Abstract
Venezuelan equine encephalitis virus (VEEV) is one of the most pathogenic members of the Alphavirus genus in the Togaviridae family. Viruses in the VEEV serocomplex continuously circulate in the Central and South Americas. The only currently available attenuated strain VEEV TC-83 is being used only for vaccination of at-risk laboratory workers and military personnel. Its attenuated phenotype was shown to rely only on two point mutations, one of which, G3A, was found in the 5′ untranslated region (5′UTR) of the viral genome. Our data demonstrate that the G3A mutation strongly affects the secondary structure of VEEV 5′UTR, but has only a minor effect on translation. The indicated mutation increases replication of the viral genome, downregulates transcription of the subgenomic RNA, and, thus, affects the ratio of genomic and subgenomic RNA synthesis. These findings and the previously reported G3A-induced, higher sensitivity of VEEV TC-83 to IFN-α/β suggest a plausible explanation for its attenuated phenotype.
INTRODUCTION
Venezuelan equine encephalitis virus (VEEV) is one of the most pathogenic members of the Alphavirus genus in the Togaviridae family. In nature, the serologically related viruses within the VEEV serocomplex persistently replicate in mosquito vectors, which infect vertebrate hosts during blood meals (Weaver and Barrett, 2004). In mammals, VEEV infection is characterized by high-titer viremia, rash and fever, followed by severe encephalitis that can result in death or neurological disorders (Dal Canto and Rabinowitz, 1981; Griffin, 2001; Johnston and Peters, 1996; Leon, 1975). Natural isolates of VEEV demonstrate some differences in the antigenic structure (Weaver and Frolov, 2005) and the severity of caused disease. In humans, the members of IAB and IC subtypes are capable of causing diffuse congestion and edema with hemorrhage in the brain, gastrointestinal (GI) tract, and lungs. Severe necrosis and vasculitis occur in lymph nodes, spleen, and the GI tract, accompanied by hepatocellular degeneration and interstitial pneumonia (de la Monte et al., 1985; Ehrenkranz and Ventura, 1974; Johnson et al., 1968). Viruses in the VEEV serocomplex continuously circulate in Central and South America, sometimes spreading into Texas (Weaver et al., 2004). They have induced epidemics in equids, and one of the recent outbreaks in Venezuela and Columbia also involved ~75,000 human cases and caused more than 20 deaths (Rivas et al., 1997; Weaver et al., 1996).
In spite of the continuous threat of VEEV epidemics, no safe and efficient vaccine, or therapeutic means have been developed for this encephalitogenic pathogen. A currently available, attenuated strain VEEV TC-83 is used only as an experimental vaccine for at-risk laboratory workers and military personnel. It is also employed as a veterinary vaccine. VEEV TC-83 strain was developed more than four decades ago by the serial passage of the wild type Trinidad donkey (TRD) strain of VEEV in guinea pig heart cells (Berge, Banks, and Tigertt, 1961). During this procedure, TC-83 accumulated 12 genomic mutations (Kinney et al., 1989), but its attenuated phenotype was shown to rely only on 2 point mutations (Kinney et al., 1993), which likely had a positive effect on virus replication in tissue culture and, thus, were the basis for the selection of the indicated strain. The T120→R mutation, located in the E2 glycoprotein, increases the protein’s positive charge, and its attenuating effect is achieved by the more efficient binding of envelope glycoprotein to heparan sulfate (Bernard, Klimstra, and Johnston, 2000). This point mutation has a strong positive effect on virus infectivity in vitro (Kamrud et al., 2008) and appears to accelerate virus entry into cultivated cells (Berge, Banks, and Tigertt, 1961; Johnston and Smith, 1988). A second attenuating mutation G3→A (G3A) was found in the 5′ untranslated region (5′UTR) of the viral genome. This mutation was shown to have no effect on virus pathogenicity in mice that exhibited a defect in IFN-α/β signaling (IFN-α/βR−/− mice) (White et al., 2001), but made the virus less virulent in adult mice, having a fully competent type I IFN system (Kinney et al., 1993; White et al., 2001). However, the effect on virulence depended on the mouse strains used in the studies. VEEV TC-83 also remained highly lethal for the immunocompetent new born mice after either s.c. or i.c. inoculation (Ludwig et al., 2001; Paessler et al., 2003). Taken together, these data and the high frequency of adverse effects in vaccinees (Alevizatos, McKinney, and Feigin, 1967) suggest to us that the currently available experimental vaccine does not meet the rigorous requirements for mass human vaccinations. Thus, the need for new, more efficient and safer VEEV-specific vaccine remains a high priority, and additional information about the molecular basis of VEEV attenuation could be very beneficial for its rational design. However, till date, the biology of this virus and its mechanism of replication are poorly understood, further dampening efforts for vaccine development.
The VEEV genome is represented by a ca. 11.5 kb-long, single-stranded RNA of positive polarity (Strauss, Rice, and Strauss, 1984), which mimics the structure of the cellular mRNAs, in that it contains a 5′ cap and poly(A)-tail at the 5′ and 3′ ends, respectively. The 5′UTR controls the translation of a 7,500-nt-long, 5′-terminal open reading frame (ORF) that is translated into viral nonstructural proteins (nsP1-4). These proteins form the enzyme complex required for the replication of viral genome and transcription of the subgenomic RNA (Barton, Sawicki, and Sawicki, 1990; Lemm and Rice, 1993). The latter RNA encodes the second polyprotein that is co- and post-translationally processed into the individual capsid, E2 and E1 proteins that form infectious viral particles (Strauss and Strauss, 1994). Thus, the 5′UTR-specific, attenuating G3A mutation in the VEEV TC-83 genome could affect the translation of viral nonstructural proteins. However, the important feature of the alphavirus genome’s 5′-terminal sequence lies in its involvement in the processes other than the mere translation of viral nsPs. The complement of this sequence at the 3′ end of the negative-strand RNA intermediate functions as a promoter for positive-strand genome synthesis (Gorchakov et al., 2004b; Niesters and Strauss, 1990a), and in the case of other alphaviruses, the 5′UTR was also suggested to function as a part of the promoter of negative-strand RNA synthesis (Frolov, Hardy, and Rice, 2001; Gorchakov et al., 2004b). Thus, it is reasonable to expect that the G3A mutation might modify the promoter activity and have an effect on the synthesis of virus-specific RNAs.
In this study, we used a combination of biochemical, biophysical, and virological approaches to further dissect the effect(s) of the 5′-terminal, TC-83-specific mutation on virus replication. Our data demonstrate that the G3A mutation strongly affects the 5′-terminal secondary structure of the VEEV genome, but has only a minor effect on the translation of viral nsPs. However, the indicated mutation strongly induced replication of the viral genome, downregulated transcription of the subgenomic RNA, and, thus, affected the ratio of genomic and subgenomic RNA synthesis. Further, these effects enhanced virus replication in vitro, and appeared to contribute to its attenuated phenotype.
