Abstract
In Podospora anserina, inactivation of the respiratory chain results in a spectacular life-span extension. This inactivation is accompanied by the induction of the alternative oxidase. Although the functional value of this response is evident, the mechanism behind it is far from understood. By screening suppressors able to reduce the life-span extension of cytochrome-deficient mutants, we identified mutations in two zinc-cluster proteins, RSE2 and RSE3, which are conserved in other ascomycetes. These mutations led to the overexpression of the genes encoding the alternative oxidase and the gluconeogenic enzymes, fructose-1, 6 biphosphatase, and pyruvate carboxykinase. Both RSE2 and RSE3 are required for the expression of these genes. We also show that, even in the absence of a respiratory deficiency, the wild-type RSE2 and RSE3 transcription factors are involved in life-span control and their inactivation retards aging. These data are discussed with respect to aging, the regulation of the alternative oxidase, and carbon metabolism.
THE filamentous fungus Podospora anserina is a model organism in which life-span control has been extensively investigated. As in other organisms, it was clear from the beginning that life span is controlled by numerous external and genetic factors. Among these factors, mitochondrial activity seems to play a determinant role (reviewed in Lorin et al. 2006). But whereas mutations that compromise mitochondrial function in humans (reviewed in Wallace 2005) and in mice (Kujoth et al., 2005, 2006, 2007; Trifunovic et al. 2004, 2005) lead to a variety of pathological life-span-shortening diseases, in P. anserina they lead to a spectacular life extension. In this organism, all wild-type cultures exhibit an unavoidable arrest of vegetative growth systematically associated with large rearrangements in the mitochondrial DNA (mtDNA). Inactivation of respiratory complex III (mutant cyc1-1) (Sellem et al. 2007) or complex IV (mutant cox5∷ble) (Dufour et al. 2000) results in an extreme increase of life span (>30-fold) associated with a reduction in reactive oxygen species (ROS) levels and an increased stability of the mtDNA. In Caenorhabditis elegans, a class of mutants (Mit mutants) with disruptions (either genetic or mediated by RNA interference) in genes essential for the mitochondrial electron transport chain (ETC) are also long lived (reviewed in Rea 2005 and Rea et al. 2007).
How does the loss of genes critical for mitochondrial activity lead to life extension in P. anserina and C. elegans? One characteristic shared by C. elegans Mit mutants and P. anserina respiratory mutants is the activation of compensatory metabolic pathways in an attempt to supplement deficits in ETC function. Such pathways could produce less toxicity, e.g., by reducing mitochondrial ROS production or activating antioxidant mechanisms. In P. anserina, inactivation of genes essential for complex III or IV activity leads to the induction of an alternative oxidase (AOX) that catalyzes the transfer of electrons directly from the ubiquinol pool to oxygen and does not couple this transfer to proton translocation (Affourtit et al. 2002; Moore et al. 2002). Some phenotypic traits of the cox5∷ble and cyc1-1 mutants can be attributed to the following characteristics: reduced growth rate, loss of fertility, and reduced ROS production. In these mutants, only complex I is conserved as a site of proton gradient formation for ATP synthesis leading to a reduction of the energy yield associated with respiration. Furthermore, the alternative oxidase is thought to have an antioxidant role, preventing over-reduction of the mitochondrial quinone pool known to favor superoxide production (Maxwell et al. 1999).
The reasons for the spectacular long-lived phenotype of these mutants are more puzzling. One hypothesis proposed that the reduction of ROS and/or ATP production might be sufficient to account for life-span extension. Another hypothesis invoked a mitochondria-to-nucleus signaling pathway as the reason for this life-span extension (Lorin et al. 2001). The AOX is encoded in the nucleus and imported into mitochondria. In fungi, AOX expression has been extensively studied in Neurospora crassa and in P. anserina. The protein is not detectable under standard growth conditions. Its expression is strongly induced by mutations or chemicals that inhibit the ETC, and significant regulation occurs at the level of transcription (Lambowitz et al. 1989; Li et al. 1996; Lorin et al. 2001; Affourtit et al. 2002; Tanton et al. 2003; Descheneau et al. 2005; Chae et al. 2007a,b). In higher plants, AOX expression depends on developmental signals, stress conditions, and inhibition of the respiratory chain (reviewed in Clifton et al. 2006). The AOX expression therefore implies the existence of one or more pathways for transducing signals from the mitochondria to the nucleus to control the expression of the gene. The mechanisms by which mitochondria communicate with the nucleus have been referred to as retrograde signaling (Butow and Avadhani 2004; Liu and Butow 2006; Rhoads and Subbaiah 2007). In Saccharomyces cerevisiae, one retrograde pathway (the RTG pathway) has been extensively studied and shown to be an important determinant of life span (Kirchman et al. 1999).
In an attempt to clarify the relationships among AOX expression, retrograde signaling, and life span in P. anserina, we isolated fast-growing, short-lived revertants from the long-lived respiratory cox5∷ble- and cyc1-1-deficient mutants. We identified three mutations localized in two zinc-cluster transcription factors that control AOX expression both in P. anserina (this work) and in N. crassa (Chae et al. 2007b). Interestingly, these two mutations activate the expression of the alternative oxidase and also of gluconeogenic genes.
MATERIALS AND METHODS
P. anserina strains, growth conditions, transformation, and genetic analysis:
Except strain TS24 that is used for positional cloning, all the strains used in this study were derived from the s wild-type strain (Rizet 1952). The gpd-aox strain contains a transgenic copy of the aox gene under the control of the strong constitutive P.anserina gpd promotor associated with a hygromycin resistance cassette (Lorin et al. 2001). The long-lived cox5∷ble and cyc1-1 strains have been described in Dufour et al. (2000) and in Sellem et al. (2007), respectively. The ΔPaKu70 strain inactivated for the KU70 mammalian ortholog provides an efficient method for producing deletion mutants (El-Khoury et al. 2008). The TS24 strain used for positional cloning was obtained from the progeny of crosses between the P. anserina S (Rizet 1952) and the P. comata T (ATCC 36713) strains. TS24 exhibits the wild-type, fertile P. anserina phenotype and retained, on chromosome III, at least 12 simple sequence repeat markers (SSR) characteristic of the P. comata strain. The germination medium contains ground corn meal (50 g/liter), agar (12.5 g/liter), and ammonium acetate (4.4 g/liter). Minimal standard medium (M2) contains 1% dextrin as the carbon source (Esser 1974). When necessary, hygromycin, phleomycin, nourseothricin, and antimycin A were added to the medium at 75 μg/ml, 10 μg/ml, 50 μg/ml, and 10 μg/ml, respectively. Transformation experiments were conducted as previously described (Berges and Barreau 1989) on protoplasts obtained by incubation with 40 mg/ml glucanex (Laffort). Genetic methods for P. anserina have been described (Esser 1974). For the construction of double-mutant strains, the appropriate single mutants of opposite mating types were crossed. The Δrse2 Δrse3 strain carrying the two alleles inactivated by the same cassette conferring nourseothricin resistance and the double-mutant strain rse2-1 rse3-1 carrying the two alleles that both confer the ability to grow without delay on a medium supplemented with antimycin A were identified by analyzing the segregation of the cassette or the resistance to antimycin A in isolated asci.
