Abstract
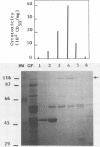
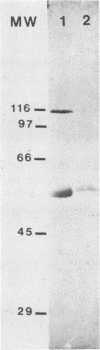
The aim of this study was to identify Escherichia coli cytotoxic necrotizing factor (CNF) by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis and to investigate the possible dissociation of CNF from hemolysin (Hly), which is often produced by CNF-producing strains. CNF was purified from cell lysates of a CNF-producing strain by using ammonium sulfate precipitation, ion-exchange chromatography, gel filtration, and preparative nondenaturing PAGE. All eluates from successive longitudinal slices of a preparative polyacrylamide gel were tested for cytoxicity and analyzed by SDS-PAGE; CNF activity was quantitatively correlated with a protein of 115 kilodaltons (kDa). This procedure increased both cytotoxicity and lethal activity (about 300-fold). We then compared SDS-PAGE protein patterns of enriched lysates from eight field and mutant E. coli strains producing both CNF and Hly, Hly alone, or neither; the 115-kDa band was present solely in CNF-producing strains, irrespective of Hly production. A neutralizing antiserum was produced against unpurified CNF from strain BM2-1 and then extensively adsorbed with cells and extracts of a CNF-defective mutant from BM2-1. The adsorbed antiserum possessed antitoxin activity and neutralized both lethal and necrotic effects of cell lysates from all the CNF-producing strains tested. In an immunoblot of enriched extract from BM2-1, the adsorbed antiserum recognized, besides the 115-kDa protein, another protein of 59 kDa, which was present in the CNF-defective mutant from BM2-1 and was not associated with cytotoxicity. We can conclude from these findings that CNF is a protein of 115 kDa associated with both cytotoxicity and in vivo toxicity, distinct from Hly, and present in all presumed CNF-producing strains tested.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bisicchia R., Ciammarughi R., Caprioli A., Falbo V., Ruggeri F. M. Toxin production and haemagglutination in strains of Escherichia coli from diarrhoea in Brescia, Italy. J Hyg (Lond) 1985 Oct;95(2):353–361. doi: 10.1017/s002217240006277x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco J., Gonzalez E. A., Blanco M., Alonso M. P., Barbadillo M. J. Toxins and serotypes of faecal non-enterotoxigenic and non-enteropathogenic Escherichia coli strains causing mannose-resistant haemagglutination: relation with haemagglutination patterns. Zentralbl Bakteriol Mikrobiol Hyg A. 1988 Jul;269(1):43–55. doi: 10.1016/s0176-6724(88)80083-x. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Caprioli A., Donelli G., Falbo V., Possenti R., Roda L. G., Roscetti G., Ruggeri F. M. A cell division-active protein from E. coli. Biochem Biophys Res Commun. 1984 Jan 30;118(2):587–593. doi: 10.1016/0006-291x(84)91343-3. [DOI] [PubMed] [Google Scholar]
- Caprioli A., Falbo V., Roda L. G., Ruggeri F. M., Zona C. Partial purification and characterization of an escherichia coli toxic factor that induces morphological cell alterations. Infect Immun. 1983 Mar;39(3):1300–1306. doi: 10.1128/iai.39.3.1300-1306.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli A., Falbo V., Ruggeri F. M., Baldassarri L., Bisicchia R., Ippolito G., Romoli E., Donelli G. Cytotoxic necrotizing factor production by hemolytic strains of Escherichia coli causing extraintestinal infections. J Clin Microbiol. 1987 Jan;25(1):146–149. doi: 10.1128/jcm.25.1.146-149.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri S. J., Bohach G. A., Snyder I. S. Escherichia coli alpha-hemolysin: characteristics and probable role in pathogenicity. Microbiol Rev. 1984 Dec;48(4):326–343. doi: 10.1128/mr.48.4.326-343.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rycke J., Guillot J. F., Boivin R. Cytotoxins in non-enterotoxigenic strains of Escherichia coli isolated from feces of diarrheic calves. Vet Microbiol. 1987 Oct;15(1-2):137–150. doi: 10.1016/0378-1135(87)90139-8. [DOI] [PubMed] [Google Scholar]
- Emödy L., Kétyi I., Kuch B., Pácsa S. Antitoxic immunity against the so-called lung toxin produced by Escherichia coli. Acta Microbiol Acad Sci Hung. 1979;26(3):233–239. [PubMed] [Google Scholar]
- Guesdon J. L., Avrameas S. Polyacrylamide-agarose beads for the preparation of effective immunoabsorbents. J Immunol Methods. 1976;11(2):129–133. doi: 10.1016/0022-1759(76)90140-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McLaren I., Wray C. Another animal Escherichia coli cytopathic factor. Vet Rec. 1986 Dec 6;119(23):576–577. [PubMed] [Google Scholar]
- O'Brien A. D., Holmes R. K. Shiga and Shiga-like toxins. Microbiol Rev. 1987 Jun;51(2):206–220. doi: 10.1128/mr.51.2.206-220.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short E. C., Kurtz H. J. Properties of the Hemolytic Activities of Escherichia coli. Infect Immun. 1971 May;3(5):678–687. doi: 10.1128/iai.3.5.678-687.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer W., Goebel W. Synthesis and secretion of hemolysin by Escherichia coli. J Bacteriol. 1980 Oct;144(1):53–59. doi: 10.1128/jb.144.1.53-59.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]