Abstract
HIV-1 variants resistant to small molecule CCR5 inhibitors such as vicriviroc (VVC) have modified Env complexes that can use both the inhibitor-bound and -free forms of the CCR5 co-receptor to enter target cells. However, entry via the inhibitor-CCR5 complex is inefficient in some, but not all, cell types, particularly cell lines engineered to express CCR5. We investigated the effect of increasing CCR5 expression, and hence the density of the inhibitor-CCR5 complex when a saturating inhibitor (VVC) concentration was present, by using 293-Affinofile cells, in which CCR5 expression is up-regulated by the transcriptional activator, ponasterone. When CCR5 expression was low, the resistant virus entered the target cells to a lesser extent when VVC was present than absent. However, at a higher CCR5 level, there was much less entry inhibition at a constant, saturating VVC concentration. We conclude that the relative decrease in entry of a VVC-resistant virus in some cell types results from its less efficient use of the VVC-CCR5 complex, and that increasing the CCR5 expression level can compensate for this inefficiency.
Introduction
The small molecule CCR5 inhibitors represent a new class of therapy for HIV-1 infection, with the first class member (Maraviroc; MVC) now a licensed drug and a second (Vicriviroc; VVC) in late-stage trials (Hammer et al., 2006; Kuhmann and Hartley, 2008). These compounds bind to the CCR5 co-receptor and prevent its use by HIV-1 during virus-cell fusion. The inhibitory mechanism is non-competitive or allosteric; insertion of the small molecule into a cavity located within the transmembrane helices disrupts the geometry of a multi-point interaction between CCR5 and the HIV-1 gp120 glycoprotein (Dragic et al., 2000; Seibert et al., 2006; Tsamis et al., 2003; Watson et al., 2005). That association involves, at a minimum, the second extracellular loop (ECL-2) and tyrosine-sulfated N-terminus (Tyr-Nt) of CCR5 binding, respectively, to elements of the gp120 V3 region and the more conserved bridging sheet that forms between the C1, C2 and C4 domains after CD4 binding has occurred (Cormier and Dragic, 2002; Huang et al., 2007).
Although MVC, VVC and related compounds do efficiently suppress HIV-1 replication in cell culture and cause substantial reductions in plasma viremia, resistant variants can arise over time both in vitro and in vivo (Marozsan et al., 2005; Ogert et al., 2008; Trkola et al., 2002; Tsibris et al., 2008; Westby et al., 2007). These escape mutants are substantially resistant to the selecting compound, and are usually cross-resistant to other members of the same class (Pugach et al., 2008), although the latter is not always observed (Westby et al., 2007). The mechanism of resistance involves acquiring the ability to use the inhibitor-CCR5 complex, in addition to the free co-receptor, so that the virus can enter its target cells whether or not an inhibitor is present (Pugach et al., 2007; Westby et al., 2007). The escape mutants tend to be stable and fit; they replicate efficiently in the presence or absence of the inhibitor, and they do not rapidly revert to sensitivity when cultured in its absence in vitro, although the re-emergence of pre-treatment genetic sequences was seen after discontinuation of therapy in one infected person (Anastassopoulou et al., 2007; Trkola et al., 2002; Tsibris et al., 2008; Westby et al., 2007). The genetic pathway to resistance is complex, but it usually involves the accumulation of sequence changes in the gp120 V3 region (Baba et al., 2007; Kuhmann et al., 2004; Ogert et al., 2008; Tsibris et al., 2008; Westby et al., 2007). However, an alternative genetic pathway to the same phenotype involves sequence alterations elsewhere in Env, without changes in the V3 sequence (Marozsan et al., 2005). How gp120 from the resistant viruses can still interact with the inhibitor-bound form of CCR5 is not yet fully understood, but is thought to involve alterations in the relative usage of the different elements of the multi-point binding interaction.
The inhibition profiles for small molecule CCR5 inhibitors against resistant viruses are unusual in form and they vary with the target cell type and virus inoculum (Ogert et al., 2008; Pugach et al., 2007; Westby et al., 2007). Irrespective of the target cell type, saturating concentrations of the inhibitors cause essentially 100% inhibition of wild-type HIV-1 isolates, clones or Env-pseudotyped viruses, allowing the determination of conventional IC50 and IC90 values. The inhibitors have little or no activity against in vitro-selected resistant viruses, when the target cells are PBMC. However, when the same inhibitors are tested against the same resistant viruses in various CCR5-expressing cell lines, they generally do cause some inhibition of entry/replication, although with an unconventional manifestation (Ogert et al., 2008; Pugach et al., 2007; Westby et al., 2007). Thus, in plots that depict how entry inhibition varies with the inhibitor concentration, the curves asymptote to a plateau. Once the plateau is reached, increasing the inhibitor concentration has no additional effect. The explanation of the “plateau effect” is that it measures the entry of the resistant virus via the inhibitor-bound CCR5 relative to the drug-free form; the more efficiently entry occurs via the inhibitor-CCR5 complex, the lower the plateau height, which is also known as the Maximum Percent Inhibition (MPI) value (Pugach et al., 2007; Westby et al., 2007). In extreme cases, entry via the inhibitor-receptor complex may even be more efficient than via the free receptor, leading to negative inhibition (Pugach et al., 2007; Tsibris et al., 2008). Understanding these issues is necessary for interpretation of clinical resistance to CCR5 inhibitors, which is usually measured using the commercial Trofile assay that relies on single-cycle infection in a CCR5-expressing, engineered cell line (Whitcomb et al., 2007). Here, we show that the amount of CCR5 expressed on a target cell line affects the plateau height and hence how resistance is manifested. We also discuss what factors might influence the efficiency with which resistant viruses use the inhibitor-CCR5 complex on different cells and in different assays.
