Abstract
Cell adhesion is the fundamental driving force that establishes complex cellular architectures, with the nervous system offering a striking, sophisticated case study. Developing neurons adhere to neighboring neurons, their synaptic partners, and to glial cells. These adhesive interactions are required in a diverse array of contexts, including cell migration, axon guidance and targeting, as well as synapse formation and physiology. Forward and reverse genetic screens in the fruit fly Drosophila have uncovered several adhesion molecules that are required for neural development, and detailed cell biological analyses are beginning to unravel how these factors shape nervous system connectivity. Here we review our current understanding of the most prominent of these adhesion factors and their modes of action.
Key words: drosophila, cell adhesion, nervous system, glia, axon, synapse
Introduction
Cell adhesion is the main force that sorts cells into distinct functional groups, a prerequisite for the establishment and maintenance of tissues and organs. In this context, adhesion plays many roles, affecting the survival and proliferation of cells through interactions with the substrate, as well as controlling the morphogenesis and assembly of the more complex cellular arrangements seen in all organs. Intriguingly, relative to our knowledge of the plethora of functions mediated by cell signaling pathways, our understanding of the mechanisms that underlie how cell adhesion influences development remains fragmentary. Here we examine our understanding of cell adhesion in the nervous system because it is a prominent example of both the power and complexity of adhesive interactions to define structure.
Neurons and their processes must navigate within a complex three-dimensional environment, where they undergo selective interactions with neighboring neurons, including both synaptic and non-synaptic partners, as well as with glia. These interactions regulate axon guidance, dendrite elaboration, fasciculation patterns, layer-specific targeting, as well as choice of synaptic partners. Furthermore, adhesive contacts also continue into adult life, and are critical for nervous system function and maintenance. As individual neurons use adhesion in multiple contexts and environments, these interactions must be both diverse and distinct. Here we outline some of this functional diversity, and discuss how adhesion might be regulated in vivo.
Drosophila provides a useful model to study cell adhesion in vivo for three reasons. First, both the anatomy, as well as the function of many individual circuits has been described. Second, a number of sophisticated somatic mosaic techniques allow targeted genetic manipulation of single cells in the context of an otherwise wild-type animal. These technologies are especially useful as many cell adhesion molecules are broadly expressed and have pleiotropic functions.1 Finally, the Drosophila brain is both stereotyped from animal to animal, and genetically “hard-wired”, making the precise regulation of adhesion mechanisms critical to normal development. Thus, genetic studies in the fruit fly have been particularly informative, and form the focus of this review.
Fundamentals of Adhesion
The role of physical interactions in shaping cells and tissues has been a subject of considerable thought for more than 90 years.2 Initial studies examined how surface tension shapes cellular geometry, and led to critical insights that continue to influence our thinking today. Foremost among these was the notion of equilibrium, or minimum free energy, in which differences in surface tension between cells could cause some cell surfaces to contract, and others to enlarge.2 However, these initial ideas arose in the absence of any knowledge of the molecules that might alter the “surface tension” between two cellular surfaces, and did not examine the complexities that become apparent when considering the elaborate geometries of cells in the nervous system. More recently, both in vitro and in vivo studies of non-neuronal cells have begun to shed light on how adhesion molecules, particularly members of the cadherin superfamily, can provide the driving forces necessary for cellular reorganizations.3 In order to minimize free energy, cells with stronger adhesive force will engage in more stable interactions with each other than with neighbors that have weaker adhesive capacity. These interactions will sort the two cell populations, causing cells with stronger interactions to be surrounded by cells with weaker interactions.3 Differences in adhesive force can be achieved either qualitatively, by expression of different adhesion factors, or quantitatively, by expression of varying levels of the same factor.4,5 Similar principles apply in vivo, where E-cadherin and N-cadherin dictate the spatial arrangement of cells in the Drosophila oocyte and the eye.6,7 These latter studies also demonstrated that by restricting N-cadherin expression to a specific cell type, differential adhesive interactions can occur amongst subsets of cells within a larger structure. This specificity allows cells expressing the same cadherins to arrange themselves by minimizing their mutual surface tensions in a highly localized way, within the in vivo environment.7 Similar interactions may play important roles in shaping the fine structure of the nervous system and we anticipate that as adhesion factors are identified and characterized in more detail, many of their actions will be attributable to differential adhesion. The challenge that we are only beginning to confront is how all of these adhesive cues might be integrated by developing neurites into “decisions” about where to project.
Adhesive Interactions Amongst Neurons in the Developing Nervous System
Many genetic screens for mutations affecting the establishment of neuronal connectivity have identified cell adhesion molecules of many different molecular families. Foremost among these are members of the cadherin superfamily, Immunoglobulin (Ig) domain containing proteins and members of the leucine-rich repeat (LRR) family. We focus on three contexts in which these molecules have been studied extensively, examining the formation of specific connections in the adult visual system and the neuromuscular junction, and discussing the use of cell adhesion in neuronal self-recognition. We further narrow our discussion to molecules that have been shown to directly mediate adhesion, without excluding the possibility that they might act as signaling molecules as well. However, at present, the signaling pathways that might be regulated by these molecules have not been studied in flies. Finally, it is important to recognize that drawing a distinction between adhesion and signaling molecules becomes somewhat arbitrary as one begins to consider that cell signaling pathways can regulate cell adhesion and vice versa.
The structure and development of the adult visual system.
The visual system has provided a facile system to examine the molecular mechanisms by which developing axons choose appropriate synaptic partners. These choices are informed by adhesive interactions between axons and their targets, and surprisingly, also amongst afferent axons themselves. Here we briefly review the anatomy of the system, and then describe these interactions, and their molecular mechanisms.
