Abstract
PTEN/MMAC1 is a tumor suppressor gene located on chromosome 10q23. Inherited PTEN/MMAC1 mutations are associated with a cancer predisposition syndrome known as Cowden’s disease. Somatic mutation of PTEN has been found in a number of malignancies, including glioblastoma, melanoma, and carcinoma of the prostate and endometrium. The protein product (PTEN) encodes a dual-specificity protein phosphatase and in addition can dephosphorylate certain lipid substrates. Herein, we show that PTEN protein induces a G1 block when reconstituted in PTEN-null cells. A PTEN mutant associated with Cowden’s disease (PTEN;G129E) has protein phosphatase activity yet is defective in dephosphorylating inositol 1,3,4,5-tetrakisphosphate in vitro and fails to arrest cells in G1. These data suggest a link between induction of a cell-cycle block by PTEN and its ability to dephosphorylate, in vivo, phosphatidylinositol 3,4,5-trisphosphate. In keeping with this notion, PTEN can inhibit the phosphatidylinositol 3,4,5-trisphosphate-dependent Akt kinase, a downstream target of phosphatidylinositol 3-kinase, and constitutively active, but not wild-type, Akt overrides a PTEN G1 arrest. Finally, tumor cells lacking PTEN contain high levels of activated Akt, suggesting that PTEN is necessary for the appropriate regulation of the phosphatidylinositol 3-kinase/Akt pathway.
Abnormalities of chromosomal region 10q23 are frequent in a number of malignancies, including prostate cancer and glioblastoma (1, 2). Recently, a candidate tumor suppressor gene PTEN/MMAC1/TEP1 (for simplicity hereafter referred to as PTEN) was cloned and mapped to this region (3–5). Somatic mutations of PTEN are found in a number of malignancies, including glioblastoma, melanoma, and carcinomas of the prostate, lung, endometrium, and head and neck (3, 4, 6–14). Germ-line mutations of the PTEN gene are associated with the development of Cowden’s disease (CD) and Bannayan–Zonana syndrome (BZS) (15–18). CD is characterized by the occurrence of multiple hamartomas in the skin, gastrointestinal tract, breast, thyroid, and central nervous system and an increased incidence of breast and thyroid cancers (18). BZS is a related syndrome in which intestinal hamartomas are accompanied by neurological abnormalities including mild mental retardation, delayed motor development, vascular malformations, and speckled penis (18).
The predicted protein product of the PTEN gene (referred to hereafter as PTEN) has homology to tensin, an actin binding protein localized to focal adhesion complexes (19); to auxilin, a protein involved in the uncoating of clatherin-coated vesicles (20); and to dual-specificity phosphatases (4, 21). Recombinant PTEN is capable of dephosphorylating both tyrosine- and threonine-phosphorylated substrates and in addition can dephosphorylate phosphatidylinositol 3,4,5-trisphosphate (PtdIns-3,4,5-P3) (22, 23). Overproduction of PTEN can suppress colony formation in certain cells, growth in soft agar, and tumor formation in nude mice (24, 25). Recent data suggest that PTEN might function, at least in part, through regulation of focal adhesion kinase and the subsequent inhibition of adhesion and migration (26). PTEN is essential for murine embryonic development beyond day 7.5. In the mouse loss of PTEN allele leads to hyperplasia and dysplasia in the skin, gastrointestinal tract, and prostate, as well as tumor formation (27).
In this study, we found that reintroduction of a PTEN cDNA into cells lacking a wild-type PTEN protein led to a cell-cycle block in G1. This function was tightly linked to the phosphatase activity of PTEN and was inactivated by tumor-derived mutations. Furthermore, a PTEN mutant, associated with CD, that retains protein phosphatase activity was defective in arresting cells in G1 and was also defective in dephosphorylating inositol 1,3,4,5-tetrakisphosphate (IP4).
These data suggested that PTEN might regulate cell-cycle progression by blocking activation of downstream targets of phosphatidylinositol 3-kinase such as the protooncogene Akt. In keeping with this notion, PTEN was capable of inhibiting wild-type Akt kinase activity in cells. Furthermore, a constitutively active form of Akt, but not wild-type Akt, overrode a PTEN-induced cell-cycle block.
MATERIALS AND METHODS
Cell Culture, Transfection, and Metabolic Labeling.
ACHN, 786-O, SAOS-2, and U2-OS cells (gifts from the Kaelin laboratory) were maintained in DMEM containing 10% Fetal Clone (HyClone), penicillin and streptomycin at 37°C. Cells were transfected with Fugene 6 (Boehringer-Mannheim) for 786-O cells or by the calcium phosphate procedure for U2-OS, ACHN, and SAOS-2 cells, as described (28, 29). Transfected 786-O cells were metabolically labeled for 3 h in 5 ml of methionine-free medium supplemented with 10% dialyzed fetal calf serum and [35S]methionine (100 μCi/ml; 1 Ci = 37 GBq).
Plasmids.