Results
The maximum extent of inhibition of a CCR5 inhibitor-resistant virus varies with the cell surface CCR5 density
Previous studies have shown that the manifestation of resistance to small molecule CCR5 inhibitors is cell type-dependent; in particular, the MPI value (plateau height) for CCR5 inhibitor-resistant viruses varies between target cell types, particularly between PBMC and cell lines engineered to express CCR5 (Ogert et al., 2008; Pugach et al., 2007; Westby et al., 2007). One plausible explanation is that the MPI is influenced by the cell surface CCR5 concentration. Because cell surface areas differ between cell types, it is difficult to compare the densities of receptors such as CCR5 based solely on expression level measurements. To address the relationship between MPI and CCR5 surface density, we therefore used the 293-Affinofile cell line in which the amount of cell surface CCR5 can be varied via application of different concentrations of the transcriptional activator, ponasterone (B. Lee, unpublished results). Under these circumstances, increases in CCR5 expression translate into increases in receptor density, as the cell surface area is unchanged. Of note is that without ponasterone induction, 293-Affinofile cells naturally express low levels of CCR5 that are, however, sufficient for infection by some HIV-1 and SIV strains (B. Lee, unpublished results).
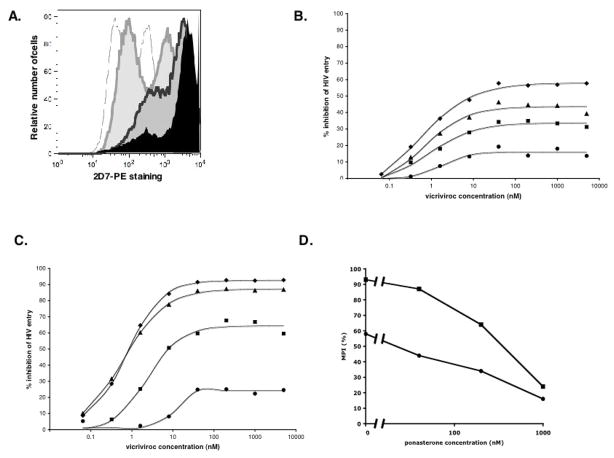
The 293-Affinofile cells were treated with three different ponasterone concentrations to induce low, intermediate or high levels of cell surface CCR5, and with a fixed (100 ng/ml) concentration of tetracycline to induce high level CD4 expression (B. Lee, unpublished results). In addition to increasing the overall expression of CCR5, ponasterone treatment also resulted in a more uniform distribution of the coreceptor across the cell population (Fig. 1A). Thus, in the absence of ponasterone, two subpopulations of cells with distinct levels of CCR5 expression were evident. The lowest ponasterone concentration increased CCR5 expression in both subpopulations, but at the higher concentrations the highest-expressing subpopulation became predominant (Fig. 1A).
Figure 1. The maximum percent inhibition of a VVC-resistant Env-pseudotyped viruses vary inversely with the cell surface CCR5 level.
293-Affinofile cells were treated for 18h with ponasterone concentrations of 0.04, 0.2 or 1 μM, to induce low, intermediate or high levels of cell surface CCR5, respectively. The concentration of tetracycline, which induces CD4 expression, was held constant at 100 ng/ml. A) The cell surface expression of CCR5 was determined by staining with PE-labeled MAb 2D7 followed by flow cytometry. Raw histogram data are shown for cells treated with medium alone (grey outline), 0.04 μM (light grey area), 0.2 μM (dark grey area) or 1 μM ponasterone (black area). B, C) Cells treated with medium alone (diamonds), 0.04 μM (triangles), 0.2 μM (squares) or 1 μM ponasterone (circles) were incubated for 1h with varying concentrations of VVC. The cells were then infected with the CC101.19 cl.7 (panel B) or D1/85.16 cl.23 (panel C) Env-pseudotyped virus and luciferase production was measured 48h later. The extent of inhibition was calculated from the level of entry observed in the absence of inhibitors at each ponasterone concentration (100% entry = 0% inhibition). The MPI values for CC101.19 and D1/85.16 were, respectively, 58% and 93% in the absence of ponasterone, 44% and 87% at 40 nM ponasterone; 34% and 64% at 200 nM ponasterone; 16% and 24% at 1 μM ponasterone. D. The MPI values derived from the above assays are expressed as a function of the ponasterone concentration for D1/85.16 cl.23 (squares) and CC101.19 cl.7 (circles).
To assess the effect of varying the CCR5 expression levels, we infected the ponasterone-treated cells with the Env-pseudotyped virus CC101.19 cl.7, in the presence of varying concentrations of VVC. The CC101.19 isolate was derived from the CC1/85 primary virus under the selection pressure of the AD101 small molecule CCR5 inhibitor, and is cross-resistant to VVC and other, similar inhibitors (Pugach et al., 2008).
The resulting MPI values for VVC inhibition of CC101.19 cl.7 varied inversely with the ponasterone concentration, i.e., with the amount of CCR5 on the cell surface (Fig. 1B). In other words, the more CCR5 was present on the target cells, the less that entry of the resistant virus was inhibited by VVC. At the highest ponasterone (i.e., CCR5) concentration, the MPI represented ~15% inhibition, similar to what we previously observed with the same test virus and inhibitors in PBMC; the virus is almost completely resistant (Kuhmann et al., 2004; Pugach et al., 2007). In contrast, in the absence of ponasterone, the MPI of ~60% was comparable to that seen with the same virus on U87-CD4/CCR5 cells (Pugach et al., 2007). The latter cells are the basis of the commercial Trofile assay for CCR5 inhibitor-resistance (Whitcomb et al., 2007). Virtually identical results were obtained when maraviroc, a different small molecule CCR5 inhibitor, was used instead of VVC (data not shown).
