Abstract
Mitochondrial complex II (succinate dehydrogenase) is part of the tricarboxylic acid cycle and the respiratory chain. Three nuclear genes encode its essential iron-sulfur subunit in Arabidopsis (Arabidopsis thaliana). One of them, SUCCINATE DEHYDROGENASE2-3 (SDH2-3), is specifically expressed in the embryo during seed maturation, suggesting that SDH2-3 may have a role as the complex II iron-sulfur subunit during embryo maturation and/or germination. Here, we present data demonstrating that three abscisic acid-responsive elements and one RY-like enhancer element, present in the SDH2-3 promoter, are involved in embryo-specific SDH2-3 transcriptional regulation. Furthermore, we show that ABSCISIC ACID INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEAFY COTYLEDON2, three key B3 domain transcription factors involved in gene expression during seed maturation, control SDH2-3 expression. Whereas ABI3 and FUS3 interact with the RY element in the SDH2-3 promoter, the abscisic acid-responsive elements are shown to be a target for bZIP53, a member of the basic leucine zipper (bZIP) family of transcription factors. We show that group S1 bZIP53 protein binds the promoter as a heterodimer with group C bZIP10 or bZIP25. To the best of our knowledge, the SDH2-3 promoter is the first embryo-specific promoter characterized for a mitochondrial respiratory complex protein. Characterization of succinate dehydrogenase activity in embryos from two homozygous sdh2-3 mutant lines permits us to conclude that SDH2-3 is the major iron-sulfur subunit of mature embryo complex II. Finally, the absence of SDH2-3 in mutant seeds slows down their germination, pointing to a role of SDH2-3-containing complex II at an early step of germination.
Succinate:ubiquinone oxidoreductase (succinate dehydrogenase [SDH]; EC 1.3.5.1), commonly referred to as mitochondrial complex II, has a central role in mitochondrial metabolism as a member of both the electron transport chain and the tricarboxylic acid (TCA) cycle. This important membrane-associated complex catalyzes the oxidation of succinate to fumarate and the reduction of ubiquinone to ubiquinol. In bacteria and heterotrophic eukaryotes, complex II is constituted by four subunits: two peripheral membrane proteins, a flavoprotein (SDH1) and an iron-sulfur protein (SDH2), and two small integral membrane proteins (SDH3 and SDH4; Lemire and Oyedotun, 2002; Yankovskaya et al., 2003). The succinate-binding site is formed by the SDH1 protein, and this flavoprotein subunit interacts with the SDH2 subunit, which contains three nonheme iron-sulfur centers acting as conductors of electrons from the flavoprotein to the membrane. The two integral membrane proteins, SDH3 and SDH4, anchor the SDH1-SDH2 subcomplex to the matrix side of the inner mitochondrial membrane and contain a b-type heme and the ubiquinone-binding site (Yankovskaya et al., 2003). Surprisingly, plant complex II may contain additional subunits of unknown function, along with the four classical subunits (Millar et al., 2004).
Complex II subunits are all encoded in the nuclear genome in Arabidopsis (Arabidopsis thaliana; Figueroa et al., 2001, 2002; Millar et al., 2004). Surprisingly, we found that three nuclear genes, designated SDH2-1 (At3g27380), SDH2-2 (At5g40650), and SDH2-3 (At5g65165), encode the iron-sulfur subunit in Arabidopsis (Figueroa et al., 2001). The three proteins would be functional as complex II iron-sulfur subunits, since they are highly conserved when compared with their homologs in other organisms and contain the Cys motifs involved in binding the three iron-sulfur clusters essential for electron transport (Figueroa et al., 2001). However, only SDH2-1 and SDH2-2 have been identified in the Arabidopsis mitochondrial proteome (Heazlewood et al., 2004). There is only one previous report describing more than one SDH2 gene in a eukaryotic organism, a sheep nematode, and this fact may be related to a switch in energy metabolism during development (Roos and Tielens, 1994). The unusual presence of three SDH2 genes in Arabidopsis raises interesting questions about their origin and function.