The Drosophila visual system comprises the retina and four optic ganglia, the lamina, medulla, lobula and lobula plate8 (Fig. 1A). The retina consists of approximately 800 subunits, ommatidia, each containing eight distinct photoreceptor neurons, designated R1–R8. Photoreceptor axons from each ommatidium form a fascicle that innervates the optic lobe in a retinotopic pattern. R1–R6 cells project to the most peripheral optic neuropil, the lamina, while the R7 and R8 cells send their projections through the lamina into the medulla8,9 (Fig. 1A). During development, R1–R6 cells initially stop between two layers of glial cells in the lamina. Subsequently, each R cell growth cone defasciculates, and extends away from the ommatidial bundle along a unique trajectory to join a neighboring column of post-synaptic targets (Fig. 1A). R1–R6 axons, together with their post-synaptic targets, then form a new fascicle, called a cartridge, and initiate synapse formation. By contrast, R7 and R8 target within distinct layers in the medulla. These targeting choices emerge in two steps: initially both R7 and R8 axons target to relatively shallow, temporary layers.9 Then, after additional medulla layers assemble, both R7 and R8 axons extend deeper to their final target layers where they form stable connections with higher order neurons.8 Thus, the adult visual system captures at least two distinct types of synaptic specificity: cartridge formation in the lamina, and layer-specific targeting in the medulla.
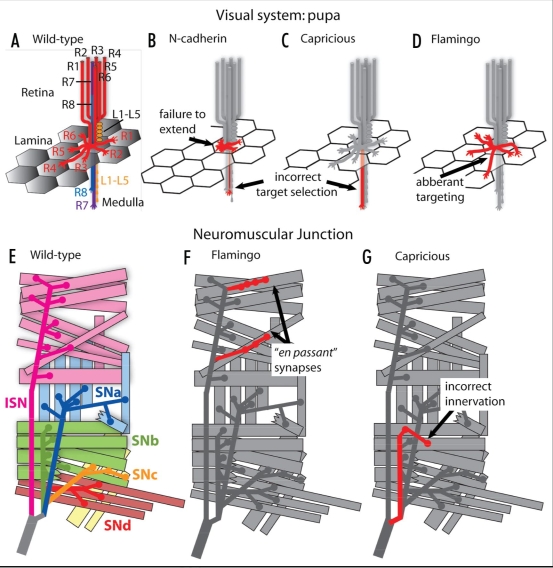
Figure 1.
Anatomy of the visual system and the neuromuscular junction. (A) Perspective diagram of R cell axons projecting out of the retina, into the lamina and medulla, during pupal development in wild type. R cells (R1–R8) and lamina neurons (L1–L5). (B) Illustrative example of R1–R6 axons failing to extend to targets in the lamina, and R7 axons innervating the inappropriate target layer in the medulla, as would be seen in N-cadherin mutants. (C) Example of R8 axons failing to choose the appropriate layer in the medulla, as seen in a capricious mutant. (D) Example of R1–R6 axons extending to aberrant targets in the lamina, as seen in flamingo mutants. (E) Anatomy of the wild-type neuromuscular junctions in a single larval hemisegment. (F) Example of en passant synapse formation, as seen in flamingo mutants. (G) Example incorrect innervation of the target field, as seen in capricious mutants.
Adhesive interactions in the developing visual system.
The molecular mechanisms that underlie the formation of these connections are incompletely understood. However, three different cell adhesion molecules, the classical cadherin N-cadherin, the leucine-rich repeat adhesion molecule Capricious, and the protocadherin Flamingo play important roles.10–12 N-cadherin is required both for cartridge formation and layer-specific targeting while Capricious regulates layer-specific targeting, and Flamingo cartridge formation. Moreover, N-cadherin and Capricious mediate interactions between R cell axons and their targets, whereas Flamingo mediates interactions only amongst R cell axons. Here we consider the functions of each one in turn.
N-cadherin is broadly expressed in the nervous system, and homozygous mutant animals display pleiotropic defects in axon fasciculation and guidance.13 On the other hand, mosaic studies demonstrated specific roles for N-cadherin in particular aspects of R cell axon targeting10,14 (Fig. 1B). These studies revealed that N-cadherin was required for R1–R6 axons to reach their appropriate lamina cartridges, as well as for R7 axons (but not R8 axons) to reach the correct layer of the medulla. Subsequent analyses revealed that N-cadherin is also required in lamina and medulla neurons for R cell axon targeting.15,16 Finally, these studies demonstrated that N-cadherin mediates homophilic, attractive interactions between R cell axons and their targets, providing the first functional evidence for such an N-cadherin mediated interaction in any context.15
N-cadherin activity is subject to sophisticated, dynamic and cell-type specific regulation of its expression level and sub-cellular localization. In the context of R1–R6 axon targeting in the lamina, N-cadherin adhesion appears to be asymmetrically regulated within the growth cone to allow for defasciculation and lateral movement towards the correct targets.17 This lateral extension is dependent upon two additional proteins, the receptor tyrosine phosphatase LAR, and the synaptic scaffolding molecule Liprin-α, both of which are required in R cells for target selection, raising the possibility that they could interact with N-cadherin.17 Additional complexity has been revealed through studies of N-cadherin in other visual neurons. In particular, N-cadherin functions in lamina neurons, controlling the targeting of their axons to specific layers in the medulla.18 Individual lamina neurons have complex and varied N-cadherin requirements, with some requiring N-cadherin cell-autonomously while others necessitate N-cadherin function in their neighbors. At least part of this complexity is achieved by dynamic regulation of N-cadherin localization to specific neuronal processes. These data support a model where cell- and stage-specific modulation of N-cadherin controls discrete targeting steps. Finally, it is also likely that N-cadherin insertion into the plasma membrane is regulated in developing R cell axons, perhaps through the actions of the exocyst complex.19 These studies underscore that molecules such as N-cadherin do not simply act as “on and off” cellular adhesives, but rather that they are under complex regulation at multiple levels. Future studies will undoubtedly uncover both additional regulatory mechanisms as well as provide molecular details regarding the known mechanisms.