To determine whether the above observations were unique to the CC101.19 strain or more generally applicable, we repeated the experiment but using an Env-pseudotyped virus cloned from the D1/85.16 isolate. The D1/85.16 virus was also derived from CC1/85 but under the selection pressure of VVC; like CC101.19 it uses the inhibitor-CCR5 complex for entry (Marozsan et al., 2005; Pugach et al., 2007). However, D1/85.16 cl.23, unlike CC101.19 cl.7, has no V3 sequence changes, and its genetic route to resistance involves the acquisition of 3 amino acid substitutions in the gp41 fusion peptide (Marozsan et al., 2005; Anastossopolou et al., 2009).
The phenotype of D1/85.16 cl.23 was broadly similar similarly to that of CC101.19 cl.7, in that its resistance to VVC was also dependent on the CCR5 expression level (Fig. 1C). However, D1/85.16 cl.23 was less resistant to VVC than CC101.19 cl.7 at low CCR5 concentrations; the MPI value was 24% with cells treated with high concentrations of ponasterone, but as high as 93% with untreated cells (Fig. 1C). For comparison, the MPI values for the parental virus were 96% at 0 nM and 40 nM ponasterone, 90% at 200 nM and 85% at 1 μM. Furthermore, the relationship between the extent of (MPI value) and the CCR5 expression level (ponasterone concentration) was steeper for D1/85.16 cl.23 than for CC101.19 cl.7 (Fig. 1D). This effect of increased CCR5 levels was mainly on the extent, and not the potency, of inhibition; thus, the half-maximal inhibitory concentrations for the two resistant viruses were in the range 1–10 nM irrespective of the ponasterone concentration (Figs, 1B, C).
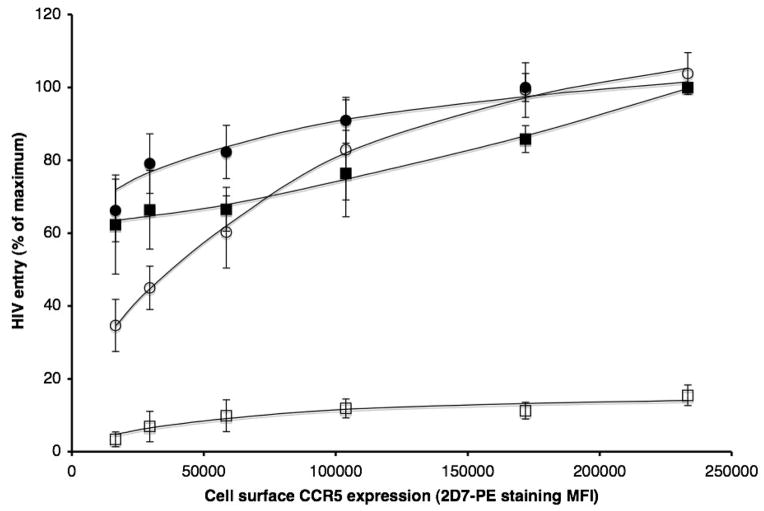
When the ponasterone concentration was titrated to vary the cell surface CCR5 expression level, the mean fluorescence intensity (MFI) values derived by staining the cells with the phycoerythrin (PE)-labeled anti-CCR5 MAb 2D7-PE ranged from ~16,000 to ~230,000. Under the same conditions, we measured entry of the parental (CC1/85 cl.6) and resistant (CC101.19 cl.7) Env-pseudotyped viruses, in the presence or absence of a saturating VVC concentration (5 μM) (Fig. 2). The entry of both viruses was only partially dependent on the CCR5 concentration in the absence of VVC, although a steady increase occurred at the higher end of the concentration range, particularly for CC101.19 cl.7. Thus even when CCR5 concentrations were at the low end of the expression range, each virus could enter the target cells fairly well (~65% of the extent seen at the highest CCR5 level).
Figure 2. Increased CCR5 concentration compensates for the reduced efficiency of cell entry by the resistant virus via the VVC-CCR5 complex.
293-Affinofile cells were treated with tetracycline (100 ng/ml) and varying ponasterone concentrations (from 0 to 1 μM). Entry of the parental CC1/85 cl.6 (squares) or the resistant CC101.19 cl.7 (circles) Env-pseudotyped virus was measured in the presence (open symbols) or absence (closed symbols) of 5 μM VVC. The extent of entry of each Env-pseudotyped virus into cells treated with 1 μM ponasterone (the highest concentration used), in the absence of VVC, was defined as 100% for normalization purposes. For each data point, an identical sample of cells was labeled with the 2D7-PE MAb and analyzed for CCR5 expression by flow cytometry. The resulting MFI value was plotted against the extent of HIV-1 entry.
When VVC was present, entry of the parental virus was, as expected, negligible, although it did increase slightly with the CCR5 expression level. However, the resistant virus was much more dependent on CCR5 levels when VVC was present; at the lowest CCR5 expression level, the extent of entry was only 35% of that seen at the highest (Fig. 2). This observation implies that entry of the resistant virus via the VVC-CCR5 complex is inefficient when the CCR5 concentration is at the low end of the titration range, but that increasing the abundance of VVC-CCR5 complexes can compensate.