RESULTS
Enzymatic analysis of the TRD- and the TC-83-specific secondary structures of the 5′UTR
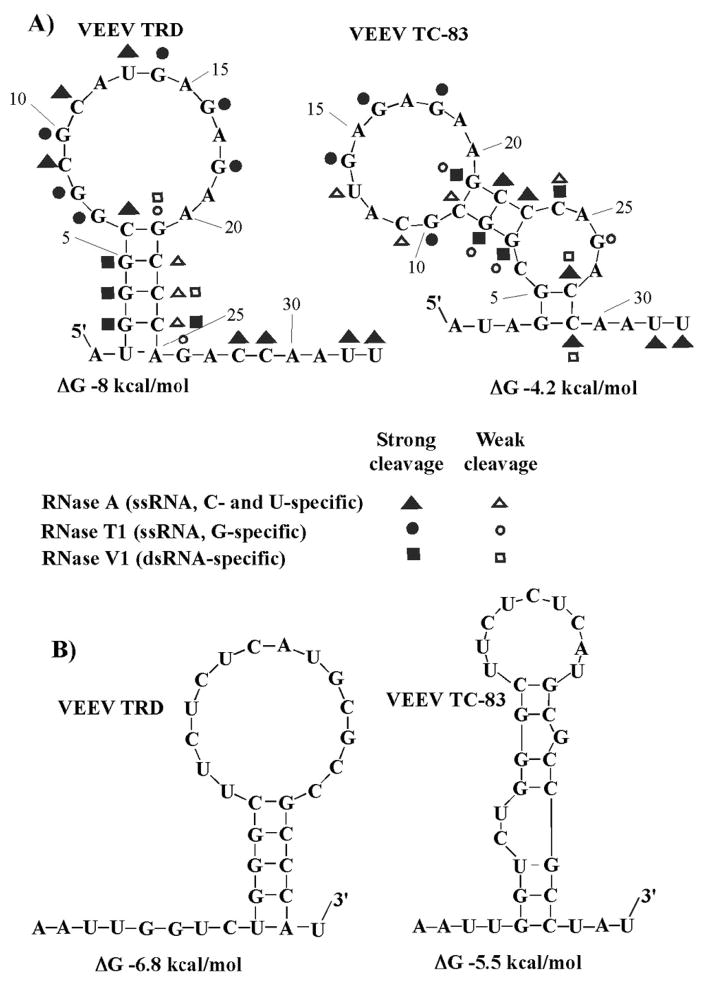
The TC-83-specific G3A mutation strongly affected the computer-predicted secondary structure of the 5′ end of the VEEV TRD genome in terms of the stability and configuration of the very 5′-terminal stem-loop (Fig. 1A). Both the M-fold (Jaeger, Turner, and Zuker, 1989) and Vienna fold (Hofacker, 2003) programs yield identical results for secondary structure prediction. The TRD-specific 5′UTR was predicted to fold into a hairpin composed of 5 base-pair-long stem and a large (14-nt-long) terminal loop. In contrast, the predicted secondary structure of the TC-83 5′UTR contained two short stems separated by an internal, 4-nt-long bulge, and had a smaller (11-nt-long) loop (Fig. 1A). The calculated overall free energy of the TC-83-specific 5′-terminal stem-loop was expected to be higher than that in the TRD strain, and thus, its stability at physiological conditions (temperature and salt concentration) was questionable. Similarly, the mutation had a strong effect on the computer-predicted folding of the 3′ terminus of the negative strand of the VEEV genome (Fig. 1B).
Fig. 1.
The effect of G3A mutation on RNA folding. (A) The computer-predicted (M-fold) secondary structures of the 5′ termini of VEEV TRD and TC-83 genomes, and the results of the RNase probing. (B) The computer-predicted secondary structures of the 3′ termini of the negative strands of viral genomes.
To verify the effect of the G3A mutation on the RNA folding, we performed enzymatic analysis using nucleotide- and dsRNA-specific RNases. The 5′-terminal 130-nt-long cDNA fragments of TRD and TC-83 genomes, comprising the 5′-terminal stem-loop and the ensuing sequence, which was predicted to fold into stable stem-loop SL2, were synthesized by PCR and cloned into plasmids under control of the promoter of the T7 DNA-dependent RNA polymerase (see Materials and Methods for details). The SL2 was left to protect the proper folding of the 5′-terminal sequences. The in vitro-synthesized, 32P-labeled RNA fragments were partially digested by RNase T1, RNase V1, and RNase A. The enzymatic probing conditions were optimized to leave at least 80% of the RNA intact. The cleavage products were then analyzed on denaturing polyacrylamide gels to reveal the cleavage patterns, which were further used for generating potential secondary structures and comparing them to those computer-predicted (Fig. 1A).
The data strongly suggested that in TRD 5′UTR the G4 and G5 are base-paired with C23 and C22 respectively, while in TC-83 they form base-pairs with C29 and C28. In TC-83, however, both C28 and C29 were cleaved strongly by RNase A in addition to being cleaved by RNase V1 suggesting that the G4-C29 and G5-C28 base pairs were not stable. Nucleotides G7 and G8 are not base-paired in TRD 5′UTR, while they form the second stem in TC-83 RNA and base-pairs with C23 and C22, respectively. All of the uridine, cytosine and guanine residues in both the TRD loop G7-A20 and the TC-83 loop G10-A20 were cleaved by RNase A or RNase T1, suggesting that they are not involved in base-pairing and are likely to be in single stranded form. RNase A and RNase T1 cleaved C24 and G26 respectively in TC-83 5′UTR, supporting the possible existence of an internal bulge. In contrast to TRD 5′UTR, the RNA fragments, predicted to form stems in TC-83-specific sequence, were partially cleaved by the single-strand-specific nuclease RNase T1 or RNase A, indicating low stability of the RNA secondary structure under the used experimental conditions. The overall results of the RNase mapping were consistent with the predicted secondary structures and supported the hypothesis that the G3A mutation changed the secondary structure and stability of the 5′-terminal stem-loop.
In the next experiments, we synthesized 31-nt-long ribo oligonucleotides representing the very 5′ terminal sequences of TRD and TC-83 genomes (see Materials and Methods for details). The thermal melting temperatures of the TRD and TC-83 5′-terminal stem-loop RNA fragments monitored by UV-absorbance, revealed that the G3A mutation decreased the melting temperature of oligonucleotide by 8.2 °C (data not shown). Next, the imino proton signals of the TRD and TC-83 5′UTR fragments were monitored by NMR spectroscopy, and the 1D NMR spectra were collected as a function of temperature. Unlike the UV-monitored melting experiments, the NMR monitoring provides information on the melting of individual base pairs. Sharp, downfield shifted NMR signals in the imino region (11.5 –14 ppm) indicate that the imino protons are base-paired and thus protected from rapid exchange with solvent (Fig. 2). At 5°C, the 1D NMR spectra of the TRD-specific 5′-terminal RNA fragment contained 6 signals in the imino proton region. Most of the signals were assigned to specific imino protons using correlations in the 2D NOESY spectra based on sequential NOE interactions (Wuthrich, 1986). Because no NOE cross-correlations were observed for some of the imino signals, they were tentatively assigned as arising from residues G7/G8 or G(s) in the loop. As the temperature was increased, signals corresponding to the G3, G4, G5 and G21 imino protons remained intact, while those of G7 and G8 disappeared. In the case of VEEV TC-83-specific RNA, the 1D spectra collected at 5°C had 5 signals in the imino proton region. As in the above-described TRD RNA-based experiments, signals were either specifically assigned from sequential cross-correlations in 2D NOESY spectra, or, if no cross-peaks were detected, were provisionally assigned to the G4, and G5, in the two-base-pair stem and G26 in the 4-nt bulge. The latter assumption was based on the fact that, G4 and G5 could base-pair with C29 and C28 at low temperatures, and this further correlated with the results of enzymatic analysis (Fig. 1A). With an increase in temperature of the TC-83-specific 31-mer, the signals corresponding to the G4, G5 and G26 imino protons disappeared, which was indicative of either very weak hydrogen bonds or a lack of base-pairing. At 25°C, TRD-specific 5′UTR had 4 imino peaks intact while, only 3 imino peaks remained for TC-83-specific 5′UTR suggesting that the two 5′UTR RNAs had different secondary structures. Further, the disappearance of stem-specific signals corresponding to G4, G5 and G26 with increase in temperature suggested that the TC-83-specific 5′UTR has a less stable secondary structure compared to that of TRD, whose G3, G4, G5 and G21 imino signals remained intact at higher temperatures.
Fig. 2.
Analysis of stability of the VEEV TRD- and VEEV TC-83-specific, 5′-terminal stem-loops by NMR. The 31-nt-long RNA molecules, representing nt 1–31 of the VEEV TRD and TC-83 genomes, were synthesized in vitro and prepared for NMR as described in Materials and Methods. The imino regions of the 1D NOESY spectra presented here were collected at 5° C, 15° C, and 25° C. The signals were assigned from the 2D NOESY spectra, using sequential NOE connectivities.