The adhesive leucine-rich repeat protein Capricious plays an instructive role in R8 cell layer-specific targeting in the medulla.11 In particular, capricious loss-of-function mutations cause targeting errors in R8 axons, with many extending laterally and others terminating in inappropriate medulla layers (Fig. 1C). In contrast, the layer-specific targeting of R7 axons is unaffected by capricious mutations. Consistent with a function restricted to R8, Capricious protein is expressed in R8 axons, and their target layer, but not in R7 cells, or their recipient layer. Remarkably, Capricious is sufficient to specify axonal targeting, as ectopic expression of Capricious in R7 cells redirects them to the Capricious-positive R8 layer. This implies that by mediating afferent-target interactions, Capricious provides a cue that is both necessary and sufficient to specify targeting to a single layer, implying that other layer-specific cues remain to be discovered.
Flamingo encodes an unusual, evolutionarily conserved member of the cadherin superfamily, containing both cadherin repeats and a seven trans-membrane domain. Initial studies of Flamingo focused on its roles in epithelial planar cell polarity and dendrite patterning in the peripheral nervous system of the embryo and larva.20–23 Flamingo also plays at least three distinct roles in photoreceptor target selection.12,24,25 First, Flamingo regulates the spacing of R cell axon bundles, establishing the local topography of cartridges and columns in the lamina and the medulla, respectively.12,24 Second, Flamingo appears to be required for layer-specific targeting. In flamingo mutants, R8 axons (but not R7 cells) fail to reach their target layer and instead terminate in more superficial layers of the medulla.24 Finally, Flamingo mediates homophilic interactions among R1–R6 axons that guide these axons to their appropriate target cartridges12,25 (Fig. 1D). Detailed mosaic analyses have provided new insight into Flamingo's mechanism of action. In particular, Flamingo acts exclusively in a cell non-autonomous fashion, mediating afferent-afferent interactions that take place prior to growth cone extension.25 These studies demonstrated that while Flamingo function in any single growth cone is dispensable for normal target selection, individual growth cones are sensitive to differences in Flamingo activity between themselves and their neighbors. In effect, individual growth cones balance Flamingo-mediated interactions with their immediate neighbors, creating a form of “opponency” where the strength of the interaction with one neighbor is opposed by an interaction with another neighbor. This simple model provides a mechanism by which differences in adhesivity between cells, perhaps reflecting differences in cell fate, can be translated into changes in growth cone orientation necessary for appropriate target selection. While Flamingo's molecular structure, containing a seven pass trans-membrane domain similar to those of G-protein coupled receptors, suggests that it functions as more than simply an adhesive factor, there is no evidence in any experimental context that it acts as a signaling receptor. In summary, the cellular opponency model that emerged from studies of Flamingo in R cell axons provides a powerful framework for considering homophilic, adhesive interactions more broadly.
The structure and development of the neuromuscular junction.
Adhesive interactions also play critical roles in patterning the connections between motor neurons and their muscle targets in the developing embryo and larva. The abdominal body wall of embryos and larvae is segmented, with each segment containing more than 30 bilaterally symmetrical muscle fibers, targeted in a precise pattern by approximately 40 identified motor neurons (Fig. 1E). Each fiber is unique, and can be defined by morphological criteria.26 Synapses between motor neurons and muscles, the neuromuscular junctions (NMJ), form during embryogenesis and reflect a developmental sequence of axon guidance decisions made at a series of so-called “choice points.” These decisions culminate in the arrival of each motor neuron at a specific muscle target, and the formation of a NMJ of unique morphology. Thus, studies of cell adhesion in this context have focused both on the roles of adhesive interactions in shaping axon guidance decisions, and regulating the strength and patterning of the NMJ.
Adhesive interactions at the neuromuscular junction.
As in the visual system, our understanding of the molecular factors that shape the NMJ are only partly understood. Here we focus on just two adhesion molecules where both gain and loss-of function studies demonstrate critical developmental roles, namely Flamingo and Capricious.
Recent work has linked Flamingo to target selection and synaptogenesis at the NMJ.27 Homozygous mutant animals display increased numbers of ectopic synapses, abnormal “en passant” synapses distal to the axon terminal, as well as pre-synaptic varicosities within normally asynaptic axonal segments (Fig. 1F). These observations suggest that Flamingo suppresses synapse formation in this context. Intriguingly, this study also described a function for Flamingo in the maintenance of axons and synapses as homozygous mutant animals display age-dependent axon degeneration and loss of muscle innervation. Thus, Flamingo acts differently at the NMJ in comparison to the visual system, specifically affecting synapse formation in one case, while affecting target selection in the other.
Capricious acts as a molecular matchmaker between pre- and post-synaptic cells at the neuromuscular junction, a function similar to its role in the visual system. In the embryo, Capricious is expressed in a small number of muscles and neurons. In one well-studied case, Capricious is expressed in both a single muscle and the motor neurons that innervate it.28 In capricious mutants, motor neurons form ectopic synaptic contacts with muscles that neighbor the appropriate target, arguing that Capricious functions to restrict neuronal innervation to a single target (Fig. 1G). Conversely, expression of Capricious in all muscles causes the formation of many ectopic synapses. Intriguingly, the intracellular domain of Capricious is required in the muscle but not the innervating neuron, suggesting an asymmetric signaling role.29 Taken together, these studies argue that Capricious mediates specific adhesive interactions between a particular muscle and its neuronal targets, and that this interaction promotes synapse formation with the target while inhibiting synapse formation on neighboring cells.
DSCAM: A new paradigm for self-recognition in the nervous system.
