Abstract
Background
The basis of hepatocellular injury and progressive fibrosis in a subset of patients with NAFLD is poorly understood. We sought to identify hepatic proteins that are differentially abundant across the histologic spectrum of NAFLD.
Methods
Hepatic protein abundance was measured in liver samples from four groups (n=10 each) of obese (body mass index >30kg/m2) patients: 1) obese normal group (normal liver histology), 2) Simple steatosis (SS), 3) NASH-mild (steatohepatitis with fibrosis stage 0–1), and 4) NASH-progressive (steatohepatitis with fibrosis stage 2–4). Hepatic peptides were analysed on an API Qstar XL quadrupole time of flight mass spectrometer using Analyst QS software. Linear trends tests were performed and used to screen for differential abundance.
Results
Nine known proteins were expressed with differential abundance between study groups. For seven proteins (albumin, hemoglobin beta, hemoglobin delta, dihydropyrimidinase, enolase, metal transport protein ATX1 and HSP gp96) differential abundance is likely to have been on the basis of known biologic effects of increased hepatic lipid content and/or inflammation. Lumican, a 40kDa keratin sulfate proteoglycan that regulates collagen fibril assembly and activates TGF-beta and smooth muscle actin, was expressed similarly in obese normal and SS but was overexpressed in a progressive manner in NASH-mild vs. SS (124%, p<0.001), NASH-progressive vs. NASH-mild (156%, p<0.001) and NASH-progressive vs. obese normal (178%, p<0.001). Fatty acid binding protein-1 (FABP-1), which is protective against the detergent effects of excess FFAs, facilitates intracellular FFA transport and is an important ligand for PPAR-mediated transcription, was overexpressed in SS when compared obese normal (128%, p<0.001), but was paradoxically underexpressed in NASH-mild vs. SS (73%, p<0.001), NASH-progressive vs. NASH-mild (81%, p<0.001) and NASH-progressive vs. obese normal (59%, p<0.001).
Conclusions
Histologically progressive NAFLD is associated with overexpression of lumican, an important mediator of fibrosis in non-hepatic tissues, while FABP-1 is paradoxically underexpressed in NASH compared to milder histologic variants of NAFLD, suggesting a new potential mechanism of lipotoxicity in NAFLD. Further studies are needed to determine the biologic basis of lumican and/of FABP-1 dysregulation in NAFLD.
Introduction
Many conditions are associated with the histologic features of non-alcoholic fatty liver disease (NAFLD). The condition most commonly associated with NAFLD is chronic overnutrition with consequent obesity and, often, insulin resistance.(1–6) NAFLD progresses to cirrhosis within 10 years in approximately 5% of patients(7) and has been projected to become the most common indication for liver transplantation in the next 10–20 years.(8–10) The biologic basis for the diverse histological spectrum that occurs in NAFLD is poorly defined.
Regardless of the etiology, the effectors of hepatic injury in NAFLD must have a protein basis. Therefore we hypothesized that analysis of the hepatic proteome will provide insights into the biology of this condition. Proteomic analyses in general have been limited by poor sensitivity in detecting low-abundance proteins as well as difficulties with maintaining sample stability and data management. Recent advances in protein separation and improvements in detection and identification of peptides and proteins have facilitated detailed characterization of complex biologic samples, including the liver. Proteomic analysis would be likely to identify hepatic proteins that contribute to the development of NASH with progressive fibrosis. Differential abundance of hepatic proteins could be confirmed and further studied in a second phase of experiments including immunohistochemical analysis of samples. We utilised a tandem mass spectrometric approach that provides simultaneous analysis of the relative expression of a large number of proteins in a sensitive manner to determine the relative expression of hepatic proteins across the histologic spectrum of NAFLD.
Methods and Materials
The hepatic proteome was measured in liver samples from four groups of obese (BMI>30kg/m2) patients:
obese normal group (normal liver histology, n=10),
Simple steatosis (SS, n=10),
NASH-mild (steatosis, lobular and/or portal inflammation grade 1 and fibrosis stage 0–1,
NASH-progressive (as for NASH-mild but with fibrosis stage 2–4).
The study was approved by the Institutional Review Board. All participants gave written informed consent for participation in medical research. Participants were recruited from patients undergoing bariatric surgery for medically complicated obesity and from obese patients undergoing resection of benign liver masses. Patients with NAFLD who had secondary causes of steatohepatitis (drugs, prior gastric surgery for obesity), and patients with other etiologies of chronic liver disease (excessive alcohol consumption, viral hepatitis (B, C), cholestatic liver disease, hemochromatosis, Wilson’s disease, drug-induced liver disease, and alpha 1-antitrypsin deficiency), were excluded from this study. The histological classification of samples was chosen a priori.
Protein labeling
A total of 100μg of proteins from each liver sample was labeled with iTRAQ according to Applied Biosystems iTRAQ protein labeling protocol (Applied Biosystems, Foster City, CA). The tryptic-digested peptides of each sample were labeled with either 114, 115, 116 or 117 iTRAQ reagents. The labeled peptides from two different samples, e.g. simple steatosis and NASH-progressive matched for age and sex, were mixed together. The combined sample was cleaned up of excess trypsin as well as iTRAQ reagents using Waters c18 Sep-Pak (Milford, MA) before mass spectrometric analysis. Although all iTRAQ reagents (114, 115, 116 or 117) were used, as is routine to avoid reporter effect, only two were present in any pair-wise analysis. Replicate analyses were performed using different iTRAQ reagents, all from the original protein extraction.
Sample preparation for Multidimensional Liquid Chromatography – Tandem mass Spectrometry (LC-MS/MS) analysis
iTRAQ-labeled samples were fractionated into 10 fractions on a strong cation exchange column, Biox SCX 300μm × 5cm (Dionex, Sunnyvale, CA) using an off-line Agilent 1100 series capillary liquid chromatography system (Wilmington, DE). LC/MS/MS analysis of the peptides in each fraction was performed on an Applied Biosystems API Qstar XL quadrupole time of flight mass spectrometer configured with a Protana nano spray ion source (Proxeon, Denmark) and with an Ultimate nano liquid chromatography system (Dionex, Sunnyvale, CA). The mass spectroscopy data was obtained via information-dependent acquisition (IDA) mode in the Analyst QS software.
