity of the strain to antibiotics and detergents, the mutations in gonococcal PilQ characterized here increase resistance provided that at least mtrR and penB are present. The pilQ2 mutation also prevents formation of an SDS-resistant PilQ oligomer, but the direct cause and effect of these two observations has yet to be established. To our knowledge, this is the first report of a mutation in PilQ from Neisseria that interferes with formation of an SDS-resistant HMM multimer, although a recent report showed that deletion of the presumed chaperone protein, PilW, also resulted in the loss of the SDS-resistant PilQ multimer (Carbonnelle et al., 2005). While the actual effect of the pilQ2 mutation on PilQ structure is unclear, gel filtration of wild-type and mutant PilQ proteins (Fig. 3D) suggests that at least a portion of the PilQ protein in the pilQ2 mutant exists as a oligomeric species (presumably, like the wild-type, a dodecamer). These results suggest that the pilin-expressing membraneous projections in FA19 pilQ2 observed by immunoelectron microscopy (Fig. 1B) may be the result of this oligomeric PilQ interacting with pili and could explain the high residual transformation competence of the pilQ2 strain (Fig. 2). However, if present these PilQ complexes are unable to facilitate the diffusion of antibiotics across the outer membrane. More knowledge about how pilQ assembles and interacts with the pilus assembly proteins will be required before a detailed molecular understanding of the effect of the effect of these mutations on the entry of DNA and antibiotics into the bacterial cell is known.
Type IV pilin biogenesis in N. gonorrhoeae requires multiple accessory subunits in addition to pilin and PilQ (Wolfgang et al., 1998; Wolfgang et al., 2000). Thus, we also examined the role of pilin, PilT, and PilP in antibiotic resistance by creating deletion mutants in SZ3, a 3rd level transformant of RM11.2 recA6 with a wild type pilQ allele (Fig. 5). The lack of a substantial increase in penicillin resistance in SZ3 following deletion of the pilE or pilT gene shows that neither assembly or disassembly of the pilus fiber alters the ability of PilQ to enable penicillin entry, even though structural modeling suggests that a pilin fiber would fully occupy the central cavity of PilQ (Collins et al., 2003). In contrast, penicillin resistance in SZ3 ΔpilP increased significantly, although not to the same level as the pilQ2 mutation. Because deletion of PilP decreased the expression of the multimeric form of PilQ without changing the levels of the monomeric form (data not shown; (Drake et al., 1997)), our data suggest that the intermediate increase in resistance is the result of a decrease in the number of stable PilQ multimers in the outer membrane. The simplest model to account for all of these observations is that only some of the PilQ complexes are dedicated to expressing pili, but that all PilQ complexes are influenced by expression of PilP. This model predicts that different complexes of pilQ are involved in different functions in the outer membrane and that the decreased multimeric form detected in gonococcal membranes of a pilP strain may explain the differential phenotypes of the pilQ2 mutation.
One of the intriguing aspects of chromosomally mediated resistance is that the resistance determinants must be transferred in a specific order. For example, our results demonstrate that pilQ2 does not increase the MIC of penicillin or tetracycline unless other resistance determinants are present (Fig. 4). Of the previously described resistance determinants, only mtrR and penB are necessary for pilQ2 to increase resistance to penicillin or tetracycline (Fig. 4, right panels). The penA gene further increases resistance, but its presence is not required for the phenotypic increase in resistance mediated by pilQ2 (Fig. 4). The fact that pilQ2 has no influence on the MIC unless other resistance determinants are present that reduce the concentration of antibiotics in the periplasm (mtrR and penB) suggests that influx of antibiotics through the PilQ multimer in susceptible strains is only a small fraction of the antibiotic influx compared to that through porin. Only when mtrR and penB combine to decrease influx through porins does the influx of antibiotics through PilQ become significant, thereby increasing resistance when PilQ is deleted or mutated (see Figure 8). Because this mutation disrupts normal piliation, increases in resistance as a result of pilQ mutations are unlikely to be observed in the clinical setting owing to the importance of pili in mucosal infections (Hamrick et al., 2001; Swanson, 1973), although pilQ mutations may have a role in resistance in disseminated infections in which the role of pili is not well established. Nonetheless, our results underscore the role of the PilQ secretin in the diffusion of antibiotics and other molecules into the periplasm of the bacterium.