Fusion mediated by mutant Env glycoproteins via the VVC-CCR5 complex
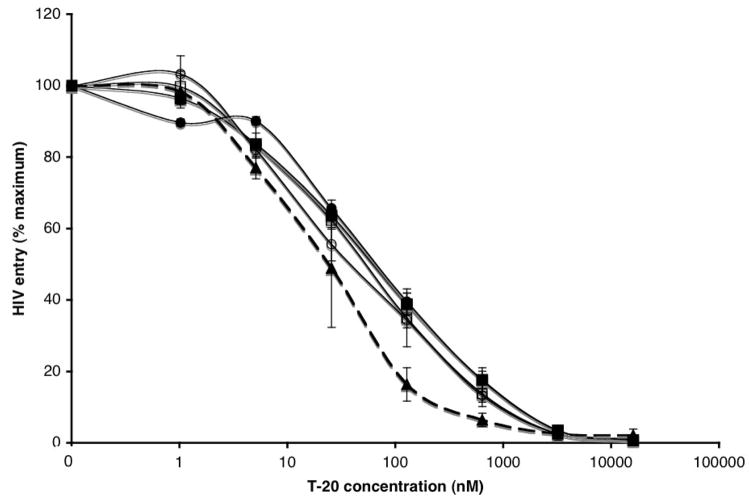
The plateau effect in infection-inhibition assays on CCR5-expressing cell lines arises because the resistant viruses use the inhibitor-CCR5 complex less efficiently than the free co-receptor; the extent of entry is therefore reduced when the inhibitor concentration is sufficient to saturate all the available CCR5 (Pugach et al., 2007; Westby et al., 2007). We showed earlier that increasing the CCR5 concentration (and hence the quantity of inhibitor-CCR5 complexes present) compensates for the reduced efficiency with which the inhibitor-CCR5 complex is used (Fig. 1B, C). But why is entry via inhibitor-bound CCR5 less efficient? HIV-1 coreceptor affinity and fusion kinetics are directly correlated with each other and inversely correlated with sensitivity to fusion inhibitors such as T-20, because the longer the “window period” when the fusion-intermediate structure of gp41 is exposed to T-20, the less T-20 is required for inhibition (Reeves et al., 2002). We therefore assessed whether the reduced efficiency of CC101.19 cl.7 Env-mediated entry in the presence of VVC affected T-20 sensitivity (Fig. 3). The CC101.19 cl.7 Env-pseudotyped virus was ~2.5-fold less sensitive to T-20 than the parental virus in the absence of VVC (IC50 = 65 nM, compared to 25 nM). This is consistent with the modest difference in T-20 sensitivity (~2-fold) we observed with the corresponding uncloned isolates in a PBMC-based replication assay (Pugach et al., 2008). When VVC was present, it did not affect the inhibition of CC101.19 cl.7 by T-20, irrespective of the CCR5 concentration on the 293-Affinofile cells (Fig. 3). Hence, even when the cells expressed low levels of CCR5, a condition when CC101.19 cl.7 entry was relatively inefficient (Fig. 2), this did not translate into an influence on T-20 sensitivity (Fig. 3). Similar results were obtained using the corresponding, fully infectious parental and resistant viruses on Tzm-bl cells (data not shown).
Figure 3. The inefficient use of the VVC-CCR5 complex by a VVC-resistant virus does not affect T-20 sensitivity.
The parental CC1/85 cl.6 (dashed line) or resistant CC101.19 cl.7 (solid lines) Env-pseudotyped viruses were incubated for 1h with varying concentrations of T-20 and then used to infect 293-Affinofile cells in the presence (open symbols) or absence (closed symbols) of 5 μM VVC. The cells had been treated medium alone (not shown, for clarity), or with ponasterone at concentrations of 0.04 μM (circles), 0.2 μM (not shown, for clarity) or 1 μM (squares) to induce varying levels of cell surface CCR5. The curves for CC1/85 cl.6 in the absence of VVC were virtually identical to each other, so only the one for the lowest ponasterone concentration is shown, for clarity. There was no detectable entry of CC1/85 cl.6 in the presence of VVC (not shown). The extent of entry of each virus is expressed relative to that observed in the absence of T-20 (= 100%). The data shown were averaged from three independent experiments.
We also varied the time of addition of a maximally effective T-20 concentration (10μM) after CC101.19 cl.7 had been allowed to bind to the Affinofile cells at 4°C and then transferred to 37°C to allow the fusion events to take place. The time taken for the virus to become resistant to T-20 (i.e., for the T-20-sensitive conformational changes in gp41 to be completed) was similar whether VVC was present or not (data not shown).
Taken together, the above experiments imply that using the VVC-CCR5 complex for entry does not lead to an increased exposure of the T-20-sensitive fusion intermediate structure of gp41, compared to use of free CCR5. Hence, the reduced entry efficiency in the presence of VVC appears not to be due to any slowing of the rate of conformation changes in gp41 subsequent to receptor binding.
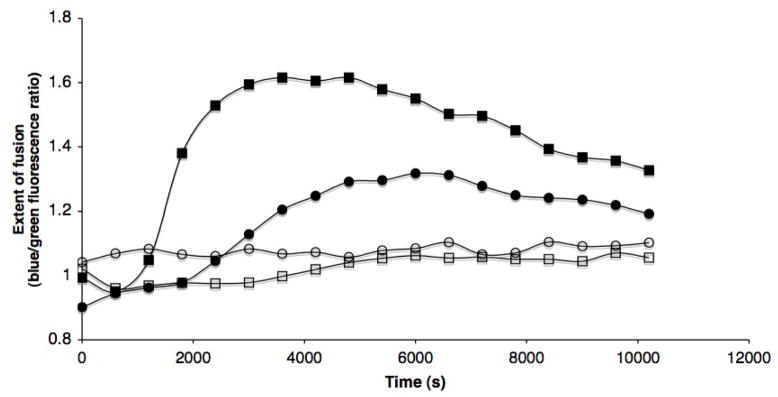
As an additional measure of the rate and extent of Env-mediated membrane fusion activity, we used a cell-cell fusion assay in which Env-expressing QT6 cells were mixed with CD4/CCR5-expressing JC53 (HeLa-derived) cells (Reeves et al., 2005). Env proteins from the parental CC1/85 cl.6 and the CCR5 inhibitor-resistant CC101.19 cl.7 viruses were expressed at similar levels on the transfected QT6 cells (data not shown). Both Env proteins mediated membrane fusion activity in the absence of VVC, but the maximal extent of fusion was 2-fold lower for the resistant Env than the parental, and it took longer for the resistant Env to drive fusion to its half-maximal level, 2400s vs 1500s for the parental Env (Fig. 4). Hence, the resistant CC101.19 cl.7 Env is less fusion-competent than the parental CC1/85 cl.6 Env in this experimental system. This difference agrees with our previous report that the replication capacity of CC101.19 cl.7 is less than that of CC1/85 cl.6 (Anastassopoulou et al., 2007).