Life-span measurements:
Life spans were measured on M2 medium on three to five subcultures derived from two to five independent spores exhibiting a given genotype. Cultures were grown in 30 ml/30 cm race tubes at 27° in the dark. The life span of a strain was defined in centimeters as the mean length (given with standard errors) of growth of parallel cultures between the point of the incubation of freshly germinated spores and the arrested edge of the dead culture. Survival curves, plotted as the percentage of surviving cultures in the course of time, also defined the life span (in days) as the time at which 50% of the cultures are still alive.
SSR markers and localization of gene rse2:
Twelve SSR markers overlapping 2 Mb on the long arm of chromosome III were found polymorphic between the P. anserina and P. comata isolates. Their characteristic markers are presented in supporting information, Table S1. To position gene rse2, crosses between (TS24) rse2+(comata origin) and (s) rse2-1 (anserina origin) parental strains provided us with a collection of monocaryotic spores for which linkage analysis was performed. The nature of the rse2 allele was determined by growth on antimycin A, and the nature of the 12 SSR markers was identified by PCR analysis. PCR amplifications were performed on rapid mini-preparations of DNA extracted from mycelium grown 24 hr after the germination of each spore.
Nucleic acid and protein manipulation:
Southern blots were done using total DNA extracted by the mini-preparation method (Lecellier and Silar 1994). Western blot analysis of the AOX protein was performed on isolated mitochondria as previously described (Sellem et al. 2007). Immunochemistry was performed with an anti-AOX mouse monoclonal antibody generated against the AOX of Sauromatum guttatum (Elthon et al. 1989). Additionally, blots were reprobed with an anti-βATPase rabbit antibody (a gift from J. Velours) as a standardization control. The bound antibodies were detected using an enhanced chemiluminescence detection system (Pierce Supersignal West picochemiluminescent substrate). Quantifications of the signal intensity that reflects the amount of protein were performed using the ImageQuant program on at least three independent blots (Molecular Dynamics, Amersham Bioscience, Piscataway, NJ).
Quantitative RT–PCR:
Total RNA from various strains grown for 48 hr on standard medium (1% dextrin) covered with cellophane disc was extracted using the RNeasy plant kit (Qiagen) with RLT buffer and DNAase I according to the manufacturer's instructions except that mycelium was broken with glass beads in a Fastprep apparatus (40 sec, intensity 6.5). For quantitative RT–PCR (qRT–PCR) analysis, 2 μg of RNA was reverse transcribed and random primed with oligo(dT)20 using the Supercript II reverse transcriptase (Invitrogen) according to the instructions of the supplier. Pairs of primers for PCR were developed for the aox (5′-GATGTCTGTTCCCCATCGAC-3′/5′-GAGGAAATGTTGGCAGTGGT-3′), gpd (5′-CACCGAGGACGAGATTGTCT-3′/5′-TCAGGGAGATACCAGCCTTG-3′), fbp (5′-CACCGGTGACTTTACGCTCC-3′/5′-GGAGAATTGGAGGGCGTGGC-3′), and pck (5′-ACCAAACCATCCGACATGC-3′/5′-GGTCTTGTTTACTGTGTTGA-3′) genes to give products of ∼40–50 bp in length. One primer of each set was designed across an exon/intron boundary to avoid amplification of any contaminating genomic DNA. The product of the first-strand cDNA reaction was diluted 10-fold before real-time PCR analysis. Amplifications were performed in duplicate in a LightCycler (Roche) using the LightCycler FastStart DNA MasterPLUS SYBR Green I kit (Roche) with no reverse transcriptase controls to estimate the contribution of contaminating DNA. Amplification efficiencies were measured for each primer pair and every set of amplification reactions. For each strain, the levels of aox, fbp, and pck transcripts were normalized using the gpd transcript level, which was used as a standard because its expression remained stable in all the strains and conditions examined. At least three independent experiments were performed from one to three different RNA preparations. For a given strain and a given gene, results are expressed as the level of expression of this gene in this strain relative to the level of expression of this gene in the wild-type strain.
Cloning of rse2 and rse3 genes:
PCR amplifications of the rse2 and rse3 mutated genes were performed using the primer pairs 5′-GGCTCGAGGACGGGAACCGGGAAG-3′/5′-GGGGACTAGTCGAAGGGGCGGCATTGTG-3′and 5′-CCCCCATGGCCGAGTAAA TACTGGATTTTG-3′/5′-CCCAGATCTGCCGCGTGACCAGGACC-3′. They were cloned in the XhoI/SpeI sites of the PBCHygro vector (Silar 1995) or in the NcoI/BglII sites of the pAPI508 vector (El-Khoury et al. 2008), respectively. Transformation of wild-type protoplasts resulted in hygromycin- or nourseothricin-resistant strains purified through genetic crosses with the wild type.
Inactivation of rse2 and rse3 genes:
Seven hundred nucleotides of the 5′ and 200 nucleotides of the 3′ region of the rse3 wild-type gene were amplified with the primers 5′-GAAAGCGGCCGCGTGACCAGGACCAAG-3′/5′-GGGCCATGGCTCTATCTGGACGG GACGGC-3′and 5′-GGGAGATCTGGAGTGCAGTTATACTTGG-3′/5′-CACGCGGCCGC TTTCGCCTCTTCTTTAAAC-3′, respectively, as described in El-Khoury et al. (2008) and cloned in the BglII/NcoI sites of the pAPI508 vector containing the nourseothricin resistance cassette. Protoplasts of the ΔKU70 strain were transformed and nourseothricin-resistant transformants were isolated and purified through a genetic cross with the wild-type. Nourseothricin resistance cosegregated with antimycin sensitivity. Following the same strategy, rse2 was inactivated using the primers 5′-AGGAAAAAAGCGGCCGCTGGGAAAGGGGAAGGAAG-3′/5′-GAAGATCGCAGTCGTTCGGCTTTGT-3′ for the 5′ region and 5′-AGGAAGCTTGGTGGGAGCATCGACAAA-3′/5′-AGGAAAAAAGCGGCCGCAAT CCGCCTCTCGGTCTT-3′ for the 3′ region and cloned in the HindIII/BglII sites of the pAPI508 vector.