Many neurons display complex axonal and dendritic architectures whose organization reflects, in part, interactions that discriminate between processes elaborated by the same cell, versus those elaborated by neighbors. The Ig superfamily member the Down Syndrome cell adhesion molecule (DSCAM) plays a central role in these interactions. Mutations in DSCAM cause defects in the development of a variety of neurons, affecting the outgrowth patterns of simple branched axons in the mushroom body, the elaboration of complex dendritic fields in the larva, and the connectivity of the olfactory system, among others.30–35 Additional analysis has provided evidence for a novel, DSCAM-mediated mechanism that supports neuronal self-recognition. DSCAM displays astonishing molecular diversity, having more than 38,000 alternatively spliced isoforms that differ in their extracellular and transmembrane domains.36 Individual neurons express multiple isoforms, with single cells of the same type expressing distinct, but overlapping subsets of the genomic repertoire.37,38 Homophilic adhesive interactions are isoform-specific, with negligible binding of different splice variants to each other.39–41 Finally, diversity per se, rather than expression of particular isoforms in specific cells, is central to DSCAM function.42 Taken together, these data support a model in which neuronal processes can distinguish self from non-self because only neurites from the same cell will contain exactly the same complement of DSCAM isoforms. Through unknown signaling pathways, these interactions then alter neurite behavior to prevent processes from the same cell from crossing one another.
Glial-Neuron Interactions in Development
The preceding studies have focused on adhesive interactions between neurons and their targets. However, this restricted view ignores an array of significant interactions between neurons and the non-neuronal constituents of the nervous system, namely glia. Glial cells form elaborate shapes in order to ensheath neuronal cell bodies and axons, and to become intimately associated with synapses43 (Fig. 2A). Although our knowledge about the functions of glia is still fragmentary, neuron-glia interactions are required during glial migration, axon guidance and targeting, as well as synapse formation and function. For many of these processes, contact-mediated, potentially adhesive interactions are particularly important and it is apparent that we have only just begun to identify the critical molecular players. Here we focus on instances where neuron-glia interactions influence migration, axon targeting, synapse formation and physiology.
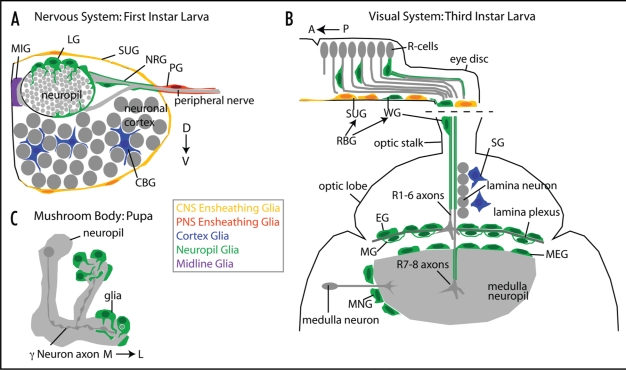
Figure 2.
Arrangement of glial types in the Drosophila nervous system. (A) Cross-section of a single hemi-segment of the embryonic ventral nerve chord. The midline is to the left. Embryonic and larval glia are: MIG, midline glia; LG, longitudinal glia; SUG, surface glia; NRG, nerve root glia; PG, peripheral glia; CBG, cell body glia. (B) Horizontal view of the eye disc and frontal view of the optic lobe of third instar larvae, depicting both the location and shapes of glial subtypes: retinal basal glia (RBG), which comprise surface glia (SUG) that ensheath the eye disc as a whole and wrapping glia (WG) that ensheath individual R cell bundles; SG, satellite glia; EG, epithelial glia; MG, marginal glia; MNG, medulla neuropil glia, MEG, medulla glia. (C) Mushroom body neuropil of the pupal CNS during axon pruning. An individual γ Neuron is highlighted. Upon an Ecdysone pulse, glial processes invade the neuropil and phagocytose degenerating axonal debris.
The roles of neuron-glial interactions in cell migration and axon extension.
Neuron-glial interactions influence both glial migration and axon extension during visual system development (Fig. 2B). During late larval stages, R cells differentiate in a wave that spreads from posterior to anterior. As they differentiate, R cell axons extend basally and exit the eye disc at the posterior, through the optic stalk. Concomitantly, retinal basal glia migrate through the optic stalk, into the eye disc, moving in the opposite direction, while staying in close contact with R cell axons.44,45 This migration is induced by a retinal signal: in mutants that lack R cells, glia do not enter the eye disc and instead accumulate in the optic stalk.46 Surprisingly, when R cells are present, but fail to extend axons, glial migration is unaffected. Thus, R cell axons are not a required substrate for glial migration. However, retinal basal glia will migrate towards large ectopic patches of differentiating R cells, suggesting that R cells provide an attractant that is sufficient for migration, but not necessary.45 Intriguingly, although glia do not need R cell axons to direct their migration, R cell axons do require glial cells as guideposts. When glial migration into eye disc is inhibited, R cell axons frequently stall and fail to exit the eye disc into the optic stalk.45,47 Similarly, in gilgamesh mutants, glia migrate too far into the eye disc, causing R cell axons to extend anteriorly rather than posteriorly.47 A related guidepost role can be found in the embryonic nervous system. There, the formation of the main axon tracts depends on longitudinal glia: when these glia are ablated, the axons of pioneer neurons, which form the first longitudinal pathways, frequently stall or extend inappropriately.43,48,49
Glia can also shape neuronal architecture by serving as intermediate targets. In the visual system, glial cells are the first to colonize the lamina. By arriving before R cell axons reach the brain, and before lamina neuron differentiation occurs, glial cells play a critical role in directing R cell axon targeting. Glial cells establish two distinct layers, designated the epithelial layer and the marginal layer, and provide a short-range signal that directs R cell axons to stop between them (Fig. 2B). When glia fail to migrate into the lamina, R1–R6 axons fail to stop, and instead project into the medulla.50,51 However, while this stop signal is very likely to be a local, possibly adhesive signal, its molecular identity is unknown.