Protein Quantification and Data Analysis
Quantification
After separate analysis of the ion exchange fractions in each iTRAQ experiment, the fraction results were grouped (ProQuant; Applied Biosystems). For each MS/MS scan, ProQuant identifies precursor m/z and charge in the TOF MS scan for that cycle. ProQuant checks the next 10 cycles for precursor m/z and charge and flags spectra that are matched for m/z and charge as part of a merged set. The program finds the peaks in the spectrum (or summed spectra) for the four signature ions derived from the iTRAQ reagents (114, 115, 116, 117). The relative contribution of each of the four samples was measured by integrating the peaks corresponding to the signature ions in the MS/MS spectrum.
Peptide/Protein identification
Results from the MS/MS were used to search the protein sequence database (KBMS human; March 2005; Celera) with the Interrogator™ Algorithm in ProQuant. The KBMS database is a compilation of several protein sequence databases (NCBI RefSeq; SwissProt; Celera; translated EMBL, and others).
Protein Abundance Comparisons
The relative protein abundances were analyzed for relationships between each pair in the iTRAQ runs (four conditions in each iTRAQ run—that is, six comparisons). Signature ion ratios (corresponding to comparisons in the experimental design) for each accession number identified during the Interrogator search were stored in an Access database. Tables for the full dataset or for subsets were created in Excel using the Access database as source. “Fold-change” was derived from ratios by use of the ratio for values ≥ 1 and −1/ratio for ratios <1. For plots around 0 (=no change) ratios were converted to [ratio-1] for ratios ≥ 1 and to [−1/ratio + 1] for ratio <1 (no change=0 of 50% =±0.5). Classification of biologic and molecular functions were derived from the Panther database (http://www.pantherdb.org/). Tests of linear trends were performed and used to determine the statistical significance of differential abundance of proteins. The corresponding p-values were used for ranking the proteins according to statistical significance. Because of the large number of proteins being tested, the value of 0.05 was not used as an absolute cut-off value for significance but rather as a screening threshold for identifying proteins of most interest for further investigation. Thus, all of the proteins that were stated to be differentially abundant in a comparison had p-values of ≤0.05. Proteins of interest that were found to be differentially abundant were further analysed with immunohistochemistry (to confirm differential protein abundance and also into protein distribution) and RT-PCR, (which also provided insight into whether the basis for differential protein abundance was on a pretranscriptional basis). The immunohistochemistry findings studies were repeated in a further set of 16 patients (4 new participants from each study group).
RNA isolation and real-time PCR
Total RNA was isolated from the same snap frozen liver biopsy samples as were used ion the proteomics portion of the analysis (RNeasy Plus kit, Qiagen, GmbH, Germany). Real time PCR was carried out on a LightCycler (Roche, San Francisco, CA, USA) using equal quantities of template cDNA using the QuantiTect SYBR Green PCR kit (Qiagen, GmbH, Germany). The primers used were: Lumican Forward, 5′ CTTCAATCAGATAGCCAGACTGC-3 ′; Lumican Reverse, 5 ′-AGCCAGTTCGTTGTGAGATAAAC-3′. Expression of 18-S rRNA was used as the internal standard (Quantum RNA 18S Internal Standards, Ambion Inc, Austin, TX, USA). Standard curves were generated for each optimized assay using known PCR copy numbers to produce a linear plot of threshold cycle (Ct) against log dilution.
Immunohistochemistry
Lumican
In brief, biopsy sections from the same samples used for the proteomic analyses were deparaffinized successively, hydrated in deionized water, and washed (DAKO, Carpenteria, CA, USA) and background blocked (SNIPER, Biocare Medical, Concord, CA, USA). Sections were then incubated with primary goat anti-human lumican at 1:1000 (R&D systems, Minneapolis, MN, USA) in a background reducing diluent for 1h at room temperature. After washing (DAKO S3006), the tissue was incubated with horseradish peroxidase-labeled anti goat antibody (Promark Goat Polymer, Biocare Medical) After further washings, sections were developed with betazoid diamobenzidine (Biocare Medical) for 10 min at room temperature and then counterstained with hematoxylin for 5 min. For negative controls, tissue sections were incubated without primary antibody in TBS and 1% BSA. All samples were analyzed by a single anatomical pathologist expert in liver pathology (SS).
FABP-1
Sections were incubated for 1 h at room temperature with primary anti-human L-FABP at 1:25 (Abcam, MA, USA). After washing (DAKO S3006), the sections were incubated with EnVision dual antibody (DAKO, USA) for 15 min. After washing, sections were developed with diamobenzidine (DAKO) for 10 min at room temperature. Sections were then counterstained with hematoxylin for 5 min. For negative controls, tissue sections were incubated without primary antibody in TBS and 1% BSA.
Digital image analysis of slides stained for Lumican and L-FABP
Quantitative analysis of tissue sections immuno-stained for lumican or L-FABP was performed using histomorphometric computer assisted analysis of their images. Image acquisition was carried out using an Axioplan-2 upright microscope and Axiocam digital color camera (Carl Zeiss Inc, Oberkochen, Germany). For each tissue section, a series of 4 images was generated at an objective magnification of 5x covering >90% of the total tissue area. All images within the specific stain group were generated at uniform settings of exposure time and neutral density filters. Analysis was performed using KS400 image analysis software package (Carl Zeiss Inc.). All staining intensities were analyzed were calculated as a percentage of the total tissue area. The student’s T test was used to evaluate statistical significance between staining among the four study groups.
Results
Clinical characteristics of Patients
The clinical characteristics of all groups are summarized in Table 1. Subjects in the NASH-progressive group were younger and had a predictably greater BMI, as well as serum levels of fasting glucose, cholesterol, triglycerides, AST, and ALT than subjects in the obese normal and simple steatosis group (p<0.05 all cases).
Table 1.