MATERIALS AND METHODS
Bacterial strains and plasmids
FA19 (Maness and Sparling, 1973) is a penicillin-susceptible laboratory strain. FA6140 is a high-level chromosomally mediated resistant strain of N. gonorrhoeae isolated in Durham in the 1970's (Danielsson et al., 1986). RM11.2 recA6 (Seifert, 1997) is a derivative of FA1090 (Dempsey et al., 1991) containing the recA gene under regulation of the lac operator/promoter. Other strains used in this work are described in Table 1. Transformations with FA19 strains were carried out exactly as described previously (Ropp et al., 2002). Transformations of RM11.2 recA6 were similar except that 1 mM IPTG was added to the cells before exposure to DNA to allow for the expression of the recA gene product required for homologous recombination (Seifert, 1997). Selection for the penA4, mtrR, and penB5 stepwise transformants in RM11.2 recA6 was carried out on GCB plates with 0.04 μg/ml penicillin G, 2 μg/ml erythromycin, and 0.28 μg/ml penicillin G respectively. SZ3 ΔpilE was constructed by transforming SZ3 with genomic DNA from a strain of FA1090 with a complete deletion in the pilE gene (kindly provided by Dr. Janne Cannon, UNC-Chapel Hill) and selecting for nonpiliated colonies.
In order to generate cell lines harboring the pilQ2 mutation, we constructed a plasmid (pUC18-pilQ2-kan) containing the last 864 bp of FA19 pilQ2 (encoding residues 445-731), the kanamycin phosphotransferase gene inserted 20 bp past the pilQ stop codon, and 280 bp of additional downstream sequence, and used this plasmid to transform the various recipient strains to kanamycin resistance. A silent NsiI site was created by changing T to G at nucleotide 1959 of the FA19 pilQ2 gene to aid in identifying transformants containing the pilQ2 mutation. To generate strains with a disrupted pilQ allele (pilQ::Ω), strains were transformed with pUNCH290 (Chen et al., 2004). pUNCH290 comprises nucleotides 434 of the FA1090 pilQ gene to 152 bp downstream of the stop codon, in which an internal HincII fragment was replaced with the Ω fragment conferring spectinomycin resistance (Prentki and Krisch, 1984). All strains were verified by PCR amplification and sequencing, and by Western blotting to verify the absence of the high molecular mass PilQ oligomer (pilQ2 strains) or PilQ reactivity (pilQ::Ω).
Growth conditions and MIC determinations
N. gonorrhoeae were grown on GCB plates with Supplements I and II (Kellogg et al., 1963) at 37°C in a humidified 5% CO2 atmosphere, and E. coli were growth on LB plates or in 2X YT media (Sambrook et al., 1989) as described (Ropp et al., 2002). MICs were determined by the spot method as described (Ropp et al., 2002). Briefly, cells were resuspended at a density of 1 × 107 cells/ml and 5 μl (50,000 colonies) were spotted onto GCB agar plates containing increasing concentrations of the appropriate antibiotics. The MIC was defined as the lowest concentration at which no more than 5 colonies grew following incubation for 24 hr. MICs were determined for multiple transformants and represent the average ± S.E.M of at least three independent experiments.