RESULTS
Mutations in genes rse2 and rse3 restore senescence in cox5∷ble and cyc1-1 contexts:
Inactivation of complex III (cyc1-1) or complex IV (cox5∷ble) leads to a spectacular increase in life span associated with several phenotypic defects: alteration in germinating mycelium; poorly colored, thin growing mycelium; reduction of the growth rate; and female sterility (Dufour et al. 2000, Sellem et al. 2007). To shed light on the parameters responsible for these different characteristics, we isolated suppressor mutations able to improve the phenotype of these mutants. Spontaneous revertants were obtained independently as sectors of aerial fast-growing mycelium from cox5∷ble and cyc1-1 cultures. Most cultures grown in race tubes led to such sectors. Two of them, sectors 2 and 3, were isolated from cox5∷ble and cyc1-1 cultures, respectively. They were crossed with wild type to test the genetic basis of the reversion and to obtain pure revertant strains (the sectors probably contain a mixture of mutant and revertant nuclei). The presence of extragenic suppressors was revealed by recovery in the progeny of the crosses of three types of ascospores: ascospores that germinate to give sparse mycelium like the original mutant, ascospores that germinate normally, and ascospores that display an intermediate germinating mycelium giving rise to a growing mycelium similar to that of the initial sectors. Genetic analysis of the two pure revertant strains revealed that the two suppressor mutations named rse2-1 and rse3-1 were unlinked to the original cox5∷ble and cyc1-1 mutations. The characteristics of the revertants and of the strains carrying the suppressor mutations dissociated from the initial respiratory mutation are shown in Figure 1 and Table 1.
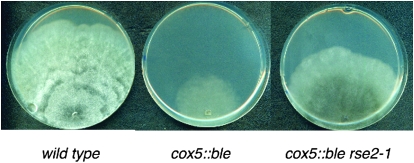
Figure 1.—
Mycelium aspect of the wild-type, cox5∷ble, and cox5∷ble rse2-1 strains. Petri plates of M2 medium were inoculated with an explant of each strain and incubated for 10 days at 27°. The wild-type strain exhibits a dense, aerial, colored mycelium and is fast growing whereas the cox5∷ble mutant exhibits a thin, poorly colored mycelium and is slow growing. The cox5∷ble rse2-1 revertant exhibits an intermediate phenotype.
TABLE 1.
Phenotypic properties of the cox5∷ble and cyc1-1 respiratory-deficient mutants and of the rse2-1 and rse3-1 suppressors
| Wild type | cox5∷ble | cyc1-1 | cox5∷ble rse2-1 | cyc1-1 rse3-1 | rse2-1 | rse3-1 | |
|---|---|---|---|---|---|---|---|
| Mycelium aspect | Aerial | Thin | Thin | Aerial | Aerial | Aerial | Aerial |
| Ascospore coloration | Black | Black | Colorlessa | Black | Colorlessa | Black | Black |
| Germination rateb | +++ | + | + | ++ | ++ | +++ | +++ |
| Growth ratec (cm/day) | 0.60 ± 0.03 | 0.21 ± 0.01 | 0.21 ± 0.01 | 0.48 ± 0.10 | 0.48 ± 0.11 | 0.50 ± 0.04 | 0.51 ± 0.04 |
| Female fertility | Fertile | Sterile | Sterile | Sterile | Sterile | Fertile | Fertile |
This phenotype exhibits a variable penetrance (Sellem et al. 2007).
Germination rate is given as +++ for wild-type thalli, + for sparse and slow-growing thalli, and ++ for thalli of intermediate phenotype on germination medium.
Growth rates are mean values ± standard deviation.
As shown in Table 1, the rse2-1 or rse3-1 mutations do not restore a wild-type phenotype to the cox5∷ble and cyc1-1 mutants. However, they considerably improve the germination of the ascospores (germinating thalli of cox5∷ble rse2-1 and cyc1-1 rse3-1 appeared more dense and grew better than germinating thalli of cox5∷ble and cyc1-1), they improve the aspect of the growing mycelium that appears more aerial and colored, and they restore a growth rate of 0.48 ± 0.1 cm/day compared with 0.21 ± 0.01 cm/day for the mutants. In contrast, they do not restore female fertility to the cox5∷ble and cyc1-1 mutants or normal ascospore pigmentation to cyc1-1. As P. anserina crosses yield dicaryotic ascospores, the recovery of heterocaryotic cox5∷ble rse2-1/cox5∷ble rse2+ and cyc1-1 rse3-1/cyc1-1 rse3+ ascospores allowed us to test the dominance/recessivity of the suppressors and to conclude that they are dominant with respect to the improved phenotypes.
Interestingly, analysis of longevity of the cox5∷ble rse2-1 and cyc1-1 rse3-1 strains revealed that the two suppressors also restored the senescence phenomenon (hence the name rse for restorator of senescence). Longevity of the revertants was ∼90 days (60 ± 10 cm) compared to ∼17 days (11.3 ± 1.6 cm) for the wild-type strain and >2 years (>300 cm) for the cox5∷ble and cyc1-1 mutants. An analysis of the mtDNA content of the senescent revertant cultures revealed the presence of mtDNA rearrangements called senDNAs as in senescent wild-type cultures (Belcour et al. 1999; Albert and Sellem 2002). However, in contrast to the wild-type strain in which senDNAα is systematically observed in a large amount, the senDNAs in the revertants originated mainly from the γ region as previously shown in other mutants (Lorin et al. 2001; Figure S3).
Recombination between cox5∷ble and rse3-1, on the one hand, and between cyc1-1 and rse2-1, on the other hand, revealed that either mutation, rse2-1 or rse3-1, is able to suppress complex III and complex IV loss-of-function mutations.