Demographic and Laboratory Data
| Obese normal (n=10) | Simple steatosis (n=10) | NASH-mild (n=10) | NASH-progressive (n=10) | |
|---|---|---|---|---|
| Age (years) | 55.8 ± 2.7* | 54.6 ± 1.9* | 49.1 ± 2.3 | 47.2 ± 2.9 |
| Gender (female/male) | 5/5 | 5/5 | 5/5 | 5/5 |
| BMI (kg/m2) | 43.7 ± 2.8* | 47.0 ± 1.4 | 46.3 ± 1.7 | 52.6 ± 3.3 |
| Triglycerides (mg/dL) | 118 ± 8* | 162 ± 28 | 129 ± 21 | 168 ± 22 |
| glucose (mg/dL) | 114 ± 15** | 113 ± 10** | 151 ± 13 | 151 ± 11 |
| Cholesterol (mg/dL) | 125 ± 19* | 181 ± 22 | 153 ± 31 | 194 ± 25 |
| AST (U/L) | 26.2 ± 2.3** | 28.5 ± 2.1** | 66.2 ± 24.4 | 71.4 ± 25.6 |
| ALT (U/L) | 22.3 ± 3.2** | 28.0 ± 1.6** | 65.5 ± 38.5 | 85.7 ± 24.6 |
| Total bilirubin (mg/dL) | 0.6 ± 0.1 | 0.4 ± 0.1 | 0.7 ± 0.1 | 0.5 ± 0.1 |
Data are the mean ± SEM.
Abbreviation: AST, aspartate transaminase; ALT, alanine aminotransferase; BMI, body mass index.
indicates P< 0.05 vs. NASH-progressive
indicates P< 0.05 vs. NASH-progressive and NASH-mild
The mean BMI was greater among participants with NASH-progressive when compared to those in the obese normal group (43.7 ± 2.8 vs. 52.6 ± 3.3 kg/m2, p=0.04). BMIs did not significantly differ between other groups.
Patient weights were measured in the two months before surgery to document similar changes between study groups. As is typical of patients in bariatric surgery programs, there was some weight loss in all groups, deemed to be clinically not significant. Mean weight losses (+/− SEM) were: Obese Normal: 3.4 + 4.5 kg, Simple Steatosis: 2.3 + 2.2 kg, NASH-mild: 1.0 + 2.0 kg and NASH-progressive: 3.5 + 4.3 kg (p=ns between all groups).
Protein Expression
A total of 1362 hepatic proteins were identified. Nine known proteins were consistently differentially abundant between study groups. Seven proteins (albumin, hemoglobin beta, hemoglobin delta, dihydropyrimidinase, enolase, metal transport protein ATX1 and HSP gp96) are likely to have been differentially abundant on the basis of known biologic effects of increased hepatic lipid content and/or inflammation.
Lumican, a 40kDa keratin sulfate proteoglycan that regulates the assembly of collagen fibrils, as well as expression of TGF-beta and smooth muscle actin, was expressed similarly in obese normal and SS. Lumican was overexpressed, however, in a dose-dependent manner in NASH-mild vs. SS (124%, p<0.001), NASH-progressive vs. NASH-mild (156%, p<0.001), and NASH-progressive vs. obese normal (178%, p<0.001). In contrast, fatty acid binding protein-1 (FABP-1) was relatively overexpressed in SS when compared obese normal (128%, p<0.001), but was underexpressed in NASH-mild vs. SS (73%, p<0.001), NASH-progressive vs. NASH-mild (81%, p<0.001) and NASH-progressive vs. obese normal (59%, p<0.001)(Table 2).
Table 2.
Relative Abundance of Lumican on Proteomic Analysis
| Disease Progression | Normal Obese | Simple Steatosis | NASH-mild | NASH-progressive |
|---|---|---|---|---|
| Normal Obese | - | 0.92 | 1.15 | 1.79 |
| Simple Steatosis | 1.09 | - | 1.24 | 1.94 |
| NASH-mild | 0.87 | 0.81 | - | 1.56 |
| NASH-progressive | 0.56 | 0.52 | 0.64 | - |
RNA isolation and real-time PCR
Gene expression of lumican, as measured by abundance of lumican mRNA, and FABP-1 were detected in all study groups. Patterns of relative abundance of gene expression of lumican was similar the patterns of relative abundance of lumican protein. Lumican mRNA abundance was 7.82, 5.19, 2.63 and 1.93 log copies/microg RNA in NASH-progressive, NASH, mild, simple steatosis and obese normal respectively. The differences in lumican gene expression between NASH-progressive and all other groups was significant (p≤0.01). FABP-1 mRNA abundance was 2.67, 2.82, 3.99 and 3.66 log copies/microg RNA in NASH-progressive, NASH, mild, simple steatosis and obese normal respectively. Although the differences in FABP-1 gene expression were directionally similar to differences in FABP-1 protein abundance seen in the proteomic analysis, the differences were not statistically different for gene expression.
Immunohistochemistry (IHC)
Lumican IHC staining patterns varied in intensity and distribution between study groups. Biopsy samples used for IHC analysis were the same as those from which samples were harvested for proteomic analyses and were confirmed in a further set of 4 participants from each of the study groups. Tables 3 and 4 present comparisons of the relative IHC staining intensity for lumican and FABP-1 between study groups. The results are very similar to those generated by the proteomic analysis.
Table 3.
Relative Abundance of Fatty Acid Binding Protein-1 on Proteomic Analysis
| Disease Progression | Normal Obese | Simple Steatosis | NASH-mild | NASH-progressive |
|---|---|---|---|---|
| Normal Obese | - | 0.86 | 0.69 | 0.68 |
| Simple Steatosis | 1.16 | - | 0.81 | 0.79 |
| NASH-mild | 1.45 | 1.24 | - | 0.99 |
| NASH-progressive | 1.47 | 1.27 | 1.02 | - |
Table 4.
Relative Protein Abundance of Lumican on IHC Analysis
| Disease Progression | Normal Obese | Simple Steatosis | Non-Alcoholic Steatohepatitis-1 | Non-Alcoholic Steatohepatitis-2 |
|---|---|---|---|---|
| Normal Obese | 1.00 | 0.83 | 0.67* | 0.60* |
| Simple Steatosis | 1.20 | 1.00 | 0.80 | 0.72* |
| Non-Alcoholic Steatohepatitis-1 | 1.50* | 1.25 | 1.00 | 0.90 |
| Non-Alcoholic Steatohepatitis-2 | 1.67* | 1.39* | 1.11 | 1.00 |
indicates p value <0.05.