A cDNA fragment encoding PTEN amino acid residues 1–403 was PCR-amplified from a 293 cDNA library (30) and ligated to vector pSG5L-HA (28) to give pSG5L-HA-PTEN;WT. An Akt-1 cDNA was amplified by reverse transcription-coupled PCR from total HeLa cell RNA and reamplified with a 5′ primer containing a Kozak sequence and sequences encoding a hemagglutinin (HA) epitope and cloned into pLNCX to give pLNCX-HA-Akt. A double-stranded oligonucleotide encoding the src myristoylation sequence was inserted 5′ of the HA tag to generate pLNCX-Myr-HA-Akt. pSG5L-HA-PTEN;C124S, pSG5L-HA-PTEN;G129R, pSG5L-HA-PTEN;G129E, pSG5L-HA-PTEN;1–274, pSG5L-HA-PTEN;1–336 and pSG5L-HA-PTEN;Δ274–342, pLNCX-HA-Akt;K179M, pLNCX-myr-HA-Akt;K179M were generated by site-directed mutagenesis or by PCR mutagenesis. Inserts from the pSG5L-HA-PTEN plasmids were cloned into pGEX2T to give the corresponding pGEX2T-PTEN plasmids. A cDNA for AKT-1 was PCR-amplified from a fetal brain library and ligated to the vector from pCDNA3-T7-VHL to give pCDNA3-T7-AKT. All plasmid inserts obtained by PCR or altered by site-directed mutagenesis were verified by sequencing. pCD19 has been previously described (31).
Antibodies.
Production of PTEN antiserum (C54) will be described elsewhere (32). HA.11, fluorescein isothiocyanate-conjugated anti-CD19, anti-T7, and anti-phospho-Akt (Ser-473) antibodies were obtained from Babco (Richmond, CA), Novagen, Caltag (South San Francisco, CA), and New England Biolabs, respectively.
Immunoprecipitations and Immunoblotting.
Preparation of whole cell extracts, immunoprecipitations, and immunoblotting conditions are as described (28). For immunoblotting, C54 antiserum was diluted 1:500 in TBS/4% milk. Secondary antibodies, alkaline phosphatase-conjugated goat anti-mouse or goat anti-rabbit (Southern Biotechnology Associates) were diluted 1:5,000. For chemiluminescent detection, horseradish peroxidase-conjugated anti-mouse antibody (Santa Cruz Biotechnology) was used at a 1:2,000 dilution and detected with the SuperSignal kit (Pierce).
Fluorescence-Activated Cell Sorting.
Cells grown on p100 plates were transfected with 4 μg of pCD19 and either 11 μg (Fugene 6 transfections) or 21 μg (calcium phosphate transfections) of the backbone pSG5L plasmid or pSG5L plasmids encoding PTEN or the indicated PTEN mutants. Cell-cycle determination of the CD19+ cells was carried out as described (28).
Protein and Inositol Phosphatase Assays.
Poly(Glu4-Tyr1) copolymer (Sigma) was phosphorylated in vitro essentially as described (22). Briefly, poly(Glu4-Tyr1) was resuspended at a final concentration of 3.3 mg/ml in 50 mM Tris⋅HCl (pH 7.4), 2 mM MnCl2, 10 mM MgCl2, and 0.1 mM ATP in a reaction mixture containing 10 μCi of [γ-33P]ATP and 100 units of β-insulin receptor kinase (Stratagene) and incubated at 30°C for 4 h. Labeled copolymer was precipitated with 100% trichloroacetic acid (TCA) washed with 20% TCA and acetone, lyophilized, and resuspended in, and dialyzed against 50 mM imidazole (pH 7.2). Protein phosphatase assays were done as described (22). Dephosphorylation of [3H]inositol 1,3,4,5- tetrakisphosphate (NEN) was performed as described by using 1 μg of the relevant glutathione S-transferase (GST) fusion proteins (23).
Akt Kinase Assays.
U2-OS cells were transfected with plasmids encoding T7-Akt-1 and pSG5L, pSG5L-HA-PTEN, or mutant derivatives. Thirty-six hours after transfection T7 immunoprecipitates were prepared from cell lysates, collected on protein A-Sepharose and incubated in a reaction mixture containing 30 mM Hepes (pH 7.5), 10 mM MgCl2, 5 mM MnCl2, 1 mM DTT, 20 μM ATP, 10 μCi of [γ-32P]ATP, and 5 μg of a GST fusion protein containing an Akt peptide substrate, for 30 min at 25°C. Radiolabeled substrate was separated from unincorporated [γ-32P]ATP by gel electrophoresis and detected by autoradiography.
RESULTS
PTEN Induces a Block in the G1 Phase of the Cell Cycle.