The mutations rse2-1 and rse3-1 are responsible for the constitutive expression of the alternative oxidase:
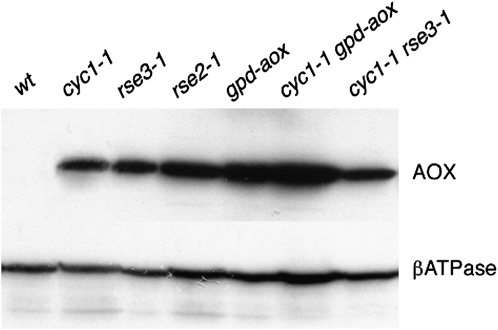
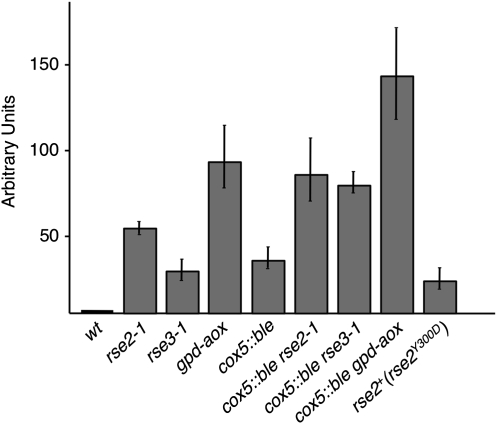
We previously showed that constitutive overexpression of the alternative oxidase in the cox5∷ble and cyc1-1 mutants improved mycelium aspect and growth rate and also restored the senescence process. This was demonstrated by expressing a fusion (gpd-aox) between the gpd promoter and the aox coding sequence in these mutants (Lorin et al. 2001; Sellem et al. 2007). Because of the similarities between the effects of the gpd-aox transgene and the rse2-1/rse3-1 mutations, the level of expression of the alternative oxidase was examined in strains carrying these mutations. The gpd-aox strain was used as a control. The Western blot analysis (Figure 2) corroborated previous results (Lorin et al. 2001) showing that AOX is undetectable in the wild type grown under standard conditions whereas it is induced in long-lived respiratory mutants (cyc1-1, for example, in Figure 2) and very strongly expressed in the gpd-aox strain. Our results show that it is also expressed in the strains carrying the rse2-1 or rse3-1 mutations. However, the AOX level is 2- to 3-fold lower in rse2-1 and rse3-1 strains than in strains carrying the gpd-aox transgene. These results were confirmed by qRT-PCR experiments shown in Figure 3. Expression levels of the aox gene were normalized to the gpd gene, and the aox mRNA copy number was given a value of 1 in the wild-type strain. Aox mRNA copy number increased ∼20-fold in rse3-1, 40-fold in rse2-1, and 60- to 80-fold in gpd-aox strains. In the cox5∷ble (and cyc1-1) strain, aox transcript levels were increased ∼20-fold compared to wild type. These levels were increased ∼3-fold in the presence of rse2-1 or rse3-1 mutations and ∼5-fold in the presence of the gpd-aox transgene. Altogether, these results are in accordance with data obtained by Western blot. The expression of the aox gene in rse2-1 and rse3-1 strains was also confirmed by testing the ability of these strains to grow on a medium containing antimycin A. Antimycin A is an inhibitor of complex III, leading to induction of the alternative oxidase in wild-type cells and allowing them to grow in the presence of the drug after a delay necessary for the induction whereas the gpd-aox, rse2-1, and rse3-1 strains grow without delay on this medium because of the constitutive expression of AOX. Heterocaryotic strains rse2-1/rse2+ and rse3-1/rse3+ also grow without a delay on medium containing antimycin A, confirming the dominance of these mutations. The double mutant rse2-1 rse3-1 was constructed by genetic cross, and it exhibited a phenotype very similar to that of each simple mutant, indicating the absence of a synergistic effect between the two mutations.
Figure 2.—
Western blot analysis of the AOX protein. Mitochondria (10 μg of mitochondrial protein) were extracted from the wild-type (wt), cyc1-1, rse3-1, rse2-1, gpd-aox, cyc1-1 gpd-aox, and cyc1-1 rse3-1 strains and loaded onto a 12% SDS–PAGE acrylamide gel. The AOX was revealed with a mouse antiserum against S. guttatum AOX provided by T. Elthon. As an internal control, the blot was reprobed with a rabbit anti βATPase provided by J. Velours.
Figure 3.—
Relative abundance of aox transcripts. For each strain, the levels of aox and gpd transcripts were determined by quantitative RT–PCR performed on one to three different RNA preparations (one to three replicates). For each experiment, the level of aox transcripts was normalized using the level of gpd transcripts as a reference. The graph shows the level of aox transcripts relative to the level of aox transcripts in the wild type for each strain. The error bars correspond to standard error. The rse2+(rse2Y300D) strain corresponds to a strain in which an ectopic copy of the mutated rse2Y300D has been integrated.
The rse2 and rse3 genes encode two zinc-cluster transcription factors:
Since the strains carrying the wild-type rse2+ and rse3+ or the mutated rse2-1 and rse3-1 alleles differ by their growth with or without delay on a medium containing antimycin A, the segregation of these alleles can be easily analyzed through crosses. Genetic analysis showed that rse2 and rse3 were localized on chromosomes III and IV, respectively, near ura5 for rse2 and near sir2 for rse3.
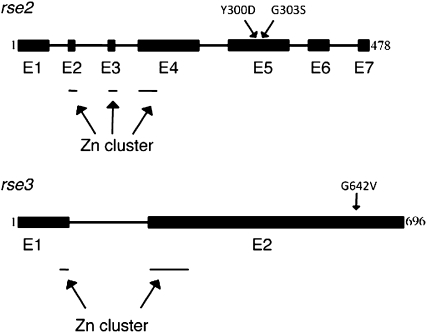
Taking advantage of the genome sequence of P. anserina (Espagne et al. 2008) and of the characterization on each of the seven chromosomes of marker polymorphisms between the geographic strains P. anserina and P. comata, segregation analysis of gene rse2 with the markers linked to gene ura5 was undertaken. A total of 198 monocaryotic spores derived from the cross rse2-1 (anserina) × rse2+(comata) were generated. The rse2-1/rse2+ segregation was determined by growth on antimycin A, and each spore was genotyped for 12 simple sequence repeat markers covering 2 Mb. All the polymorphic markers and the rse2 gene segregated in a 1:1 ratio. Of the 12 molecular markers, 2 always remained in parental association with the rse2 alleles. They covered ∼250 kb containing 67 predicted open reading frames (ORFs) and three encoding putative transcription factors of which one (Pa_3_6340) is a zinc-cluster transcription factor containing the canonical motif CX2CX6CX5-12CX2CX6-8C. Genomic DNA was prepared from rse2-1 and wild-type strains. PCR products that contained the ORF Pa_3_6340 were generated and sequenced. The rse2-1 gene was shown to contain a single T-to-G substitution changing a Tyr into an Asp codon at position 300. Sequence comparisons revealed that this protein is homologous to the recently reported AOD2 protein from N. crassa, which acts synergistically with another transcription factor of the zinc-cluster family, AOD5 (Chae et al. 2007b). This prompted us to search by BLAST the homolog of this protein in the P. anserina genome. The protein most related to AOD5 was Pa_4_8760 (54.7% identity) located on chromosome IV to which the rse3-1 mutation was genetically assigned. Sequencing of the corresponding gene in wild-type and rse3-1 strains revealed a G-to-T substitution changing a Gly into a Val codon at position 642 in the rse3-1 strain. The structure of the two genes and the position of the mutations are shown in Figure 4.
Figure 4.—
Structure of the rse2 and rse3 genes. The amino acids mutated in the rse2 gene (top) and the rse3 gene (bottom) are indicated. Exons (E) are shown as solid boxes and introns as solid lines. The first and the last amino acids of each protein are indicated. Sites within exons that contain a motif identified as a possible zinc cluster are indicated by thin lines below the genes.