Neuroglian mediates neuron-glial interactions.
While many neuron-glial interactions are local, the molecular identities of the factors involved are largely unknown. The only known adhesion molecule that appears to mediate homophilic interactions between neurons and glia during axon guidance is Neuroglian. Neuroglian mutant animals display defects in both neuronal and glial morphogenesis: sensory axons form ectopic branches and glia fail to ensheath them.52 This ectopic branching phenotype could only be rescued when Neuroglian was expressed in both neurons and glia, suggesting that Neuroglian acts as a homophilic adhesion molecule. These axon branching defects were not caused by defects in glial ensheathment, suggesting that Neuroglian plays distinct roles in neurons and glia. Intriguingly, Neuroglian is differentially spliced, with one form being restricted to neurons, while another form is expressed by neurons, glia and epithelial cells.53–55 These splice variants have identical extracellular but different intracellular domains, suggesting that Neuroglian-mediated interactions could activate distinct signaling pathways in neurons and glia. Finally the role of Neuroglian as a mediator between axons and glia appears to be conserved, as its vertebrate homolog, Neurofascin 155, mediates neuron-glia contacts in myelinated axons.56
Glial functions in synapse development.
The best understood requirement for neuron-glial interactions at the developing synapse occurs during axon pruning, a synaptic remodeling process that takes place during metamorphosis. In one prominent brain structure, the mushroom body, γ neurons initially project two branches, one medially and one dorsally (Fig. 2C). Both branches degenerate during early metamorphosis;57 at later stages only the medial branch re-extends. This axonal degeneration is triggered by the steroid hormone Ecdysone and relies on interactions between γ neurons and glia. Indeed, pruning is implemented by glial processes that invade the mushroom body neuropil to phagocytose degenerating axonal debris.57–59 These events are mediated by the scavenger receptor Draper.60 Blocking glial phagocytosis either by ectopic expression of a dominant-negative Dynamin, or by removal of Draper, results in failure of axon degeneration and persistence of ectopic axon branches.59,60 Finally, while Ecdysone signaling within γ neurons triggers both cytoskeletal degeneration in the axon, and induces glial invagination, it is also required in the glia themselves to upregulate Draper expression.59,60 Thus, one signal activates complementary pathways in neurons and glia, which then act in synchrony to prune axons.
Glial activities at mature synapses.
Anatomical studies of the NMJ and the visual system demonstrate that glial processes are in close proximity to synapses, forming highly specialized contacts that must surely be regulated by adhesion.61,62 In addition, glial cells regulate synaptic function by establishing and maintaining extracellular homeostasis. For example, glia form the blood-brain barrier to protect neurons from the high potassium and glutamate concentration of the hemolymph.63–65 Glia also express ion channels required for neurotransmitter recycling, such as the cysteine-glutamate exchanger Genderblind and the glutamate transporter dEAAT1, both of which remove excess glutamate from the synapse.66,67 Moreover, glia modify biogenic amines using the β-alanyl-biogenic amine synthase Ebony.68–70 This function is best understood at R cell synapses, where Ebony conjugates the neurotransmitter histamine to β-alanine, generating carcinine.68,69 Carcinine is thought to be transported back into R cell terminals, where another enzyme, Tan, hydrolyses it back to histamine.71 These observations suggest a tightly coupled neuron-glial biosynthetic cycle. At this synapse, the close association of neurons and glia is morphologically apparent, as glial processes, called capitate projections, invaginate into the pre-synaptic terminal.62 These structures are thought to be highly dynamic and show differential localization of Dynamin and Endophilin, suggesting that they are the sites of vesicle endocytosis and recycling.72 The Ig-domain containing molecule Basigin regulates capitate projection formation and basigin mutants display reduced numbers of capitate projections and defective synaptic transmission.73 However, the causal relationship between defects in capitate projections and synaptic transmission are unclear. Finally, although Basigin is expressed both by R cells and glia, it binds the Laminin receptor Integrin.74 Thus while morphological and functional data demonstrate intimate neuron-glial interactions, the molecules that actually hold these cells together remain largely unknown.
Concluding Remarks
These studies highlight three fundamental themes. First, it is striking how diverse the functions of adhesion molecules truly are; in no sense can the actions of these proteins be attributed to something akin to a simple “glue” acting to hold cell surfaces together. While it is unclear whether this functional diversity reflects differences between the molecules themselves, or between the cellular contexts in which they act, understanding how it arises presents a critical challenge. Second, it is apparent that both the timing and precise levels of expression of each adhesion molecule are critical for their function: manipulations that moderately alter expression often cause dramatic phenotypes. Thus we need to understand the regulatory mechanisms that control the activities of adhesion molecules, and have better models that define how cells read and interpret differences in adhesivity. Finally, while we have made some progress in identifying and characterizing the adhesive molecules that play critical roles during development, our understanding of adhesive interactions amongst neurons, and between neurons and glia in the adult nervous system remains incomplete. A systematic study of the mechanisms responsible for the long-term maintenance of brain architecture will likely lead us to new insights into cell adhesion.