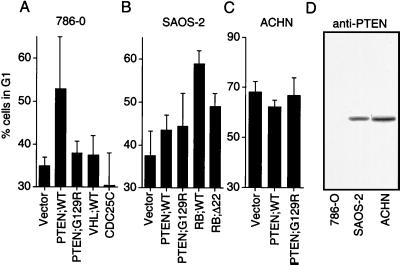
Attempts at stable expression of PTEN in PTEN-null 786-O renal carcinoma and A172 glioblastoma cell lines failed to yield clonal lines that produced detectable HA-PTEN (data not shown). Next, transient assays were used to determine whether PTEN might be capable of altering cell-cycle progression. 786-O renal carcinoma cells, which lack PTEN protein (Fig. 1D), were transiently transfected with either empty vector or plasmids encoding either HA-tagged PTEN (PTEN;WT) or a tumor-derived PTEN catalytic domain mutant (PTEN;G129R) (4), along with a plasmid encoding the cell surface marker CD19 (pCD19). After 40 h, the DNA content of the successfully transfected cells was determined by staining cells with fluorescein isothiocyanate-conjugated anti-CD19 and propidium iodide followed by fluorescence-activated cell sorting. Wild-type PTEN reproducibly induced an increase in the percentage of cells in G1 when compared with the vector alone or to PTEN;G129R (Fig. 1A). In contrast, reintroduction of plasmids encoding either the tumor suppressor proteins pRB or VHL [which is defective in 786-O cells (30)] or the dual-specificity phosphatase cdc25C failed to induce a G1 arrest in these cells (Fig. 1A and data not shown). Production of HA-PTEN in two cell lines that retain endogenous PTEN protein (SAOS-2 and ACHN) did not alter the cell-cycle distribution of these cells (Fig. 1 B–D), but, as a positive control in pRB-null SAOS-2 cells, reintroduction of a plasmid encoding pRB did effect a G1 arrest (Fig. 1B). Under these same experimental conditions, production of PTEN protein in 786-O cells did not lead to an increase in the percentage of cells harboring a sub-2N DNA content, suggesting that PTEN did not induce apoptosis in these cells (Table 1). Thus, PTEN specifically induced a G1 block in 786-O cells, which lack PTEN.
Figure 1.
PTEN induces a G1 block. (A) PTEN, but not pVHL or cdc25C, induced a G1 block in 786-O cells. 786-O cells were transiently cotransfected with a plasmid encoding CD19 (pCD19) along with plasmids encoding the indicated proteins. Forty hours after transfection, cells were fixed and the cell-cycle distribution of the successfully transfected cells was determined by fluorescence-activated cell sorting analysis. The mean and SEM of two experiments are shown. (B) PTEN does not alter the cell-cycle profile of SAOS-2(RB−/−) cells. SAOS-2 cells were transiently transfected with pCD19 and the plasmids encoding the indicated proteins and analyzed as in Fig. 2A. The mean and SEM of two experiments are shown. (C) PTEN does not alter the cell-cycle profile of ACHN cells. ACHN cells were transiently transfected with pCD19 or plasmids encoding the indicated proteins and analyzed as in A. The mean and SEM of two experiments are shown. (D) Immunoblot detection of PTEN protein in 786-O, SAOS-2, and ACHN cells. C54 anti-PTEN antiserum was used to detect PTEN by immunoblot analysis of protein extracts from the indicated cell lines.
Table 1.
Percentage of CD19+ 786-O cells with sub-2N DNA content
| Exp. | Vector | PTEN;WT | PTEN;G129R |
|---|---|---|---|
| 1 | 4.6 | 5.1 | 5.8 |
| 2 | 2.4 | 1.5 | 1.4 |
| 3 | 1.0 | 1.3 | 1.0 |
786-O cells were transiently transfected with pCD19 and the indicated pSGL-HA expression plasmids. After 36 h, cells were harvested and processed as in Fig. 1A.
Tumor-Derived Mutants Inactivate PTEN Phosphatase Activity and Cell-Cycle Control.
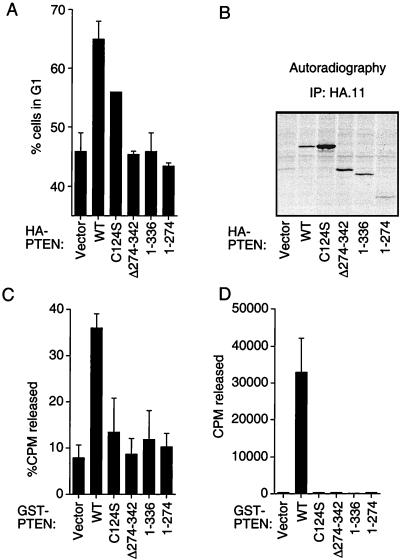
A number of tumor-derived PTEN mutations have been reported that lie outside of the predicted phosphatase and tensin–auxilin homology domains. Three such tumor-derived mutants, PTEN;1–274, PTEN;1–336, and PTEN;Δ274–342 were tested and were defective in the cell-cycle assay (Fig. 2A). With the exception of PTEN;1–274, these mutant proteins were produced to levels similar to that of wild-type PTEN in 786-O cells (Fig. 2B). Two biochemical properties have been ascribed to PTEN. PTEN can dephosphorylate certain protein substrates containing either phosphotyrosine or phosphothreonine (33). In addition, PTEN can dephosphorylate PtdIns-3,4,5-P3 (23). We next asked whether the three tumor-derived mutants were defective for either of these functions. When produced as GST fusion proteins, all three mutant proteins were defective in catalyzing the release of phosphate from either a 33P-labeled poly(Glu4-Tyr1) substrate or [3H]inositol 1,3,4,5-tetrakisphosphate ([3H]IP4) (Fig. 2 C and D).
Figure 2.