SDH2-1 and SDH2-2 genes likely arose via a relatively recent duplication event, while separation with SDH2-3 would be more ancient (Figueroa et al., 2001). This is supported by the completely different exon-intron structure of SDH2-3, which encodes a protein only 67% similar to SDH2-1 and SDH2-2. Moreover, whereas SDH2-1 and SDH2-2 have similar expression patterns, being expressed in all organs from adult plants (Figueroa et al., 2001; Elorza et al., 2004), SDH2-3 is highly expressed in the embryo during the maturation phase of seed development and SDH2-3 transcripts are abundant in dry seeds and decline during germination (Elorza et al., 2006). These data suggest that SDH2-1 and SDH2-2 are probably redundant and that SDH2-3 may have a specific role as the complex II iron-sulfur protein-coding gene during embryo maturation and/or germination.
Analysis of Arabidopsis plants carrying SDH2-3 promoter fusions to the GUS reporter gene allowed us to show that SDH2-3 expression is regulated at the transcriptional level during seed development (Elorza et al., 2006). In silico analysis of the promoter revealed the presence of three potential abscisic acid (ABA)-responsive elements (ABREs), characterized by the consensus sequence YACGTGGC containing the ACGT core (Busk and Pagès, 1998; Leung and Giraudat, 1998), and a RY-like enhancer element (Nambara and Marion-Poll, 2003). The seed-specific expression of SDH2-3 overlaps with that of genes encoding abundant seed storage proteins (SSPs; e.g. At2S3) and late embryogenesis abundant proteins (LEAs; e.g. AtEm1; Parcy et al., 1994). Since ABRE and RY elements have been implicated in the expression of these genes (Busk and Pagès, 1998; Nambara and Marion-Poll, 2003), we mutated them in the SDH2-3 promoter and show here that they are involved in the high embryo expression of SDH2-3.
ABSCISIC ACID INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEAFY COTYLEDON2 (LEC2) encode related plant-specific transcription factors containing the conserved B3 DNA-binding domain (Giraudat et al., 1992; Luerssen et al., 1998; Stone et al., 2001) and act in concert to regulate key pathways during seed maturation (Santos-Mendoza et al., 2008). They are interlocked in a cross-regulated network, resulting in overlapping or specific functions. For instance, abi3, fus3, and lec2 mutants share common phenotypes in reduced expression of SSP but exhibit specific phenotypes, such as ABA insensitivity (abi3), early germination of immature embryos (fus3), lack of chlorophyll degradation (abi3 and lec2), desiccation intolerance (abi3 and fus3), or leafy cotyledons (fus3 and lec2; To et al., 2006). Here, we demonstrate that ABI3, FUS3, and LEC2 are also involved in the regulation of SDH2-3 expression.
A wide range of mutant, antisense, or silenced plants with deficient expression of enzymes from the TCA cycle have been described, including citrate synthase (Landschütze et al., 1995), aconitase (Carrari et al., 2003), malate dehydrogenase (Nunes-Nesi et al., 2005), fumarase (Nunes-Nesi et al., 2007), succinyl CoA ligase (Studart-Guimarães et al., 2007), and NAD+-dependent isocitrate dehydrogenase (Lemaitre et al., 2007). These studies have shown that modifications in the TCA cycle can modulate photosynthetic performance and, in the case of potato (Solanum tuberosum) citrate synthase, lead to a specific disintegration of the ovary tissues of flower. Nevertheless, they have not analyzed the expression of TCA cycle genes during seed development, nor have phenotypic alterations been reported during seed maturation or germination.
To gain insight into the physiological role of complex II and to explore the function of the multiple genes encoding the same SDH subunit, our group has undertaken a reverse genetic analysis of the SDH genes (León et al., 2007). Here, we report the analysis of two insertional mutants in SDH2-3 and show that SDH2-3 is the major iron-sulfur subunit of embryo complex II and plays a role during germination.