G3A mutation has only a minor positive effect on protein translation
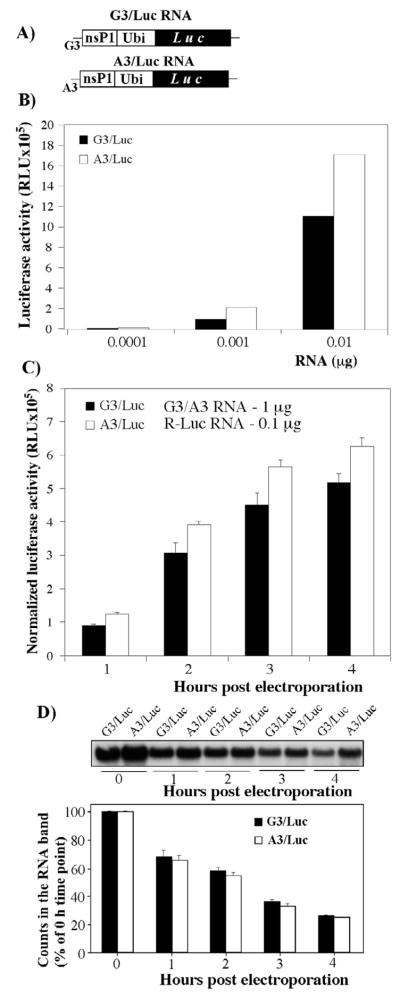
The biochemical data demonstrated that the G3A mutation destabilized the 5′-terminal stem-loop in the VEEV genome and, thus, could have a strong effect on the translation of the encoded ns proteins. To test this possibility, we designed reporter, luciferase-expressing constructs (Fig. 3A) containing either TRD or TC-83-specific 5′UTR, and compared the efficiency of translation of the encoded proteins both in vitro and in vivo.
Fig. 3.
Analysis of the effect of G3A mutation on the RNA translation efficiency. (A) The schematic representation of the firefly luciferase constructs used to evaluate the effect of G3A mutation on the template translation. The detailed description of the constructs is presented in Materials and Methods. (B) Luciferase activities in the RRL reaction mixtures, supplemented with different amounts of in vitro-synthesized G3/Luc and A3/Luc capped template RNAs. (C) Firefly luciferase expression in BHK-21 cells transfected with the in vitro-synthesized, capped G3/Luc or A3/Luc RNA. One μg of each template was mixed with 0.1 μg of Renilla luciferase-encoding RNA, and electroporated into BHK-21 cells, as described in Materials and Methods. Equal aliquots of transfected cells were seeded into 35-mm dishes, and luciferase activities were determined at indicated time points using a Dual-Luciferase system (Promega). The activity of Renilla luciferase was used to normalize the data. (D) Analysis of the effect of the G3A mutation on RNA stability. Cells were transfected with similar amounts of 32P-labeled G3/Luc or A3/Luc labeled RNAs. At indicated time points, total RNAs were isolated and analyzed by agarose gel electrophoresis under denaturing conditions. Radioactivity in the RNA bands was evaluated on a phosphorimager. The experiments presented in panels B and C were repeated 2 and 3 times, respectively. One of the highly reproducible experiments is presented.
The 5′-terminal, 191-nt-long fragments of the VEEV genome, encoding either TC-83 or TRD 5′UTRs and 49 aa of nsP1, were cloned under control of the SP6 promoter. The protein-coding sequence contained both SL2 and the 51nt-long conserved sequence element (51-nt CSE) to preserve the secondary structure architecture of the 5′ termini. The nsP1-coding sequence was fused in frame with the firefly luciferase gene using the ubiquitine gene. This design was aimed at expressing the luciferase in a free form, because fusion proteins might demonstrate lower enzymatic activities. To preserve the coding strategy of the VEEV genome, the cassettes also contained VEEV-specific 3′UTR, followed by the 25-nt-long poly(A) tail.
In the initial experiments, the in vitro-synthesized, capped RNAs were translated in the rabbit reticulocyte lysate (RRL) in vitro translation system (see Materials and Methods for details). To avoid saturation of the in vitro translation system with tested RNAs, the experiments were performed using different, low concentrations of the templates. In all of the experiments, the RNAs having the TC-83-specific 5′UTR, demonstrated a higher efficiency of luciferase expression. This suggested to us that the G3A mutation that destabilizes the 5′-terminal stem-loop has a detectable positive effect on RNA translation (see the results of one of the reproducible experiments in Fig. 3B). However, the effect was moderate and mostly detectable early in the translation reaction.
To make the data more biologically relevant, we transfected equal amounts of the same RNAs into BHK-21 cells by electroporation, and assessed luciferase expression at different times post transfection (Fig. 3C). The second in vitro-synthesized RNA that encoded the Renilla luciferase was included in the transfection mixture as a standard for normalization of the data. However, the electroporation procedure was found to be very reproducible, and no normalizing was necessary. In all of the experiments, RNA template encoding the TRD-specific 5′UTR produced luciferase almost as efficiently as did the template with the TC-83 5′UTR (Fig. 3C). Considering the possibility that either the 5′UTR sequence or secondary structure could determine the stability of the RNAs, we also transfected BHK-21 cells with the same RNAs, labeled in vitro with 32P. Total cellular RNA was isolated at 0, 1, 2 and 4 h post transfection, and analyzed by electrophoresis in denaturing conditions, followed by assessment of the radioactivity in the RNA bands on a phosphorimager (Fig. 3D). Both RNAs exhibited the same rates of degradation. Thus, taken together, the results of the in vivo and in vitro studies suggest that the TC-83-specific G3A mutation has a very modest effect on the translation of the encoded proteins.
Effect of G3A mutation on the synthesis of virus-specific RNAs
The results of our previous studies and those of other research groups demonstrated that the 5′ terminus of the alphavirus genome and its complement in the negative-strand RNA of the replicative intermediate function as part of the promoters for negative- and positive-strand RNA synthesis, respectively (Frolov, Hardy, and Rice, 2001; Gorchakov et al., 2004b; Niesters and Strauss, 1990a; Niesters and Strauss, 1990b). Therefore, the indicated G3A point mutation could have a significant effect on viral RNA replication. To test this possibility, we designed infectious cDNA clones for two recombinant alphavirus genomes (Fig. 4A), in which the subgenomic RNA encoded Sindbis virus-specific structural proteins. All of the nonstructural protein genes and cis-acting RNA elements, including the 5′ and 3′ UTRs, 51-nt CSE and the subgenomic promoter, were derived from the VEEV genome. The genomes of G3/VEE/SINV and A3/VEE/SINV differed only in the nt3, which was either TRD- or TC-83-specific (G3 or A3, respectively). The benefit of using VEEV/SINV chimeras was in their highly attenuated phenotype, resulting from the lack of VEEV structural protein genes (Petrakova et al., 2005). The replacement of the VEEV capsid by a SINV-specific counterpart was particularly important, because this protein is one of the determinants of VEEV pathogenesis in vivo and cytopathogenicity in vitro (Atasheva et al., 2008; Garmashova et al., 2007a; Garmashova et al., 2007b). Thus, the experiments with these chimeric viruses did not require high biocontainment conditions.
Fig. 4.
Synthesis of virus-specific RNAs in the cells infected with A3/VEE/SINV and G3/VEE/SINV. (A) The schematic representation of chimeric virus genomes. Open boxes indicate VEEV-specific sequences and filled boxes indicate SINV-specific structural genes. (B) BHK-21 cells were infected with G3/VEE/SINV and A3/VEE/SINV at an MOI of 10 PFU/cell. RNAs were labeled with [3H]uridine in the presence of ActD for 4 hours from 2, 4 and 6 h post infection, then isolated and analyzed by agarose gel electrophoresis under denaturing conditions as described in Materials and Methods. Bands corresponding to genomic and subgenomic RNAs were visualized by autoradiography. G and SG indicate positions of the viral genomic and subgenomic RNAs, respectively. (C) RNA bands were excised, and radioactivity in the genome and subgenomic RNA was measured by liquid scintillation counting. One of three reproducible experiments is presented.
Upon transfection into BHK-21 cells of the in vitro-synthesized RNAs (see Materials and Methods for details), the designed chimeras demonstrated low cytopathogenicity, but the released viruses were capable of developing plaques in BHK-21 cells under agarose cover in the presence of low concentrations of serum. The in vitro-synthesized RNAs exhibited the same infectivity as did VEEV TC-83 RNA in the infectious center assay (5–10×105 PFU/μg of transfected RNA) and universal plaque sizes (data not shown), which indicated that no adaptive mutations were required for the viability of the chimeras. Virus stocks having titers above 109 PFU/ml were harvested at 24 h post transfection. Next, we evaluated the synthesis of viral genomic and subgenomic RNAs at different times post infection by their metabolic labeling with [3H]uridine in the presence of ActD. The isolated RNAs were analyzed by agarose gel electrophoresis (Fig. 4B), and radioactivity in the bands, corresponding to virus-specific RNAs, was assessed (Fig. 4C). The results presented in Fig. 4 demonstrated that the G3A mutation had a positive effect on the replication of viral genome and, at the same time, affected subgenomic RNA synthesis (Figs. 4B and C), resulting in an almost 6-fold decrease in the molar ratio of the subgenomic to genomic (SG:G) RNA synthesis.