Substrate trapping variants of PTEN induce a G1 block but tumor-derived mutants do not. (A) Comparison of wild-type, substrate-trapping and C-terminal mutant forms of PTEN in the cell-cycle assay. 786-O cells were cotransfected with pCD19 and plasmids encoding the indicated proteins and analyzed as in Fig. 1A. (B) Expression of PTEN proteins in 786-O cells. 786-O cells were transfected with plasmids encoding the indicated proteins. Forty hours after transfection anti-HA immunoprecipitates of protein extracts prepared from metabolically labeled cells were separated by gel electrophoresis and subjected to fluorography. (C) Inositol phosphatase activity of GST-PTEN and mutant derivatives. The indicated GST-PTEN fusion proteins were used to dephosphorylate [3H]IP4. (D) Protein tyrosine phosphatase activity of GST-PTEN and mutant derivatives. The indicated GST-PTEN fusion proteins were used to dephosphorylate 33P-labeled poly(Glu4-Tyr1) copolymers. (A, C, and D) The mean and SEM of two experiments are shown.
Two catalytically inert PTEN variants were created (PTEN;C124S and PTEN;D92A). Mutations of the corresponding residues in other phosphatases results in loss of enzymatic activity but allows for preservation of substrate binding and have been termed “substrate trapping” (34). As predicted, and as previously published (22, 23), the PTEN substrate-trapping mutants lack catalytic activity (Fig. 2 C and D and data not shown). Nonetheless, both PTEN;C124S and PTEN;D92A retained a partial ability to induce cells to accumulate in G1 (Fig. 2A and data not shown). These data suggest that sequestration of phosphorylated substrates might be sufficient for cell-cycle inhibition by PTEN. PTEN;C124S can induce an accumulation of PtdIns-3,4,5-P3 in cells, suggesting that there may be stable binding to this substrate (23). On the other hand the C124S mutant is defective in regulating cell motility (26). Thus, these data raised the possibility that the ability of PTEN to induce a G1 block was distinct from cell motility control and suggested a relationship between the ability of PTEN to interact with a PtdIns substrate and the ability to arrest cells in G1.
Protein Phosphatase Activity Is Not Sufficient for Induction of a G1 Block by PTEN.
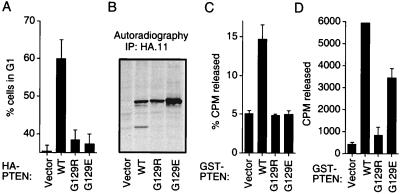
We next compared the G129R mutant to a second mutant in which the same codon is affected (G129E). G129E is encoded by a PTEN allele found in the germ-line PTEN gene of two families afflicted with CD (35). Others have found that protein phosphatase activity of PTEN;G129E is equivalent to the wild-type protein (22) and that PTEN;G129E is comparable to wild-type PTEN in its ability to inhibit cell spreading (26). Indeed, in our assays we likewise see retention of protein phosphatase activity (Fig. 3D). In five preparations of GST-PTEN;G129E, activity varied from 30% to 70% of the wild-type activity (Fig. 3D and data not shown). However, when produced in 786-O cells, this PTEN mutant did not induce a G1 block (Fig. 3A). Thus, both G129E and G129R failed to induce a G1 block, even though the former retains protein phosphatase activity (Fig. 3D). We next asked whether the G129E mutant might be defective in dephosphorylating [3H]IP4, as a measure of lipid phosphatase activity. Indeed, although PTEN;WT catalyzed the dephosphorylation of [3H]IP4, neither G129R nor G129E had measurable activity in this assay (Fig. 3C). Thus, the ability of PTEN to induce a cell-cycle block correlated best with its ability to dephosphorylate a lipid substrate, and preservation of protein phosphatase activity, as measured in vitro, was not sufficient for the induction of a G1 block.
Figure 3.
CD mutant G129E preserves protein phosphatase activity but lacks inositol phosphatase activity and is incapable of inducing a G1 block. (A) Comparison of wild-type PTEN and the mutants G129R and G129E in the cell-cycle assay. 786-O cells were cotransfected with pCD19 and plasmids encoding the indicated proteins and analyzed as in Fig. 1A. (B) Comparison of the expression of wild-type PTEN and the mutants G129R and G129E in 786-O cells. 786-O cells were transfected with plasmids encoding the indicated proteins. Anti-HA immunoprecipitates of protein extracts prepared from metabolically labeled cells were separated by gel electrophoresis and subject to fluorography. (C) Inositol phosphatase activity of GST-PTEN and mutant derivatives. The indicated GST-PTEN fusion proteins were analyzed as in Fig. 2C. (D) Protein tyrosine phosphatase activity of GST-PTEN and mutant derivatives. The indicated GST-PTEN fusion proteins were analyzed as in Fig. 2D. (A, C, and D) The mean and SEM of two experiments are shown.
Akt Is Downstream of PTEN.
