Abstract
The epistatic interactions among mutations have a large effect on the evolution of populations. In this article we provide a formalism under which epistatic interactions among pairs of mutations have a distribution whose mean can be modulated. We find that the mean epistasis is correlated to the effect of mutations or genetic robustness, which suggests that such formalism is in good agreement with most in silico models of evolution where the same pattern is observed. We further show that the evolution of epistasis is highly dependant on the intensity of drift and of how complex the organisms are, and that either positive or negative epistasis could be selected for, depending on the balance between the efficiency of selection and the intensity of drift.
THE long-term evolutionary fate of a population relies on the shape of its adaptive landscape. The adaptive landscape is the precise map that associates a fitness value to any possible genotype or phenotype. While a perfect knowledge of the adaptive landscape is out of reach, even for the simplest organisms, understanding its statistical properties can be very valuable to decipher the different selective pressures acting on an organism and its genetic system (e.g., its mutation rate or its recombination rate). Among such statistical properties, the mean epistasis among mutations is one that has been broadly studied in population genetics (Wolf et al. 2000). Epistasis refers to the existence of interactions between mutations: the effect of a mutation depends on the genetic background in which it appears. The spread of a mutation in a population depends on its effect on fitness and as epistasis affects fitness it can potentially influence the evolution of a population. This is why we focus here on the effect of mutations and their interactions on fitness.
Epistasis has been studied widely because it was demonstrated that depending on its sign selection could favor or not the evolution of recombination (Kondrashov 1993). Since that study was published, much more convincing models have been developed on the evolution of sex (Otto and Barton 2001; Poon and Chao 2004; Barton and Otto 2005; de Visser and Elena 2007). Nevertheless, the early model generated a strong interest for the experimental study of epistasis among the population genetics community (de Visser et al. 1996, 1997; Elena and Lenski 1997; Bonhoeffer et al. 2004; Burch and Chao 2004; Sanjuan et al. 2005; Beerenwinkel et al. 2007; Jasnos and Korona 2007)
There are several closely related ways to calculate epistasis (Wolf et al. 2000); in this article epistasis between two mutations is defined as
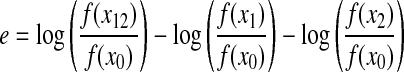 |
(1) |
In this definition e measures the deviation of the log-fitness for a double mutant, log(f(x12)/f(x0)), from that expected if the fitness effects of each individual mutation were multiplicative. (x0 is the ancestral genotype, x1 and x2 are the genotypes having mutation 1 or mutation 2, and x12 is the double mutant). With this definition epistasis is a relative deviation from single-mutant effects and is therefore not directly connected to the amplitude of the effect of single mutations on fitness. A positive epistasis refers to the case in which a combination of mutations has a higher fitness than the one expected from independent mutations. Negative epistasis refers to the opposite: double mutants have lower fitness than expected.
Epistasis is a key component of genomic architecture. Two main approaches, which can be traced back to the Wright vs. Fisher debate, have been used to study epistasis. In a Wrightian approach epistasis dominates the adaptive landscape such that the landscape is defined only by the epistatic interactions among mutations. Depending on the amount of epistasis and its (a priori imposed) distribution, smooth or rugged fitness landscapes can emerge (Goodnight 1988; Kauffman 1993; Hansen and Wagner 2001). The epistasis of the genotypes present in the population evolves then as the population wanders in such genetic space. In a Fisherian view (Fisher 1930), the adaptive landscape has a single peak, and epistasis either is included to modulate how fitness decays with the distance to the optimum (usually in terms of Hamming distance) or emerges as a consequence of the phenotype to fitness map (Wolf et al. 2000) (see later). In this article we focus on the Fisherian view of epistasis. It corresponds to the case in which a population is well adapted and evolves close to an optimum of fitness, staying in a region far from other local optima.
Because epistasis has a large effect on the spread of mutations, both deleterious and beneficial, within populations, it has recently been suggested that epistasis could be under selection itself as it has been shown for the mutation rate (Tenaillon et al. 1999; Denamur and Matic 2006) and the recombination rate (de Visser and Elena 2007). Lines of evidence supporting these ideas come (among others) from the recent studies of epistasis in artificial systems. For instance, it was shown in a model of regulatory networks that epistasis could evolve and even change sign in the presence of genetic exchange (Azevedo et al. 2006). In several systems, such as RNA folding or artificial life (Wilke and Adami 2001), it was found that epistasis was correlated with the mean effect of mutations. On the basis of these observations, analytical models have been developed in which a locus affects the epistatic interactions between mutations at other loci (Liberman and Feldman 2005, 2006, 2008; Desai et al. 2007; Liberman et al. 2007). Our present work extends these theoretical works on modifiers of epistasis to the case of a continuum of mutations effects. For the sake of simplicity we focus here exclusively on asexual populations, in which it was predicted that epistasis was neutral (Desai et al. 2007).
These recent models studying the evolution of a modifier of epistasis considered loci in which some deleterious mutations of fixed effect could appear; moreover, all interactions among mutations presented the same epistasis value (the discrete value imposed by the modifier). Although such assumptions allow one to develop simplified models, they suffer some limitations. For example, considering mutations of a single type of mutational effect on fitness can substantially affect some of the conclusions on the accumulation of deleterious mutations such as Muller's ratchet: while in small populations, the accumulation of deleterious mutations is stopped in the presence of synergistic epistasis when the effects of mutations are discrete (Kondrashov 1994), such an accumulation is endless if mutations have a continuous distribution of effects (Butcher 1995). Moreover, many experiments have demonstrated that both the fitness effect of mutations on fitness and their epistatic interactions have a large variability (Elena and Lenski 1997; Sanjuan et al. 2005; Jasnos and Korona 2007), so assuming they are not variable is an important limitation.
It has been suggested for a long time (Fisher 1958; Wolf et al. 2000) that epistasis could emerge from the phenotype to fitness relation in an integrated model of phenotypic adaptation with continuous effects, referred to as the Fisher geometric model. Fisher's geometric model of adaptation, a quite old model (Fisher 1930), has received a lot of attention recently (Fisher 1930; Kimura 1962; Orr 1998, 1999, 2000, 2006; Welch and Meselson 2001; Waxman and Welch 2005; Martin and Lenormand 2006) as it provides an interesting integrated model of adaptation that appears to be in agreement with experimental data, both qualitatively and quantitatively (Burch and Chao 1999; Orr 1999; Martin and Lenormand 2006; Silander et al. 2007; Tenaillon et al. 2007). Recently, the properties of the distribution of epistasis were characterized in this framework (Martin et al. 2007) and the emerging distribution appeared to be in good agreement with experimental observations (Martin et al. 2007). Fisher's model stipulates that organisms are characterized by a number of independent trait values. Each trait is under stabilizing selection, which means that a unique optimal value of the trait exists. This results in the existence of an optimal combination of traits that provides the best possible fitness (Figure 1A). This simplified model is also interesting because it has only a few parameters as follows:
The number of independent traits, which in geometric terms is the number of dimensions of the space: We mention this parameter later as “the phenotypic complexity.”
The way fitness declines for each trait given the distance to the optimal trait value: In this article, we use the same fitness decay function for all traits, which produces circular fitness isoclines. We decided to use a fitness function in which epistasis could be affected by a single parameter, Q. In agreement with previous studies, the function we use is f(x) = exp(−xQ) (Wilke 2001; Tenaillon et al. 2007), with x being the distance to the optimum and Q an epistasis parameter (see later).
The way mutations affect traits: In such a model a mutation is a vector that moves one organism defined by its position in the phenotypic space to a new position. In the present analysis the direction of the vector is drawn from a uniform distribution and its norm is drawn from a centered normal distribution with standard deviation
 .
.
Figure 1.—
(A) Representation of Fisher's model in two dimensions. Here an organism is defined by a combination of its traits' x and y values; its fitness is a function of the distance of its phenotype (x1, x2) to the optimum genotype (0, 0), and therefore fitness isoclines are circular. Mutations move an organism from one point of the phenotypic space to another. (B) Impact of parameter Q on fitness decline. For distances to the optimum >1, low values of Q have a higher fitness than high values, while the opposite is true for distances <1. Q takes values 0.5, 1, 1.5, 2, 2.5, and 3 with low values in light gray and higher values in darker gray. (C) Schematic representation of the effect of a mutation for several values of Q: log-fitness as a function of the phenotype (x1, x2) in two dimensions for Q = 0, Q = 0.5, Q = 1, Q = 2, Q = 3, Q = 10. The thick black line represents the mean displacement caused by a mutation at the optimum, whereas the thin black line represents the mean displacement when away from the optimum [at the position denoted by the white (Q = 0, 0.5, 1, 2) or black dot (Q = 3, 10)]. The dotted white (Q = 0, 0.5, 1, 2) or gray (Q = 3, 10) line is the fitness isocline at this distance. Mutations tend to be more deleterious when Q increases. Note that for Q = 0, log-fitness is −1 away from the optimum and 0 at the optimum, as in models with a single-fit “master sequence.” The case Q = 10 (or Q → ∞) corresponds to a flat neutral space (for a distance smaller than unity) and lethal genotypes (for a distance greater than unity).
In a one-dimensional discrete model, the parameter Q refers intuitively to epistasis. If Q = 1, then the logarithm of fitness decays linearly with the Hamming distance to the optimum. If Q > 1, there is a synergistic effect of the distance to the optimum on fitness, the further away the bigger the effect on fitness, and the opposite (diminishing return, or antagonistic effect) is seen if Q < 1. It is, however, unclear how Q influences epistasis in multiple dimensions and with continuous mutations; for example, if Q = 2 no average epistasis is found (Martin et al. 2007).