Figure 4. Cells expressing VVC-resistant Env do not fuse efficiently in the presence of VVC.
Env-transfected, β-lactamase-expressing QT6 cells were incubated with CCF2-AM-loaded, CD4- and CCR5-expressing JC53 cells. Membrane fusion by the parental CC1/85 cl.6 (squares) or the resistant CC101.19 cl.7 (circles) Env glycoproteins was measured in the presence (open symbols) or absence (closed symbols) of 5 μM VVC. The extent of fusion over time is shown as the blue/green fluorescence ratio normalized for background by dividing by the ratio obtained with control effector cells that were transfected with a vector that does not express Env.
When the JC53 target cells were treated with 5 μM VVC, the rate and extent of cell-cell fusion was reduced to negligible levels, for both the parental and the resistant Envs. The CC101.19 cl.7 Env is, therefore, resistant to VVC in a virus-entry assay, but sensitive to it in a cell-cell fusion assay; under the conditions of the fusion assay, the CC101.19 cl.7 Env glycoproteins use CCR5 inefficiently and the VVC-CCR5 complex even more so.
Discussion
We previously reported that the manifestation of resistance to small molecule CCR5 inhibitors varies with the cell type used to detect and quantify it (Pugach et al., 2007). Here, we show that CCR5 density is one of the relevant host cell factors that account for these differences. The resistant viruses have evolved a way to recognize the inhibitor-bound conformation of CCR5 while not losing the ability to interact with the free receptor (Pugach et al., 2007; Westby et al., 2007). Hence they are not “drug-dependent”, merely “drug-tolerant”. The acquisition of resistance appears to involve an alteration in how gp120 binds to CCR5. In engineered cell lines, the inhibitor-bound and –free forms of CCR5 are not utilized equally well by resistant viruses (the inhibitor-bound form is, of course, not recognized at all by wild type strains). Usually, the inhibitor-bound form is the less effective receptor configuration, but there are some rare examples of the converse situation, in which an inhibitor modestly increases entry of a resistant virus (Pugach et al., 2007; Tsibris et al., 2008).
The differential recognition of the two receptor configurations underlies the plateau effect, the partial inhibition that arises when the resistant viruses are tested using cell lines such as the U87-CD4/CCR5 cells that form the basis of the commercial Trofile assay (Pugach et al., 2007; Westby et al., 2007; Whitcomb et al., 2007). Here, the height of the plateau (MPI value) is a quantitative representation of the differential use of the two CCR5 configurations; the lower the MPI, the more efficiently the inhibitor-CCR5 complex is used and the greater the extent of resistance (Pugach et al., 2007; Westby et al., 2007). We now show that the MPI value is an inverse function of the CCR5 cell-surface density. Specifically, high-level resistance (a low plateau or low MPI value) is observed only when the target cells express high levels of CCR5. Presumably, the increased amount of the inhibitor-CCR5 complex compensates for the decreased entry efficiency via this complex compared to the free co-receptor, allowing the resistant virus to enter cells efficiently in the presence of the inhibitor. Overall, these observations should further help to understand the resistance profiles generated in the commercial Trofile assay (Pugach et al., 2007; Westby et al., 2007; Whitcomb et al., 2007).
CCR5 cell surface levels are highly variable in the population and depend on multiple genetic factors, including polymorphisms in the CCR5 promoter and the expression of its chemokine ligands (Martin et al., 1998; Trkola et al., 1996). Low CCR5 and high chemokine levels are associated with slower HIV-1 disease progression rates (Gonzalez et al., 2005; Reynes et al., 2000). Variation in CCR5 and chemokine expression may also have implications for the use of CCR5 inhibitors as therapy, including from the perspective of resistance development. Our observations suggest that individuals with low levels of free CCR5 (low CCR5 and/or high chemokine) may be less likely to develop CCR5 inhibitor-resistant variants, because the less effective use of the inhibitor-CCR5 complex when CCR5 expression is low would reduce their replicative fitness. Paradoxically, however, this factor might increase the chances of resistance developing by what is the normally unfavorable process of co-receptor switching to CXCR4 use. Clinical studies of CCR5 inhibitors should therefore take into account CCR5 and chemokine population genetics when assessing not just the effectiveness of therapy, but also the rapidity and nature of resistance development. While what we have observed using engineered, CCR5-expressing cell lines may not extrapolate completely to the different setting of natural lymphoid cells, a similar relationship between the extent of resistance and the level of CCR5 expression was found in recent studies that used rapamycin to down-regulate CCR5 in PBMCs (Heredia et al., 2008). By extension, VVC-resistant strains may not be capable of efficiently infecting macrophages in the presence of VVC, since CCR5 density is expressed at only low levels on these cells (Lee et al., 1999). Indeed, neither CC101.19 nor D1/85 can infect macrophages at a quantifiable level whether VVC is present or not (Pugach et al., 2004) (P. Pugach and J. P. Moore, unpublished results).