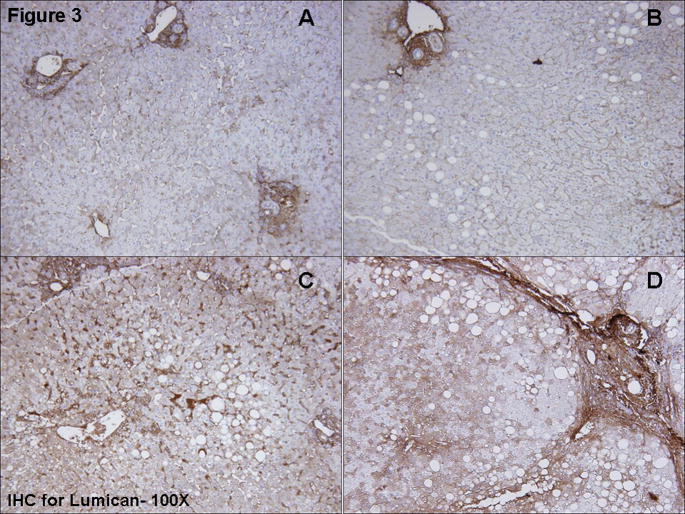
Lumican (Figure 3)
Figure 3.
Immunohistochemical staining for lumican. Moderate stromal staining was seen in portal areas. Staining intensity for lumican in hepatocytes varied between study groups, with more intense staining occurring in NASH-mild (3c) and NASH-severe (3d) than in simple steatosis (3b) or obese normal (3a) biopsies. Hepatocyte staining was cytoplasmic and was apparent throughout the parenchyma but most notable in zones 2–3 in a mosaic pattern. Mild sinusoidal staining was seen. The pattern of lumican staining closely paralleled the fibrosis.
IHC staining for lumican in biopsies from subjects from the Obese Normal and Simple Steatosis groups (Figure 3a and b), was mild to moderate in stromal and sinusoidal regions, with moderately intense cytoplasmic staining in a mosaic pattern in 25–50% of hepatocytes. Sinusoidal staining intensity was increased in NASH-mild and NASH-severe (Figure 3 c and d). Lumican staining was moderately intense in 50–75% of hepatocytes in NASH-severe, for which staining was throughout the parenchyma with accentuation in zone 2–3.
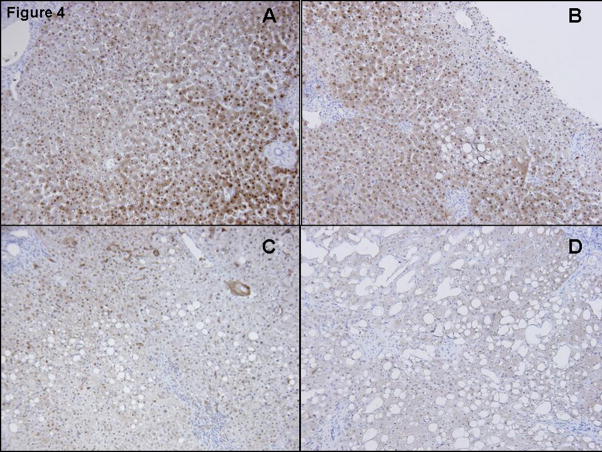
FABP-1 (Figure 4)
Figure 4. Immunohistochemical staining for FABP-1.
Immunohistology for FABP-1 revealed moderately intense abundance within the cytoplasm of hepatocytes in normal obese (Fig 4A) and in simple steatosis (Fig 4B). L-FABP staining was less intense in mild (Fig 4C) and severe forms of NASH (Fig 4D).
Staining for FABP-1 was apparent in 90–100% of hepatocytes for participants in the Obese Normal amnd Simple Steatosis groups (Figure 4a and b), with cytoplasmic staining in zones 1–3 and no sinusoidal or portal staining. Staining distribution was similar in subjects in the NASH-mild group but with less intensity than ON and SS. Little or no FABP-1 staining was seen in biopsies from the NASH-severe group.
Discussion
The number of obese individuals has been increasing relentlessly throughout most of the World, with an attendant increase in the number of people at risk for NAFLD. Patients who are at seemingly similarly risk for histologically progressive NAFLD can have disparate findings on liver biopsy, despite similar duration and severity of obesity.(11–13) Our analysis of the hepatic proteome has produced two novel observations that may be important in understanding the biologic basis of the histologic diversity of NAFLD. The first was that lumican is expressed in the human liver and is overexpressed in histologically progressive NAFLD. The second important observation is that FABP-1 is paradoxically underexpressed in histologically progressive NAFLD.
One of the principal findings is that lumican, a highly biologically active 40kDa keratin sulfate proteoglycan member of the leucine-rich repeat protein superfamily, is expressed differentially across the progressive stages of NAFLD. Lumican was similarly abundant in obese patients with normal liver histology and obese patients with simple steatosis but was overexpressed in NASH-mild when compared to simple steatosis, NASH-progressive vs. NASH-mild and NASH-progressive vs. obese normal. Although lumican expression in human liver has not previously been described, lumican is known to be expressed in the extracellular matrix of several other mesenchymal tissues, including skin,(14) lung,(15) cornea,(16, 17) and intestine(18). To confirm that lumican was expressed in human liver, we performed RT-PCR and lumican-specific immunohistochemical staining of the same liver biopsies that were used for the proteomic portion of the analysis. The RT-PCR studies demonstrated that the basis of the differential abundance of lumican across the histological spectrum of NAFLD is likely to be pre-transcriptional. Secondly, lumican detected in the proteomic analyses is likely to be hepatic in origin, rather than accumulated in the liver as a circulating protein, (although lumican circulation and accumulation may also occur). The directional relationship of lumican and mRNA for lumican across study groups was similar in the RT-PCR and proteomic analyses. That the magnitude of the differential expression of lumican and mRNA for lumican was different is not unexpected given the differences in the methods and biology of genes and gene products. Immunohistochemical staining with quantitative analysis demonstrated the same relative abundances of lumican in the various study groups as the proteomic analysis (Figures 1–4), confirming the relative overexpression of lumican in more histologically severe NAFLD.