In this article, we further extend the study of epistasis in Fisher's model by studying, first, the distribution of epistasis when Q ≠ 2 and, second, the selective pressures acting on Q.
MODEL AND RESULTS
Impact of Q on the mean epistasis:
Along the same line as Martin et al. (2007), we study the mean epistasis among pairs of mutations. For an individual at position x = (x1, x2, … , xn) in the space of phenotypic complexity n, its Wright–Fisher fitness is defined as
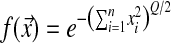 |
(Figure 1).
It is worth noting that the relation between fitness and distance in the phenotypic space is arbitrary and could be modulated by the addition of a parameter without loss of generality by defining instead 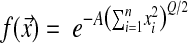 . The parameter A can scale the average effect of mutations on fitness; however, A can be absorbed in a change of units for the xi, so we set A = 1 in the rest of the article.
. The parameter A can scale the average effect of mutations on fitness; however, A can be absorbed in a change of units for the xi, so we set A = 1 in the rest of the article.
For mutations, all traits are treated in the same way: each phenotype is changed by a centered normal deviate of standard deviation σ.
Before we proceed to our calculations, we can intuitively guess the effect of Q on epistasis: since phenotype mutations are normally distributed with mean 0 and standard deviation σ, each mutation entails a move in the phenotypic space of the order of σ such that fitness is modified by a factor of the order of Exp(−σQ) so log-fitness changes are of order σQ. Epistasis measures the difference between the effect on log-fitness of a double mutation and the sum of the effects of two single mutations. A double mutation is the sum of two independent random variables with standard deviation σ such that its standard deviation is 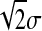 . Thus the effect on log-fitness of a double mutation is of the order of
. Thus the effect on log-fitness of a double mutation is of the order of  . Thus epistasis is of the order of
. Thus epistasis is of the order of 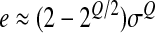 . Thus epistasis should be on average positive if Q < 2, negative if Q > 2, and null if Q = 2. We also see that e, which is homogeneous to a log-fitness effect, is approximately proportional to σQ, i.e., approximately proportional to the average log-fitness effect of single mutations.
. Thus epistasis should be on average positive if Q < 2, negative if Q > 2, and null if Q = 2. We also see that e, which is homogeneous to a log-fitness effect, is approximately proportional to σQ, i.e., approximately proportional to the average log-fitness effect of single mutations.
We now describe more rigorously the precise form of epistasis.
At the optimum:
Let us first consider that the organism is at the optimum (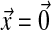 ). Epistasis among two mutations originating from the optimum is
). Epistasis among two mutations originating from the optimum is
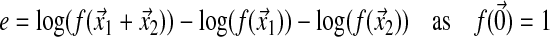 |
in which 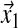 and
and  are two random mutations having each of their components drawn from a normal distribution with mean 0 and standard deviation
are two random mutations having each of their components drawn from a normal distribution with mean 0 and standard deviation 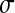 . We can rewrite epistasis as
. We can rewrite epistasis as
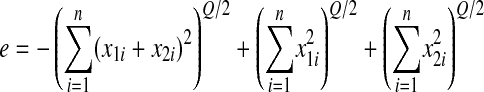 |
and then
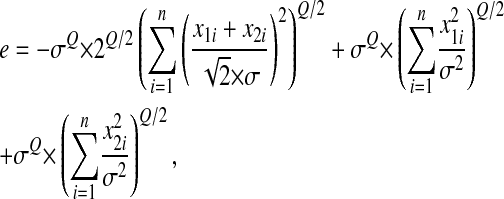 |
which makes appear in parentheses the sum of squared centered, reduced normal deviates (the sum of two independent Gaussians being a Gaussian).
Hence the mean epistasis at the optimum is
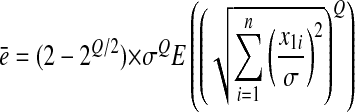 |
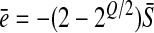 |
(2) |
with
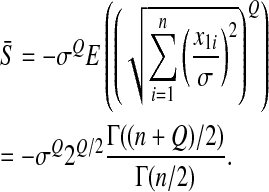 |
(3) |
Note that the mean effect of mutations on the logarithm of fitness is proportional to σQ as expected. For this last derivation we used the fact that the term in parentheses is the (generalized) Qth moment of a chi distribution with n degrees of freedom.
Hence, in our model, epistasis at the fitness optimum has a mean directly linked to the parameter Q (Figure 2, A and B). As expected, when Q = 2, E(e) = 0. If Q > 2, the mean epistasis at the optimum is negative. Since all mutations are deleterious, this means that they act synergistically, a double mutation being worse less than expected from the single effects. The opposite is true (positive or antagonistic epistasis) when Q < 2.
Figure 2.—
(A) Mean log-fitness effect of mutations as a function of the distance to the optimum. From up to down: Q = 0.5, 1, 1.5, 2, 2.5, 3. (B) The distribution of epistasis among pairs of mutations for several values of Q and of the number of dimensions. The mean effect of mutation on logarithm of fitness was normalized to be equal to −0.2 in all conditions. (C) Impact of parameter Q on the average epistasis among pairs of mutations as a function of the mean effect of mutations on logarithm of fitness. Q takes values 0.5, 1, 1.5, 2, 2.5, and 3 with low values in light gray and higher values in darker gray (D) Impact of the distance (translated in log-fitness) to the optimum on the relationship between epistasis and effect of mutations for various values of Q. Black line, Q = 1.5, z = 0; dashed black line, Q = 1.5, z = −0.1; dotted black line, Q = 1.5, z = −0.2; gray line, Q = 2.5, z = 0; dashed gray line, Q = 2.5, z = −0.1; dotted gray line, Q = 2.5, z = −0.2.
Away from the optimum:
Let us now consider that we are away from the optimal fitness, at position  having fitness f0 and at a distance z0 from the optimum.
having fitness f0 and at a distance z0 from the optimum.
Before we detail how mutations interact, let us first study the mean effect of mutations at any distance z0 from the optimum:
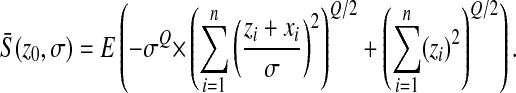 |
The first term is  times the (generalized) raw moment of order Q of a reduced, noncentered chi distribution with n degrees of freedom, which can be written as
times the (generalized) raw moment of order Q of a reduced, noncentered chi distribution with n degrees of freedom, which can be written as
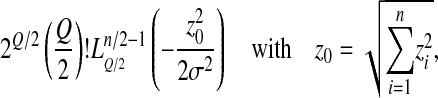 |
with L being a generalized Laguerre function.
Thus
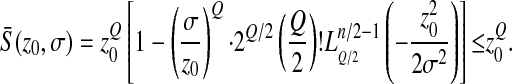 |
(4) |
The mean effect of mutations on log-fitness at a distance  from the optimum is thus the sum of a positive term
from the optimum is thus the sum of a positive term 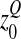 , the maximal beneficial log-fitness effect (i.e., that of a mutation that moves the genotype exactly to the optimum), and a negative term (“deleterious contribution”). If this deleterious contribution is large (
, the maximal beneficial log-fitness effect (i.e., that of a mutation that moves the genotype exactly to the optimum), and a negative term (“deleterious contribution”). If this deleterious contribution is large ( ≫ 1) such that mutations become mostly deleterious as is the case at the optimum, we find again the mean log-fitness effect at the optimum:
≫ 1) such that mutations become mostly deleterious as is the case at the optimum, we find again the mean log-fitness effect at the optimum:
 |
The behavior of 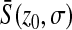 as z0 increases is depicted in Figure 2A. For Q = 2, since
as z0 increases is depicted in Figure 2A. For Q = 2, since 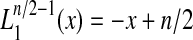 , we have
, we have  , which is independent of
, which is independent of 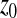 . For Q > 2, on average, mutations are also deleterious (
. For Q > 2, on average, mutations are also deleterious (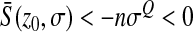 ) and when
) and when 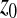 increases, the absolute value of
increases, the absolute value of  increases: mutations are more deleterious when one is far from the optimum than close to it.
increases: mutations are more deleterious when one is far from the optimum than close to it.
If 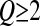 , since mutations are deleterious on average, a first mutation tends to increase the distance from the optimum, where mutations are more deleterious (Q > 2) or as deleterious (Q = 2) as in the previous position. Thus, we again observe a negative mean epistasis if Q > 2, but a null mean epistasis if Q = 2. When Q > 2,
, since mutations are deleterious on average, a first mutation tends to increase the distance from the optimum, where mutations are more deleterious (Q > 2) or as deleterious (Q = 2) as in the previous position. Thus, we again observe a negative mean epistasis if Q > 2, but a null mean epistasis if Q = 2. When Q > 2, 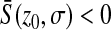 decreases more rapidly (mutations becoming on average more deleterious) as z0 increases if σ is large than if σ is small (not shown); thus, when mutational effect increases, the absolute value of epistasis should also increase (epistasis becoming more negative).
decreases more rapidly (mutations becoming on average more deleterious) as z0 increases if σ is large than if σ is small (not shown); thus, when mutational effect increases, the absolute value of epistasis should also increase (epistasis becoming more negative).