The sensitivity of HIV-1 strains to T-20 correlates inversely with their fusion kinetics and capacity to interact with coreceptors expressed at low levels (Abrahamyan et al., 2005; Platt et al., 2007; Reeves et al., 2002; Taylor et al., 2008). Given that CC101.19 cl.7 enters cells less efficiently at low cell-surface CCR5 levels when VVC is present, we investigated whether this virus would become more sensitive to T-20 in the presence of VVC, especially at low CCR5 expression levels. However, we found that VVC had no such effect on the sensitivity of CC101.19 cl.7 to T-20 at any of the CCR5 levels tested (Fig. 3, and data not shown). One possible explanation for this observation is that the resistance mutations affect a step(s) in the fusion process that precedes the T-20-sensitive refolding of Env. For example, the minimum number of Env-CCR5 complexes required to form a fusion pore may be greater for the resistant Env, or the recruitment of sufficient CCR5 molecules into a fusion complex may occur more slowly; the resistance mutations could affect such Env functions both when VVC is present and absent, creating differences between the parental and resistant viruses. Five to eight Env-receptor complexes have been proposed to be involved in HIV-1 entry (Klasse, 2007; Kuhmann et al., 2000; Magnus et al., 2009; Sougrat et al., 2007); it is conceivable that this number might be greater for CCR5 inhibitor-resistant variants. Variation in how CCR5 is recruited into fusion complexes could also explain the different dependences of the parental and resistant viruses on CCR5 density. It is also possible that parental and resistant viruses preferentially use different subsets of CCR5 conformational variants for entry, and that these CCR5 forms differ in abundance between cell types or, in the case of the 293-Affinofile cells, with the ponasterone concentration (Fig. 1A) (Anastassopoulou et al., 2009).
CCR5 inhibitor resistance was not recapitulated in Env-mediated cell-cell fusion assays. Thus, VVC significantly reduced the membrane fusion activity of Env glycoproteins from the resistant CC101.19 cl.7 virus. Overall, we conclude that, under the conditions of the fusion assay, the CC101.19 cl.7 Env glycoproteins must use the VVC-CCR5 complex very inefficiently. This contrasts with the resistance of the same Env proteins in infectivity assays using different cell types, although the extent of resistance was weak at the lowest CCR5 expression levels. Differences in the total amount or forms of CCR5 and/or Env, or in their localization in rafts and other membrane areas, between the different assay systems might underlie these observations. The requirements for the lateral diffusion of CCR5 proteins and their recruitment into fusion pores, may also differ markedly between virus-cell and cell-cell fusion systems as well as between the sensitive and resistant viruses (see above).
We conclude that the interaction between inhibitor-resistant HIV-1 Env and CCR5 is more sensitive to decreases in CCR5 expression levels when an inhibitor such as VVC is present, than not. The resistant virus does not use the inhibitor-CCR5 complex as efficiently as it uses the free coreceptor. However, under some conditions, for example in a PBMC-based, multi-cycle virus replication assay, the loss of efficiency does not reduce the maximum degree of entry, which can even be enhanced. Hence the resistant viruses are fit and do not revert rapidly to sensitivity when cultured in PBMC in the absence of the inhibitor (Anastassopoulou et al., 2007; Trkola et al., 2002; Westby et al., 2007). Complete resistance, as seen in a PBMC assay, occurs only when CCR5 and Env are present at a high enough density, or are in suitable configurations, to compensate for less efficient use of the inhibitor-CCR5 complex; under other circumstances, such as in the Trofile assay, resistance is incomplete, leading to distinct plateaus of inhibition.
Methods
Reagents and cell lines
The CCR5 inhibitors VVC and MVC were gifts from Dr. Julie Strizki (Schering-Plough Research Institute, Kenilworth, NJ) and Dr. Chris Hitchcock (Pfizer, Sandwich, UK), respectively. T-20 was provided by Dr. William Olson (Progenics Pharmaceuticals Inc., Tarrytown, NY). Tetracycline and ponasterone were purchased from Sigma (St. Louis, Mo), Blasticidin from Invitrogen (Carlsbad, CA). 293-Affinofile cells were maintained in DMEM (Invitrogen), 10% fetal bovine serum (FBS, Invitrogen) and 50ug/ml blasticidin (B. Lee, unpublished results). Tzm-bl cells were obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. Cells were cultured at 37°C in an atmosphere containing 5% CO2.
Flow cytometry
293-Affinofile cells were detached with versene 18h after treatment with ponasterone (Invitrogen). Samples (1 × 106 cells) were then stained with PE-labeled 2D7 antibody (BD-Biosciences, San Jose, CA) for 30 minutes at 4°C in the cytometry buffer (phosphate-buffered saline with 10% FBS). After two washes, the antibody-labeled cells were analyzed using an LSRII digital cytometer (BD Biosciences). Overlay of the histograms was performed using the FlowJo software (Tree Star, Inc., Ashland, OR).
HIV-1 entry assays
293-Affinofile cells were seeded in white 96-well plates at a density of 5000 cells per well, in 100 μl of medium, 18h prior to infection. At this point, 293-Affinofile cells were treated with varying concentrations of ponasterone and tetracycline. VVC and MVC (50 μl) were added to the cells 1h prior to infection. Env-pseudotyped viruses were made by cotransfection of the pCI-env and pNLluc-AM plasmids into 293T cells at a 3:1 ratio, using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The pCI-env and pNLluc-AM plasmids were constructed as described previously (Pugach et al., 2007). Two days post-transfection, supernatants were passed through a 0.45 μm filter and immediately used to infect the 293-Affinofile cells. Luciferase production was quantified after 48 hours, as described previously (Pugach et al., 2007). To assess T-20 sensitivity, Env-pseudotyped viruses were incubated for 1h at 37°C with varying concentrations of T-20 and then used to infect 293-Affinofile cells.