Cloning the pilQ2 gene
Chromosomal DNA from PR100 penC (Ropp et al., 2002) was digested with ClaI, separated on a 0.7% agarose gel, and ten fractions ranging in size from 12 kb to 300 bp were tested for their ability to transform PR100 to penicillin resistance on GCB with 1.2 μg/ml penicillin. Fraction 2, comprising fragments from ∼ 8 to 10 kb, was enriched in the pilQ2 gene and was used to create a mini-library in ClaI-digested pBeloBac 11 bacmid vector (Kim et al., 1996). Initial attempts to clone the gene into the medium copy-number plasmid pACYC184 were unsuccessful, necessitating the use of the single copy-number bacmid vector. Three clones were found to transform PR100 to penicillin resistance with high frequency. Each clone harbored an identical 10.6 kb insert containing seven genes: the 3′-end of pilQ, aroK, aroB, yafJ, and three hypothetical genes of unknown function. The location of the penC gene was narrowed down by amplifying each gene and testing the fragments in the transformation assay. Only the fragments containing the pilQ gene transformed PR100 to higher penicillin resistance with high frequency, demonstrating that the penC mutation maps to the 3′-end of the pilQ gene (bp 1333-2196 encoding residues 445-731). All three clones revealed the identical basepair mutation.
Immunoelectron microscopy of pilin fibers
Analysis of the piliation states of gonococcal strains was determined exactly as described previously (Chen et al., 2004). The C-terminal domain of FA19 PilQ differs from FA1090 PilQ by two amino acid differences (G523R and N620S). To ensure that sequences of PilQ had no bearing on the piliation state of the gonococci, the C-terminal domain of pilQ from RM11.2 recA6 (an FA1090 dertivative) was also exchanged with the corresponding region from FA19. A plasmid identical to the one used to transfer the pilQ2 allele into strains (see above), except that it contained the wild type sequence from FA19, was used to construct RM11.2 recA6 pilQFA19.
Transformation efficiency
Strains were passaged on GCB agar plates, resuspended in GCB with supplements I and II, 20 mM NaHCO3, 10 mM MgCl2 and 1 mM IPTG (GCB+), and diluted to OD600 = 0.18 in the same medium. Twenty μl of cells and 100 ng of pSY6 were added to 180 μl of GCB+ and incubated at 37°C in 5% CO2/95% air for 15 min. GCB+ (1.8 ml) was then added and the cells were incubated for a further 5 hrs, followed by serial 10-fold dilutions in GCB+. A portion (50 μl) of the appropriate dilution was plated in duplicate on GCB + 2 μg/ml nalidixic acid (NAL) plates. For determination of total CFU, 50 μl of a 1 × 104 dilution were plated on GCB plates.
Electrophoresis and Western blotting
For SDS-PAGE and Western blotting of whole cell lysates, cells were swabbed from GCB plates, resuspended in GCB+ at an OD560 of 0.18, and 1 ml was removed to a microfuge tube. After pelleting the cells and removal of the medium, 100 μl of SDS-PAGE loading buffer was added and the cells were dispersed. Three to ten μl of the samples were loaded onto SDS-polyacrylamide gels (8 or 10%) and separated by electrophoresis. The proteins in the gel were then electrotransferred to nitrocellulose filters overnight in 25 mM Tris, 192 mM glycine, and 10% methanol. The lower methanol appeared to increase the efficiency of transfer of the high molecular mass PilQ oligomer. Following transfer, the filter was blocked with 5% dry milk in phosphate-buffered saline with 0.1% Tween (PBS-T), then incubated with a 1:10,000 dilution of a PilQ antibody (kindly provided by Charles E. Wilde III, Indiana University School of Medicine) for 2 hrs at 25°C. The filter was washed 3 times with PBS-T, and incubated for another 1 hr with HRP-conjugated mouse anti-rabbit secondary antibody (Amersham Biosciences, Piscataway, NJ). After washing the filter, PilQ bands were visualized with SuperSignal West Pico (Pierce Chemical Co., Rockford, IL).
For pilin blots, cells were prepared as described above, checked for protein levels to ensure equal loading, and separated on 15% SDS-polyacrylamide gels. Proteins were transferred from gels to PVDF membranes, and membranes were blocked with 5% dry milk in Tris-buffered saline with 0.1% Tween. The membranes were probed with the pilin monoclonal antibody 1E8/G8 (gift from M. Koomey and M. Blake), washed, incubated with HRP-conjugated goat anti-mouse secondary antibody (Roche Molecular Biochemicals), and washed again. The ECL Plus Western Blot kit (Amersham Biosciences) was used to detect pilin bands.