Footnotes
Previously published online as a Cell Adhesion & Migration E-publication: http://www.landesbioscience.com/journals/celladhesion/article/6918
References
- 1.Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting and tissue morphogenesis. Genes Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- 2.Thompson DAW. On growth and form. Cambridge, UK: The University Press; 1917. [Google Scholar]
- 3.Steinberg MS. Differential adhesion in morphogenesis: a modern view. Curr Opin Genet Dev. 2007;17:281–286. doi: 10.1016/j.gde.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Steinberg MS, Takeichi M. Experimental specification of cell sorting, tissue spreading and specific spatial patterning by quantitative differences in cadherin expression. Proc Natl Acad Sci USA. 1994;91:206–209. doi: 10.1073/pnas.91.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duguay D, Foty RA, Steinberg MS. Cadherin-mediated cell adhesion and tissue segregation: qualitative and quantitative determinants. Dev Biol. 2003;253:309–323. doi: 10.1016/s0012-1606(02)00016-7. [DOI] [PubMed] [Google Scholar]
- 6.Godt D, Tepass U. Drosophila oocyte localization is mediated by differential cadherin-based adhesion. Nature. 1998;395:387–391. doi: 10.1038/26493. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi T, Carthew RW. Surface mechanics mediate pattern formation in the developing retina. Nature. 2004;431:647–652. doi: 10.1038/nature02952. [DOI] [PubMed] [Google Scholar]
- 8.Meinertzhagen IA, Hanson TE, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 9.Ting CY, Yonekura S, Chung P, Hsu SN, Robertson HM, Chiba A, Lee CH. Drosophila N-cadherin functions in the first stage of the two-stage layer-selection process of R7 photoreceptor afferents. Development. 2005;132:953–963. doi: 10.1242/dev.01661. [DOI] [PubMed] [Google Scholar]
- 10.Lee CH, Herman T, Clandinin TR, Lee R, Zipursky SL. N-cadherin regulates target specificity in the Drosophila visual system. Neuron. 2001;30:437–450. doi: 10.1016/s0896-6273(01)00291-4. [DOI] [PubMed] [Google Scholar]
- 11.Shinza-Kameda M, Takasu E, Sakurai K, Hayashi S, Nose A. Regulation of layer-specific targeting by reciprocal expression of a cell adhesion molecule, capricious. Neuron. 2006;49:205–213. doi: 10.1016/j.neuron.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Lee RC, Clandinin TR, Lee CH, Chen PL, Meinertzhagen IA, Zipursky SL. The protocadherin Flamingo is required for axon target selection in the Drosophila visual system. Nature neuroscience. 2003;6:557–563. doi: 10.1038/nn1063. [DOI] [PubMed] [Google Scholar]
- 13.Iwai Y, Usui T, Hirano S, Steward R, Takeichi M, Uemura T. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 14.Iwai Y, Hirota Y, Ozaki K, Okano H, Takeichi M, Uemura T. DN-cadherin is required for spatial arrangement of nerve terminals and ultrastructural organization of synapses. Mol Cell Neurosci. 2002;19:375–388. doi: 10.1006/mcne.2001.1081. [DOI] [PubMed] [Google Scholar]
- 15.Prakash S, Caldwell JC, Eberl DF, Clandinin TR. Drosophila N-cadherin mediates an attractive interaction between photoreceptor axons and their targets. Nature neuroscience. 2005;8:443–450. doi: 10.1038/nn1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yonekura S, Xu L, Ting CY, Lee CH. Adhesive but not signaling activity of Drosophila N-cadherin is essential for target selection of photoreceptor afferents. Dev Biol. 2007;304:759–770. doi: 10.1016/j.ydbio.2007.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choe KM, Prakash S, Bright A, Clandinin TR. Liprin-alpha is required for photoreceptor target selection in Drosophila. Proc Natl Acad Sci USA. 2006;103:11601–11606. doi: 10.1073/pnas.0601185103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nern A, Zhu Y, Zipursky SL. Local N-cadherin interactions mediate distinct steps in the targeting of lamina neurons. Neuron. 2008;58:34–41. doi: 10.1016/j.neuron.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehta SQ, Hiesinger PR, Beronja S, Zhai RG, Schulze KL, Verstreken P, et al. Mutations in Drosophila sec15 reveal a function in neuronal targeting for a subset of exocyst components. Neuron. 2005;46:219–232. doi: 10.1016/j.neuron.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 20.Usui T, Shima Y, Shimada Y, Hirano S, Burgess RW, Schwarz TL, et al. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell. 1999;98:585–595. doi: 10.1016/s0092-8674(00)80046-x. [DOI] [PubMed] [Google Scholar]
- 21.Kimura H, Usui T, Tsubouchi A, Uemura T. Potential dual molecular interaction of the Drosophila 7-pass transmembrane cadherin Flamingo in dendritic morphogenesis. J Cell Sci. 2006;119:1118–1129. doi: 10.1242/jcs.02832. [DOI] [PubMed] [Google Scholar]
- 22.Gao FB, Brenman JE, Jan LY, Jan YN. Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev. 1999;13:2549–2561. doi: 10.1101/gad.13.19.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao FB, Kohwi M, Brenman JE, Jan LY, Jan YN. Control of dendritic field formation in Drosophila: the roles of flamingo and competition between homologous neurons. Neuron. 2000;28:91–101. doi: 10.1016/s0896-6273(00)00088-x. [DOI] [PubMed] [Google Scholar]
- 24.Senti KA, Usui T, Boucke K, Greber U, Uemura T, Dickson BJ. Flamingo regulates R8 axon-axon and axon-target interactions in the Drosophila visual system. Curr Biol. 2003;13:828–832. doi: 10.1016/s0960-9822(03)00291-4. [DOI] [PubMed] [Google Scholar]