For Q < 2, the behavior of  is more complicated and can be positive (i.e., mutations can be advantageous on average) or negative. However, it seems that for
is more complicated and can be positive (i.e., mutations can be advantageous on average) or negative. However, it seems that for 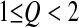 , as the log-fitness is still a concave function of the distance to the optimum (Figure 1, B and C), the mean effect of advantageous mutations (decreasing the distance to the optimum) is smaller than the mean effect of deleterious mutations (increasing the distance to the optimum). Since the proportion of deleterious mutations is higher than the proportion of beneficial ones (Fisher 1930), mutations stay deleterious on average. Nevertheless in contrast to what happens when Q > 2, mutations are on average less deleterious far from the optimum than close to it, so that we can expect epistasis among pairwise mutations to be positive on average. For
, as the log-fitness is still a concave function of the distance to the optimum (Figure 1, B and C), the mean effect of advantageous mutations (decreasing the distance to the optimum) is smaller than the mean effect of deleterious mutations (increasing the distance to the optimum). Since the proportion of deleterious mutations is higher than the proportion of beneficial ones (Fisher 1930), mutations stay deleterious on average. Nevertheless in contrast to what happens when Q > 2, mutations are on average less deleterious far from the optimum than close to it, so that we can expect epistasis among pairwise mutations to be positive on average. For 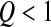 , the log-fitness function is convex (Figure 1, B and C) and, depending on the distance to the optimum and the standard deviation σ, mutations can be either deleterious (close to the optimum) or beneficial (far from the optimum) on average (Figure 2A).
, the log-fitness function is convex (Figure 1, B and C) and, depending on the distance to the optimum and the standard deviation σ, mutations can be either deleterious (close to the optimum) or beneficial (far from the optimum) on average (Figure 2A).
We can also see in Figure 2A that the curvature of the mean log-fitness effect of mutations decreases when the distance to the optimum increases: epistasis should be more pronounced close to the optimum than far from the optimum.
The epistasis between two random mutations can be written as
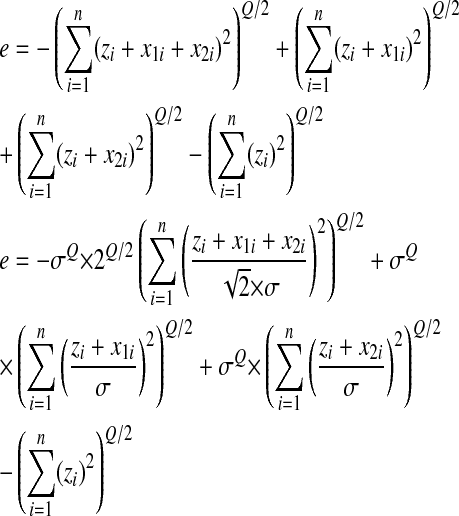 |
and the mean epistasis among pairs of mutations as
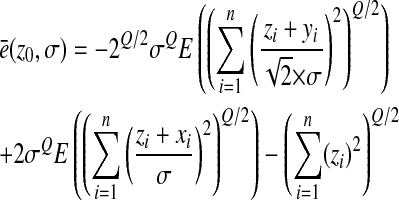 |
with yi = x1,i + x2,i. And
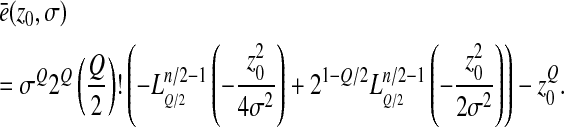 |
(5) |
Note that for Q = 2, since 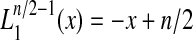 , E(e) = 0.
, E(e) = 0.
To make appear more clearly the dependency of the mean epistasis on 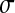 , we can write
, we can write
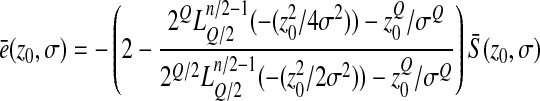 |
or
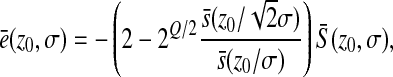 |
where the reduced variable  depends only on the ratio z0/σ.
depends only on the ratio z0/σ.
This last expression shows that, as expected, the mean epistasis is homogeneous to a mutational effect on log-fitness, i.e., homogeneous to 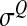 .
.
We also see that when  converges to 0, the mean epistasis increases linearly with the mean effect of mutation on log-fitness as found around the optimum value as
converges to 0, the mean epistasis increases linearly with the mean effect of mutation on log-fitness as found around the optimum value as
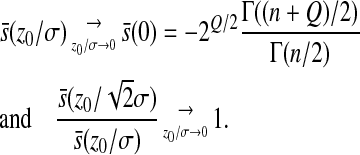 |
Finally, by writing
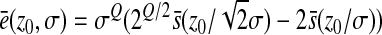 |
we clearly see the contribution of each factor on epistasis: the difference in parentheses is the difference between the effect of the double mutation ( is the standard deviation of the distribution of the double mutation, which makes a factor
is the standard deviation of the distribution of the double mutation, which makes a factor 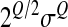 appear because of the form of the log-fitness with a Q exponent) and the sum of each single mutational effect.
appear because of the form of the log-fitness with a Q exponent) and the sum of each single mutational effect.
Using parametric Equations 4 and 5 with σ as a moving parameter, we can plot the average epistasis and the average effect of mutations on log-fitness as a function of Q and of the distance to the optimum (Figure 2D). While the number of dimensions affects the mean effect of mutations, it has no impact on its relationship with the average epistasis; i.e., for any phenotypic complexity the relationship between the average effect of mutations on the logarithm of fitness and the average epistasis between pairs of mutations is unchanged (data not shown). As expected, around Q = 2, the mean pairwise epistasis is negative if Q > 2 and positive if Q < 2.
We can further see that, as expected in this model, around the optimum, the more distant an organism is from the optimum, the smaller the mean epistasis is among its mutations (Figure 2D).
Evolution of the parameter Q:
Now that we have shown how epistasis is related to the parameter Q, we can study the selective pressure acting on Q. We now assume that in addition to their position in the phenotypic space, organisms have a genetically encoded value of Q that is itself subject to mutations. We are therefore in the theories of modifiers, in which we study how a trait controlling the parameter Q evolves. We once again concentrate on Fisher's model and therefore consider haploid asexual organisms.
Mutation–selection–fixed-drift equilibrium theory:
We recently studied Fisher's model at mutation–selection–fixed-drift equilibrium (MSFDE) with low mutation rate and showed that the equilibrium fitness depends only on the population size, the number of dimensions, and the parameter Q (Tenaillon et al. 2007). We can use a similar formalism to study the evolution of Q and its distribution at equilibrium. In such formalism, n is the phenotypic complexity, f the fitness, and v a population size parameter such that v = 2N − 2 in the haploid case with N being the population size in a Wright–Fisher model.
It is important to note that the framework considers only transiently dimorphic populations: a resident population, fully homogeneous, in which a single mutant occurs, either affecting the phenotypes or affecting the modifier allele, appears and gets either fixed or lost according to its fitness relative to that of the first resident.
In such a process, the probability that a mutation gets fixed depends on both the fitness of the resident and the fitness of the mutant (and the intensity of drift). If it gets fixed, the mutant then becomes the new resident. However, as this process goes on and mutations get successively fixed, this probability converges to a stable distribution that does not depend on the fitness of the first resident (i.e., the beginning of the process) anymore. Thus after several fixations of mutations, the probability that the current resident has a given fitness becomes constant: this is the fitness distribution at MSFDE.
If the mutations affecting the modifier Q of epistasis are rare, the population will reach the MSFDE before Q mutates. We can then compute the fitness distribution at MSFDE given a fixed value of Q (Sella and Hirsh 2005; Tenaillon et al. 2007),
 |
or, translated into the distance z to the optimum (the probability distribution of distance to the optimum),
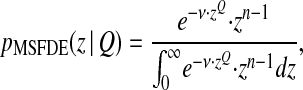 |
which gives for the mean fitness and the mean log-fitness (a “*” in the following formula indicates a mean taken with respect to the equilibrium distribution)
 |
and
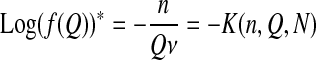 |
and a mean distance to the optimum
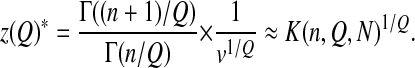 |
The parameter K reflects the mean log-fitness at equilibrium and reflects therefore an intensity of drift; the higher K is, the lower the fitness. Note that it allows for the definition of an effective population size Ne that sums up the intensity of drift in this model. Taking as a reference a fitness landscape with no epistasis (Q = 2) and the smallest complexity (n = 1), we define
 |
such that
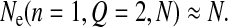 |
Ne quantifies the intensity of drift since
 |
i.e., the smaller Ne is, the higher the intensity of drift and the higher the fixed-drift load.
We note that these results are strictly valid when the mutation rate is very small such that the effect on fitness of the recurrent production of the deleterious mutant is neglected. For only small mutation rates these values have to be modified by a factor e−U, with U being the mutation rate [e.g., 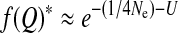 ].
].
Given that the distance to the optimum of the resident is z we can now calculate how a slight change from Q to Q′ should be selected for. The effect on log-fitness of a mutation changing Q into Q′ is given by
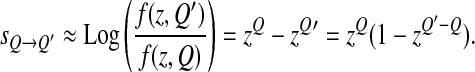 |
Thus if z < 1, z(Q′−Q) < 1 if Q′ > Q: a mutation increasing Q should be selected for. On the contrary if z > 1, Q should decrease upon selection.