HIV-1 Env-mediated fusion assay
Fusion mediated by the parental and CCR5 inhibitor-resistant Env proteins was determined using a β-lactamase reporter cell-cell fusion assay, as described previously (Lineberger et al., 2002; Reeves et al., 2005). Briefly, effector QT6 cells were cotransfected with Env and codon-optimized β-lactamase expressing plasmids and infected with a T7 polymerase-encoding vaccinia virus. At 4°C, effector cells were added to Hela/CD4/CCR5 cells (JC53) loaded with CCF2-AM (an acetoxymethylester derivative of CCF2 that has a donor fluorophore (coumarin) linked to an acceptor (fluorescein) by a β-lactam ring), and then the temperature was shifted immediately to 37°C. Cell-cell fusion in this assay can be detected as a shift from green to blue fluorescence, which indicates cleavage of CCF2 by β-lactamase. Fluorescence was detected using a fluorometer (FLUOstar OPTIMA, BMG Labtech). The extent of fusion is expressed as the blue/green fluorescence ratio, which was normalized for background by dividing by the ratio obtained with control effector cells that were transfected with a vector that does not express Env.
Acknowledgments
We thank Shawn Kuhmann for his many contributions to the evolution of this project. We appreciate the discussions we have had with Rogier Sanders, Reem Berro and Cleo Anastassopoulou on resistance to CCR5 inhibitors and fusion inhibition. This work was funded by NIH grants R01 AI41420, T32 AI07621 (PP) and R01 AI40880.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahamyan LG, Mkrtchyan SR, Binley J, Lu M, Melikyan GB, Cohen FS. The cytoplasmic tail slows the folding of human immunodeficiency virus type 1 Env from a late prebundle configuration into the six-helix bundle. J Virol. 2005;79 (1):106–15. doi: 10.1128/JVI.79.1.106-115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassopoulou CG, Ketas TJ, Klasse PJ, Moore JP. Resistance to CCR5 inhibitors caused by sequence changes in the fusion peptide of HIV-1 gp41. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0811713106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassopoulou CG, Marozsan AJ, Matet A, Snyder AD, Arts EJ, Kuhmann SE, Moore JP. Escape of HIV-1 from a small molecule CCR5 inhibitor is not associated with a fitness loss. PLoS Pathog. 2007;3 (6):e79. doi: 10.1371/journal.ppat.0030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Miyake H, Wang X, Okamoto M, Takashima K. Isolation and characterization of human immunodeficiency virus type 1 resistant to the small-molecule CCR5 antagonist TAK-652. Antimicrob Agents Chemother. 2007;51(2):707–15. doi: 10.1128/AAC.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier EG, Dragic T. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J Virol. 2002;76 (17):8953–7. doi: 10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragic T, Trkola A, Thompson DA, Cormier EG, Kajumo FA, Maxwell E, Lin SW, Ying W, Smith SO, Sakmar TP, Moore JP. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc Natl Acad Sci U S A. 2000;97 (10):5639–44. doi: 10.1073/pnas.090576697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, Murthy KK, Rovin BH, Bradley W, Clark RA, Anderson SA, O’Connell RJ, Agan BK, Ahuja SS, Bologna R, Sen L, Dolan MJ, Ahuja SK. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307 (5714):1434–40. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- Hammer SM, Saag MS, Schechter M, Montaner JS, Schooley RT, Jacobsen DM, Thompson MA, Carpenter CC, Fischl MA, Gazzard BG, Gatell JM, Hirsch MS, Katzenstein DA, Richman DD, Vella S, Yeni PG, Volberding PA. Treatment for adult HIV infection: 2006 recommendations of the International AIDS Society--USA panel. Top HIV Med. 2006;14(3):827–43. [PubMed] [Google Scholar]
- Heredia A, Latinovic O, Gallo RC, Melikyan G, Reitz M, Le N, Redfield RR. Reduction of CCR5 with low-dose rapamycin enhances the antiviral activity of vicriviroc against both sensitive and drug-resistant HIV-1. Proc Natl Acad Sci U S A. 2008;105 (51):20476–81. doi: 10.1073/pnas.0810843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317 (5846):1930–4. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse PJ. Modeling how many envelope glycoprotein trimers per virion participate in human immunodeficiency virus infectivity and its neutralization by antibody. Virology. 2007;369(2):245–62. doi: 10.1016/j.virol.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhmann SE, Hartley O. Targeting chemokine receptors in HIV: a status report. Annu Rev Pharmacol Toxicol. 2008;48:425–61. doi: 10.1146/annurev.pharmtox.48.113006.094847. [DOI] [PubMed] [Google Scholar]
- Kuhmann SE, Platt EJ, Kozak SL, Kabat D. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J Virol. 2000;74(15):7005–15. doi: 10.1128/jvi.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhmann SE, Pugach P, Kunstman KJ, Taylor J, Stanfield RL, Snyder A, Strizki JM, Riley J, Baroudy BM, Wilson IA, Korber BT, Wolinsky SM, Moore JP. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J Virol. 2004;78 (6):2790–807. doi: 10.1128/JVI.78.6.2790-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Quantification of CD4, CCR5 and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci U S A. 1999;96 (9):5215–20. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lineberger JE, Danzeisen R, Hazuda DJ, Simon AJ, Miller MD. Altering expression levels of human immunodeficiency virus type 1 gp120-gp41 affects efficiency but not kinetics of cell-cell fusion. J Virol. 2002;76 (7):3522–33. doi: 10.1128/JVI.76.7.3522-3533.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus C, Rusert P, Bonhoeffer S, Trkola A, Regoes RR. Estimating the stoichiometry of human immunodeficiency virus entry. J Virol. 2009;83 (3):1523–31. doi: 10.1128/JVI.01764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marozsan AJ, Kuhmann SE, Morgan T, Herrera C, Rivera-Troche E, Xu S, Baroudy BM, Strizki J, Moore JP. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D) Virology. 2005;338 (1):182–99. doi: 10.1016/j.virol.2005.04.035. [DOI] [PubMed] [Google Scholar]