- 25.Chen PL, Clandinin TR. The cadherin Flamingo mediates level-dependent interactions that guide photoreceptor target choice in Drosophila. Neuron. 2008;58:26–33. doi: 10.1016/j.neuron.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keshishian H, Broadie K, Chiba A, Bate M. The Drosophila neuromuscular junction: a model system for studying synaptic development and function. Annu Rev Neurosci. 1996;19:545–575. doi: 10.1146/annurev.ne.19.030196.002553. [DOI] [PubMed] [Google Scholar]
- 27.Bao H, Berlanga ML, Xue M, Hapip SM, Daniels RW, Mendenhall JM, et al. The atypical cadherin flamingo regulates synaptogenesis and helps prevent axonal and synaptic degeneration in Drosophila. Mol Cell Neurosci. 2007;34:662–678. doi: 10.1016/j.mcn.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shishido E, Takeichi M, Nose A. Drosophila synapse formation: regulation by transmembrane protein with Leu-rich repeats, CAPRICIOUS. Science. 1998;280:2118–2121. doi: 10.1126/science.280.5372.2118. [DOI] [PubMed] [Google Scholar]
- 29.Taniguchi H, Shishido E, Takeichi M, Nose A. Functional dissection of Drosophila capricious: its novel roles in neuronal pathfinding and selective synapse formation. J Neurobiol. 2000;42:104–116. doi: 10.1002/(sici)1097-4695(200001)42:1<104::aid-neu10>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 30.Hughes ME, Bortnick R, Tsubouchi A, Baumer P, Kondo M, Uemura T, et al. Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron. 2007;54:417–427. doi: 10.1016/j.neuron.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, et al. Dendrite self-avoidance is controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 32.Soba P, Zhu S, Emoto K, Younger S, Yang SJ, Yu HH, et al. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron. 2007;54:403–416. doi: 10.1016/j.neuron.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu H, Hummel T, Clemens JC, Berdnik D, Zipursky SL, Luo L. Dendritic patterning by Dscam and synaptic partner matching in the Drosophila antennal lobe. Nat Neurosci. 2006;9:349–355. doi: 10.1038/nn1652. [DOI] [PubMed] [Google Scholar]
- 34.Hummel T, Vasconcelos ML, Clemens JC, Fishilevich Y, Vosshall LB, Zipursky SL. Axonal targeting of olfactory receptor neurons in Drosophila is controlled by Dscam. Neuron. 2003;37:221–231. doi: 10.1016/s0896-6273(02)01183-2. [DOI] [PubMed] [Google Scholar]
- 35.Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, et al. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–686. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 36.Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 37.Neves G, Chess A. Dscam-mediated self- versus non-self-recognition by individual neurons. Cold Spring Harbor symposia on quantitative biology. 2004;69:485–488. doi: 10.1101/sqb.2004.69.485. [DOI] [PubMed] [Google Scholar]
- 38.Chen BE, Kondo M, Garnier A, Watson FL, Puettmann-Holgado R, Lamar DR, et al. The molecular diversity of Dscam is functionally required for neuronal wiring specificity in Drosophila. Cell. 2006;125:607–620. doi: 10.1016/j.cell.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 39.Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meijers R, Puettmann-Holgado R, Skiniotis G, Liu JH, Walz T, Wang JH, et al. Structural basis of Dscam isoform specificity. Nature. 2007;449:487–491. doi: 10.1038/nature06147. [DOI] [PubMed] [Google Scholar]
- 41.Wojtowicz WM, Wu W, Andre I, Qian B, Baker D, Zipursky SL. A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell. 2007;130:1134–1145. doi: 10.1016/j.cell.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hattori D, Demir E, Kim HW, Viragh E, Zipursky SL, Dickson BJ. Dscam diversity is essential for neuronal wiring and self-recognition. Nature. 2007;449:223–227. doi: 10.1038/nature06099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito KU J, Technau GM. Distribution, classification and development of Drosophila glial cells in the late embryonic and early larval ventral nerve chord. Roux's Arch Dev Biol. 1995;204:284–307. doi: 10.1007/BF02179499. [DOI] [PubMed] [Google Scholar]
- 44.Chotard C, Salecker I. Glial cell development and function in the Drosophila visual system. Neuron Glia Biol. 2007;3:17–25. doi: 10.1017/S1740925X07000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rangarajan R, Gong Q, Gaul U. Migration and function of glia in the developing Drosophila eye. Development. 1999;126:3285–3292. doi: 10.1242/dev.126.15.3285. [DOI] [PubMed] [Google Scholar]
- 46.Choi KW, Benzer S. Migration of glia along photoreceptor axons in the developing Drosophila eye. Neuron. 1994;12:423–431. doi: 10.1016/0896-6273(94)90282-8. [DOI] [PubMed] [Google Scholar]
- 47.Hummel T, Attix S, Gunning D, Zipursky SL. Temporal control of glial cell migration in the Drosophila eye requires gilgamesh, hedgehog and eye specification genes. Neuron. 2002;33:193–203. doi: 10.1016/s0896-6273(01)00581-5. [DOI] [PubMed] [Google Scholar]
- 48.Hidalgo A, Booth GE. Glia dictate pioneer axon trajectories in the Drosophila embryonic CNS. Development. 2000;127:393–402. doi: 10.1242/dev.127.2.393. [DOI] [PubMed] [Google Scholar]
- 49.Hidalgo A, Urban J, Brand AH. Targeted ablation of glia disrupts axon tract formation in the Drosophila CNS. Development. 1995;121:3703–3712. doi: 10.1242/dev.121.11.3703. [DOI] [PubMed] [Google Scholar]
- 50.Poeck B, Fischer S, Gunning D, Zipursky SL, Salecker I. Glial cells mediate target layer selection of retinal axons in the developing visual system of Drosophila. Neuron. 2001;29:99–113. doi: 10.1016/s0896-6273(01)00183-0. [DOI] [PubMed] [Google Scholar]