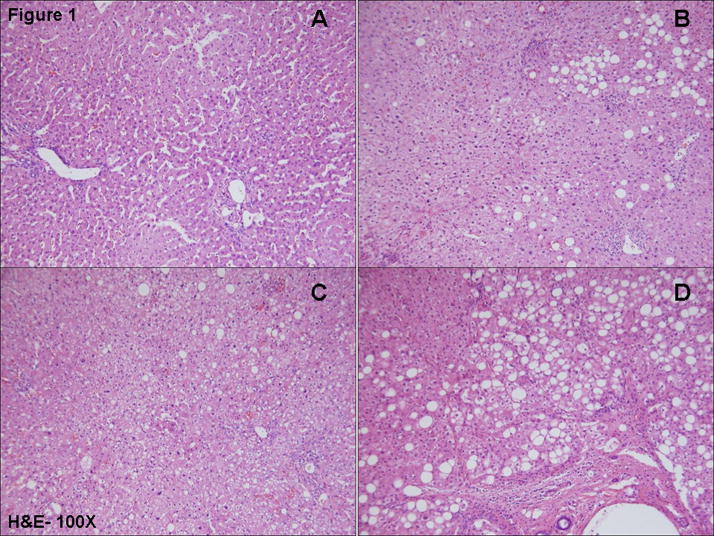
Figure 1.
Hematoxylin and eosin staining of (A)-Normal Obese; (B)-Simple Steatosis; (C)-NASH-mild and (D)- NASH-progressive is shown. Proteomic analysis was carried out on the same samples used for histological (H & E and Masson’s trichrome) and immunohistological analysis.
The implications of the identification of lumican within the human liver merits detailed consideration. Lumican has diverse biologic roles but has been thought to be involved primarily in fibrosis of the extracellular matrix through the binding of collagen fibrils and regulation of their lateral growth.(19, 20) Lumican has been studied most extensively in corneal tissue, where it appears to control the polymerization of collagen into small diameter fibrils.(20, 21) In addition to its role in regulating collagen fibrillogenesis, lumican participates in the injury-related transition of epithelial cells to mesenchymal cells,(16) and to exert a proapoptotic effect on stromal cells.(22, 23) Lumican expression is also increased with progression of hepatic fibrosis in rats.(24) Moreover, lumican promotes Fas-Fas ligand (FasL)-mediated cytokine signaling, probably through the presentation of Fas ligand to Fas ligand receptor.(22) Control of lumican expression is poorly understood, but has been shown to be stimulated by oxidative stress, (25, 26) and is at least partly mediated by NF-κB.(27) Decreased sulforylation of lumican side chains stimulates macrophage adhesion and the cellular inflammatory response,(28, 29) suggesting that changes in the structure of lumican may promote the inflammatory process that precedes and enhances collagen deposition during the process of hepatic fibrosis. Our finding that lumican was overexpressed in early stages of NAFLD, and not just in patients with moderate to advanced fibrosis, raises the possibility that overexpression of hepatic lumican might be an early marker of a pro-fibrotic state in patients with NAFLD at risk for histologically progressive disease. Our immunohistochemical studies show that lumican is abundant in the parenchyma, with mesenchymal staining also apparent. The immunohistochemical studies suggest that hepatocytes are the predominant source of lumican expression. It remains to be determined whether increased lumican synthesis is specific for NAFLD or plays a broader role in the progression of hepatic fibrosis, independent of the underlying etiology of disease. Lumican abundance in serum has not been reported to date, although because proteoglycans are typically quite soluble, it may be detectable in serum. It also remains to be determined whether increased lumican synthesis is specific for NAFLD or that it plays a broader role in the progression of hepatic fibrosis, independent of the underlying disease. Heparan sulfate proteoglycans have been shown to mediate the clearance of hepatic lipoproteins, which might suggest an additional role of proteoglycans in fatty liver disease.(30)
The other major finding of this analysis was that fatty acid binding protein-1 (FABP-1 or L-FABP) was (appropriately) relatively overexpressed in simple steatosis when compared obese normal (128%, p<0.001); but as the stages of NAFLD progressed FABP-1 was paradoxically underexpressed in NASH-mild vs. simple steatosis (73%, p<0.001), NASH-progressive vs. NASH-mild (81%, p<0.001) and NASH-progressive vs. obese normal (59%, p<0.001). FABPs are small, abundantly expressed cytoplasmic proteins that reversibly bind saturated and unsaturated long chain fatty acids, eicosanoids, and other lipids.(31–33) FABPs have multiple biologic functions, including roles in intracellular fatty acid transport, storage, and export, as well as cholesterol and phospholipid metabolism.(31–37) FABP-1 also plays an important facilitative role in hepatic fatty acid oxidation.(38, 39) The effects of FABP-1 deficiency have been described in null mice, which develop features of the metabolic syndrome, including hypertriglyceridemia, when fed a high-fat/high-sugar diet fed.(40) FABP-1 deficiency is also associated increased lipogenesis, hepatic insulin resistance and triglyceride secretion in mice.(37) Potentially more importantly, by binding fatty acids, FABP-1 is protective against the detergent effects of excess FFAs and is an important ligand for PPAR-mediated transcription. FABP-1 deficiency may thus contribute to many of the metabolic abnormalities associated with NAFLD and NASH.