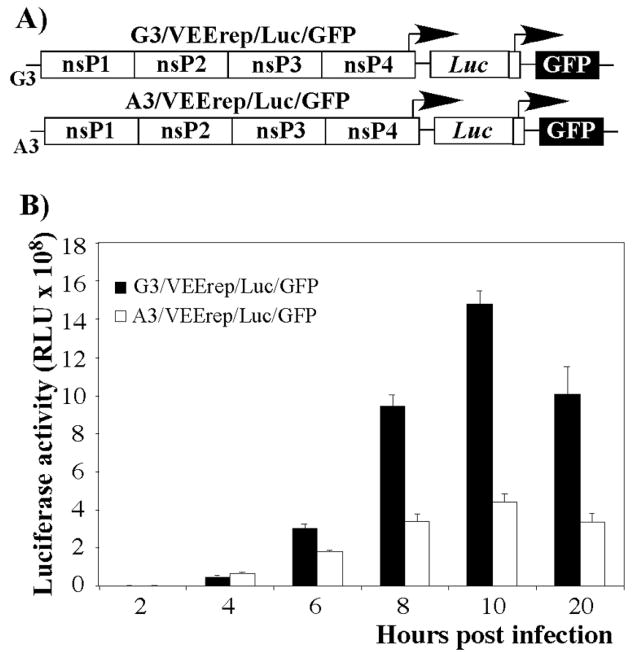
To confirm the effect of the mutation on the subgenomic RNA synthesis, we designed VEEV replicons A3/VEErep/Luc/GFP and G3/VEErep/Luc/GFP, encoding firefly luciferase and GFP under the control of the subgenomic promoters (Fig. 5A). Both replicons were packaged into infectious virus particles by using capsid- and glycoprotein-producing helpers. After determining the titers, cells were infected at the same multiplicity, and luciferase activity was determined at different times post infection. In multiple reproducible experiments, replicons with TC-83-specific 5′UTR produced lower levels of luciferase from 6 h post infection, compared to the replicons that encoded the TRD-specific 5′UTR. A significant, 3-fold difference was observed by 10 and 12 h post transfection. Similar results were obtained by measuring GFP fluorescence levels by flow cytometry (data not shown), as we detected a 3-fold difference in the mean fluorescence of GFP by 8 h post infection.
Fig. 5.
Expression of heterologous genes encoded by subgenomic RNA of VEEV replicons having either VEEV TRD- or VEEV TC-83-specific 5′UTRs. (A) The schematic representation of the replicons’ genomes. (B) BHK-21 cells were infected with packaged A3/VEErep/Luc/GFP and G3/VEErep/Luc/GFP replicons at an MOI of 10 inf.u/cell and the expression of luciferase was measured at the indicated time points.
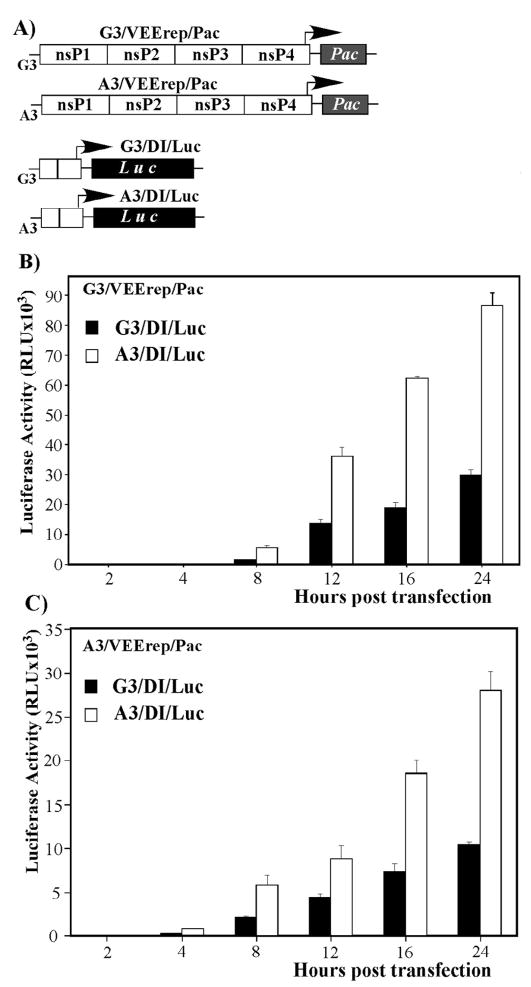
A higher level of genomic RNA synthesis by virus harboring G3A mutation could have multiple explanations. Therefore, to further understand the effect of the mutation on virus replication, we designed i) VEEV replicons A3/VEErep/Pac and G3/VEErep/Pac and ii) defective viral genomes A3/DI/Luc and G3/DI/Luc (Fig. 6A). The VEEV replicons encoded the viral nonstructural proteins and the Pac gene, which is unrelated to viral infection, under the subgenomic promoter; while the DI genomes encoded the 5′-terminal 519 nucleotides of VEEV genome and the firefly luciferase, cloned under control of the subgenomic promoter (Fig. 6A). The DI RNAs contained no nonstructural genes, were incapable of self-replication, and, thus, required trans-complementation with nsPs. Upon delivery into the same cell, replicon genomes were expected to produce VEEV nsPs for both their own and DI RNA synthesis. This resulted in a competition between the DI RNAs and the replicon genomes for the latter proteins. The efficiency of competition was determined by the promoter sequences, and, ultimately, it defined the level of luciferase expression.
Fig. 6.
Replication of the DI RNAs, encoding VEEV TRD- and VEEV TC-83-specific 5′UTRs, in the presence of VEEV replicons. (A) The schematic representation of the DI RNA and VEEV replicon genomes used in the study. (B and C) BHK-21 cells were co-transfected with 2μg of indicated replicons and 2 μg of indicated DI RNAs. Equal numbers of electroporated cells were seeded into 35-mm dishes and incubated at 37°C in 5% CO2. At the indicated time points, cells were lysed, and luciferase activity was measured as described in Materials and Methods.
Different combinations of the in vitro-synthesized replicon and DI genomes were transfected into BHK-21 cells (see Materials and Methods for details), and the luciferase activity was examined at different times post electroporation. In all of the experiments, the A3/DI/Luc RNA, having TC-83-derived 5′UTR, produced more luciferase than did G3/DI/Luc (Figs. 6B and C) and, thus, was more efficient in utilizing the VEEV replicative enzymes supplied in trans. Notably, the A3/DI/Luc RNA replication was likely even higher than one might assume from the data presented in Fig. 6B, as the DI genomes containing a vaccine strain-specific G3A mutation were less efficient in the transcription of the subgenomic RNA (Fig. 4), and still demonstrated higher levels of luciferase expression.
These results indicated that the more efficient replication of viral genomes (Fig. 4), having the TC-83-specific 5′-terminal sequence, was not the result of the positive effect of G3A mutation on translation efficiency, but rather depended on the modification of the RNA promoter element, leading to its more efficient functioning in genome replication. The indicated mutation also made the promoter of positive-strand RNA synthesis more competitive with the subgenomic promoter for the replicative enzymes, and, thus, reduced the molar ratio of SG:G RNA synthesis.
Virus replication
To evaluate the effect of the G3A mutation on virus replication, we assessed the replication rates of both A3/VEE/SINV and G3/VEE/SINV variants in BHK-21 and NIH 3T3 cells. As indicated above, both viruses were poorly cytopathic, but demonstrated efficient replication in both cell types, albeit at lower rates than did VEEV TC-83 (Fig. 7). The more efficient replication of TC-83 was not surprising, because formation of infectious viral particles was determined by the interaction of the genome with heterologous, SINV-derived capsid protein. Our previous studies strongly indicated that the VEEV ns polyprotein-coding sequence contains RNA elements, so-called packaging signals (PS), that promote genome packaging into the nucleocapsid (Volkova, Gorchakov, and Frolov, 2006). Therefore, it is highly unlikely that the SINV capsid is capable of recognizing the VEEV-specific PSs, and, based on very low levels of the nucleotide sequence identity, it was impossible to expect presence of SINV-specific RCs in the VEEV-specific sequence. Efficient formation of viral particles was rather determined either by close compartmentalization of the capsid and newly synthesized RNA or by the presentation of the latter RNA by the replicative complexes for packaging. In multiple repeat experiments, the replication of A3/VEE/SINV was always detectably higher than that of G3/VEE/SINV. This increase correlated with the more efficient synthesis of viral genome RNA, suggesting its critical role in virus production.