The effect of drift is to impede the population from approaching the optimum. When drift is intense, z should be on average >1. Whereas when selection is efficient, z < 1 and the population is closer to the optimum. The further evolution of Q will therefore depend on this balance between drift and selection. The effect of a mutation that changes Q, taking into account this balance, can be measured by computing the mean selective effect of a mutation of Q at MSFDE, i.e., taking into account the probability that the distance from the optimum of the resident is z:
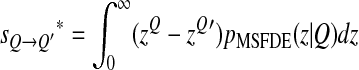 |
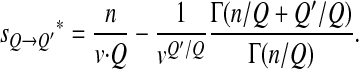 |
If n ≫ Q (e.g., n/Q > 5 or 10, and in the following we assume this condition holds true), then as Q′ should be of the order of Q, n/Q ≫ Q′/Q and
 |
thus
 |
(6) |
and
 |
If K > 1 (intense drift), then Q′ < Q is selected for. The inverse is true for K < 1 (strong selection). Selection will favor high or low values of Q depending on the intensity of drift. Note that usually Q is of the order of 1, such that K is of order n/N and the direction of selection on Q is generally set by n/N: when selection is strong, i.e., when n/N is small, selection will favor an increase of Q, while when drift dominates the process, selection will favor a small Q (Figure 3).
Figure 3.—
(A) Impact of population size on the average value of Q at MSFDE for several phenotypic complexities. Solid line, phenotypic complexity, n = 100; dashed line, n = 30; dotted line, n = 10. (B) Impact of phenotypic complexity on the average value of Q at MSFDE for several population sizes. Solid line, population size N = 100; dashed line, N = 30; dotted line, N = 10.
Is this selection efficient? In other words do we have  ? If K ≫ 1 and Q′ < Q, then
? If K ≫ 1 and Q′ < Q, then  whereas if Q′ > Q, then
whereas if Q′ > Q, then 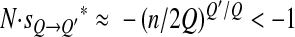 ; in other words, when K ≫ 1, Q′ < Q is selected for, whereas Q′ > Q is selected against. The opposite is true when K ≪ 1. In the intermediate case, when K is of the order of 1,
; in other words, when K ≫ 1, Q′ < Q is selected for, whereas Q′ > Q is selected against. The opposite is true when K ≪ 1. In the intermediate case, when K is of the order of 1,  , which can be small if
, which can be small if  ∼
∼  . For N not too small, around the threshold value K = 1, a slight change (i.e., >1/N) of the value of K can result in a sharp change in the mean direction of evolution of epistasis (i.e., Q = 2 is “unstable”).
. For N not too small, around the threshold value K = 1, a slight change (i.e., >1/N) of the value of K can result in a sharp change in the mean direction of evolution of epistasis (i.e., Q = 2 is “unstable”).
Hence we see that a change of Q can be selected for in populations and that depending on the intensity of drift (parameter K, or 1/4Ne, in our model) selection will either favor high or low values of Q. It is important to note that an intense drift does not mean here that there is no selection at all. When drift is intense (K ≫ 1), the population tends, on average, to be delocalized from the optimum and is in a region where a mutation that changes Q can have a substantial effect on fitness and therefore selection for or against this change can be strong. Interestingly, the intensity of drift depends on Q: 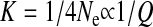 . Thus a high value of Q (e.g., large negative epistasis) tends to increase selection for still higher values of Q (more negative epistasis) if n/N ≪ 1 and to reduce selection for small values of Q if n/N ≫ 1, whereas a small value of Q (e.g., positive epistasis) tends to increase selection for still smaller Q (more positive epistasis) if n/N ≫ 1 and to decrease selection for high values of Q if n/N ≪ 1. The precise value of Q could also be decisive in the special situation where n/N ∼ 1.
. Thus a high value of Q (e.g., large negative epistasis) tends to increase selection for still higher values of Q (more negative epistasis) if n/N ≪ 1 and to reduce selection for small values of Q if n/N ≫ 1, whereas a small value of Q (e.g., positive epistasis) tends to increase selection for still smaller Q (more positive epistasis) if n/N ≫ 1 and to decrease selection for high values of Q if n/N ≪ 1. The precise value of Q could also be decisive in the special situation where n/N ∼ 1.
On a longer term the equilibrium will be reached both by the phenotypes and by the modifier of epistasis. Importantly, if mutations of the modifier Q are not rare (i.e., as frequent as those affecting the classical traits), this equilibrium does not need a longer time to be reached. The evolutionary tendency of Q at this new equilibrium will reflect the one at the first equilibrium. Especially in cases where selection for increasing Q (strong selection) or decreasing Q (intense drift) is efficient, Q will rapidly reach its maximal value Qmax or its minimal value Qmin and stay for a long time at this value. However, when selection for either large or small values of Q is not very strong, the evolution of Q will mostly depend on the dynamics of the system, i.e., on the mutational spectrum, the mutation rate, and the initial conditions.
At a long timescale, the probability density of Q can be written as
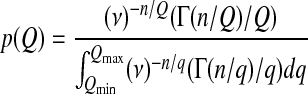 |
(7) |
such that the unconditioned mean fitness is
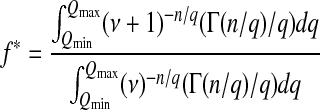 |
and the mean Q is
 |
(8) |
These functions cannot be integrated but can nevertheless be explored numerically. First of all it appears, as expected, that when drift is strong (i.e., small population size, large phenotypic complexity), Q evolves to its minimal value, favoring positive epistasis. On the opposite, when selection is stronger than drift (large population size, small phenotypic complexity), Q evolves to its maximum value, hence favoring negative epistatic interactions among pairs of mutations. This is illustrated in Figure 3, A and B, in which we can see that for a given population size as phenotypic complexity increases Q evolves from the maximum value to the minimum, and similarly at a given phenotypic complexity, as population size increases, Q evolves from the minimum value to the maximum. Hence depending on the interplay between population size and the phenotypic complexity, selection should favor either strong negative epistasis or strong positive epistasis among pairs of mutations.
It is interesting to note that if we assume that the modifier of Q affects only a fraction m of the dimensions, such that fitness can be written as 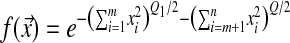 , the modifier affecting only Q1, then the equilibrium value of Q1 is the same as in the previous model with a phenotypic complexity of m. As noted before (Tenaillon et al. 2007), at equilibrium each dimension behaves as if it were independent from the others (independent traits).
, the modifier affecting only Q1, then the equilibrium value of Q1 is the same as in the previous model with a phenotypic complexity of m. As noted before (Tenaillon et al. 2007), at equilibrium each dimension behaves as if it were independent from the others (independent traits).
Simulations:
The derivations we used assume that populations have converged to the MSFDE and are therefore to be used in the limit of infinite time. They are also based upon the hypothesis of a small mutation rate. We therefore decided to implement some simulations to see how valid they are. An individual-based model of evolution was developed [as previously described (Silander et al. 2007)] in which the parameter Q was encoded for each individual. Along the evolutionary process the mean Q of the population was monitored to find its equilibrium value after 100 million generations. We chose different initial values of Q, the mutation rate, and the mean mutation size in the phenotypic space to determine the impact of those parameters on the final equilibrium. The parameter Q evolved through mutations 1000 times less frequently than mutations affecting the phenotypes and was then drawn from a uniform distribution between its minimal and maximal value.
From such simulations it appeared that the theoretical model was qualitatively very accurate for various mutation rates and mean effects of mutations, but that the switching point, where Q moves from maximal value to minimal, is not precisely estimated (Figure 4C). Indeed, we could find three regimes, two in which both theories and simulations agree fully and one in which they do not.
Figure 4.—
(A) Fitness as a function of the distance to the optimum and parameter Q, and distance distributions at MSFDE. Top panel: log-fitness as a function of the distance to the optimum for Qmax = 2.5 (solid line) and Qmin = 0.8 (shaded line). Second, third, and fourth panels: density distribution of the distance to the optimum at MSFDE (“short-term equilibrium”) for a given Q: Qmax (solid line) and Qmin (shaded line). Second panel: strong selection (Kmax < 1, N = 100, n = 25), an increase of Q is always advantageous. Third panel: intermediate selection (Kmin < 1 < Kmax, N = 100, n = 200). Fourth panel: intense drift (Kmin > 1, N = 100, n = 700), a decrease if Q is always advantageous. (B) Distribution of Q at MSFDE as a function of phenotypic complexity. For a population size of 30, and phenotypic complexity of 10 (solid line), 15, 20, 25, 30, 40, 50, and 100 (dotted line), we see that for intermediate values of n, high and low values of Q have a high probability, which explains the dependency on initial conditions observed in simulations. (C) Impact of phenotypic complexity on the average value of Q for population sizes of 30 in the simulation model, with several initial values of Q. The average size of a mutation was 10. Each point is the average of 10 simulations.
When population sizes are big and phenotypic complexity is small (K ≪ 1), even at the smallest value of Q the population at MSFDE is at a distance from the optimum <1 [Kmax = n/(2N − 2)Qmin, < 1; Figure 4A, second panel]. Hence, whatever the initial value of Q or the initial fitness of the population is, selection will bring the population toward a smaller than unity distance from the optimum. For an organism whose distance to the optimum is <1, a mutation increasing Q is always beneficial as it directly increases its fitness (see Figure 1). Hence for all populations Q will evolve to its maximal value as predicted by theory (Figure 4A).
Similarly when populations are small and/or phenotypic complexity is large (K ≫ 1), even at the highest value of Q, the distance to the optimum is >1 [Kmin = n/(2N − 2)Qmax, >1; Figure 4A, bottom panel]. Hence, whatever the initial conditions are on the initial fitness or the initial value of Q of the population, drift will bring the population to a bigger than unity distance to the optimum. For an organism whose distance to the optimum is >1, a mutation decreasing Q is always beneficial as it directly increases it fitness (see Figure 1). Hence for all populations Q will evolve to the minimal value of Q as predicted by theory (Figure 4A).