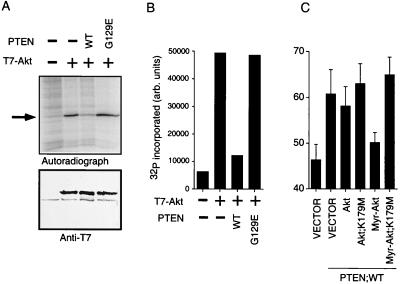
Thus, our data suggested that the ability of PTEN to regulate cell-cycle progression was dependent upon its ability to dephosphorylate PtdIns-3,4,5-P3 and raised the possibility that the regulation of downstream targets of phosphatidylinositol 3-kinase might be critical for PTEN-mediated cell-cycle control. One such effector is the protein product of the protooncogene AKT. We next sought to determine whether PTEN might down-regulate Akt kinase activity. U2-OS cells were transiently transfected with an empty vector plasmid or a plasmid encoding T7-epitope tagged Akt along with the backbone vector plasmid or plasmids encoding HA-PTEN or PTEN;G129E. Akt was recovered by anti-T7 immunoprecipitation and used to phosphorylate a polypeptide substrate in the presence of [γ-32P]ATP. PTEN efficiently down-regulated Akt kinase activity, whereas PTEN;G129E did not (Fig. 4 A and B). Identical results were obtained when this experiment was carried out in 786-O cells, which lack PTEN protein (data not shown). Thus, PTEN can negatively regulate Akt kinase activity.
Figure 4.
PTEN inhibits Akt kinase activity and an activated form of Akt can override a PTEN induced G1 block. (A) Inhibition of Akt kinase by wild-type PTEN. U2-OS cells were transfected with plasmids encoding the indicated proteins. After transfection, anti-T7 immunoprecipitates were prepared and used to phosphorylate a GST-peptide substrate in vitro. Autoradiography (Upper) and anti-T7 immunoblot (Lower) of the same membrane are shown. The black arrow indicates the position of the substrate. Results are representative of two experiments. (B) Quantitation of 32P incorporation in A with a PhosphoImager. (C) Myr-Akt overrides a PTEN-induced G1 block. 786-O cells were transiently cotransfected with pCD19 and plasmids encoding PTEN with empty vector (pLNCX) or with plasmids encoding the indicated Akt proteins. After transfection the cells were analyzed as in Fig. 1A. The mean and SEM of two experiments are shown.
We next asked whether Akt or a myristoylated form of Akt could override a PTEN-induced G1 block. 786-O cells were transiently transfected with pCD19 and plasmids encoding either empty vector or wild-type PTEN and either empty pLNCX vector or pLNCX plasmids encoding Akt or the indicated derivatives. Although wild-type Akt had a minimal effect on the PTEN-induced G1 block, a myristoylated form of Akt that is targeted to the membrane independently of PtdIns-3,4,5-P3 overcame a PTEN block. In contrast, kinase-inactive versions of both Akt and Myr-Akt were unable to override PTEN (Fig. 4C). PTEN levels were unchanged by overproduction of Akt or the indicated derivatives (data not shown). These data suggest that PTEN-mediated cell-cycle inhibition depends on negative regulation of the phosphatidylinositol 3-kinase/Akt signaling pathway.
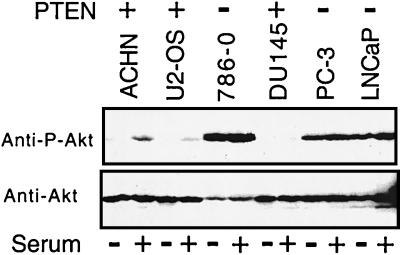
If Akt is a critical downstream target of PTEN tumor suppressor function, tumors or cell lines that lack PTEN protein might be predicted to harbor elevated levels of Akt kinase activity. As a measure of endogenous Akt activity, we subjected cell extracts to immunoblotting with an antibody specific to phospho-Ser-473 on Akt. Phosphorylation of this residue along with Thr-308 is required for Akt kinase activity (36). Protein extracts from duplicate plates of a panel of either PTEN+ or PTEN− cells were prepared after serum deprivation or after 90 min of serum stimulation and immunoblotted with the phospho-specific antibody. Protein extracts prepared from cells that contain PTEN protein by immunoblot, ACHN, DU145, and U2-OS (Fig. 1D and data not shown) were found to have little phosphorylated Akt (Fig. 5 Upper). In contrast, cells that lack PTEN protein, 786-O, LNCaP, and PC-3 (Fig. 1D and data not shown), had increased levels of phosphorylated Akt that was not down-regulated by serum withdrawal (Fig. 5 Upper). Levels of total Akt-1 protein in these cells were comparable (Fig. 5 Lower). Thus, loss of PTEN, in these tumor cell lines, was specifically correlated with a deregulation of the phosphorylated form of Akt.
Figure 5.
Cells lacking PTEN protein contain high levels of Akt, phosphorylated on Ser-473. The indicated cells lines were serum-deprived for 24 h in duplicate. One p100 plate from each pair was refed with fresh medium containing 10% fetal calf serum for 90 min, after which protein extracts were prepared and subject to immunoblot analysis with an antibody directed at Akt phospho-Ser-473 (Upper) or an antibody that recognizes Akt independent of its phosphorylation status (Lower).
DISCUSSION
Reintroduction of PTEN into 786-O cells led to an increase in the proportion of cells in G1. Whether this represents a block in G1 or a prolongation of G1 awaits further study. Every human derived PTEN mutation we have tested to date has been defective in this cell-cycle assay. Thus, it is clear that the G1 arrest is a reflection of a critical function of PTEN that is tightly linked to its function as a tumor suppressor protein.