Fig. 7.
Analysis of virus replication in BHK-21 and NIH-3T3 cells. Cells were infected with G3/VEE/SINV and A3/VEE/SINV at an MOI of 10 PFU/cell. Media was replaced at the indicated time points, and virus titers were analyzed by plaque assay on BHK-21 cells. The experiments were repeated three times with very high reproducibility. Figure represents one of the repeated experiments.
DISCUSSION
In the infected cells, alphaviruses produce only a handful of proteins, which function in replication of the viral genome and assembly of infectious virions. In addition to their role in virus replication and assembly, these proteins also determine other aspects of virus-host cell interactions, such as the development of a cytopathic effect and downregulation of the cell response to virus replication (Fazakerley et al., 2002; Frolova et al., 2002; Gorchakov, Frolova, and Frolov, 2005; Gorchakov et al., 2008a; Gorchakov et al., 2004a). The level of RNA replication required for efficient packaging and assembly of virions depends on the functioning of the promoter elements that are encoded by the virus genome and negative-strand RNA intermediate (Strauss and Strauss, 1994). The very 5′ end of the VEEV genome has been shown to contain two functional elements (Gorchakov et al., 2004b; Michel et al., 2007), one of which is located in the 5′UTR, and the second is present in the nsP1-coding sequence. The latter cis-acting element, the 51-nt CSE, is a replication enhancer that appears to function in VEEV RNA replication by positioning of the replicative enzymes in the initiation of the RNA synthesis (Michel et al., 2007). The negative effect of the mutations in the VEEV and SINV genome 51-nt CSEs can be compensated by the mutations in nsP2 and nsP3 (Fayzulin and Frolov, 2004; Michel et al., 2007). The 5′UTR (more precisely, its complement in the negative strand of the viral genome) was defined as a core promoter, and some of the mutations in this sequence or its replacement by the 5′UTR derived from other alphaviruses, have deleterious effects on RNA replication (Gorchakov et al., 2004b; Niesters and Strauss, 1990a). Our previously published data and those of other research groups demonstrated that the G3A mutation in the VEEV 5′UTR strongly affects the pathogenicity (Kinney et al., 1993; White et al., 2001) of the virus and the ability of the VEEV-specific replicons to cause CPE (Petrakova et al., 2005). Interestingly, there was no correlation between these two characteristics: the replication of the VEEV TC-83-specific replicon, which lacks the structural genes (A3/VEErep/Pac), is more cytopathic (Petrakova et al., 2005). Cells, transfected with A3/VEErep/Pac replicons form fewer PurR foci and demonstrate dramatically slower growth rates compared to those transfected with the replicons having the same nucleotide sequence but G3 in the 5′UTR. On the other hand, G3A mutation in the VEEV TRD background makes the virus less pathogenic in mice (Kinney et al., 1993; White et al., 2001), albeit this effect appears to be mouse strain- and age-dependent.
Our new data provide a plausible explanation for the variances in the cytopathogenicity of the replicons differing only in the nt3 of the 5′UTR and suggest that the secondary structure of this RNA fragment plays crucial role(s) in virus replication. The TC-83-specific G3A mutation changed folding of the 5′UTR and, based on computer predictions, the secondary structure of the 3′ end of the negative strand of VEEV genome as well. Its effect on the RNA secondary structure was further confirmed by enzymatic probing, UV-melting experiments and NMR studies. Our results clearly demonstrate that the indicated mutation reduced the stability of the very 5′-terminal stem-loop and, most likely, caused a similar change in the 3′end of the negative-strand intermediate. Accordingly, this mutation could affect multiple processes of virus replication. The effect of this mutation on the secondary structure of the negative strand is not very clear. This uncertainty comes from the fact that, to date, it has not been unambiguously demonstrated whether the 3′ terminus of the negative strand is annealed to the complementary sequence at the 5′UTR of viral genome, or if it remains in free form and is capable of folding into the stem-loop during virus replication. The experimental data from our previous studies indicate that the replicative RNA intermediate is a dsRNA (Gorchakov et al., 2008b); however, the negative-strand RNA could function in both genomic and subgenomic RNA synthesis in vitro (Li and Stollar, 2004; Li and Stollar, 2007), which suggests that dsRNA formation, at least in the promoter sites, is not necessary. In addition to these findings, the in vitro experiments have shown that the 3′ end of the negative-strand genome can bind a number of cellular proteins (Pardigon, Lenches, and Strauss, 1993). Moreover, the genomic RNA synthesis is partially terminated in close proximity to another (SG) promoter (Wielgosz and Huang, 1997), and, in the replicative intermediate, this sequence is also sensitive to the RNase digestion (Simmons and Strauss, 1972). The latter phenomena lead us to suggest that in the dsRNA intermediate, the RNA promoter elements might be present in the ss form. Based on these data, we hypothesized that the G3A mutation-specific change in the RNA secondary structure and stability could result both in modification of the translation efficiency of the RNA and affect the function of the 5′UTR-associated promoter(s) in the RNA synthesis.
It was surprising to find that the G3A mutation had almost no effect on the 5′UTR activity in translation of the encoded proteins, because stable stem-loops at the 5′ terminus are generally believed to strongly affect the translation efficiency of the templates (Kozak, 2005). However, some of the available data suggest that this is also the case for another alphavirus, Sindbis virus. The pathogenicity of the neuroinvasive strain of SINV was shown to be determined by the mutation of nt8 in the 5′UTR (Dubuisson et al., 1997). This mutation stabilized the 5′-terminal stem, but did not alter the translation efficiency of the RNA in vitro. The pathogenic and nonpathogenic strains of SINV also differ in nt5. As in the case of VEEV G3A mutation, the G5A mutation, which is present in tissue culture-adapted strain of SINV and in the widely used SINV Toto1101, is also predicted to destabilize the 5′-terminal stem (McKnight et al., 1996), but it caused only a minor increase in the efficiency of RNA translation (Nickens and Hardy, 2008).
The G3A mutation in the VEEV genome mediated a higher level of RNA replication and downregulated transcription of the subgenomic RNA. Analysis of the level of viral RNA synthesis in the infected cells and the DI RNA competition experiments strongly suggested that the promoters of the positive-strand genome synthesis and the transcription of the subgenomic RNA appear to compete for the replicative enzymes. In the case of the TC-83-specific 5′UTR, the balance is shifted towards genomic RNA synthesis. Based on the available data on SINV replication (Gorchakov et al., 2004b; Nickens and Hardy, 2008), it is possible that the G3A mutation might also affect the negative-strand RNA synthesis due to the interaction of the 3′ and 5′ ends of the viral genome. Nevertheless, this additional effect, on the other hand, would not affect the current hypothesis that the nt3 mutation in VEEV TC-83 changes RNA and virus replication by favoring viral genomic RNA synthesis.
The ability of VEEV to cause a disease was previously shown to depend on multiple factors and, first of all, on the amino acid sequence of E2 glycoprotein. A single amino acid change in this protein led to profound attenuation of the virus (Kinney et al., 1993). Secondly, the VEEV capsid protein was found to interfere with nuclear-cytoplasmic trafficking and, ultimately, to inhibit the transcription of cellular messenger and ribosomal RNAs. Modifications of the defined 35-aa-long capsid-specific peptide or its replacement, with one derived from the SINV capsid, made the virus dramatically less cytopathic and strongly attenuated (Garmashova et al., 2007a). In this study, we have demonstrated that the TC-83-specific, 5′UTR mutation strongly affects the secondary structure of the 5′ terminus of the viral genome. The G3A mutation has no significant effect on the RNA translation efficiency, but increases replication of both viral genome and virus itself in tissue culture. Similar G5A mutation was previously described for the cell culture-adapted SINV strain. Thus, higher RNA replication rates of both VEEV and SINV might affect the intimate balance between the virus-induced cell response and the ability of the virus to interfere or escape its development (Nickens and Hardy, 2008; White et al., 2001). This, in combination with the previously described, G3A-induced higher sensitivity to IFN-α/β (White et al., 2001), appears to form a strong basis for the attenuated phenotype of VEEV TC-83.