- 51.Suh GS, Poeck B, Chouard T, Oron E, Segal D, Chamovitz DA, et al. Drosophila JAB1/CSN5 acts in photoreceptor cells to induce glial cells. Neuron. 2002;33:35–46. doi: 10.1016/s0896-6273(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto M, Ueda R, Takahashi K, Saigo K, Uemura T. Control of axonal sprouting and dendrite branching by the Nrg-Ank complex at the neuron-glia interface. Curr Biol. 2006;16:1678–1683. doi: 10.1016/j.cub.2006.06.061. [DOI] [PubMed] [Google Scholar]
- 53.Hortsch M, Bieber AJ, Patel NH, Goodman CS. Differential splicing generates a nervous system-specific form of Drosophila neuroglian. Neuron. 1990;4:697–709. doi: 10.1016/0896-6273(90)90196-m. [DOI] [PubMed] [Google Scholar]
- 54.Hortsch M, Wang YM, Marikar Y, Bieber AJ. The cytoplasmic domain of the Drosophila cell adhesion molecule neuroglian is not essential for its homophilic adhesive properties in S2 cells. J Biol Chem. 1995;270:18809–18817. doi: 10.1074/jbc.270.32.18809. [DOI] [PubMed] [Google Scholar]
- 55.Genova JL, Fehon RG. Neuroglian, Gliotactin, and the Na+/K+ ATPase are essential for septate junction function in Drosophila. J Cell Biol. 2003;161:979–989. doi: 10.1083/jcb.200212054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhat MA. Molecular organization of axo-glial junctions. Curr Opin Neurobiol. 2003;13:552–559. doi: 10.1016/j.conb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 57.Lee T, Marticke S, Sung C, Robinow S, Luo L. Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron. 2000;28:807–818. doi: 10.1016/s0896-6273(00)00155-0. [DOI] [PubMed] [Google Scholar]
- 58.Watts RJ, Schuldiner O, Perrino J, Larsen C, Luo L. Glia engulf degenerating axons during developmental axon pruning. Curr Biol. 2004;14:678–684. doi: 10.1016/j.cub.2004.03.035. [DOI] [PubMed] [Google Scholar]
- 59.Awasaki T, Ito K. Engulfing action of glial cells is required for programmed axon pruning during Drosophila metamorphosis. Curr Biol. 2004;14:668–677. doi: 10.1016/j.cub.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 60.Awasaki T, Tatsumi R, Takahashi K, Arai K, Nakanishi Y, Ueda R, et al. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron. 2006;50:855–867. doi: 10.1016/j.neuron.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 61.Sepp KJ, Schulte J, Auld VJ. Developmental dynamics of peripheral glia in Drosophila melanogaster. Glia. 2000;30:122–133. doi: 10.1002/(sici)1098-1136(200004)30:2<122::aid-glia2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 62.Stark WS, Carlson SD. Ultrastructure of capitate projections in the optic neuropil of Diptera. Cell Tissue Res. 1986;246:481–486. doi: 10.1007/BF00215187. [DOI] [PubMed] [Google Scholar]
- 63.Auld VJ, Fetter RD, Broadie K, Goodman CS. Gliotactin, a novel transmembrane protein on peripheral glia, is required to form the blood-nerve barrier in Drosophila. Cell. 1995;81:757–767. doi: 10.1016/0092-8674(95)90537-5. [DOI] [PubMed] [Google Scholar]
- 64.Baumgartner S, Littleton JT, Broadie K, Bhat MA, Harbecke R, Lengyel JA, et al. A Drosophila neurexin is required for septate junction and blood-nerve barrier formation and function. Cell. 1996;87:1059–1068. doi: 10.1016/s0092-8674(00)81800-0. [DOI] [PubMed] [Google Scholar]
- 65.Schwabe T, Bainton RJ, Fetter RD, Heberlein U, Gaul U. GPCR signaling is required for blood-brain barrier formation in Drosophila. Cell. 2005;123:133–144. doi: 10.1016/j.cell.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 66.Grosjean Y, Grillet M, Augustin H, Ferveur JF, Featherstone DE. A glial amino-acid transporter controls synapse strength and courtship in Drosophila. Nature neuroscience. 2008;11:54–61. doi: 10.1038/nn2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rival T, Soustelle L, Cattaert D, Strambi C, Iche M, Birman S. Physiological requirement for the glutamate transporter dEAAT1 at the adult Drosophila neuromuscular junction. J Neurobiol. 2006;66:1061–1074. doi: 10.1002/neu.20270. [DOI] [PubMed] [Google Scholar]
- 68.Borycz J, Borycz JA, Loubani M, Meinertzhagen IA. tan and ebony genes regulate a novel pathway for transmitter metabolism at fly photoreceptor terminals. J Neurosci. 2002;22:10549–10557. doi: 10.1523/JNEUROSCI.22-24-10549.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Richardt A, Rybak J, Stortkuhl KF, Meinertzhagen IA, Hovemann BT. Ebony protein in the Drosophila nervous system: optic neuropile expression in glial cells. J Comp Neurol. 2002;452:93–102. doi: 10.1002/cne.10360. [DOI] [PubMed] [Google Scholar]
- 70.Suh J, Jackson FR. Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron. 2007;55:435–447. doi: 10.1016/j.neuron.2007.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wagner S, Heseding C, Szlachta K, True JR, Prinz H, Hovemann BT. Drosophila photoreceptors express cysteine peptidase tan. J Comp Neurol. 2007;500:601–611. doi: 10.1002/cne.21138. [DOI] [PubMed] [Google Scholar]
- 72.Fabian-Fine R, Verstreken P, Hiesinger PR, Horne JA, Kostyleva R, Zhou Y, et al. Endophilin promotes a late step in endocytosis at glial invaginations in Drosophila photoreceptor terminals. J Neurosci. 2003;23:10732–10744. doi: 10.1523/JNEUROSCI.23-33-10732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Curtin KD, Wyman RJ, Meinertzhagen IA. Basigin/EMMPRIN/CD147 mediates neuronglia interactions in the optic lamina of Drosophila. Glia. 2007;55:1542–1553. doi: 10.1002/glia.20568. [DOI] [PubMed] [Google Scholar]
- 74.Curtin KD, Meinertzhagen IA, Wyman RJ. Basigin (EMMPRIN/CD147) interacts with integrin to affect cellular architecture. J Cell Sci. 2005;118:2649–2660. doi: 10.1242/jcs.02408. [DOI] [PubMed] [Google Scholar]