In between these two cases [for Qmin < n/(2N − 2) < Qmax or Kmin < 1 < Kmax] lies the situation in which the equilibrium distance of the largest Q is smaller than unity and the equilibrium distance at the minimal value of Q is larger than unity. In this range, there are two attractors, and depending on the initial conditions, on the frequency of mutations acting on Q and, on their effect on Q, a population will either evolve to a distance smaller than unity and therefore evolve toward the maximal Q or evolve to bigger than unity distance favoring minimal values of Q (Figure 4A, third panel). We confirmed this by initiating different populations at their equilibrium distance from the optimum with different values of Q, and, as expected depending on their positions, they favored either the minimal or the maximal value of Q (Figure 4C).
The study of the average Q at MSFDE does not predict clearly this instability (since at equilibrium—when time goes to infinity—the initial values do not matter); however, with a closer look at the distribution of Q at MSFDE, we can see that high probabilities are observed at both high and low values of Q in the region of instability we described previously (Figure 4B). Hence the discrepancy between simulations and theory lies in the fact that MSFDE reflects very long-term equilibrium while simulations might get trapped in some local optimum and infinite time would be required for the theoretical limit to be reached.
In the simulations presented previously, no clear effect of the average size of mutations was observed on the patterns described; whether the average mutation size was 0.01 or 100, the transition of Q between extreme values was observed at the same ratio of phenotypic complexity to population sizes (data not shown).
Impact of the mutation rate:
In the simulations presented before, the genomic mutation rate was assumed to be quite low, i.e., 0.001 per generation, which ensured that the simulation was fast and in conditions compatible with the formalism used for MSFDE. However, we wanted to know how robust such results are in the face of increased mutation rate. To do so we increased the mutation rate to 0.1 and followed in a population size of 100 the evolution of Q. Similarly to what has been observed before, a shift occurred as a function of phenotypic complexity, arising at different phenotypic complexities for various initial conditions of Q. The value at which the shift occurred was, however, affected by the mutation rate. It arose at lower values of phenotypic complexity compared to simulations with a lower mutation rate (Figure 5).
Figure 5.—
Impact of the mutation rate on the evolution of Q. The mutation rate was set either to 0.001 or to 0.1. The position of the switch from maximal to minimal value of Q was decreased when mutation rate increased. Here population size was 100, and several effects of mutation size gave the same result.
This shift is somehow expected as we know that an increased mutation rate will have two consequences. First, it will allow the population to explore more the genotypic space and therefore enhance the chances to find the best solution, either low or high Q predicted by theory. Hence increased mutation rate should favor a transition in Q closer to the one predicted by theory. Second, an increase in mutation rate reduces the average fitness as many deleterious mutants are present within the population (Haigh 1978) and is also equivalent to a reduction in effective population size (Charlesworth et al. 1993). This effect results in an increased distance to the optimum and therefore a faster transition to low values of Q as phenotypic complexity increases. The first and second effects described act synergistically when the transition of Q occurs at higher than expected phenotypic complexity and result in a transition occurring much closer to the theoretical value (Figure 5). However, when the transition occurs at a lower than expected value of phenotypic complexity, the effects are antagonistic and result in a moderate displacement of the transition point (Figure 5).
DISCUSSION
Since the “omic” era (genomic, transcriptomic, proteomic, metabolomic), the study of interactions between an organism and its environment or interactions within an organism's genome is becoming more feasible experimentally, generating a renewed interest for the theories aiming at understanding how selection should shape the genomic architecture of organisms. While it is clear that all kinds of interactions could exist within a genome, one of the goals of population genetics is to see whether such interactions could be captured by some summary statistics that would on the one hand describe the genomic architecture of the organism and on the other hand be predictive of the various selective pressures acting on such an organism. In genetics, epistasis refers to the existence of some genetic interactions between some loci; in population genetics it refers more precisely to how such interactions affect phenotypes and fitness (Wolf et al. 2000). Generally two kind of epistasis are opposed: antagonistic epistasis in which the combination of mutations has a fitness closer to the ancestral fitness than what is expected from their individual effects and synergistic epistasis in which the combination of mutations has an increased fitness effect (more distant from the ancestral fitness) compared to that expected. The other formalism that we use here refers to positive or negative epistasis. Positive epistasis refers to the fact that combinations of mutations that are beneficial, deleterious, or a mixture of both have fitness values higher than the ones expected from the product of the fitness effects of single mutants. Negative epistasis refers to the opposite situation in which mutations in combination have a lower than expected fitness effect.
To study how epistasis influences population evolution and how it can evolve, one can study a locus controlling epistasis, its impact on the population, and the selective pressures acting on it. One classical framework to do so is to assume a single peak fitness landscape in which epistasis can modulate how fitness will decay as organisms accumulate deleterious mutations. Epistasis is then chosen to be homogenous among loci, being either positive or negative. With this framework it was shown that epistasis could largely influence the way populations would respond to selection (Kimura and Maruyama 1966; Charlesworth 1990; Kauffman 1993; Hansen and Wagner 2001; de Visser and Elena 2007), and some recent studies have shown how natural selection shapes epistasis (Liberman and Feldman 2005, 2006, 2008; Desai et al. 2007; Liberman et al. 2007). However, all experimental data, to date, have found that both kinds of epistasis were present simultaneously (Elena and Lenski 1997; Sanjuan et al. 2005; Jasnos and Korona 2007), and therefore such theories should be revisited to see how robust they are to the presence of both kinds of epistasis (Kouyos et al. 2006, 2007). To go further in such a direction, one needs a proper framework in which epistasis can be distributed with both positive and negative interactions, with positive or negative averages (Jasnos and Korona 2007), and with a distribution that has potentially some biological meaning. In this article, we present a theoretical framework that on the one hand provides a distributed epistasis with nonzero mean and on the other hand allows us to study how epistasis evolves.
The framework we use is the one first briefly introduced by Fisher (1930) and presented briefly in Figure 1A. In the last decade a large number of theoretical studies and experimental works have used such a framework and showed its interest and validity to study evolution (Burch and Chao 1999; Orr 1999; Martin and Lenormand 2006; Silander et al. 2007; Tenaillon et al. 2007). Two lines of investigations have been studied mainly: the study of the effect of mutations (Fisher 1930; Kimura 1962; Orr 1998, 1999, 2000, 2006; Welch and Meselson 2001; Waxman and Welch 2005; Martin and Lenormand 2006) and the study of MSFDE (Wagner and Gabriel 1990; Hartl and Taubes 1996, 1998; Poon and Otto 2000; Tenaillon et al. 2007). For our purposes, we use both aspects.
A direct link between mutation effects on fitness and epistasis:
Using mutation models, we study here the epistatic interactions among pairs of mutations. First, as shown before (Martin et al. 2007), we find that the model provides a whole distribution of possible epistatic interactions among pairs of mutations (Figure 2B). Such a distribution can have both positive and negative epistatic interactions as it has been observed in biological organisms. Moreover we show that the average epistasis is nonzero as long as Q is different from 2 (Figure 2C). Interestingly we find a tight link between epistasis and the mean effect of mutations on fitness logarithm. This observation is very interesting, as it agrees with the analysis of various in silico models in which a correlation between the effect of mutations and epistasis was found, and therefore suggests that such a model could be analyzed under Fisher's model framework. Hence, we show that while the parameter Q is directly related to epistasis, it is also directly related to the effect of mutations on fitness, something that we could refer to as genetic robustness, i.e., how stable the fitness of an organism is in the face of mutations. Hence, as suggested before in a simple discrete model (Wilke and Adami 2001), our formalism suggests that epistasis and genetic robustness might be intimately linked, without involving any selection mechanism: the correlation between the two appears as an emerging mutational property of the adaptive landscape and not a property resulting from the action of natural selection.
The parallel with previous definitions of epistasis in discrete models can be pushed further. In genotypic models fitness is often defined as  , or
, or 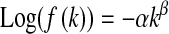 , with k being the number of mutations [f(0) being the optimal fitness],
, with k being the number of mutations [f(0) being the optimal fitness], 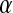 the mean effect of the first mutation on logarithm of fitness, and
the mean effect of the first mutation on logarithm of fitness, and 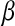 the epistasis parameter. Using our formalism, if
the epistasis parameter. Using our formalism, if 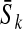 denotes the average log-fitness of individuals with k mutations, we have, for all k,
denotes the average log-fitness of individuals with k mutations, we have, for all k,
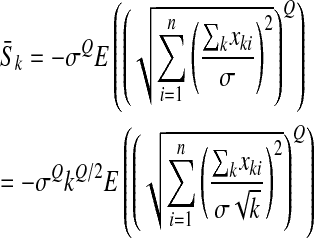 |
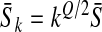 |
such that we can write  for all k with
for all k with 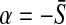 and
and 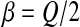 . So a direct parallel between
. So a direct parallel between 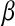 and Q/2 can be made. Interestingly, Wilke and Adami (Wilke 2001) predicted that α and β should be negatively correlated. In our framework, we have
and Q/2 can be made. Interestingly, Wilke and Adami (Wilke 2001) predicted that α and β should be negatively correlated. In our framework, we have  if fitness is defined as
if fitness is defined as  . Thus
. Thus  or
or 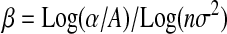 . As β > 0 (mutations are deleterious on average), if α < A, then
. As β > 0 (mutations are deleterious on average), if α < A, then 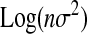 < 0 and α and β are negatively correlated, β being approximately a log function of α as seems to be the case in Wilke and Adami's (2001) study. It would be interesting to compare this result to their data, as the complexity of their system could then be estimated.