MATERIALS AND METHODS
Cell cultures
BHK-21 cells were kindly provided by Dr. Sondra Schlesinger (Washington University, St Louis, MO). NIH 3T3 cells were obtained from the American Type Tissue Culture Collection (Manassas, Va). BHK-21 and NIH 3T3 cells were propagated in Alpha MEM supplemented with 10% fetal bovine serum (FBS) and vitamins.
Plasmid constructs
The parental plasmid with VEEV TC-83 genome was described elsewhere (Petrakova et al., 2005). It contained a cDNA of the viral genome under the control of an SP6 DNA-dependent RNA polymerase promoter. pG3/VEE/SINV, encoding the genome of G3/VEE/SINV chimeric virus genome was described elsewhere as VEE/SINV (Garmashova et al., 2007b; Petrakova et al., 2005). In this report, the name of the virus was changed to make it consistent with other constructs. This plasmid encoded 5′UTR of the VEEV TRD strain, 3′ UTR, subgenomic promoter and ns polyprotein-coding sequence from VEEV TC-83 and structural polyprotein-coding sequence from SINV Toto1101 (Rice et al., 1987). pA3/VEE/SINV differed only by one nucleotide (A3), specific for the VEEV TC-83 5′UTR. In both genomes, the poly(A) sequence was followed by a MluI restriction site. pA3/VEErep/Pac and pG3/VEErep/Pac replicons had viral structural genes replaced by a puromycin acetyltransferase (Pac) sequence, and were described elsewhere (Petrakova et al., 2005).
All other plasmids encoding modified VEEV genomes and luciferase were constructed by standard PCR-based mutagenesis and cloning methods. After cloning into the plasmids, all of the PCR fragments were sequenced to exclude the possibility of spontaneous mutations. The details of the cloning procedures and sequences can be provided upon request. pG3/Luc and pA3/Luc encoded the promoter for the SP6 DNA-dependent RNA polymerase, followed by nt 1–191 of the viral genome, fused with the entire ubiquitin gene and firefly luciferase-coding sequence. This protein-coding sequence was followed by VEEV TC-83-specific 3′UTR, poly(A) and MluI restriction site, required for linearization of the plasmid before in vitro transcription reaction.
pG3/DI/Luc and pA3/DI/Luc plasmids encoded defective viral genomes under control of the SP6 promoter. These genomes contained the 5′ terminal 519 nt derived from VEEV TRD or VEEV TC-83 genomes, respectively, followed by nt 7291–7564 (encoding VEEV subgenomic promoter), a firefly luciferase gene, 3′ terminal nt 11202–11446 of VEEV genome, poly(A) tail and MluI restriction site.
RNA transcriptions
All of the plasmids were purified by centrifugation in CsCl gradients. Prior to transcription, they were lineraized by MluI, and RNAs were synthesized in vitro by SP6 RNA polymerase in the presence of cap analog (Rice et al., 1987) under the conditions recommended by the manufacturer (Invitrogen). The yield and integrity of the transcripts were monitored by gel electrophoresis under nondenaturing conditions, followed by analysis of the RNA concentration on a FluorChem imager (Alpha Innotech). For virus rescue and analysis of DI RNA replication, the appropriate volumes of reaction mixtures were directly used for electroporation. For comparative studies of translation efficiencies of the Luc-coding RNAs, the transcripts were additionally purified using RNeasy columns (Qiagen), and RNA concentration was measured as described above.
RNA transfections
BHK-21 cells were electroporated with 2 μg of in vitro-synthesized, viral genome RNA using previously described conditions (Liljeström et al., 1991), and cells were seeded into 100-mm dishes. The released viruses were harvested 24 h post transfection. Titers were determined by a plaque assay on BHK-21 cells. For analysis of DI RNA replication, replicons and DI RNAs were co-electroporated as described in the figure legends. Equal amounts of cells were seeded into 35-mm dishes, and luciferase activity was measured at different times post transfection by using a luciferase assay kit according to the manufacturer’s instructions (Promega). To assess the translation efficiency of the templates, equal amounts of A3/Luc and G3/Luc RNAs were electroporated into BHK-21 cells, equal aliquots of the cells were seeded into 35-mm dishes, and luciferase activity was measured at different times post transfection. To determine the stability of the RNAs, similar amounts of 32P-labeled, in vitro-synthesized A3/Luc and G3/Luc RNAs were electroporated into BHK-21 cells, equal aliquots of the cells were seeded into 35-mm dishes. At the indicated times, RNAs were isolated by using TRizol reagent as recommended by the manufacturer (Invitrogen), followed by electrophoresis in agarose gel and autoradiography. Dried gels were further analyzed on a Storm phosphorimager (Molecular Dynamics). The radioactivity detected in the RNA bands, was normalized on that found in the 0 h samples isolated immediately after RNA transfection. Replicons were packaged into infectious viral particles by co-electroporation of the in vitro-synthesized replicon and two helper RNAs (Volkova, Gorchakov, and Frolov, 2006) into BHK-21 cells. Media were harvested 24 h post electroporation, and titers of the packaged replicons were determined by infecting BHK-21 cells with different dilutions of the samples and measuring the numbers of infected, GFP-expressing cells after 8 h of incubation at 37°C in a CO2 incubator.
In vivo translation assay
Different amounts of in vitro-synthesized A3/Luc and G3/Luc RNAs, presented in the Fig. 3, were translated in 10 μl of the rabbit reticulocyte lysate (RRL) system according to the manufacturer’s instructions (Promega). Aliquots of the reaction mixtures were taken after 1 h incubation at 30°C, and luciferase activity was measured by a luciferase assay kit according to the manufacture’s protocol (Promega).
Analysis of virus replication
5 ×105 BHK-21 or NIH 3T3 cells were seeded into 35-mm dishes. After 4 h of incubation at 37°C in 5% CO2, monolayers were infected at the multiplicities of infection (MOI) indicated in the legend to Fig. 7, for 1 h, washed three times with phosphate-buffered saline (PBS), and overlaid with 1 ml of complete medium. At the indicated times post infection, media were replaced by fresh media, and virus titers in the harvested samples were determined by plaque assay on BHK-21 cells as previously described (Lemm et al., 1990).
RNA analysis
BHK-21 cells were infected with chimeric viruses at an MOI of 10 PFU/cell. At the indicated times post infection, the intracellular RNAs were labeled with [3H]uridine (20 μCi/ml), in the presence of 1 μg/ml of dactinomycin (ActD)/ml for 4 h at 37°C in 5% CO2. RNAs were isolated from the cells using TRIzol reagent according to the manufacturer’s instructions (Invitrogen). The RNAs were denatured with glyoxal in dimethyl sulfoxide and analyzed by agarose gel electrophoresis as previously described (Bredenbeek et al., 1993). For quantitative analysis, the RNA bands were excised from the 2,5-diphenyloxazole (PPO)-impregnated gels, and the radioactivity was measured by liquid scintillation counting.
Enzymatic analysis of the RNA secondary structure
RNA secondary structure mapping experiments were carried out using a protocol and reagents obtained from Ambion. Briefly, the in vitro-synthesized RNA fragments were purified by gel electrophoresis, eluted, dephosphorylated and 5′ labeled using T4 polynucleotide kinase from the KinaseMax kit (Ambion) and [γ-32P] ATP (Amersham). Labeled RNAs were purified by denaturing 12% polyacrylamide gel, and eluted into SDS-containing buffer, followed by ethanol precipitation. In some experiments, 32P-labeled RNA transcripts were digested in the buffer containing 10 mM Tris–HCl pH 7.0, 100 mM KCl, 10mM MgCl2), supplemented with 1 μg of yeast tRNA. Digestions were performed by either RNase T1 (0.01 or 0.001U/μl), or RNase A (0.01 or 0.001 μg/ml), or RNase V1 (0.001 or 0.0001U/μl) at 25° C for 10 min. Samples were precipitated, re-suspended in 10μl of denaturing loading buffer, and subjected to electrophoresis in 12% or 20% sequencing gels. Alkaline hydrolysis ladders were obtained by treatment of 5′ end-labeled transcripts in the alkaline buffer (Ambion) at 95 °C for 5min.