To confirm that the Y300D mutation in the rse2-1 strain and the G642V mutation in the rse3-1 strain are responsible for the constitutive expression of the AOX and the restoration of senescence in strains deficient for the III/IV respiratory complex, we took advantage of the dominance of the two suppressor mutations. The mutated genes were cloned in plasmids pBCHygro (Silar et al. 1995) and pAPI508 (El-Khoury et al. 2008) and introduced into a wild-type strain by transformation. Hygromycin- and nourseothricin-resistant transformants resulting from a nonhomologous integration were selected for each transformation. These rse2+(rse2Y300D) and rse3+(rse3G642V) transformants, which carried an endogenous wild-type allele and an ectopic mutant allele, showed constitutive overexpression of the alternative oxidase [see rse2+(rse2Y300D) in Figure 3], revealing that the introduction of the Y300D mutation in the rse2 gene, or of the G642V mutation in the rse3 gene, is sufficient to cause this phenotype. Furthermore, strains with the cox5∷ble rse2+(rseY300D) genotype obtained by genetic cross exhibited the same growth phenotype and longevity as cox5∷ble rse2-1 (90 days compared to >2 years for cox5∷ble rse2+).
The rse2 and rse3 genes of eight other revertants derived from cox5∷ble or cyc1-1 cultures were sequenced. One revertant had a single base-pair substitution changing a glycine to a serine at position 303 in the rse2 gene, only three amino acids from the rse2Y300D mutation (Figure 4). The seven other revertants carried wild-type alleles for rse2 and rse3.
The rse2 and rse3 gene products also activate the expression of gluconeogenic genes:
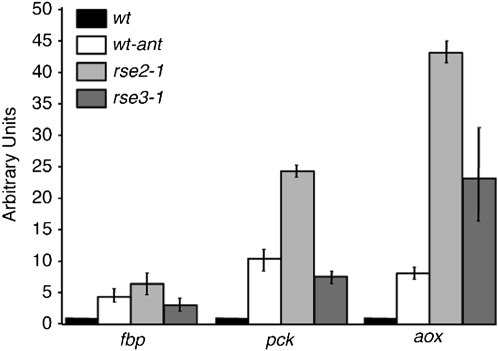
The rse2 and rse3 gene products are conserved in several ascomycetes (see Figure S1 and Figure S2). RSE2 is homologous to AOD2 from N. crassa (53% identity), AcuM from Aspergillus nidulans (34% identity), and RDS2 from S. cerevisiae (28.8% identity). Recently, it was shown that RDS2 in S. cerevisiae (Soontorngun et al. 2007) and AcuM and AcuK in A. nidulans (Hynes et al. 2007) act as activators of the expression of genes encoding central enzymes in the gluconeogenic pathway, in particular phosphoenolpyruvate carboxykinase (PCK) and fructose-1,6-biphosphatase (FBPase). PCK catalyzes an early step in gluconeogenesis and converts oxaloacetate to phosphoenolpyruvate, and FBPase catalyzes the final step in hexose monophosphate formation by dephosphorylating fructose-1,6-biphosphate to yield fructose-6-phosphate. This prompted us to examine the expression of the genes encoding these two enzymes in rse2Y300D, rse3G642V, and wild-type strains by quantitative RT–PCR. Two coding sequences potentially encoding PCK (Pa_4_3160) and FBPase (Pa_4_ 9360) were identified by homology searches. RNA was isolated from the rse2Y300D, rse3G642V, and wild-type strains grown on standard medium (containing 1% dextrin), and the abundance of transcript levels of the genes encoding Pa_4_ 9360 (Papck) and Pa_4_ 9360 (Pafbp) were examined in the three strains. The expression of the aox gene was determined in parallel and the level of transcripts expressed from the constitutive gpd gene was used as a reference control. Figure 5 shows the expression level of each gene in a given strain compared to the expression level of the gpd gene in the wild-type strain. These experiments corroborate the results shown in Figure 3: an increase of aox mRNAs levels of ∼40-fold in rse2Y300D and of ∼20-fold in rse3G642V. They also reveal that the expression of Papck and Pafbp is significantly increased in the mutant stains: for Papck ∼20-fold in rse2Y300D and ∼5-fold in rse3G642V and for Pafbp ∼5-fold in rse2Y300D and ∼2-fold in rse3G642V.
Figure 5.—
Quantification of fbp, pck, and aox expression. Total RNA was extracted from cultures of the wild-type strain grown under normal conditions (solid) or in the presence of antimycin A (10 μg/ml) (open) and from cultures of the rse2Y300D (light shading) and rse3G642V (dark shading) mutants grown under normal conditions. qPCR reactions were performed on cDNA to quantify the level of fbp, pck, aox, and gpd transcripts in each strain. Experiments were performed at least three times. As in Figure 3, in each experiment, the level of fbp, pck, and aox transcripts was normalized using the level of gpd transcripts as a reference. The diagram shows the level of fbp, pck, and aox transcripts in the different strains and culture conditions relative to the level of these transcripts in the wild type. The error bars correspond to standard error.
To be sure that the activation of the Papck and Pafbp genes in the mutant strains does not result from a qualitative change of the properties of the mutated transcription factors, the expression level of the two genes and of the aox gene was also tested in a wild-type strain grown on antimycin A. As shown in Figure 5, the expression of the aox, Papck, and Pafbp genes is increased 5-fold, 10-fold, and 3-fold, respectively, in the wild-type strain grown on antimycin A, indicating that Papck and Pafbp are indeed coregulated with the aox gene.
Genes rse2 and rse3 are nonessential in respiratory-competent strains grown in standard conditions but both are essential for induction of the alternative oxidase:
To determine the function of these transcription factors more clearly, strains deleted for the rse2 and rse3 genes were constructed by replacement with a cassette conferring resistance to nourseothricin (El-Khoury et al. 2008). The correct replacement was verified by Southern blot analysis (data not shown). Using rse2∷nat (Δrse2) and rse3∷nat (Δrse3) as parents in genetic crosses, we subsequently isolated the double-deleted strain Δrse2 Δrse3. All these strains were viable and displayed no impairment of growth, pigmentation, or fertility on standard synthetic M2 medium (the same mycelium aspect as the wild-type strain; cf. Figure 1). These results demonstrate that rse2 and rse3 are nonessential genes in respiratory-competent strains grown in standard conditions. However, none of these strains, including the simple Δrse2 and Δrse3 deletions, are able to induce the aox gene. This was demonstrated in two ways. First, none of the deleted strains is able to grow on a medium containing antimycin A even after the lag necessary for the wild type to begin growth. Second, the association by genetic cross of mutations cox5∷ble and Δrse2 or cox5∷ble and Δrse3 led to lethal spores that were unable to germinate. These results indicate that the inactivation of either of the two genes, rse2 and rse3, prevents the expression of the aox gene under inducing conditions and therefore that both proteins RSE2 and RSE3 are required for aox induction. Finally, we constructed the Δrse2 rse3G642V and rse2Y300D Δrse3 strains in which the deleted allele of one gene is associated with the mutated allele of the other. None of these strains was able to grow on a medium containing antimycin A, demonstrating that even when one of the two proteins is present in its mutant form, the other protein is still required to induce the alternative oxidase.