We recently reported that dehydroepiandrostenedione (DHEA) levels are low in patients with more histologically severe NAFLD,(41) raising the question of whether our proteomic results support this observation. DHEA and its interchangeable sulfated form, DHEA-S, are, of course steroid hormones and will not have been detectable in a proteomic analysis. The mechanism of hepatic regulation of lumican expression is entirely unknown, as hepatic lumican expression has not previously been described. We are currently examining hepatic regulation of lumican expression in vivo and in vitro, including the effects of DHEA. With respect to FABP-1, DHEA is known to inhibitPPAR-gamma expression, an important regulator of FABP expression.(42)
There are some important caveats to consider when interpreting the results of any proteomic analysis. When investigating associations between protein expression and a disease, it is likely that proteins will be highlighted by chance. The risk of random associations can be reduced by restricting the analysis to proteins that have been identified a priori based on an hypothesis or the known biology of the disease. A disadvantage of limiting the scope of the proteins that are analysed is the greatly decreased likelihood of identifying novel pathways. For this reason, we did not limit our proteomics analysis to pre-selected proteins. To reduce the likelihood of random associations of differential protein abundance we used, instead, a strict statistical selection strategy and were very careful only to select those proteins which were consistently over- or under expressed across the different disease stages. Moreover, we confirmed our proteomics findings by RT-PCR and immunohistochemistry. Both RT-PCR and immunohistochemistry analyses supported the proteomic findings. We also did not perform a proteomic analysis of serum, which may well have yielded entirely different results. We did not have samples suitable for proteomic analysis of serum and, in any case, serum will contain proteins produced by many sources other than the liver. Our primary interest was in comparing the hepatic proteome between study groups. A further consideration is whether or not our findings are applicable to a wider population of patients with NAFLD and whether medications may have played a role in the observed differences in hepatic protein expression. We matched patients in terms of clinical and biochemical characteristics as far as we were able to (Table 1). As all patients had medically complicated obesity, as defines a bariatric surgery population, many patients were receiving prescription medications at the time of surgery. To minimize the impact of medications between the pairs of patients (all analyses were pair-wise), we required the protein expression differences to be present all pairs, e.g different between all subjects within one group when compared to paired subjects from another group. No two patients in or between groups were on the same medications. This stringent requirement greatly narrowed the number of proteins that were seen to be differentially expressed. Medication effects thus may have caused us to miss differential abundance for some proteins but it is highly unlikely to have accounted for the uniform differential protein abundance that was seen for the proteins we did identify.
In summary, more histologically progressive NAFLD is associated with overexpression of lumican, an important mediator of fibrosis in non-hepatic tissues, in association with more severe/advanced NAFLD. In contrast, FABP-1 is paradoxically underexpressed in NASH when compared to milder histologic variants of NAFLD, suggesting a new potential mechanism of lipotoxicity in NAFLD. Further studies are needed to determine the biologic basis of the dysregulation of lumican and/of FABP-1 expression in NAFLD.
Supplementary Material
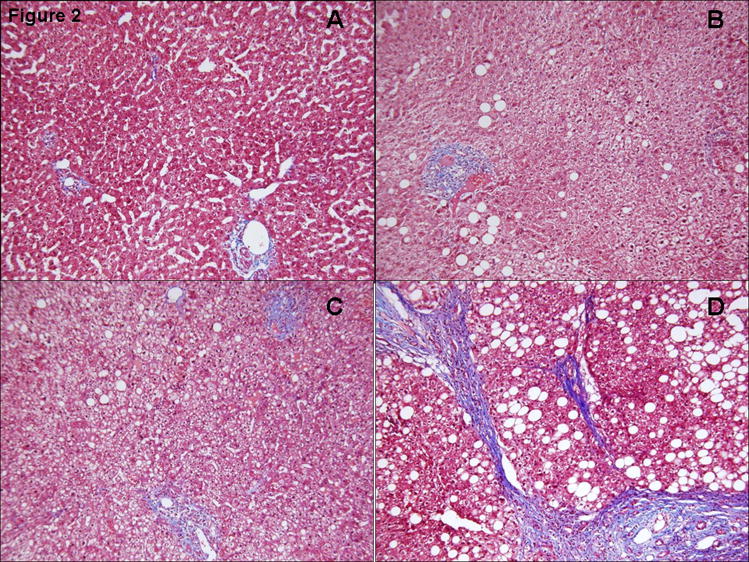
Figure 2.
Masson’s trichrome staining of (A)-Normal Obese; (B)-Simple Steatosis; (C)-NASH (mild); (D)- NASH-progressive is shown.
Table 5.
Relative Protein Abundance of FABP-1 on IHC Analysis
| Disease Progression | Normal Obese | Simple Steatosis | Non-Alcoholic Steatohepatitis-1 | Non-Alcoholic Steatohepatitis-2 |
|---|---|---|---|---|
| Normal Obese | 1.00 | 1.33 | 1.43 | 2.89* |
| Simple Steatosis | 0.75 | 1.00 | 1.08 | 2.18* |
| Non-Alcoholic Steatohepatitis-1 | 0.7 | 0.93 | 1.00 | 2.02* |
| Non-Alcoholic Steatohepatitis-2 | 0.35* | 0.46* | 0.49* | 1.00 |
indicates p value <0.05.
Acknowledgments
This work has been supported by Public Health Service grant NIDDK RO1 DK069757-01 and GCRC RR00585.
Footnotes
No conflicts of interest exist.