Gel Filtration of PilQ proteins
Liter cultures of FA19 (wild-type PilQ) and FA19 pilQ2 (PilQ-E666K) were grown in GCB broth with Supplements B, I and II, and 10 mM sodium bicarbonate. Cells were pelleted, lysed with an EmulsiFlex-C5 homogenizer (Avestin, Ottawa, Canada), and membranes were isolated by centrifugation at 100,000g. The membrane pellets were dounce-homogenized in Buffer A (20 mM Tris, 150 mM NaCl, 2 mM EDTA, pH 8.0), mixed with an equal volume of 40 mM Tris, 4% SB-10, 3 M NaCl, 2 mM EDTA, pH 8, and incubated on ice for 1 h. The membranes were pelleted at 15,000g for 30 min, and the supernatants containing the extracted PilQ proteins were filtered through 0.45μm filters. Aliquots (300 μl) were submitted to gel filtration on a Sephacryl S-500 column (16 mm × 70 cm) in 20 mM Tris, pH 8, 500 mM NaCl, 1 mM EDTA, 1.2% SB-10. Twenty μl from the indicated fractions were processed for SDS-PAGE and Western blotting, and the resulting bands were imaged on a Bio-Rad Fluoro-S system and quantitated with QuantityOne software (Bio-Rad, Hercules, CA).
Site-directed and site-saturation mutagenesis of pilQ
To generate a library of pilQ fragments in which the codon for Glu-666 was randomized, we utilized overlap-extension PCR (Ho et al., 1989). The sense mutagenic primer (5′- TGAATACGCAGGCTATGGTTNNNAACGGCGGCACTTTGATTG-3′) comprised 20 bp at the 5′ end of codon-666, three random bases, and 19 bp at the 3′ end of codon-666, while the antisense mutagenic primer (5′-AACCATAGCCTGCGTATTCA-3′) comprised the reverse complement of the first 20 bp of the sense primer. The template was a plasmid vector (pUC18) containing a portion of the pilQ gene starting at 1333 bp to 300 bp downstream of the stop codon. In the first set of reactions, a sense (relative to the direction of pilQ) vector primer and the antisense mutagenic primer or the sense mutagenic primer and an antisense vector primer were used to amplify the same template in separate amplifications. The resulting fragments were isolated, combined, and the full-length pilQ gene with a randomized codon at position-666 was generated in a second PCR reaction with only the vector primers. The mutagenized PCR fragments were purified on a Marligen (Gaithersburg, MD) PCR purification column and used to transform PR100 to penicillin resistance. Resistant colonies were passaged, examined by western blotting to determine clones expressing only the monomeric PilQ subunit, and the pilQ gene from those clones were amplified from genomic DNA and sequenced to determine the identity of the mutation. Site-directed mutagenesis was carried out with the same template described above using the QuickChange kit from Stratagene (La Jolla, CA).
Supplementary Material
Figure 7. Model showing why the pilQ2 and pilQ::Ω alleles increase resistance only in strains also harboring mtrR and penB mutations.
The two panels show a schematic representation of the cell envelope of N. gonorrhoeae. Left panel, FA19. In this strain the entry of penicillin through porin is much greater than the entry through the PilQ complex. The low level of the MtrC-MtrD-MtrE efflux pump is represented by light shading. Right panel, PR100. This strain has acquired penA, mtrR, and penB resistance determinants, resulting in the expression of altered forms of PBP 2 and porin IB and overexpression of the MtrC-MtrD-MtrE efflux pump. The mtrR and penB determinants function to lower the influx of penicillin through porin such that entry of penicillin through the PilQ complex becomes a significant portion of the total influx. Only in PR100 will functional deletion of the PilQ complex (due to pilQ2 or pilQ::Ω) lead to increase resistance to penicillin. See text for details.