- Martin MP, Dean M, Smith MW, Winkler C, Gerrard B, Michael NL, Lee B, Doms RW, Margolick J, Buchbinder S, Goedert JJ, O’Brien TR, Hilgartner MW, Vlahov D, O’Brien SJ, Carrington M. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282(5395):1907–11. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- Ogert RA, Wojcik L, Buontempo C, Ba L, Buontempo P, Ralston R, Strizki J, Howe JA. Mapping resistance to the CCR5 co-receptor antagonist vicriviroc using heterologous chimeric HIV-1 envelope genes reveals key determinants in the C2-V5 domain of gp120. Virology. 2008;373 (2):387–99. doi: 10.1016/j.virol.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Platt EJ, Durnin JP, Shinde U, Kabat D. An allosteric rheostat in HIV-1 gp120 reduces CCR5 stoichiometry required for membrane fusion and overcomes diverse entry limitations. J Mol Biol. 2007;374 (1):64–79. doi: 10.1016/j.jmb.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugach P, Kuhmann SE, Taylor T, Marozsan AJ, Snyder A, Ketas T, Wolinsky SM, Korber BT, Moore JP. The prolonged culture of human immunodeficiency virus type 1 in primary lymphocytes increases its sensitivity to neutralization by soluble CD4. Virology. 2004;321:8–22. doi: 10.1016/j.virol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Pugach P, Ketas TJ, Michael E, Moore JP. Neutralizing antibody and anti-retroviral drug sensitivities of HIV-1 isolates resistant to small molecule CCR5 inhibitors. Virology. 2008;377 (2):401–7. doi: 10.1016/j.virol.2008.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugach P, Marozsan AJ, Ketas TJ, Landes EL, Moore JP, Kuhmann SE. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology. 2007;361 (1):212–28. doi: 10.1016/j.virol.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves JD, Gallo SA, Ahmad N, Miamidian JL, Harvey PE, Sharron M, Pohlmann S, Sfakianos JN, Derdeyn CA, Blumenthal R, Hunter E, Doms RW. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc Natl Acad Sci U S A. 2002;99 (25):16249–54. doi: 10.1073/pnas.252469399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves JD, Lee FH, Miamidian JL, Jabara CB, Juntilla MM, Doms RW. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J Virol. 2005;79 (8):4991–9. doi: 10.1128/JVI.79.8.4991-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynes J, Portales P, Segondy M, Baillat V, Andre P, Reant B, Avinens O, Couderc G, Benkirane M, Clot J, Eliaou JF, Corbeau P. CD4+ T cell surface CCR5 density as a determining factor of virus load in persons infected with human immunodeficiency virus type 1. J Infect Dis. 2000;181 (3):927–32. doi: 10.1086/315315. [DOI] [PubMed] [Google Scholar]
- Seibert C, Ying W, Gavrilov S, Tsamis F, Kuhmann SE, Palani A, Tagat JR, Clader JW, McCombie SW, Baroudy BM, Smith SO, Dragic T, Moore JP, Sakmar TP. Interaction of small molecule inhibitors of HIV-1 entry with CCR5. Virology. 2006;349 (1):41–54. doi: 10.1016/j.virol.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Sougrat R, Bartesaghi A, Lifson JD, Bennett AE, Bess JW, Zabransky DJ, Subramaniam S. Electron tomography of the contact between T cells and SIV/HIV-1: implications for viral entry. PLoS Pathog. 2007;3(5):e63. doi: 10.1371/journal.ppat.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BM, Foulke JS, Flinko R, Heredia A, DeVico A, Reitz M. An alteration of human immunodeficiency virus gp41 leads to reduced CCR5 dependence and CD4 independence. J Virol. 2008;82 (11):5460–71. doi: 10.1128/JVI.01049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384(6605):184–7. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- Trkola A, Kuhmann SE, Strizki JM, Maxwell E, Ketas T, Morgan T, Pugach P, Xu S, Wojcik L, Tagat J, Palani A, Shapiro S, Clader JW, McCombie S, Reyes GR, Baroudy BM, Moore JP. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc Natl Acad Sci U S A. 2002;99 (1):395–400. doi: 10.1073/pnas.012519099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsamis F, Gavrilov S, Kajumo F, Seibert C, Kuhmann S, Ketas T, Trkola A, Palani A, Clader JW, Tagat JR, McCombie S, Baroudy B, Moore JP, Sakmar TP, Dragic T. Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J Virol. 2003;77 (9):5201–8. doi: 10.1128/JVI.77.9.5201-5208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsibris AM, Sagar M, Gulick RM, Su Z, Hughes M, Greaves W, Subramanian M, Flexner C, Giguel F, Leopold KE, Coakley E, Kuritzkes DR. In vivo emergence of vicriviroc resistance in an HIV-1 subtype C-infected subject. J Virol. 2008;82 (16):8210–4. doi: 10.1128/JVI.00444-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson C, Jenkinson S, Kazmierski W, Kenakin T. The CCR5 receptor-based mechanism of action of 873140, a potent allosteric noncompetitive HIV entry inhibitor. Mol Pharmacol. 2005;67 (4):1268–82. doi: 10.1124/mol.104.008565. [DOI] [PubMed] [Google Scholar]
- Westby M, Smith-Burchnell C, Mori J, Lewis M, Mosley M, Stockdale M, Dorr P, Ciaramella G, Perros M. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J Virol. 2007;81(5):2359–71. doi: 10.1128/JVI.02006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcomb JM, Huang W, Fransen S, Limoli K, Toma J, Wrin T, Chappey C, Kiss LD, Paxinos EE, Petropoulos CJ. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob Agents Chemother. 2007;51 (2):566–75. doi: 10.1128/AAC.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]