What are the underlying mechanisms by which PTEN might exert control over progression through G1? First, this function appears to be quite distinct from PTEN-mediated inhibition of cell spreading and cell motility. PTEN inhibition of cell motility and adhesion was observed in cells containing endogenous PTEN, was observed in cells only when grown on a fibronectin matrix, and was not observed with the PTEN;C124S mutant. In contrast, PTEN overexpression induced a G1 block in cells lacking, but not in cells producing, PTEN protein, could be induced in cells plated on poly-(l-lysine)-coated plates, and was partially induced by the C124S mutant. Finally, PTEN;G129E clearly separates these two functions, because this mutant inhibits cell spreading comparably to the wild-type protein (26) but is incapable of arresting 786-O cells in G1 (Fig. 3). Thus, these data suggest that the alterations in cell-cycle profile are not an indirect consequence of inhibition of cellular adhesion. Furthermore, that PTEN;G129E preserves protein phosphatase activity and can negatively regulate cell spreading yet is linked to the development of CD suggests that these functions are not sufficient for suppression of the CD phenotype. On the other hand, PTEN;G129E is defective in the cell-cycle assay. Thus, this biological assay might be indicative of PTEN interactions with critical physiological substrates, regulation of which might be essential for preventing the onset of CD. Indeed, this mutant is defective in catalyzing the dephosphorylation of [3H]IP4, suggesting that in vivo dephosphorylation of PtdIns-3,4,5-P3 is a critical requirement for PTEN-mediated suppression of CD. Similar results were recently reported by two groups (37, 38). Thus, we propose that the development of CD is linked to a loss of PTEN lipid phosphatase activity and to the subsequent deregulation of the cell cycle.
In support of this notion, mice that have only a single intact PTEN allele develop a syndrome not unlike CD and are phenotypically characterized by hyperplasia and dysplasia of the skin, gastrointestinal tract, and prostate (27). Notably, in the prostate of PTEN+/− mice both Ki-67 staining and the mitotic index indicated a significant increase in the proportion of cells that were proliferating when compared with wild-type mice. These data indicate that in murine prostate epithelial cells, the cell-cycle distribution has been altered by PTEN loss (27). In addition, wide-spread increased bromodeoxyuridine incorporation is found in PTEN mutant embryos at day 7.5 to 8.5 when compared with wild-type embryos (39). Finally, PTEN-mediated growth inhibition was recently linked to induction of a G1 arrest rather than induction of apoptosis in glioblastoma cell lines (37).
Thus, our data and the data describing proliferative abnormalities in PTEN+/− mice suggest that PTEN plays a role in cell-cycle control. This function in turn appears to require lipid phosphatase activity. These data led us to ask whether Akt, a known downstream target of phosphatidylinositol 3-kinase that is activated by PtdIns-3,4,5-P3, could act downstream of PTEN. First, we found that wild type but not the G129E mutant of PTEN could inhibit Akt kinase activity, suggesting again that PTEN protein phosphatase activity is not sufficient for inhibition of Akt. We next asked whether Akt could override a PTEN-mediated cell-cycle block. In keeping with the role of PTEN in limiting the availability of PtdIns-3,4,5-P3, wild-type Akt was ineffective in this assay, whereas expression of a myristoylated form of Akt led to an override of the PTEN cell-cycle block. These data support the notion that Akt is an important downstream target of PTEN regulation. Similar conclusions have been reached by studying the effect of PTEN loss on murine fibroblasts (39).
Inducible expression of phosphatidylinositol 3-kinase can induce DNA synthesis in the absence of serum and is thus sufficient for initiation of this process (40). Akt and phosphatidylinositol 3-kinase can activate a number of downstream targets that may be involved in the regulation cell proliferation (41). The ribosomal protein p70S6K regulates the increased translation of a subset of mRNA species thought to be important for cell-cycle progression and, indeed, inactivation of p70S6K function leads to an arrest of cells in G1 (42, 43). Likewise, Akt can induce phosphorylation of 4E-BP1, an event that leads to 4E-BP1 dissociation from the eukaryotic initiation factor eIF4E and a subsequent “disinhibition” of translation (44). eIF4E when overproduced transforms NIH 3T3 cells and, thus, like Akt, is an attractive tumor suppressor target (45). Which component(s) in the phosphatidylinositol 3-kinase/Akt pathway are rate-limiting for S phase entry is not known.
Akt is also an important component of a cell-survival signaling pathway and in this capacity can phosphorylate and inactivate the Bcl-2 family member BAD, rendering it incapable of blocking Bcl-2 or Bcl-XL activity (41, 46, 47). Recent data has shown that PTEN−/− cells are resistant to apoptotic stimuli and that regulation of Akt is abnormal in PTEN-deficient fibroblasts (39). To date, we have not seen an increase in apoptosis upon reintroduction of PTEN into 786-O cells (Table 1). Among many possibilities, these data may indicate that additional inactivating mutations have been sustained in this cell line, rendering it incapable of responding to a variety of apoptotic signals. Finally, increases in phosphatidylinositol 3-kinase activity do not uniformly result in protection from apoptosis. In certain circumstances, phosphatidylinositol 3-kinase activity can induce cell-cycle progression and, in fact, promote apoptosis (40) (R. Narsimhan and T.M.R., unpublished data). The notion that PTEN may regulate both cell-cycle and cell-survival functions is strikingly similar to the tumor suppressor functions imputed to p53. Thus PTEN, like p53, may serve to coordinate these activities, although possibly in response to different sets of signals (48).