ACKNOWLEDGEMENTS
This work was supported by NIH grants R01 AI36901 (RAN) and R01 AI055977 (HSS). We thank Sohban Nandi and Josh Tomberg for helpful discussions. We also thank Janne Cannon, Fred Sparling, and Ching-ju Chen for generous access to reagents and stimulating discussions.
REFERENCES
- Brannigan JA, Tirodimos IA, Zhang Q-Y, Dowson CG, Spratt BG. Insertion of an extra amino acid is the main cause of the low affinity of penicillin-binding protein 2 in penicillin-resistant strains of Neisseria gonorrhoeae. Mol Microbiol. 1990;4(6):913–919. doi: 10.1111/j.1365-2958.1990.tb00664.x. [DOI] [PubMed] [Google Scholar]
- Cannon JG, Sparling PF. The genetics of the gonococcus. Ann Rev Genet. 1984;38:111–133. doi: 10.1146/annurev.mi.38.100184.000551. [DOI] [PubMed] [Google Scholar]
- Carbonnelle E, Helaine S, Prouvensier L, Nassif X, Pelicic V. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Mol Microbiol. 2005;55:54–64. doi: 10.1111/j.1365-2958.2004.04364.x. [DOI] [PubMed] [Google Scholar]
- Chen CJ, Tobiason DM, Thomas CE, Shafer WM, Seifert HS, Sparling PF. A mutant form of the Neisseria gonorrhoeae pilus secretin protein PilQ allows increased entry of heme and antimicrobial compounds. J Bacteriol. 2004;186:730–739. doi: 10.1128/JB.186.3.730-739.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RF, Davidsen L, Derrick JP, Ford RC, Tonjum T. Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J Bacteriol. 2001;183:3825–3832. doi: 10.1128/JB.183.13.3825-3832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RF, Ford RC, Kitmitto A, Olsen RO, Tonjum T, Derrick JP. Three-dimensional structure of the Neisseria meningitidis secretin PilQ determined from negative-stain transmission electron microscopy. J Bacteriol. 2003;185:2611–2617. doi: 10.1128/JB.185.8.2611-2617.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RF, Frye SA, Kitmitto A, Ford RC, Tonjum T, Derrick JP. Structure of the Neisseria meningitidis Outer Membrane PilQ Secretin Complex at 12 Å Resolution. J Biol Chem. 2004;279:39750–39756. doi: 10.1074/jbc.M405971200. [DOI] [PubMed] [Google Scholar]
- Danielsson D, Faruki H, Dyer D, Sparling PF. Recombination near the antibiotic resistance locus penB results in antigenic variation of gonococcal outer membrane protein I. Infect Immun. 1986;52:529–533. doi: 10.1128/iai.52.2.529-533.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey JA, Litaker W, Madhure A, Snodgrass TL, Cannon JG. Physical map of the chromosome of Neisseria gonorrhoeae FA1090 with locations of genetic markers, including opa and pil genes. J Bacteriol. 1991;173:5476–5486. doi: 10.1128/jb.173.17.5476-5486.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty TJ. Genetic analysis and penicillin-binding protein alterations in Neisseria gonorrhoeae with chromosomally mediated resistance. Antimicrob Agents Chemother. 1986;30:649–652. doi: 10.1128/aac.30.5.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson CG, Jephcott AE, Gough KR, Spratt BG. Penicillin-binding protein 2 genes of non-ß-lactamase-producing, penicillin-resistant strains of Neisseria gonorrhoeae. Mol Microbiol. 1989;3:35–41. doi: 10.1111/j.1365-2958.1989.tb00101.x. [DOI] [PubMed] [Google Scholar]
- Drake SL, Sandstedt SA, Koomey M. PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular-mass multimer. Mol Microbiol. 1997;23:657–668. doi: 10.1046/j.1365-2958.1997.2511618.x. [DOI] [PubMed] [Google Scholar]