The RSE2 and RSE3 transcription factors are involved in life-span control even in the absence of mitochondrial dysfunction:
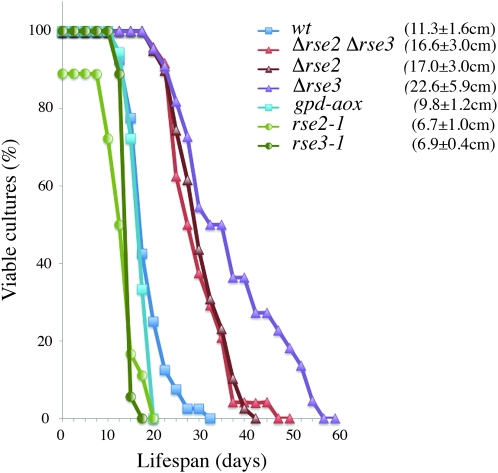
As stated above, the modification of the transcriptional pattern in the rse2Y300D and rse3G642V strains revealed that the suppressor mutations modify the expression or the activity of the RSE2 and RSE3 products. We examined the abundance of rse2 and rse3 transcript levels in the wild-type, rse2Y300D, and rse3G642V strains by quantitative RT–PCR experiments. Transcripts of both genes were virtually unchanged in the three strains (data not shown). This strongly supports the hypothesis of a modification of the activity and not of the abundance of the RSE2 and RSE3 products in the rse2Y300D and rse3G642V strains. In a respiratory-deficient context, cox5∷ble or cyc1-1, the rse2Y300D and rse3G642V mutations are responsible for severe life-span reduction. We investigated whether, in respiratory-competent strains, the rse2Y300D and rse3G642V mutations and the resulting gene expression modifications also lead to a modified life span. As shown in Figure 6, the rse2Y300D and rse3G642V strains displayed a decreased life span compared to wild type (∼12 vs. 17 days) whereas deletion of either of the rse genes results in an increased life span (∼30 days for Δrse2 and 35 days for Δrse3). These results suggest that the RSE products contribute to shortening life span and that the gene expression modifications due to the rse2Y300D and rse3G642V mutations accentuate this effect. It is very unlikely that the decreased life span of the rse2Y300D and rse3G642V strains results from the overexpression of the AOX, since as previously shown (Lorin et al. 2001 and Figure 6), we confirmed that the gpd-aox strain carrying the gpd-aox transgene displays the same longevity as the wild type.
Figure 6.—
Life-span analyses. For each genotype, at least 18 subcultures (representing the two mating types) were grown on M2 medium at 27° in race tubes. Data were plotted as the cumulative survival in time using Kaplan–Meier estimates. The mean longevity in centimeters ± standard error is in parentheses.
DISCUSSION
Characterization of mutations in two conserved zinc-cluster proteins that control the expression of the alternative oxidase and gluconeogenic genes:
We report in this study the characterization of mutations in two transcription factor genes, rse2 and rse3, each encoding a zinc-cluster protein controlling the induction of the alternative oxidase and the expression of gluconeogenic genes. These mutations were selected in a screen for suppressors of the long-lived respiratory mutants cox5∷ble and cyc1-1. They partially suppress the detrimental effects and restore the senescence process in these mutants. The three mutations reported here are located in two different genes; several other suppressors nonallelic to rse2 and rse3 were obtained but have not yet been studied in detail. Thus, the screen was not exhaustive. The two gene products contain Zn(2) Cys(6) binuclear cluster DNA-binding domains. Database searches and recent published data reveal that these genes are present in other ascomycetes. RSE2 corresponds to AOD2 from N. crassa, AcuM from A. nidulans, and RDS2 from S. cerevisiae. RSE3 corresponds to AOD5 from N. crassa and AcuK from A. nidulans.
We show here that the mutations rse2Y300D and rse3G642V confer higher levels of mRNA of the alternative oxidase and gluconeogenic genes compared to the wild-type strain and are dominant, strongly suggesting that they correspond to gain-of-function mutations. The mutations rse2Y300D and rse2G303S are not located in a conserved predicted functional domain (Schjerling and Holmberg 1996); however, their clustering pinpoints one region in RSE2 with potential significance for the function of this transcription factor and indicates that the integrity of this region is necessary to keep the transcription factor in a less-active form. Both the affected residues Y300 and G303 are conserved in aod2 (N. crassa), RDS2 (S. cerevisiae), and acuK (A. nidulans) and lie in a region of the protein highly conserved between the four organisms. In the same way, the G642 of rse3 is conserved in aod5 (N. crassa) and acuK (A. nidulans) and belongs to a short region that is conserved among the three organisms (see Figure S1 and Figure S2). The question how these single amino acid substitutions modulate the activity of the RSE2 and RSE3 transcription factors is unresolved. Although the levels of rse2 and rse3 mRNA in the mutant strains are unchanged, hypotheses such as an increased protein stability conferred by the mutations cannot be excluded. However, some other interesting possibilities can be proposed. For example, the rse3G642V mutation in the C-ter of RSE3 could make the transactivation domain more accessible or increase its intrinsic activation properties (Schjerling and Holmberg 1996). The mutations rse2Y300D and rse2G303S in RSE2 could reveal a latent activation domain or could change an interaction with an inhibitory protein. The hypothesis in which conserved motifs in zinc-cluster proteins could have an inhibitory role is based on several studies in S. cerevisiae showing that deletion of these motifs renders these proteins constitutively active (MacPherson et al. 2006).
Regarding the relationships between RSE2 and RSE3, the observation that the Δrse2 rse3+, Δrse3 rse2+, Δrse2 rse3G642, and rse2Y300D Δrse3 strains are unable to induce the alternative oxidase unambiguously demonstrate that both proteins are required for this induction even when one of them is present in a mutated form. This result agrees with observations reported for N. crassa in which neither the aod2 nor the aod5 mutants are able to induce AOX (Descheneau et al. 2005) and in A. nidulans in which neither the acuK nor the acuM mutants are able to induce PCK (Hynes et al. 2007). In N. crassa, electrophoretic mobility shift assays showed that AOD2 and AOD5 act synergistically to bind an alternative oxidase induction motif (AIM) present in the promoter of the aod-1 gene, which encodes the alternative oxidase. These data support the hypothesis that the two proteins interact with each other (Chae et al. 2007b). The AIM motif consists of one pair of CGG repeats separated by 7 bp and is essential for the inducible expression of the aod-1 gene. It is present in the upstream sequence of the aox gene of P. anserina and other Sordariales (Chae et al. 2007a); however, it is absent from the 900-bp upstream coding sequence of the Papck and Pafbp genes whose expression is also controlled by RSE2 and RSE3. Several explanations can be proposed for this observation. One possibility is that there is a cryptic motif that we have not spotted in the promoter of these genes. Another one is that activation of these genes requires other factors interacting with RSE2 and RSE3 and determining DNA-binding specificity. A third hypothesis is that RSE2 and RSE3 are indirect activators of the gluconeogenic genes by regulating the production of an inducing molecule.