< 0 and α and β are negatively correlated, β being approximately a log function of α as seems to be the case in Wilke and Adami's (2001) study. It would be interesting to compare this result to their data, as the complexity of their system could then be estimated.
Finally, we show here that even with a definition of epistasis that should be independent of the effect of mutations (Equation 1) as it measures a relative change in effect, in our model, epistasis is directly linked to the average effect of mutations on log-fitness. Hence comparative studies on the intensity of epistasis (Sanjuan and Elena 2006) should take this parameter into account.
Evolution of the parameter Q depends on drift:
Previous studies have analyzed the evolution of epistasis (Liberman and Feldman 2005, 2006, 2008; Desai et al. 2007; Liberman et al. 2007). However, they focused on modifiers of epistasis acting on two loci or on modifiers of epistasis acting on many loci having mutations of similar effects on fitness. Our framework allows the study of a modifier that has an impact on a whole distribution of epistatic interactions, which is somehow more realistic. However, as there are many different effects of mutations, an unavoidable consequence is that the modifier affects not only epistasis but also the average fitness effects of mutations. Hence our analysis unravels, at least in this model, a mixed effect of the selection acting on a modifier affecting epistasis. What we may lose in theoretical clarity, we gain in realism as we know that mutations in nature do not have a single effect and that epistasis appears to be distributed. Hence any global modifier of epistasis should be submitted to a selective pressure similar to the ones acting on Q.
In previous studies (Liberman and Feldman 2005, 2006, 2008; Desai et al. 2007; Liberman et al. 2007), the theoretical framework used was that of a very large population size and fixed effect of mutations. In such a context, the population remains at the fitness optimum. Epistasis in such models affects only the fitness of genotypes having two deleterious mutations. However, in such large asexual populations deleterious mutants having a single or even more deleterious mutations are doomed to disappear and do not contribute to the long-term fate of the population. Hence, any modifier affecting the effect of single [modifier of genetic robustness (Gardner and Kalinka 2006)] or double mutations [modifier of epistasis (Desai et al. 2007)] will be neutral. However, in sexual populations through recombination, individual having deleterious mutations can contribute to the production of mutation-free individuals and therefore epistasis is under selection. Therefore, previous studies have focused on the impact of recombination on the evolution of epistasis.
In contrast with those theoretical predictions, in asexual populations, evolution of genetic robustness has been observed (van Nimwegen et al. 1999). This has been mainly studied theoretically through the framework of neutral networks, in which fitness of an organism is either maximal or null (corresponding to an infinite value of Q in our model). In such a framework, the genotypes having the highest fraction of neutral neighbors are selected for because the fitness of the populations is linked not only to mutation rate as Haldane proposed in the absence of neutral mutations, but also to the fraction of neutral neighbors, the so-called neutrality. Indeed, as soon as the population is not doomed to stay at a single genotype of maximal fitness, selection can act on modifiers affecting the fitness of the explored genotypes. This was illustrated on the evolution of robustness by Krakauer and Plotkin (2002), who showed that a high mutation rate coupled with drift could lead to selection for genetic robustness. It is worth noting that they kept the assumption of high mutation rate, but that such an assumption is not necessary (Gros and Tenaillon 2009). For a model like Fisher's model, in which mutations can have arbitrarily small effects, some deleterious mutations will be fixed regardless of the population size. A modifier of epistasis that affects the fitness of those fixed mutations can therefore be under selection even if the mutation rate is almost null. Hence to study this selection we can first assume a near vanishing mutation rate to see how epistasis would evolve: this is the main reason why we choose to study epistasis in the MSFDE framework.
We have applied the MSFDE theory to Fisher's model to study how the parameter Q would evolve (if evolvable). In MSFDE theory, a population is always monomorphic unless a single mutant appears. Depending on its effect, either on the phenotypes or on Q, such a mutant will either invade and become the new resident genotype or disappear. The interesting feature of Fisher's model on the MSFDE side is twofold. First, the results concerning the evolution of Q are independent of the mutational effects (cf. Equations 6 and 7) and therefore robust to the different distributions of mutation effects and to their mean; these parameters influence only the speed of convergence to the equilibrium. Second, the theory combines phenotypic complexity, epistasis, and population size in a global parameter to measure the efficiency of selection vs. drift. It can be shown that at MSFDE the mean logarithm of fitness is  . Hence fitness at MSFDE decreases with phenotypic complexity (as it is harder to optimize simultaneously many traits); it is also affected, as expected, by the population size, and finally by the epistasis parameter Q (the higher Q is, the higher the fitness at MSFDE): when mutations are mostly deleterious, increasing Q makes mutations more deleterious on average and they are more easily selected against. However, this property makes the evolution of a modifier of epistasis different from that of a noncostly modifier of genetic robustness [for instance, the parameter σ or a parameter A modulating the selection intensity if fitness is defined as
. Hence fitness at MSFDE decreases with phenotypic complexity (as it is harder to optimize simultaneously many traits); it is also affected, as expected, by the population size, and finally by the epistasis parameter Q (the higher Q is, the higher the fitness at MSFDE): when mutations are mostly deleterious, increasing Q makes mutations more deleterious on average and they are more easily selected against. However, this property makes the evolution of a modifier of epistasis different from that of a noncostly modifier of genetic robustness [for instance, the parameter σ or a parameter A modulating the selection intensity if fitness is defined as 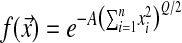 ]. The latter has no effect on mean fitness at equilibrium while a modifier of Q does. It appears therefore interesting to study the evolution of Q in Fisher's model.
]. The latter has no effect on mean fitness at equilibrium while a modifier of Q does. It appears therefore interesting to study the evolution of Q in Fisher's model.
Using MSFDE theory we find that the evolution of parameter Q is influenced by the ratio of complexity to population size (Figure 3). When selection is strong, i.e., when n/N is small, selection will favor an increase of Q, while when drift dominates the process, selection will favor a small Q. This was suggested by the theory and confirmed by simulations. Interestingly while our theory is based on the assumption of low mutation rate, simulations show that its range of validity includes large mutation rates (Figure 5). However, it is worth noting that while the model predicts very well the behavior of the population in the strong drift or strong selection regimes, it is better to use local selective pressure acting on Q rather than long-term equilibrium predictions in the intermediate regime. A precise estimate of the time needed to reach equilibrium is not available here. However, this timeframe should depend mainly on the mutation rate of the modifier of Q as well as on the mean effect of a mutation acting on Q. Increasing the mutation rate of the modifier should shorten this relaxation time as it increases the probability to escape the local optimum. The effect of the size of mutations on this modifier is more complex: a reduction in this size can increase the probability to escape a local optimum (since mutations are less deleterious) while more mutations may then be needed to fully escape this optimum. An increase in the size may also increase the probability that a mutation passes through the valley between the two local optima.
As we have seen, the intensity of drift is determined mainly by the ratio n/N of phenotypic complexity and population size. Therefore an estimate of both parameters is essential to decipher whether negative or positive epistasis should evolve. The main difficulty may probably come from the estimate of n as we have to date no idea of what precisely determines phenotypic complexity. So far, the only study that precisely addressed this question experimentally is that of Tenaillon et al. (2007) for the vesicular stomatite virus (VSV) and the bacteriophage PhiX174. They found, respectively, n/Q ∼10 and ∼45 for these two viruses coding, respectively, 5 and 11 genes. Intuitively the number of genes, the number of genetic interactions, and the diversity of environmental conditions may be parts of the determinants of this complexity. We have therefore no precise estimate of phenotypic complexity for other organisms but it is likely that it spans several orders of magnitude among life diversity, as the viruses studied previously are among the simplest life forms. Hence it is highly likely that some species may be subjected to selection for negative epistasis whereas others may be subjected to selection for positive epistasis.
Why should drift favor the evolution of antagonistic epistasis? When drift is strong, populations will not be clustered around the optimum but at some distance, natural selection failing to improve all phenotypic traits simultaneously. As positive epistasis improves the fitness of distant organisms (Figure 1B), it is hence favored under such conditions. In our model drift is just a pressure that pushes the populations away from the optimum.
Link with previous models:
In large populations, other models have shown that positive epistasis was selected for in the presence of recombination; here, positive epistasis is selected for when drift is strong. In infinite sexual populations, populations are polymorphic and recombination “moves” alleles in different genotypes, both optimal and nonoptimal, even if the populations keep some optimal genotypes present in the populations. Our analysis somehow suggests that this situation has some similarity with the evolution of finite asexual populations in which the best genotype is strongly drifting around the optimal genotype. A parallel can be made between the fraction of time spent by beneficial alleles in genotypes loaded with deleterious mutations in the presence of recombination and the time spent by the asexual monomorphic population with a fixed deleterious mutation.
Some more interplay between our result and the existence of selection for recombination can be suggested. First, our model shows that when selection is efficient compared to drift, negative epistasis is favored. This could be interesting for the evolution of sex as the existence of negative epistasis is one of the conditions that favor selection for recombination (de Visser and Elena 2007). Second, in the simulations we performed, we saw that under some ranges of parameters, the system can transiently (compared with infinite time) evolve to the maximal or the minimal value of Q. In such a regime, any additional parameter that favors the efficiency of selection could lead to a switch toward a negative epistasis. This could be one of the bases of the evolution of the epistasis observed in regulatory networks in which the addition of sexual exchanges leads to a more efficient selection and concomitantly to a change of sign of epistasis toward negative epistasis (Azevedo et al. 2006). It would therefore be interesting to see how the presence of genetic exchange in polymorphic populations could affect MSFDE and affect the evolution of epistasis.