- Faruki H, Sparling PF. Genetics of resistance in a non-ß-lactamase-producing gonococcus with relatively high-level penicillin resistance. Antimicrob Agents Chemother. 1986;30:856–860. doi: 10.1128/aac.30.6.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill MJ, Simjee S, Al-Hattawi K, Robertson BD, Easmon CS, Ison CA. Gonococcal resistance to beta-lactams and tetracycline involves mutation in loop 3 of the porin encoded at the penB locus. Antimicrob Agents Chemother. 1998;42:2799–2803. doi: 10.1128/aac.42.11.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman KE, Pan W, Spratt BG, Balthazar JT, Judd RC, Shafer WM. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995;141:611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- Hagman KE, Lucas CE, Balthazar JT, Snyder L, Nilles M, Judd RC, Shafer WM. The MtrD protein of Neisseria gonorrhoeae is a member of the resistance/nodulation/division protein family constituting part of an efflux system. Microbiology. 1997;143:2117–2125. doi: 10.1099/00221287-143-7-2117. [DOI] [PubMed] [Google Scholar]
- Hamrick TS, Dempsey JA, Cohen MS, Cannon JG. Antigenic variation of gonococcal pilin expression in vivo: analysis of the strain FA1090 pilin repertoire and identification of the pilS gene copies recombining with pilE during experimental human infection. Microbiology. 2001;147:839–849. doi: 10.1099/00221287-147-4-839. [DOI] [PubMed] [Google Scholar]
- Hansen MV, Wilde CE., 3rd Conservation of peptide structure of outer membrane protein-macromolecular complex from Neisseria gonorrhoeae. Infect Immun. 1984;43:839–845. doi: 10.1128/iai.43.3.839-845.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckels JE. The surface of Neisseria gonorrhoeae: isolation of the major components of the outer membrane. J Gen Microbiol. 1977;99:333–341. doi: 10.1099/00221287-99-2-333. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Kellogg DS, Peacock WL, Deacon WE, Browh L, Perkle CI. Neisseria gonorrhoeae. I. Virulence genetically linked to colonial variation. J Bacteriol. 1963;85:1274–1279. doi: 10.1128/jb.85.6.1274-1279.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim UJ, Shizuya H, Kang HL, Choi SS, Garrett CL, Smink LJ, Birren BW, Korenberg JR, Dunham I, Simon MI. A bacterial artificial chromosome-based framework contig map of human chromosome 22q. Proc Natl Acad Sci U S A. 1996;93:6297–6301. doi: 10.1073/pnas.93.13.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CD, Madraswala RN, Seifert HS. Comparisons between colony phase variation of Neisseria gonorrhoeae FA1090 and pilus, pilin, and S-pilin expression. Infect Immun. 1998;66:1918–1927. doi: 10.1128/iai.66.5.1918-1927.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CD, Hayes SF, van Putten JP, Harvey HA, Apicella MA, Seifert HS. Modulation of gonococcal piliation by regulatable transcription of pilE. J Bacteriol. 2001;183:1600–1609. doi: 10.1128/JB.183.5.1600-1609.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CD, Tobiason DM, Lazio MP, Kline KA, Seifert HS. Low-level pilin expression allows for substantial DNA transformation competence in Neisseria gonorrhoeae. Infect Immun. 2003;71:6279–6291. doi: 10.1128/IAI.71.11.6279-6291.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B, Potter L, So M, Long CD, Seifert HS, Sheetz MP. Single pilus motor forces exceed 100 pN. Proc Natl Acad Sci U S A. 2002;99:16012–16017. doi: 10.1073/pnas.242523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maness MJ, Sparling PF. Multiple antibiotic resistance due to a single mutation in Neisseria gonorrhoeae. J Infect Dis. 1973;128:321–330. doi: 10.1093/infdis/128.3.321. [DOI] [PubMed] [Google Scholar]
- Marciano DK, Russel M, Simon SM. An aqueous channel for filamentous phage export. Science. 1999;284:1516–1519. doi: 10.1126/science.284.5419.1516. [DOI] [PubMed] [Google Scholar]