RSE2, RSE3, and the control of longevity in P. anserina:
We have previously shown that in P. anserina, inactivation of genes encoding components of the cytochrome pathway leads to the induction of the alternative oxidase and to a spectacular increase of life span (Dufour et al. 2000; Sellem et al. 2007). We have also shown that the introduction of the gpd-aox transgene in the long-lived cox5∷ble and cyc1-1 leads to increased expression of the alternative oxidase in comparison with induced expression of this enzyme in the nontransgenic cox5∷ble mutant and to the restoration of senescence associated with an improvement of the phenotype (Lorin et al. 2001; Sellem et al. 2007). We show here that, in response to a block of the cytochrome pathway, there is induction of the aox gene but also of the pck and fbp gluconeogenic genes and that this induction is under the control of the two zinc-cluster proteins RSE2 and RSE3. These results therefore question the reasons for the very great life span of the respiratory-deficient mutants of P. anserina and the role of the different pathways that are induced in the control of longevity. The observation that gain-of-function mutations of genes rse2 and rse3 lead to a decreased life span whereas deletion of these genes, in particular of rse3, results in an increased life span, strongly suggests that some (direct or indirect) targets of the RSE2 and RSE3 proteins contribute to shortening life span. Although the aox gene is greatly induced in the rse2Y300D and rse3G642V mutants, it seems unlikely that the reduction of the life span of these mutants results from this induction because the gpd-aox transgenic strains show no reduction of life span. The aox gene is therefore probably not involved in the control of life span in a respiratory-competent context. In contrast, in a cox5∷ble or cyc1-1 context, we found a correlation among the mycelium phenotype, longevity, and AOX levels. Increased aox gene expression leads to a reduction in life extension and counters the detrimental phenotypic effects due to the cox5∷ble or cyc1-1 mutations. This positive correlation between the amount of AOX and the improvement of the phenotype of cox5∷ble and cyc1-1 mutants supports our proposed mechanism that increasing the electron flow through the alternative pathway is accompanied by increased oxygen consumption and increased ATP formation at the first coupling site (Lorin et al. 2001, 2006).
Many studies have demonstrated the central role of metabolic regulation in the aging process. While it is impossible to highlight all such studies, it is worth noting that a simple reduction of available glucose in the media results in life extension in P. anserina (Maas et al. 2004) and in yeast (Lin et al. 2000). More relevant to our discussion is that, in S. cerevisiae, a metabolic shift from glucose metabolism and fermentation toward respiration plays a central part in this life extension (Lin et al. 2002). In the same way, caloric restriction induces a metabolic reprogramming characterized by a transcriptional shift toward energy metabolism and upregulation of gluconeogenesis in mouse skeletal muscle (Lee et al. 1999). Recently, transcript profiling data from C. elegans dauer larvae and long-lived daf-2 mutant adults revealed increased expression of genes encoding gluconeogenic enzymes (McElwee et al. 2006). It is therefore possible that gluconeogenesis is a conserved pathway in the control of longevity in a wide spectrum of organisms.
A block of the respiratory cytochrome pathway is expected to lead to a wide spectrum of transcriptional changes in the cell. Our study shows that the two zinc-cluster proteins RSE2 and RSE3 are involved in this transcriptional response by activating the expression of the genes encoding the alternative oxidase and major enzymes of gluconeogenis. To identify the other genes whose expression is regulated by RSE2 and RSE3, transcriptome profiling of the strains carrying the different alleles (wild type, gain of function, deleted) of genes rse2 and rse3 will be conducted. This should allow us to gain insights into the physiological role of these zinc-cluster proteins and especially their role in the cellular response to a defect in respiratory function. We are convinced that these data will clarify the parameters involved in the control of the life span in P. anserina.
Acknowledgments
We thank Eric Migeon for help with the positional localization of the gene rse2 and Antoine Boivin and Ryad El-Khoury for help with the DNA constructions for deletion of the genes rse2 and rse3 and for useful discussion. We acknowledge Chris Herbert, Marc Maas, and Linda Sperling for critical reading of the manuscript. This work was supported by grants from the Association Française contre les Myopathies and grants from the European Community's Sixth Framework programme (LSHM-CT-200-512020).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.100834/DC1.
References
- Affourtit, C., M. S. Albury, P. G. Crichton and A. L. Moore, 2002. Exploring the molecular nature of alternative oxidase regulation and catalysis. FEBS Lett. 510 121–126. [DOI] [PubMed] [Google Scholar]
- Albert, B., and C. H. Sellem, 2002. Dynamics of the mitochondrial genome during Podospora anserina aging. Curr. Genet. 40 365–373. [DOI] [PubMed] [Google Scholar]
- Belcour, L., A. Sainsard-Chanet, C. Jamet-Vierny and M. Picard, 1999. Stability of the mitochondrial genome of Podospora anserina and its genetic control, pp. 209–228 in Mitochondrial Diseases, edited by P. Lestienne. Springer-Verlag, Berlin/Heidelberg, Germany/New York.
- Berges, T., and C. Barreau, 1989. Heat shock at an elevated temperature improves transformation efficiency of protoplasts from Podospora anserina. J. Gen. Microbiol. 135 601–604. [DOI] [PubMed] [Google Scholar]
- Butow, R. A., and N. G. Avadhani, 2004. Mitochondrial signaling: the retrograde response. Mol. Cell 14 1–15. [DOI] [PubMed] [Google Scholar]
- Chae, M. S., C. C. Lin, K. E. Kessler, C. E. Nargang, L. L. Tanton et al., 2007. a Identification of an alternative oxidase induction motif in the promoter region of the aod-1 gene in Neurospora crassa. Genetics 175 1597–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae, M. S., C. E. Nargang, I. A. Cleary, C. C. Lin, A. T. Todd et al., 2007. b Two zinc-cluster transcription factors control induction of alternative oxidase in Neurospora crassa. Genetics 177 1997–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton, R., A. H. Millar and J. Whelan, 2006. Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim. Biophys. Acta 1757 730–741. [DOI] [PubMed] [Google Scholar]
- Descheneau, A. T., I. A. Cleary and F. E. Nargang, 2005. Genetic evidence for a regulatory pathway controlling alternative oxidase production in Neurospora crassa. Genetics 169 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour, E., J. Boulkay, V. Rincheval and A. Sainsard-Chanet, 2000. A causal link between respiration and senescence in Podospora anserina. Proc. Natl. Acad. Sci. USA 97 4138–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khoury, R., C. H. Sellem, E. Coppin, A. Boivin, M. F. Maas et al., 2008. Gene deletion and allelic replacement in the filamentous fungus Podospora anserina. Curr. Genet. 53 249–258. [DOI] [PubMed] [Google Scholar]
- Elthon, T. E., R. L. Nickels and L. McIntosh, 1989. Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol. 89 1311–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espagne, E., O. Lespinet, F. Malagnac, C. Da Silva, O. Jaillon et al., 2008. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 9 R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser, K., 1974. Podospora anserina, pp. 531–551 in Handbook of Genetics, edited by R. C. King. Plenum Press, New York.