References
- 1.Ludwig J, Viggiano TR, McGill DB, Oh BJ. Nonalcoholic steatohepatitis: Mayo Clinic experiences with a hitherto unnamed disease. Mayo Clinic Proceedings. 1980;55:434–438. [PubMed] [Google Scholar]
- 2.Wanless IR, Lentz JS. Fatty liver hepatitis (steatohepatitis) and obesity: an autopsy study with analysis of risk factors. Hepatology. 1990;12:1106–1110. doi: 10.1002/hep.1840120505. [DOI] [PubMed] [Google Scholar]
- 3.Adler M, Schaffner F. Fatty liver hepatitis and cirrhosis in obese patients. American Journal of Medicine. 1979;67:811–816. doi: 10.1016/0002-9343(79)90740-x. [DOI] [PubMed] [Google Scholar]
- 4.Diehl AM, Goodman Z, Ishak KG. Alcohol like liver disease in nonalcoholics. A clinical and histologic comparison with alcohol-induced liver injury. Gastroenterology. 1988;95:1056–1062. [PubMed] [Google Scholar]
- 5.Sanyal AJ, Campbell-Sargent C, Mirshahi F, Rizzo WB, Contos MJ, Sterling RK, Luketic VA, et al. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology. 2001;120:1183–1192. doi: 10.1053/gast.2001.23256. [DOI] [PubMed] [Google Scholar]
- 6.Marchesini G, Bugianesi E, Forlani G, Cerrelli F, Lenzi M, Manini R, Natale S, et al. Nonalcoholic fatty liver, steatohepatitis, and the metabolic syndrome. 2003;37:917–923. doi: 10.1053/jhep.2003.50161. [erratum appears in Hepatology 2003 Aug;38(2):536] [DOI] [PubMed] [Google Scholar]
- 7.Adams LA, Lymp JF, St S, Sanderson SO, Lindor KD, Feldstein A, Angulo P. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. 2005;129:113–121. doi: 10.1053/j.gastro.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 8.Charlton M. Nonalcoholic fatty liver disease: a review of current understanding and future impact. Clinical Gastroenterology and Hepatology. 2004;2(12):1048–58. doi: 10.1016/s1542-3565(04)00440-9. [DOI] [PubMed] [Google Scholar]
- 9.Kim WR, Poterucha JJ, Porayko MK, Dickson ER, Steers JL, Wiesner RH. Recurrence of nonalcoholic steatohepatitis following liver transplantation. Transplantation. 1996;62:1802–1805. doi: 10.1097/00007890-199612270-00021. [DOI] [PubMed] [Google Scholar]
- 10.Angulo P. Nonalcoholic fatty liver disease and liver transplantation. NEJM. 2006;12:523–534. doi: 10.1002/lt.20738. [DOI] [PubMed] [Google Scholar]
- 11.Frantzides CT, Carlson MA, Moore RE, Zografakis JG, Madan AK, Puumala S, Keshavarzian A. Effect of body mass index on nonalcoholic fatty liver disease in patients undergoing minimally invasive bariatric surgery. Journal of Gastrointestinal Surgery. 2004;8(7):849–55. doi: 10.1016/j.gassur.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Silvestre V, Ruano M, Garcia-Lescun MC, Aguirregoicoa E, Criado L, Rodriguez A, Marco A, et al. Morbid obesity, non-alcoholic fatty liver disease, metabolic syndrome and bariatric surgery. Nutr Hosp. 2007;22:602–606. [PubMed] [Google Scholar]
- 13.Solga SF, Clark JM, Alkhuraishi AR, Torbenson M, Tabesh A, Schweitzer M, Diehl AM, et al. Race and comorbid factors predict nonalcoholic fatty liver disease histopathology in severely obese patients. Surg Obes Relat Dis. 2005;1:6–11. doi: 10.1016/j.soard.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Brezillon S, Venteo L, Ramont L, D’Onofrio MF, Perreau C, Pluot M, Maquart FX, et al. Expression of lumican, a small leucine-rich proteoglycan with antitumour activity, in human malignant melanoma. Clin Exp Dermatol. 2007;32:405–416. doi: 10.1111/j.1365-2230.2007.02437.x. [DOI] [PubMed] [Google Scholar]
- 15.Dolhnikoff M, Morin J, Roughley PJ, Ludwig MS. Expression of lumican in human lungs. Am J Respir Cell Mol Biol. 1998;19:582–587. doi: 10.1165/ajrcmb.19.4.2979. [DOI] [PubMed] [Google Scholar]
- 16.Saika S, Miyamoto T, Tanaka S, Tanaka T, Ishida I, Ohnishi Y, Ooshima A, et al. Response of lens epithelial cells to injury: role of lumican in epithelial-mesenchymal transition. Invest Ophthalmol Vis Sci. 2003;44:2094–2102. doi: 10.1167/iovs.02-1059. [DOI] [PubMed] [Google Scholar]
- 17.Saika S, Shiraishi A, Liu CY, Funderburgh JL, Kao CW, Converse RL, Kao WW. Role of lumican in the corneal epithelium during wound healing. J Biol Chem. 2000;275:2607–2612. doi: 10.1074/jbc.275.4.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blochberger TC, Vergnes JP, Hempel J, Hassell JR. cDNA to chick lumican (corneal keratan sulfate proteoglycan) reveals homology to the small interstitial proteoglycan gene family and expression in muscle and intestine. J Biol Chem. 1992;267:347–352. [PubMed] [Google Scholar]
- 19.Chakravarti S. Functions of lumican and fibromodulin: lessons from knockout mice. Glycoconj J. 2002;19:287–293. doi: 10.1023/A:1025348417078. [DOI] [PubMed] [Google Scholar]
- 20.Chakravarti S, Magnuson T, Lass JH, Jepsen KJ, LaMantia C, Carroll H. Lumican regulates collagen fibril assembly: skin fragility and corneal opacity in the absence of lumican. J Cell Biol. 1998;141:1277–1286. doi: 10.1083/jcb.141.5.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corpuz LM, Dunlevy JR, Hassell JR, Conrad AH, Conrad GW. Molecular cloning and relative tissue expression of decorin and lumican in embryonic quail cornea. Matrix Biol. 2000;19:699–704. doi: 10.1016/s0945-053x(00)00117-7. [DOI] [PubMed] [Google Scholar]
- 22.Vij N, Roberts L, Joyce S, Chakravarti S. Lumican regulates corneal inflammatory responses by modulating Fas-Fas ligand signaling. Invest Ophthalmol Vis Sci. 2005;46:88–95. doi: 10.1167/iovs.04-0833. [DOI] [PubMed] [Google Scholar]