- Merz AJ, So M, Sheetz MP. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- Newhall WJ, Wilde CE, 3rd, Sawyer WD, Haak RA. High-molecular-weight antigenic protein complex in the outer membrane of Neisseria gonorrhoeae. Infect Immun. 1980;27:475–482. doi: 10.1128/iai.27.2.475-482.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouwen N, Ranson N, Saibil H, Wolpensinger B, Engel A, Ghazi A, Pugsley AP. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc Natl Acad Sci U S A. 1999;96:8173–8177. doi: 10.1073/pnas.96.14.8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesky M, Hobbs M, Nicholas RA. Identification and analysis of amino acid mutations in porin IB that mediate intermediate-level resistance to penicillin and tetracycline in Neisseria gonorrhoeae. Antimicrob Agents Chemother. 2002;46:2811–2820. doi: 10.1128/AAC.46.9.2811-2820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W, Spratt BG. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol Microbiol. 1994;11:769–775. doi: 10.1111/j.1365-2958.1994.tb00354.x. [DOI] [PubMed] [Google Scholar]
- Prentki P, Krisch HM. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Ropp PA, Nicholas RA. Cloning and characterization of the ponA gene encoding penicillin-binding protein 1 from Neisseria gonorrhoeae and Neisseria meningitidis. J Bacteriol. 1997;179:2783–2787. doi: 10.1128/jb.179.8.2783-2787.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropp PA, Hu M, Olesky M, Nicholas RA. Mutations in ponA, the gene encoding penicillin-binding protein 1, and a novel locus, penC, are required for high-level chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother. 2002;46:769–777. doi: 10.1128/AAC.46.3.769-777.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M. Mutants at conserved positions in gene IV, a gene required for assembly and secretion of filamentous phages. Mol Microbiol. 1994;14:357–369. doi: 10.1111/j.1365-2958.1994.tb01296.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual-2nd edition. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1989. [Google Scholar]
- Sarubbi FAJ, Blackman E, Sparling PF. Genetic mapping of linked antibiotic resistance loci in Neisseria gonorrhoeae. J Bacteriol. 1974;120:1284–1292. doi: 10.1128/jb.120.3.1284-1292.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert HS. Insertionally inactivated and inducible recA alleles for use in Neisseria. Gene. 1997;188:215–220. doi: 10.1016/s0378-1119(96)00810-4. [DOI] [PubMed] [Google Scholar]
- Sparling PF, Sarubbi FAJ, Blackman E. Inheritance of low-level resistance to penicillin, tetracycline, and chloramphenicol in Neisseria gonorrhoeae. J Bacteriol. 1975;124:740–749. doi: 10.1128/jb.124.2.740-749.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein DC, Danaher RJ, Cook TM. Characterization of a gyrB mutation responsible for low-level nalidixic acid resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother. 1991;35:622–626. doi: 10.1128/aac.35.4.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J. Studies on gonococcus infection. IV. Pili: their role in attachment of gonococci to tissue culture cells. J Exp Med. 1973;137:571–589. doi: 10.1084/jem.137.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonjum T, Koomey M. The pilus colonization factor of pathogenic neisserial species: organelle biogenesis and structure/function relationships--a review. Gene. 1997;192:155–163. doi: 10.1016/s0378-1119(97)00018-8. [DOI] [PubMed] [Google Scholar]
- Veal WL, Nicholas RA, Shafer WM. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J Bacteriol. 2002;184:5619–5624. doi: 10.1128/JB.184.20.5619-5624.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D, Kolenbrander PE, Kaiser D. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J Bacteriol. 1999;181:24–33. doi: 10.1128/jb.181.1.24-33.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M, Park HS, Hayes SF, van Putten JP, Koomey M. Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc Natl Acad Sci U S A. 1998;95:14973–14978. doi: 10.1073/pnas.95.25.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M, van Putten JP, Hayes SF, Dorward D, Koomey M. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. Embo J. 2000;19:6408–6418. doi: 10.1093/emboj/19.23.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.