- Hynes, M. J., E. Szewczyk, S. L. Myrray, Y. Suzuki, M. A. Davis et al., 2007. Transcriptional control of gluconeogenesis in Aspergillus nidulans. Genetics 176 139–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman, P. A., S. Kim, C. Y. Lai and S. M. Jazwinski, 1999. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics 152 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth, G. C., A. Hiona, T. D. Pugh, S. Someya, K. Panzer et al., 2005. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309 481–484. [DOI] [PubMed] [Google Scholar]
- Kujoth, G. C., C. Leeuwenburgh and T. A. Prolla, 2006. Mitochondrial DNA mutations and apoptosis in mammalian aging. Cancer Res. 66 7386–7389. [DOI] [PubMed] [Google Scholar]
- Kujoth, G. C., P. C. Bradshaw, S. Haroon and T. A. Prolla, 2007. The role of mitochondrial DNA mutations in mammalian aging. PLoS Genet. 3 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz, A. M., J. R. Sabourin, H. Bertrand, R. Nickels and L. McIntosh, 1989. Immunological identification of the alternative oxidase of Neurospora crassa mitochondria. Mol. Cell. Biol. 9 1362–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecellier, G., and P. Silar, 1994. Rapid methods for nucleic acids extraction from petri dish-grown mycelia. Curr. Genet. 25 122–123. [DOI] [PubMed] [Google Scholar]
- Lee, C. K., R. G. Klopp, R. Weindruch and T. A. Prolla, 1999. Gene expression profile of aging and its retardation by caloric restriction. Science 285 1390–1393. [DOI] [PubMed] [Google Scholar]
- Li, Q., R. G. Ritzel, L. L. McLean, L. McIntosh, T. Ko et al., 1996. Cloning and analysis of the alternative oxidase gene of Neurospora crassa. Genetics 142 129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. J., P. A. Defossez and L. Guarente, 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289 2126–2128. [DOI] [PubMed] [Google Scholar]
- Lin, S. J., M. Kaeberlein, A. A. Andalis, L. A. Sturtz, P. A. Defossez et al., 2002. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418 344–348. [DOI] [PubMed] [Google Scholar]
- Liu, Z., and R. A. Butow, 2006. Mitochondrial retrograde signaling. Annu. Rev. Genet. 40 159–185. [DOI] [PubMed] [Google Scholar]
- Lorin, S., E. Dufour, J. Boulay, O. Begel, S. Marsy et al., 2001. Overexpression of the alternative oxidase restores senescence and fertility in a long-lived respiration-deficient mutant of Podospora anserina. Mol. Microbiol. 42 1259–1267. [DOI] [PubMed] [Google Scholar]
- Lorin, S., E. Dufour and A. Sainsard-Chanet, 2006. Mitochondrial metabolism and aging in the filamentous fungus Podospora anserina. Biochim. Biophys. Acta 1757 604–610. [DOI] [PubMed] [Google Scholar]
- Maas, M. F. P. M., H. J. De Boer, A. J. M. Debets and R. F. Hoekstra, 2004. The mitochondrial plasmid pAL2–1 reduces calorie restriction mediated life span extension in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 41 865–871. [DOI] [PubMed] [Google Scholar]
- MacPherson, S., M. Larochelle and B. Turcotte, 2006. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70 583–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell, D. P., Y. Wang and L. McIntosh, 1999. The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. USA 96 8271–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee, J. J., E. Schuster, E. Blanc, J. Thornton and D. Gems, 2006. Diapause-associated metabolic traits reiterated in long-lived daf-2 mutants in the nematode Caenorhabditis elegans. Mech. Ageing Dev. 127 458–472. [DOI] [PubMed] [Google Scholar]
- Moore, A. L., M. S. Albury, P. G. Crichton and C. Affourtit, 2002. Function of the alternative oxidase: Is it still a scavenger? Trends Plant Sci. 7 478–481. [DOI] [PubMed] [Google Scholar]
- Rea, S. L., 2005. Metabolism in the Caenorhabditis elegans Mit mutants. Exp. Gerontol. 40 841–849. [DOI] [PubMed] [Google Scholar]
- Rea, S. L., N. Ventura and T. E. Johnson, 2007. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 5 e259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads, D. M., and C. C. Subbaiah, 2007. Mitochondrial retrograde regulation in plants. Mitochondrion 7 177–194. [DOI] [PubMed] [Google Scholar]
- Rizet, G., 1952. Les phénomènes de barrage chez Podospora anserina. I. Analyse génétique des barrages entres les souches S et s. Rev. Cytol. Biol. Veget. 13 51–92. [Google Scholar]
- Schjerling, P., and S. Holmberg, 1996. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 24 4599–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellem, C. H., S. Marsy, A. Boivin, C. Lemaire and A. Sainsard-Chanet, 2007. A mutation in the gene encoding cytochrome c1 leads to a decreased ROS content and to a long-lived phenotype in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 44 648–658. [DOI] [PubMed] [Google Scholar]
- Silar, P., 1995. Two new easy to use vectors for transformation. Fungal Genet. Newsl. 42 73. [Google Scholar]
- Soontorngun, N., M. Larochelle, S. Drouin, F. Robert and B. Turcotte, 2007. Regulation of gluconeogenesis in Saccharomyces cerevisiae is mediated by activator and repressor functions of Rds2. Mol. Cell. Biol. 27 7895–7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanton, L. L., C. E. Nargang, K. E. Kessler, Q. Li and F. E. Nargang, 2003. Alternative oxidase expression in Neurospora crassa. Fungal Genet. Biol. 39 176–190. [DOI] [PubMed] [Google Scholar]
- Trifunovic, A., A. Wredenberg, M. Falkenberg, J. N. Spelbrink, A. T. Rovio et al., 2004. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429 417–423. [DOI] [PubMed] [Google Scholar]
- Trifunovic, A., A. Hansson, A. Wredenberg, A. T. Rovio, E. Dufour et al., 2005. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc. Natl. Acad. Sci. USA 102 17993–17998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, D. C., 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39 359–407. [DOI] [PMC free article] [PubMed] [Google Scholar]