NMR spectroscopy
The 31-mer RNA molecules, corresponding to residues 1 – 31 of the 5′UTRs of VEEV TRD and VEEV TC-83 genomes (see Fig. 1A for details), were synthesized by in vitro transcription of the DNA template using the T7 DNA-dependent RNA polymerase. The HPLC-purified DNA templates were obtained from IDT and additionally tested by electrophoresis in a polyacrylamide gel. After transcription, RNA molecules were precipitated by ethanol and purified on 20% polyacrylamide gels supplemented with 8 M urea. The RNAs were extracted from the gel by electroelution, recovered by ethanol precipitation and concentrated by centrifugation using a Centricon-3 concentrator (Millipore). The samples were dialyzed against 10 mM Na-phosphate buffer (pH 6.5), supplemented with 10 mM KCl and 0.05 mM EDTA, and lyophilized. The quality of the samples was confirmed by gel electrophoresis. For the experiments involving exchangeable hydrogen or non-exchangeable hydrogen, the lyophilized RNA samples were then dissolved in 300μl of either 90% H2O/10% D2O or 99.96% D2O, respectively. In order to facilitate stem-loop formation the RNA samples were heated to 95°C for three minutes followed by rapid cooling on ice to reduce duplex RNA formation. RNA concentrations in the samples used for the NMR experiments were 0.25mM.
All of the NMR spectra were acquired on Varian Direct Drive type spectrometers (Varian Associates, Palo Alto, CA), operating at either 750 MHz or 800 MHz, equipped with pulsed field gradients and direct-drive architecture. Melting of the RNA molecule was studied by monitoring the imino proton signals as a function of temperature. 1D NOESY spectra were collected at 5° C, 10° C, 15° C, 20° C, and 25° C using spectral widths of 16000 Hz and 1024 complex data points. To identify base-pairing patterns and NOE interactions between exchangeable hydrogens, 2D NOESY spectra (with 250 ms and 400 ms mixing times) were acquired at 5° C and 25°C, spectral widths of 7600 Hz and 8000 Hz and 2048 complex data points, using samples dissolved in 90%H2O/10% D2O. Additional NOESY spectra, using identical NMR parameters, were acquired with the samples in 99.96% D2O. Acquired NMR data were processed by using VNMRJ software. HOD signal at 4.76 ppm was used for referencing the 1D and 2D spectra.
Acknowledgments
The authors would like to thank Edward Nikonowicz for suggestions and discussions related to NMR experiments and David Volk for critical reading of the manuscript. This work was supported by Public Health Service grant AI070207 to IF, NHLBI grant N01HV28184, and the Welch Foundation grant H1296 to DG. RKS was supported in part by the McLaughlin Endowment Predoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alevizatos AC, McKinney RW, Feigin RD. Live, attenuated Venezuelan equine encephalomyelitis virus vaccine. I Clinical effects in man Am J Trop Med Hyg. 1967;16(6):762–8. [PubMed] [Google Scholar]
- Atasheva S, Garmashova N, Frolov I, Frolova E. Venezuelan equine encephalitis virus capsid protein inhibits nuclear import in Mammalian but not in mosquito cells. J Virol. 2008;82(8):4028–41. doi: 10.1128/JVI.02330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton DJ, Sawicki DL, Sawicki SG. Association of alphavirus replication with the cytoskeletal framework and transcription in vitro in the absence of membranes. In: Brinton MA, Heinz FX, editors. New aspects of positive-strand RNA viruses. American Society for Microbiology; Washington, D.C.: 1990. pp. 75–79. [Google Scholar]
- Berge TO, Banks IS, Tigertt WD. Attenuation of Venezuelan equine encephalomyelitis virus by in vitro cultivation in guinea pig heart cells. Am J Hyg. 1961;73:209–218. [Google Scholar]
- Bernard KA, Klimstra WB, Johnston RE. Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology. 2000;276 (1):93–103. doi: 10.1006/viro.2000.0546. [DOI] [PubMed] [Google Scholar]
- Bredenbeek PJ, Frolov I, Rice CM, Schlesinger S. Sindbis virus expression vectors: Packaging of RNA replicons by using defective helper RNAs. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Canto MC, Rabinowitz SG. Central nervous system demyelination in Venezuelan equine encephalomyelitis infection. J Neurol Sci. 1981;49 (3):397–418. doi: 10.1016/0022-510x(81)90030-7. [DOI] [PubMed] [Google Scholar]
- de la Monte SM, Castro F, Bonilla NJ, de Urdaneta AG, Hutchins GM. The systemic pathology of Venezuelan equine encephalitis virus infection in humans. Am J Trop Med Hyg. 1985;34(1):194–202. doi: 10.4269/ajtmh.1985.34.194. [DOI] [PubMed] [Google Scholar]
- Dubuisson J, Lustig S, Akov Y, Rice CM. Genetic determinants of Sindbis virus neuroinvasiveness. J Virol. 1997;71:2636–2646. doi: 10.1128/jvi.71.4.2636-2646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenkranz NJ, Ventura AK. Venezuelan equine encephalitis virus infection in man. Annu Rev Med. 1974;25:9–14. doi: 10.1146/annurev.me.25.020174.000301. [DOI] [PubMed] [Google Scholar]
- Fayzulin R, Frolov I. Changes of the secondary structure of the 5′ end of the Sindbis virus genome inhibit virus growth in mosquito cells and lead to accumulation of adaptive mutations. J Virol. 2004;78 (10):4953–64. doi: 10.1128/JVI.78.10.4953-4964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazakerley JK, Boyd A, Mikkola ML, Kaariainen L. A single amino acid change in the nuclear localization sequence of the nsP2 protein affects the neurovirulence of Semliki Forest virus. J Virol. 2002;76 (1):392–6. doi: 10.1128/JVI.76.1.392-396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I, Hardy R, Rice CM. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. Rna. 2001;7(11):1638–51. doi: 10.1017/s135583820101010x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova EI, Fayzulin RZ, Cook SH, Griffin DE, Rice CM, Frolov I. Roles of nonstructural protein nsP2 and Alpha/Beta interferons in determining the outcome of Sindbis virus infection. J Virol. 2002;76 (22):11254–64. doi: 10.1128/JVI.76.22.11254-11264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmashova N, Atasheva S, Kang W, Weaver SC, Frolova E, Frolov I. Analysis of Venezuelan equine encephalitis virus capsid protein function in the inhibition of cellular transcription. J Virol. 2007a;81 (24):13552–65. doi: 10.1128/JVI.01576-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmashova N, Gorchakov R, Volkova E, Paessler S, Frolova E, Frolov I. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J Virol. 2007b;81 (5):2472–84. doi: 10.1128/JVI.02073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R, Frolova E, Frolov I. Inhibition of transcription and translation in Sindbis virus-infected cells. J Virol. 2005;79 (15):9397–409. doi: 10.1128/JVI.79.15.9397-9409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R, Frolova E, Sawicki S, Atasheva S, Sawicki D, Frolov I. A new role for ns polyprotein cleavage in Sindbis virus replication. J Virol. 2008a;82 (13):6218–31. doi: 10.1128/JVI.02624-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R, Frolova E, Williams BR, Rice CM, Frolov I. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J Virol. 2004a;78 (16):8455–67. doi: 10.1128/JVI.78.16.8455-8467.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R, Garmashova N, Frolova E, Frolov I. Different types of nsP3-containing protein complexes in sindbis virus-infected cells. J Virol. 2008b;82 (20):10088–101. doi: 10.1128/JVI.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R, Hardy R, Rice CM, Frolov I. Selection of functional 5′ cis-acting elements promoting efficient sindbis virus genome replication. J Virol. 2004b;78 (1):61–75. doi: 10.1128/JVI.78.1.61-75.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin DE. Alphaviruses. In: Knipe DM, Howley PM, editors. Fields’ Virology. 4. Lippincott, Williams and Wilkins; New York: 2001. pp. 917–962. [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31 (13):3429–31. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger JA, Turner DH, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KM, Shelokov A, Peralta PH, Dammin GJ, Young NA. Recovery of Venezuelan equine encephalomyelitis virus in Panama. A fatal case in man Am J Trop Med Hyg. 1968;17 (3):432–440. doi: 10.4269/ajtmh.1968.17.432. [DOI] [PubMed] [Google Scholar]