If Akt is a critical downstream target of PTEN, tumor cells that lack PTEN might be predicted to harbor excessive Akt activity. Immunoblots of protein extracts from cells that were characterized as PTEN− or PTEN+ (Fig. 1D and data not shown) were found to have a marked increase in the amount of Akt phosphorylated on Ser-473 (Fig. 5). This inverse correlation between PTEN and activated Akt again argues for a role for PTEN in Akt regulation in tumors. In this regard, inhibition of Akt activity by a dominant negative form of Akt blocks BCR-ABL transformation of murine myeloid cells and stable expression of antisense Akt2 reduces tumorigenicity of pancreatic cancer cell lines known to have amplified Akt (49, 50). Thus, inhibition of Akt family members in these settings does demonstrate antitumor activity. Whether activated Akt is necessary for the transformed phenotype of PTEN null tumors is a critical question. Nonetheless, our data raise the possibility that PTEN null tumors may be susceptible to Akt inhibition as a cancer treatment strategy.
Acknowledgments
We thank William G. Kaelin, Peter Adams, Mark Ewen, Myles Brown, Kornelia Polyak, and David Livingston for hours of thoughtful discussion and for the critical reading of this manuscript. This work was supported by the U.S. Army Medical Research Institute of Infectious Diseases–Prostate Cancer Research Program PC970221 and the Claudia–Adams Barr Program (W.R.S).
ABBREVIATIONS
- IP4, inositol 1
3,4,5-tetrakisphosphate
- PtdIns
phosphatidylinositol 3,4,5-trisphosphate
- CD
Cowden’s disease
- HA
hemagglutinin
- GST
glutathione S-transferase
References
- 1.Gray I C, Phillips S M, Lee S J, Neoptolemos J P, Weissenbach J, Spurr N K. Cancer Res. 1995;55:4800–4803. [PubMed] [Google Scholar]
- 2.Rasheed B K, McLendon R E, Friedman H S, Friedman A H, Fuchs H E, Bigner D D, Bigner S H. Oncogene. 1995;10:2243–2246. [PubMed] [Google Scholar]
- 3.Steck P A, Pershouse M A, Jasser S A, Yung W K, Lin H, Ligon A H, Langford L A, Baumgard M L, Hattier T, Davis T, et al. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang S I, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 5.Li D M, Sun H. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 6.Rasheed B K, Stenzel T T, McLendon R E, Parsons R, Friedman A H, Friedman H S, Bigner D D, Bigner S H. Cancer Res. 1997;57:4187–4190. [PubMed] [Google Scholar]
- 7.Liu W, James C D, Frederick L, Alderete B E, Jenkins R B. Cancer Res. 1997;57:5254–5257. [PubMed] [Google Scholar]
- 8.Guldberg P, thor Straten P, Birck A, Ahrenkiel V, Kirkin A F, Zeuthen J. Cancer Res. 1997;57:3660–3663. [PubMed] [Google Scholar]
- 9.Cairns P, Okami K, Halachmi S, Halachmi N, Esteller M, Herman J G, Jen J, Isaacs W B, Bova G S, Sidransky D. Cancer Res. 1997;57:4997–5000. [PubMed] [Google Scholar]
- 10.Teng D H, Hu R, Lin H, Davis T, Iliev D, Frye C, Swedlund B, Hansen K L, Vinson V L, Gumpper K L, et al. Cancer Res. 1997;57:5221–5225. [PubMed] [Google Scholar]
- 11.Tashiro H, Blazes M S, Wu R, Cho K R, Bose S, Wang S I, Li J, Parsons R, Ellenson L H. Cancer Res. 1997;57:3935–3940. [PubMed] [Google Scholar]
- 12.Kohno T, Takahashi M, Manda R, Yokota J. Genes Chromosomes Cancer. 1998;22:152–156. doi: 10.1002/(sici)1098-2264(199806)22:2<152::aid-gcc10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 13.Kong D, Suzuki A, Zou T T, Sakurada A, Kemp L W, Wakatsuki S, Yokoyama T, Yamakawa H, Furukawa T, Sato M, et al. Nat Genet. 1997;17:143–144. doi: 10.1038/ng1097-143. [DOI] [PubMed] [Google Scholar]
- 14.Okami K, Wu L, Riggins G, Cairns P, Goggins M, Evron E, Halachmi N, Ahrendt S A, Reed A L, Hilgers W, et al. Cancer Res. 1998;58:509–511. [PubMed] [Google Scholar]
- 15.Liaw D, Marsh D J, Li J, Dahia P L, Wang S I, Zheng Z, Bose S, Call K M, Tsou H C, Peacocke M, Eng C, Parsons R. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 16.Nelen M R, van Staveren W C, Peeters E A, Hassel M B, Gorlin R J, Hamm H, Lindboe C F, Fryns J P, Sijmons R H, Woods D G, et al. Hum Mol Genet. 1997;6:1383–1387. doi: 10.1093/hmg/6.8.1383. [DOI] [PubMed] [Google Scholar]
- 17.Tsou H C, Ping X L, Xie X X, Gruener A C, Zhang H, Nini R, Swisshelm K, Sybert V, Diamond T M, Sutphen R, Peacocke M. Hum Genet. 1998;102:467–473. doi: 10.1007/s004390050723. [DOI] [PubMed] [Google Scholar]