- 23.Vij N, Roberts L, Joyce S, Chakravarti S. Lumican suppresses cell proliferation and aids Fas-Fas ligand mediated apoptosis: implications in the cornea. Exp Eye Res. 2004;78:957–971. doi: 10.1016/j.exer.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Krull NB, Gressner AM. Differential expression of keratan sulphate proteoglycans fibromodulin, lumican and aggrecan in normal and fibrotic rat liver. FEBS Lett. 1992;312:47–52. doi: 10.1016/0014-5793(92)81407-d. [DOI] [PubMed] [Google Scholar]
- 25.Xie L, Tsaprailis G, Chen QM. Proteomic identification of insulin-like growth factor-binding protein-6 induced by sublethal H2O2 stress from human diploid fibroblasts. Mol Cell Proteomics. 2005;4:1273–1283. doi: 10.1074/mcp.M500032-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Winokur ST, Barrett K, Martin JH, Forrester JR, Simon M, Tawil R, Chung SA, et al. Facioscapulohumeral muscular dystrophy (FSHD) myoblasts demonstrate increased susceptibility to oxidative stress. Neuromuscul Disord. 2003;13:322–333. doi: 10.1016/s0960-8966(02)00284-5. [DOI] [PubMed] [Google Scholar]
- 27.Wu F, Chakravarti S. Differential expression of inflammatory and fibrogenic genes and their regulation by NF-kappaB inhibition in a mouse model of chronic colitis. J Immunol. 2007;179:6988–7000. doi: 10.4049/jimmunol.179.10.6988. [DOI] [PubMed] [Google Scholar]
- 28.Funderburgh JL, Mitschler RR, Funderburgh ML, Roth MR, Chapes SK, Conrad GW. Macrophage receptors for lumican. A corneal keratan sulfate proteoglycan. Invest Ophthalmol Vis Sci. 1997;38:1159–1167. [PubMed] [Google Scholar]
- 29.Wu F, Vij N, Roberts L, Lopez-Briones S, Joyce S, Chakravarti S. A novel role of the lumican core protein in bacterial lipopolysaccharide-induced innate immune response. J Biol Chem. 2007;282:26409–26417. doi: 10.1074/jbc.M702402200. [DOI] [PubMed] [Google Scholar]
- 30.MacArthur JM, Bishop JR, Stanford KI, Wang L, Bensadoun A, Witztum JL, Esko JD. Liver heparan sulfate proteoglycans mediate clearance of triglyceride-rich lipoproteins independently of LDL receptor family members. J Clin Invest. 2007;117:153–164. doi: 10.1172/JCI29154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cistola DP, Sacchettini JC, Banaszak LJ, Walsh MT, Gordon JI. Fatty acid interactions with rat intestinal and liver fatty acid-binding proteins expressed in Escherichia coli. A comparative 13C NMR study. J Biol Chem. 1989;264:2700–2710. [PubMed] [Google Scholar]
- 32.Woodford JK, Behnke WD, Schroeder F. Liver fatty acid binding protein enhances sterol transfer by membrane interaction. Mol Cell Biochem. 1995;152:51–62. doi: 10.1007/BF01076463. [DOI] [PubMed] [Google Scholar]
- 33.Cistola DP, Walsh MT, Corey RP, Hamilton JA, Brecher P. Interactions of oleic acid with liver fatty acid binding protein: a carbon-13 NMR study. Biochemistry. 1988;27:711–717. doi: 10.1021/bi00402a033. [DOI] [PubMed] [Google Scholar]
- 34.Newberry EP, Kennedy SM, Xie Y, Sternard BT, Luo J, Davidson NO. Diet-induced obesity and hepatic steatosis in L-FABP−/− mice is abrogated with saturated but not polyunsaturated fat feeding and attenuated following cholesterol supplementation. Am J Physiol Gastrointest Liver Physiol. 2007 doi: 10.1152/ajpgi.00377.2007. [DOI] [PubMed] [Google Scholar]
- 35.Atshaves BP, McIntosh AM, Lyuksyutova OI, Zipfel W, Webb WW, Schroeder F. Liver fatty acid-binding protein gene ablation inhibits branched-chain fatty acid metabolism in cultured primary hepatocytes. J Biol Chem. 2004;279:30954–30965. doi: 10.1074/jbc.M313571200. [DOI] [PubMed] [Google Scholar]
- 36.Martin GG, Danneberg H, Kumar LS, Atshaves BP, Erol E, Bader M, Schroeder F, et al. Decreased liver fatty acid binding capacity and altered liver lipid distribution in mice lacking the liver fatty acid-binding protein gene. J Biol Chem. 2003;278:21429–21438. doi: 10.1074/jbc.M300287200. [DOI] [PubMed] [Google Scholar]
- 37.Newberry EP, Xie Y, Kennedy S, Han X, Buhman KK, Luo J, Gross RW, et al. Decreased hepatic triglyceride accumulation and altered fatty acid uptake in mice with deletion of the liver fatty acid-binding protein gene. J Biol Chem. 2003;278:51664–51672. doi: 10.1074/jbc.M309377200. [DOI] [PubMed] [Google Scholar]
- 38.Veerkamp JH, van Moerkerk HT. Fatty acid-binding protein and its relation to fatty acid oxidation. Mol Cell Biochem. 1993;123:101–106. doi: 10.1007/BF01076480. [DOI] [PubMed] [Google Scholar]
- 39.Kaikaus RM, Sui Z, Lysenko N, Wu NY, Ortiz de Montellano PR, Ockner RK, Bass NM. Regulation of pathways of extramitochondrial fatty acid oxidation and liver fatty acid-binding protein by long-chain monocarboxylic fatty acids in hepatocytes. Effect of inhibition of carnitine palmitoyltransferase I. J Biol Chem. 1993;268:26866–26871. [PubMed] [Google Scholar]
- 40.Newberry EP, Xie Y, Kennedy SM, Luo J, Davidson NO. Protection against Western diet-induced obesity and hepatic steatosis in liver fatty acid-binding protein knockout mice. Hepatology. 2006;44:1191–1205. doi: 10.1002/hep.21369. [DOI] [PubMed] [Google Scholar]
- 41.Charlton M, Angulo P, Chalasani N, Merriman R, Viker K, Charatcharoenwitthaya P, Sanderson S, et al. Low circulating levels of dehydroepiandrosterone in histologically advanced nonalcoholic fatty liver disease. Hepatology. 2008;47:484–492. doi: 10.1002/hep.22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashida K, Goto K, Zhao Y, Okabe T, Yanase T, Takayanagi R, Nomura M, et al. Dehydroepiandrosterone negatively regulates the p38 mitogen-activated protein kinase pathway by a novel mitogen-activated protein kinase phosphatase. Biochim Biophys Acta. 2005;1728:84–94. doi: 10.1016/j.bbaexp.2005.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.