- Johnston RE, Peters CJ. Alphaviruses. In: Fields BN, Knipe DM, Howley PM, editors. Virology. 3. Lippincott-Raven; New York: 1996. pp. 843–898. [Google Scholar]
- Johnston RE, Smith JF. Selection for accelerated penetration in cell culture coselects for attenuated mutants of Venezuelan equine encephalitis virus. Virology. 1988;162(2):437–43. doi: 10.1016/0042-6822(88)90484-9. [DOI] [PubMed] [Google Scholar]
- Kamrud KI, Alterson KD, Andrews C, Copp LO, Lewis WC, Hubby B, Patel D, Rayner JO, Talarico T, Smith JF. Analysis of Venezuelan equine encephalitis replicon particles packaged in different coats. PLoS ONE. 2008;3(7):e2709. doi: 10.1371/journal.pone.0002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney RM, Chang GJ, Tsuchiya KR, Sneider JM, Roehrig JT, Woodward TM, Trent DW. Attenuation of Venezuelan equine encephalitis virus strain TC-83 is encoded by the 5′-noncoding region and the E2 envelope glycoprotein. J Virol. 1993;67(3):1269–77. doi: 10.1128/jvi.67.3.1269-1277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney RM, Johnson BJB, Welch JB, Tsuchiya KR, Trent DW. The full-length nucleotide sequences of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and its attenuated vaccine derivative, strain TC-83. Virology. 1989;170:19–30. doi: 10.1016/0042-6822(89)90347-4. [DOI] [PubMed] [Google Scholar]
- Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Lemm JA, Durbin RK, Stollar V, Rice CM. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J Virol. 1990;64:3001–3011. doi: 10.1128/jvi.64.6.3001-3011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm JA, Rice CM. Assembly of functional Sindbis virus RNA replication complexes: Requirement for coexpression of P123 and P34. J Virol. 1993;67:1905–1915. doi: 10.1128/jvi.67.4.1905-1915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon CA. Sequelae of Venezuelan equine encephalitis in humans: a four year follow-up. Int J Epidemiol. 1975;4 (2):131–40. doi: 10.1093/ije/4.2.131. [DOI] [PubMed] [Google Scholar]
- Li ML, Stollar V. Identification of the amino acid sequence in Sindbis virus nsP4 that binds to the promoter for the synthesis of the subgenomic RNA. Proc Natl Acad Sci U S A. 2004;101 (25):9429–34. doi: 10.1073/pnas.0400995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ML, Stollar V. Distinct sites on the Sindbis virus RNA-dependent RNA polymerase for binding to the promoters for the synthesis of genomic and subgenomic RNA. J Virol. 2007;81(8):4371–3. doi: 10.1128/JVI.02672-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeström P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig GV, Turell MJ, Vogel P, Kondig JP, Kell WK, Smith JF, Pratt WD. Comparative neurovirulence of attenuated and non-attenuated strains of Venezuelan equine encephalitis virus in mice. Am J Trop Med Hyg. 2001;64(1–2):49–55. doi: 10.4269/ajtmh.2001.64.49. [DOI] [PubMed] [Google Scholar]
- McKnight KL, Simpson DA, Lin SC, Knott TA, Polo JM, Pence DF, Johannsen DB, Heidner HW, Davis NL, Johnston RE. Deduced consensus sequence of Sindbis Virus strain AR339: mutations contained In laboratory strains which affect cell culture and in vivo phenotypes. J Virol. 1996;70(3):1981–1989. doi: 10.1128/jvi.70.3.1981-1989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel G, Petrakova O, Atasheva S, Frolov I. Adaptation of Venezuelan equine encephalitis virus lacking 51-nt conserved sequence element to replication in mammalian and mosquito cells. Virology. 2007;362 (2):475–87. doi: 10.1016/j.virol.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickens DG, Hardy RW. Structural and functional analyses of stem-loop 1 of the Sindbis virus genome. Virology. 2008;370(1):158–72. doi: 10.1016/j.virol.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Niesters HGM, Strauss JH. Defined mutations in the 5′ nontranslated sequence of Sindbis virus RNA. J Virol. 1990a;64:4162–4168. doi: 10.1128/jvi.64.9.4162-4168.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesters HGM, Strauss JH. Mutagenesis of the conserved 51 nucleotide region of Sindbis virus. J Virol. 1990b;64:1639–1647. doi: 10.1128/jvi.64.4.1639-1647.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paessler S, Fayzulin RZ, Anishchenko M, Greene IP, Weaver SC, Frolov I. Recombinant sindbis/Venezuelan equine encephalitis virus is highly attenuated and immunogenic. J Virol. 2003;77 (17):9278–86. doi: 10.1128/JVI.77.17.9278-9286.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardigon N, Lenches E, Strauss JH. Multiple binding sites for cellular proteins in the 3′ end of Sindbis alphavirus minus-sense RNA. J Virol. 1993;67 (8):5003–11. doi: 10.1128/jvi.67.8.5003-5011.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrakova O, Volkova E, Gorchakov R, Paessler S, Kinney RM, Frolov I. Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in Mammalian cells. J Virol. 2005;79 (12):7597–608. doi: 10.1128/JVI.79.12.7597-7608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice CM, Levis R, Strauss JH, Huang HV. Production of infectious RNA transcripts from Sindbis virus cDNA clones: Mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987;61 (12):3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas F, Diaz LA, Cardenas VM, Daza E, Bruzon L, Alcala A, De la Hoz O, Caceres FM, Aristizabal G, Martinez JW, Revelo D, De la Hoz F, Boshell J, Camacho T, Calderon L, Olano VA, Villarreal LI, Roselli D, Alvarez G, Ludwig G, Tsai T. Epidemic Venezuelan equine encephalitis in La Guajira, Colombia, 1995. J Infect Dis. 1997;175 (4):828–32. doi: 10.1086/513978. [DOI] [PubMed] [Google Scholar]
- Simmons DT, Strauss JH. Replication of Sindbis virus. II Multiple forms of double-stranded RNA isolated from infected cells J Mol Biol. 1972;71:615–631. doi: 10.1016/s0022-2836(72)80027-5. [DOI] [PubMed] [Google Scholar]
- Strauss EG, Rice CM, Strauss JH. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984;133:92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkova E, Gorchakov R, Frolov I. The efficient packaging of Venezuelan equine encephalitis virus-specific RNAs into viral particles is determined by nsP1-3 synthesis. Virology. 2006;344 (2):315–27. doi: 10.1016/j.virol.2005.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Barrett AD. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol. 2004;2 (10):789–801. doi: 10.1038/nrmicro1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Ferro C, Barrera R, Boshell J, Navarro JC. Venezuelan equine encephalitis. Annu Rev Entomol. 2004;49:141–74. doi: 10.1146/annurev.ento.49.061802.123422. [DOI] [PubMed] [Google Scholar]
- Weaver SC, Frolov I. Togaviruses. In: Mahy BWJ, Meulen Vt, editors. Virology. 2. ASM Press; Salisbury, UK: 2005. pp. 1010–1024. [Google Scholar]
- Weaver SC, Salas R, Rico-Hesse R, Ludwig GV, Oberste MS, Boshell J, Tesh RB. Re-emergence of epidemic Venezuelan equine encephalomyelitis in South America. VEE Study Group Lancet. 1996;348 (9025):436–40. doi: 10.1016/s0140-6736(96)02275-1. [DOI] [PubMed] [Google Scholar]
- White LJ, Wang JG, Davis NL, Johnston RE. Role of Alpha/Beta Interferon in Venezuelan Equine Encephalitis Virus Pathogenesis: Effect of an Attenuating Mutation in the 5′ Untranslated Region. J Virol. 2001;75 (8):3706–18. doi: 10.1128/JVI.75.8.3706-3718.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielgosz MM, Huang HV. A novel viral RNA species in Sindbis virus-infected cells. J Virol. 1997;71 (12):9108–17. doi: 10.1128/jvi.71.12.9108-9117.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuthrich K. NMR of Proteins and Nucleic Acids. John Wiley & Sons; New York: 1986. pp. 203–255. [Google Scholar]