Impact of the evolution of Q on the mean fitness effect of mutations:
In the analysis presented before, we showed independently how Q influences the mean epistasis and how Q should evolve. If we assume that mutations affecting Q are very rare, then populations will reach MSFDE and then mutate Q step by step (in a process similar to the process used in adaptive dynamics). If we assume that the population is exactly at the equilibrium distance from the optimum, we can then see how epistasis evolves as the parameter Q evolves. As stated previously, the evolution of Q does not depend on the size of mutations in the phenotypic space, but the mean epistasis and the mean mutational effect on fitness do. We can therefore study the impact of the evolution of Q on both epistasis and fitness effects of mutations for several values of the mean distance that a mutation travels in the phenotypic space (parameter 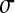 in our formalism). When drift dominates the process, whatever the “size” of mutations, Q decreases and so does the mean effect of mutations (Figure 6A). Hence selection for lower values of Q is consistent with selection for robustness under a strong drift regime. It is, however, worth noting that in such conditions fitness at MSFDE decreases with Q, something similar to “the paradox of robustness” suggested in the evolution of robustness (Frank 2007) but involving in addition a fitness decline (Gros and Tenaillon 2009).
in our formalism). When drift dominates the process, whatever the “size” of mutations, Q decreases and so does the mean effect of mutations (Figure 6A). Hence selection for lower values of Q is consistent with selection for robustness under a strong drift regime. It is, however, worth noting that in such conditions fitness at MSFDE decreases with Q, something similar to “the paradox of robustness” suggested in the evolution of robustness (Frank 2007) but involving in addition a fitness decline (Gros and Tenaillon 2009).
Figure 6.—
(A) Impact of Q on the mean effect of mutation on log-fitness at MSFDE when selection favors low values of Q. Populations are assumed to be at the mean distance for their value of Q at MSFDE. For all sizes of mutations in phenotypic space, the average effect of mutation on fitness decreases with decreasing Q. n = 10, N = 100, and the standard deviations of the mutation size in phenotypic space are 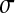 = 0.05 (solid line),
= 0.05 (solid line), 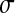 = 0.1,
= 0.1, 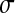 = 0.2,
= 0.2,  = 0.3, and
= 0.3, and 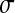 = 0.5 (dotted line). (B) Impact of Q on the mean effect of mutation on log-fitness at MSFDE when selection favors high values of Q. Depending on the size of mutations in phenotypic space, the average effect of mutation on fitness increases or decreases with increasing Q. n = 100, N = 30, and the standard deviations of mutation size in phenotypic space are
= 0.5 (dotted line). (B) Impact of Q on the mean effect of mutation on log-fitness at MSFDE when selection favors high values of Q. Depending on the size of mutations in phenotypic space, the average effect of mutation on fitness increases or decreases with increasing Q. n = 100, N = 30, and the standard deviations of mutation size in phenotypic space are 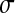 = 0.05 (solid line),
= 0.05 (solid line),  = 0.1,
= 0.1, 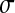 = 0.2,
= 0.2, 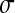 = 0.3, and
= 0.3, and  = 0.5 (dotted line).
= 0.5 (dotted line).
When selection is efficient and favors a high value of Q, we can see that the mean mutational effect on fitness either increases or decreases depending on the size of mutations (Figure 6B). If mutations are “small,” their effect on fitness will decrease with increasing Q, while when they are big the opposite is true. This is not expected if selection for epistasis is strictly equivalent to selection acting on the mean effect of mutation or genetic robustness. In fact, in such a long-term process, it is not the average effect of mutations on fitness that matters, but rather the fraction of mutations that could contribute to the population evolutionary fate, i.e., mutations that could be fixed. In this sense the evolution of the parameter Q always favors a decreasing effect on fitness of small size mutations.
Hence our analysis suggests that care should be taken in analyzing experimental data, as the mean effect of mutations that will be often used as the measure of robustness and used for epistasis might be a bad indicator of what has been selected for in populations.
Impact of the evolution of Q on the mean epistasis between pairs of mutations:
While the value of Q (compared to 2) reflects the sign of epistasis, the evolution of Q does not reflect how the intensity of epistasis evolves because epistasis also depends on the effects of mutations and on position in the phenotypic space. Similarly to what was done in the previous paragraph we can study how the intensity of epistasis evolves as Q moves toward its maximal or minimal value. When low values of Q are selected for, depending on the minimal value of Q, either strong positive epistasis will emerge or almost null epistasis for very low values of Q (Figure 7A). When selection favors high values of Q, it appears that strong negative epistasis is selected upon when mutations have large effects and almost no detectable epistasis is observed for small-effect mutations (Figure 7B).
Figure 7.—
(A) Impact of Q on the mean epistasis among mutations at MSFDE when selection favors low values of Q (same conditions as in Figure 6A). (B) Impact of Q on the mean epistasis among mutations at MSFDE when selection favors high values of Q (same conditions as in Figure 6B).
Interestingly, in a very large range of conditions, the model predicts that the intensity of epistasis should evolve to be significantly different from zero. Hence, under this asexual model of evolution, strong interactions among mutations should emerge in either direction depending on the intensity of drift. With such an increase in the interactions between mutations, our model suggests that the evolution of modularity, which would favor the lack of interactions between mutations, is unlikely in asexual populations. This observation is in good agreement with in silico models of evolution that showed that populations tend not to favor the evolution of modularity unless genetic exchanges (Misevic et al. 2006) or fluctuating environments (Kashtan and Alon 2005) are present.
Multiple modifiers of epistasis:
We previously focused our analysis on a global modifier of epistasis that would affect all traits simultaneously. Our formalism, however, allows us to study many independent modifiers of epistasis that would each affect independently m different subgroups of nk traits such that fitness would be defined as  . In such a system, each group of traits and its modifier evolve independently, according to its number of traits or phenotypic complexity. Small subgroups of traits will tend to evolve toward the maximal value of Q, as the effective drift is reduced for such parameters while a modifier of a large number of traits will tend to favor minimal values of Q. Hence a complex composite epistasis could emerge, depending on the range of action of modifiers (parameter
. In such a system, each group of traits and its modifier evolve independently, according to its number of traits or phenotypic complexity. Small subgroups of traits will tend to evolve toward the maximal value of Q, as the effective drift is reduced for such parameters while a modifier of a large number of traits will tend to favor minimal values of Q. Hence a complex composite epistasis could emerge, depending on the range of action of modifiers (parameter 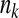 ).
).
Interestingly for a protein, it was shown that an increased stability results in a decreased effect of mutations and an increased negative epistasis as the first mutation is less likely to disrupt the protein function while pairs of mutations could still do it (Bloom et al. 2005; Bershtein et al. 2006). Hence it appears first that a local control of epistasis can be achieved by acting on protein stability and second that the observed stability of proteins suggests the existence of negative epistasis. A global control of epistasis could occur through the induction of chaperones. Once a badly folded protein is present, chaperones are induced to allow its proper refolding. This results in a strong positive epistasis (Maisnier-Patin et al. 2005), as once a deleterious mutation induces an increased level of chaperones, these helper proteins can also buffer the deleterious effects of additional mutations in other proteins. Hence, as suggested in our model, it appears that single-trait modifiers of epistasis favor negative epistasis while global regulators favor positive epistasis.
Concluding remarks:
The model we present here provides an interesting framework to study the distribution of epistasis, its influence on population evolution, and the selective pressure acting on epistasis. We have shown the existence of a strong link between epistasis and the mean effect of mutations on the logarithm of fitness and shown that at MSFDE, drift would largely influence the evolution of epistasis. More should be done to study how robustness, recombination, and mutation rates would be affected by the form of epistasis present in this model and reciprocally how they influence the evolution of epistasis.
Acknowledgments
We thank Erick Denamur, Guillaume Martin, Olivier Martin, and Olin Silander for helpful discussions. Simulations were performed at the “Centre de Calcul” of Institut Fédératif de Recherche (IFR) 02 in the Paris 7 Denis Diderot Medical School. This work was funded by the Agence Nationale de la Recherche (ANR-05-JCJC-0136-01). P.-A.G. was funded by the Délégation Générale à l'Armement.