- 18.Marsh D J, Dahia P L, Coulon V, Zheng Z, Dorion-Bonnet F, Call K M, Little R, Lin A Y, Eeles R A, Goldstein A M, et al. Genes Chromosomes Cancer. 1998;21:61–69. doi: 10.1002/(sici)1098-2264(199801)21:1<61::aid-gcc8>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 19.Chuang J Z, Lin D C, Lin S. J Cell Biol. 1995;128:1095–1109. doi: 10.1083/jcb.128.6.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ungewickell E, Ungewickell H, Holstein S E, Lindner R, Prasad K, Barouch W, Martin B, Greene L E, Eisenberg E. Nature (London) 1995;378:632–635. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]
- 21.Tonks N K, Neel B G. Cell. 1996;87:365–368. doi: 10.1016/s0092-8674(00)81357-4. [DOI] [PubMed] [Google Scholar]
- 22.Myers M P, Stolarov J, End C, Li J, Wang S I, Wigler M H, Parsons R, Tonks N. Proc Natl Acad Sci USA. 1997;94:9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maehama T, Dixon J E. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 24.Furnari F B, Lin H, Huang H S, Cavenee W K. Proc Natl Acad Sci USA. 1997;94:12479–12484. doi: 10.1073/pnas.94.23.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheney I W, Johnson D E, Vaillancourt M T, Avanzini J, Morimoto A, Demers G W, Wills K N, Shabram P W, Bolen J B, Tavtigian S V, Bookstein R. Cancer Res. 1998;58:2331–2334. [PubMed] [Google Scholar]
- 26.Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada K M. Science. 1998;280:1614–1617. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 27.Di Cristafano A, Pesce B, Cordon-Cardo C, Pandolfi P P. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 28.Sellers W R, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G., Jr Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iliopoulos O, Kibel A, Gray S, Kaelin W G., Jr Nat Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 31.Tedder T F, Isaacs C M. J Immunol. 1989;143:712–717. [PubMed] [Google Scholar]
- 32.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning:A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 33.Myers M P, Tonks N K. Am J Hum Genet. 1997;61:1234–1238. doi: 10.1086/301659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flint A J, Tiganis T, Barford D, Tonks N K. Proc Natl Acad Sci USA. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsh D J, Coulon V, Lunetta K L, Rocca-Serra P, Dahia P L, Zheng Z, Liaw D, Caron S, Duboue B, Lin A Y, et al. Hum Mol Genet. 1998;7:507–515. doi: 10.1093/hmg/7.3.507. [DOI] [PubMed] [Google Scholar]
- 36.Alessi D R, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings B A. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 37.Furnari F B, Huang H J, Cavenee W K. Cancer Res. 1998;58:5002–5008. [PubMed] [Google Scholar]
- 38.Myers M P, Pass I, Batty I H, Van der Kaay J, Stolarov J P, Hemmings B A, Wigler M H, Downes C P, Tonks N K. Proc Natl Acad Sci USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stambolic V, Suzuki A, de la Pompa J L, Brothers G M, Mirtsos C, Sasaki T, Ruland J, Penninger J M, Siderovski D P, Mak T W. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 40.Klippel A, Escobedo M A, Wachowicz M S, Apell G, Brown T W, Giedlin M A, Kavanaugh W M, Williams L T. Mol Cell Biol. 1998;18:5699–5711. doi: 10.1128/mcb.18.10.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Downward J. Curr Opin Cell Biol. 1998;10:262–267. doi: 10.1016/s0955-0674(98)80149-x. [DOI] [PubMed] [Google Scholar]
- 42.Peterson R T, Schreiber S L. Curr Biol. 1998;8:R248–R250. doi: 10.1016/s0960-9822(98)70152-6. [DOI] [PubMed] [Google Scholar]
- 43.Lane H A, Fernandez A, Lamb N J, Thomas G. Nature (London) 1993;363:170–172. doi: 10.1038/363170a0. [DOI] [PubMed] [Google Scholar]
- 44.Gingras A C, Kennedy S G, O’Leary M A, Sonenberg N, Hay N. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lazaris-Karatzas A, Montine K S, Sonenberg N. Nature (London) 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 46.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 47.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 48.Evan G, Littlewood T. Science. 1998;281:1317–1321. doi: 10.1126/science.281.5381.1317. [DOI] [PubMed] [Google Scholar]
- 49.Skorski T, Bellacosa A, Nieborowska-Skorska M, Majewski M, Martinez R, Choi J K, Trotta R, Wlodarski P, Perrotti D, Chan T O, et al. EMBO J. 1997;16:6151–6161. doi: 10.1093/emboj/16.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng J Q, Ruggeri B, Klein W M, Sonoda G, Altomare D A, Watson D K, Testa J R. Proc Natl Acad Sci USA. 1996;93:3636–3641. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]