References
- Azevedo, R. B., R. Lohaus, S. Srinivasan, K. K. Dang and C. L. Burch, 2006. Sexual reproduction selects for robustness and negative epistasis in artificial gene networks. Nature 440 87–90. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., and S. P. Otto, 2005. Evolution of recombination due to random drift. Genetics 169 2353–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerenwinkel, N., L. Pachter, B. Sturmfels, S. F. Elena and R. E. Lenski, 2007. Analysis of epistatic interactions and fitness landscapes using a new geometric approach. BMC Evol. Biol. 7 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershtein, S., M. Segal, R. Bekerman, N. Tokuriki and D. S. Tawfik, 2006. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature 444 929–932. [DOI] [PubMed] [Google Scholar]
- Bloom, J. D., J. J. Silberg, C. O. Wilke, D. A. Drummond, C. Adami et al., 2005. Thermodynamic prediction of protein neutrality. Proc. Natl. Acad. Sci. USA 102 606–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhoeffer, S., C. Chappey, N. T. Parkin, J. M. Whitcomb and C. J. Petropoulos, 2004. Evidence for positive epistasis in HIV-1. Science 306 1547–1550. [DOI] [PubMed] [Google Scholar]
- Burch, C. L., and L. Chao, 1999. Evolution by small steps and rugged landscapes in the RNA virus phi6. Genetics 151 921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch, C. L., and L. Chao, 2004. Epistasis and its relationship to canalization in the RNA virus phi 6. Genetics 167 559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher, D., 1995. Muller's ratchet, epistasis and mutation effects. Genetics 141 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., 1990. Mutation-selection balance and the evolutionary advantage of sex and recombination. Genet. Res. 55 199–221. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., M. T. Morgan and D. Charlesworth, 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser, J. A., and S. F. Elena, 2007. The evolution of sex: empirical insights into the roles of epistasis and drift. Nat. Rev. Genet. 8 139–149. [DOI] [PubMed] [Google Scholar]
- de Visser, J. A. G. M., R. F. Hoekstra and H. van den Ende, 1996. The effect of sex and deleterious mutations on fitness in Chlamydomonas. Proc. R. Soc. Lond. Ser. B Biol. Sci. 263 193–200. [Google Scholar]
- de Visser, J. A. G. M., R. F. Hoekstra and H. van den Ende, 1997. An experimental test for synergistic epistasis and its application in Chlamydomonas. Genetics 145 815–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denamur, E., and I. Matic, 2006. Evolution of mutation rates in bacteria. Mol. Microbiol. 60 820–827. [DOI] [PubMed] [Google Scholar]
- Desai, M. M., D. Weissman and M. W. Feldman, 2007. Evolution can favor antagonistic epistasis. Genetics 177 1001–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena, S. F., and R. E. Lenski, 1997. Test of synergistic interactions among deleterious mutations in bacteria. Nature 390 395–398. [DOI] [PubMed] [Google Scholar]
- Fisher, R. A., 1930. The Genetical Theory of Natural Selection. Oxford University Press, Oxford.
- Fisher, R. A. (Editor), 1958. The Genetical Theory of Natural Selection. Dover, New York.
- Frank, S. A., 2007. Maladaptation and the paradox of robustness in evolution. PLoS ONE 2 e1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, A., and A. T. Kalinka, 2006. Recombination and the evolution of mutational robustness. J. Theor. Biol. 241 707–715. [DOI] [PubMed] [Google Scholar]
- Goodnight, C. J., 1988. Epistasis and the effect of founder events on the additive genetic variance. Evolution 42 441–454. [DOI] [PubMed] [Google Scholar]
- Gros, P. -A. and O. Tenaillon, 2009. Selection for chaperone-like mediated genetic robustness at low mutation rate: impact of drift, epistasis and complexity. Genetics (in press). [DOI] [PMC free article] [PubMed]
- Haigh, J., 1978. The accumulation of deleterious genes in a population–Muller's ratchet. Theor. Popul. Biol. 14 251–267. [DOI] [PubMed] [Google Scholar]
- Hansen, T. F., and G. P. Wagner, 2001. Epistasis and the mutation load: a measurement-theoretical approach. Genetics 158 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl, D. L., and C. H. Taubes, 1996. Compensatory nearly neutral mutations: selection without adaptation. J. Theor. Biol. 182 303–309. [DOI] [PubMed] [Google Scholar]
- Hartl, D. L., and C. H. Taubes, 1998. Towards a theory of evolutionary adaptation. Genetica 102–103 525–533. [PubMed] [Google Scholar]
- Jasnos, L., and R. Korona, 2007. Epistatic buffering of fitness loss in yeast double deletion strains. Nat. Genet. 39 550–554. [DOI] [PubMed] [Google Scholar]
- Kashtan, N., and U. Alon, 2005. Spontaneous evolution of modularity and network motifs. Proc. Natl. Acad. Sci. USA 102 13773–13778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman, S., 1993. The Origin of Order. Oxford University Press, New York.
- Kimura, M., 1962. On the probability of fixation of mutant genes in a population. Genetics 47 713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., and T. Maruyama, 1966. The mutation load with epistatic gene interactions in fitness. Genetics 54 1337–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, A. S., 1993. Classification of hypotheses on the advantage of amphimixis. J. Hered. 84 372–387. [DOI] [PubMed] [Google Scholar]
- Kondrashov, A. S., 1994. Muller's ratchet under epistatic selection. Genetics 136 1469–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouyos, R. D., S. P. Otto and S. Bonhoeffer, 2006. Effect of varying epistasis on the evolution of recombination. Genetics 173 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouyos, R. D., O. K. Silander and S. Bonhoeffer, 2007. Epistasis between deleterious mutations and the evolution of recombination. Trends Ecol. Evol. 22 308–315. [DOI] [PubMed] [Google Scholar]
- Krakauer, D. C., and J. B. Plotkin, 2002. Redundancy, antiredundancy, and the robustness of genomes. Proc. Natl. Acad. Sci. USA 99 1405–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman, U., and M. W. Feldman, 2005. On the evolution of epistasis I: diploids under selection. Theor. Popul. Biol. 67 141–160. [DOI] [PubMed] [Google Scholar]
- Liberman, U., and M. W. Feldman, 2006. Evolutionary theory for modifiers of epistasis using a general symmetric model. Proc. Natl. Acad. Sci. USA 103 19402–19406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman, U., and M. Feldman, 2008. On the evolution of epistasis III: the haploid case with mutation. Theor. Popul. Biol. 73 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman, U., A. Puniyani and M. W. Feldman, 2007. On the evolution of epistasis II: a generalized Wright-Kimura framework. Theor. Popul. Biol. 71 230–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisnier-Patin, S., J. R. Roth, A. Fredriksson, T. Nystrom, O. G. Berg et al., 2005. Genomic buffering mitigates the effects of deleterious mutations in bacteria. Nat. Genet. 37 1376–1379. [DOI] [PubMed] [Google Scholar]
- Martin, G., and T. Lenormand, 2006. A general multivariate extension of Fisher's geometrical model and the distribution of mutation fitness effects across species. Evol. Int. J. Org. Evol. 60 893–907. [PubMed] [Google Scholar]
- Martin, G., S. F. Elena and T. Lenormand, 2007. Distributions of epistasis in microbes fit predictions from a fitness landscape model. Nat. Genet. 39 555–560. [DOI] [PubMed] [Google Scholar]
- Misevic, D., C. Ofria and R. E. Lenski, 2006. Sexual reproduction reshapes the genetic architecture of digital organisms. Proc. Biol. Sci. 273 457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1998. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution 52 935–949. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 1999. The evolutionary genetics of adaptation: a simulation study. Genet. Res. 74 207–214. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 2000. Adaptation and the cost of complexity. Evolution 54 13–20. [DOI] [PubMed] [Google Scholar]
- Orr, H., 2006. The distribution of fitness effects among beneficial mutations in Fisher's geometric model of adaptation. J. Theor. Biol. 238 279–285. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., and N. H. Barton, 2001. Selection for recombination in small populations. Evolution 55 1921–1931. [DOI] [PubMed] [Google Scholar]
- Poon, A., and L. Chao, 2004. Drift increases the advantage of sex in RNA bacteriophage Phi6. Genetics 166 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon, A., and S. P. Otto, 2000. Compensating for our load of mutations: freezing the meltdown of small populations. Evol. Int. J. Org. Evol. 54 1467–1479. [DOI] [PubMed] [Google Scholar]
- Sanjuan, R., and S. F. Elena, 2006. Epistasis correlates to genomic complexity. Proc. Natl. Acad. Sci. USA 103 14402–14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan, R., J. M. Cuevas, A. Moya and S. F. Elena, 2005. Epistasis and the adaptability of an RNA virus. Genetics 170 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella, G., and A. E. Hirsh, 2005. The application of statistical physics to evolutionary biology. Proc. Natl. Acad. Sci. USA 102 9541–9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silander, O. K., O. Tenaillon and L. Chao, 2007. Understanding the evolutionary fate of finite populations: the dynamics of mutational effects. PLoS Biol. 5 e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, O., B. Toupance, H. Le Nagard, F. Taddei and B. Godelle, 1999. Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria. Genetics 152 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, O., O. K. Silander, J. P. Uzan and L. Chao, 2007. Quantifying organismal complexity using a population genetic approach. PLoS ONE 2 e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nimwegen, E., J. P. Crutchfield and M. Huynen, 1999. Neutral evolution of mutational robustness. Proc. Natl. Acad. Sci. USA 96 9716–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, G. P., and W. Gabriel, 1990. Quantitative variation in finite parthenogenetic populations: What stops Muller's ratchet in the absence of recombination? Evolution 44 715–731. [DOI] [PubMed] [Google Scholar]
- Waxman, D., and J. J. Welch, 2005. Fisher's microscope and Haldane's ellipse. Am. Nat. 166 447–457. [DOI] [PubMed] [Google Scholar]
- Welch, D. B., and M. S. Meselson, 2001. Rates of nucleotide substitution in sexual and anciently asexual rotifers. Proc. Natl. Acad. Sci. USA 98 6720–6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke, C. O., 2001. Selection for fitness versus selection for robustness in RNA secondary structure folding. Evol. Int. J. Org. Evol. 55 2412–2420. [DOI] [PubMed] [Google Scholar]
- Wilke, C. O., and C. Adami, 2001. Interaction between directional epistasis and average mutational effects. Proc. R. Soc. Lond. Ser. B Biol. Sci. 268 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, J. B., E. D. Brodie, III and M. J. Wade, 2000. Epistasis and the Evolutionary Process. Oxford University Press, New York.