RESULTS
ABRE and RY Elements Are Required for SDH2-3 Promoter Activity
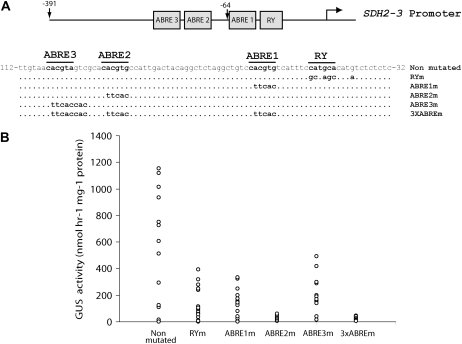
The 223 bp upstream of the SDH2-3 transcription start site are sufficient to confer high expression of the GUS reporter gene in mature seeds (Elorza et al., 2006) and have three potential ABRE elements (ABRE1, ABRE2, and ABRE3) and a RY-like enhancer element (Fig. 1A). Furthermore, removal of the region between −223 and −65 comprising ABRE2 and ABRE3 drastically reduced GUS expression (Elorza et al., 2006). To evaluate the function of these elements, constructs containing substitution mutations were made (Fig. 1A) and GUS activity was determined in mature T2 seeds from transgenic plants carrying the wild-type or mutated SDH2-3 promoters fused to GUS. Mutating any of the three ABRE elements or the RY element caused significant reduction of GUS expression, mutation of ABRE2 producing the most drastic reduction of activity (Fig. 1B).
Figure 1.
Mutation analysis of ABRE and RY elements in the SDH2-3 promoter. A, Structure of the mutant constructs. Motifs targeted by in vitro mutagenesis are boxed. Numbers are in relation to the transcription initiation site, indicated by a curved arrow. The sequence of the wild-type promoter is shown (Non mutated), and the mutated nucleotides in each construct are indicated below. B, GUS activity was measured in duplicate seed extracts from 12 to 20 independent transgenic lines carrying the wild-type or mutant promoters fused to GUS. Each point represents one transgenic line, and sd values have been omitted for clarity.
ABI3, FUS3, and LEC2 Are Involved in the Regulation of SDH2-3 Expression
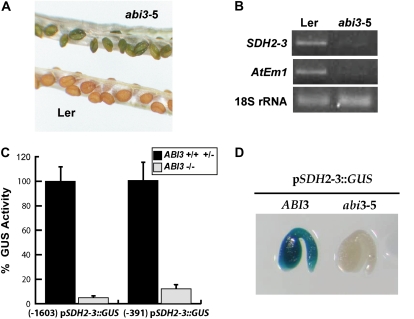
ABI3, FUS3, and LEC2 are considered master regulators of seed maturation (Santos-Mendoza et al., 2008). Accordingly, we analyzed SDH2-3 expression in a severe abi3-5 mutant allele (Ooms et al., 1993). Homozygous abi3-5 mutant seeds fail to degrade chlorophyll and are thus identified by their green color (Fig. 2A). The accumulation of SDH2-3 mRNA was dramatically reduced in abi3-5 seeds, as was the control LEA gene AtEm1 (Fig. 2B).
Figure 2.
ABI3 regulates SDH2-3 expression in seeds. A, Homozygous abi3-5 and wild-type (Ler) siliques. B, SDH2-3 expression is drastically reduced in abi3-5 seeds. RT-PCR analyses of SDH2-3 and AtEm1 transcripts were performed on total RNA from wild-type and abi3-5 seeds. Thirty-five amplification cycles were used for SDH2-3 and AtEm1, and 15 cycles were used for the 18S rRNA load control. C, SDH2-3 promoter activity is reduced in abi3-5 seeds. GUS activities were measured in protein extracts from brown seeds (black bars), carrying at least one wild-type ABI3 allele, and green homozygous abi3-5 seeds (gray bars). A value of 100% activity corresponded to 452 ± 63 nmol 4-methylumbelliferone h−1 mg−1 protein for the long SDH2-3 promoter∷GUS construct and 141 ± 25 nmol 4-methylumbelliferone h−1 mg−1 protein for the short SDH2-3 promoter∷GUS construct. Four biological replicates were performed. D, The SDH2-3 promoter is inactive in abi3-5 embryos. GUS staining was performed on embryos extracted from yellow and green seeds prior to complete desiccation.
The reduction of SDH2-3 mRNA levels in abi3-5 seeds is probably due to a decrease in promoter activity, since abi3-5 plants crossed with homozygous plants carrying either 1.6 or 0.4 kb of the SDH2-3 promoter:GUS fusions showed a dramatic reduction of GUS activity (Fig. 2C). Furthermore, no GUS staining was observed in any isolated embryo from ABI3-deficient green seeds, whereas embryos containing wild-type ABI3 alleles showed strong staining.
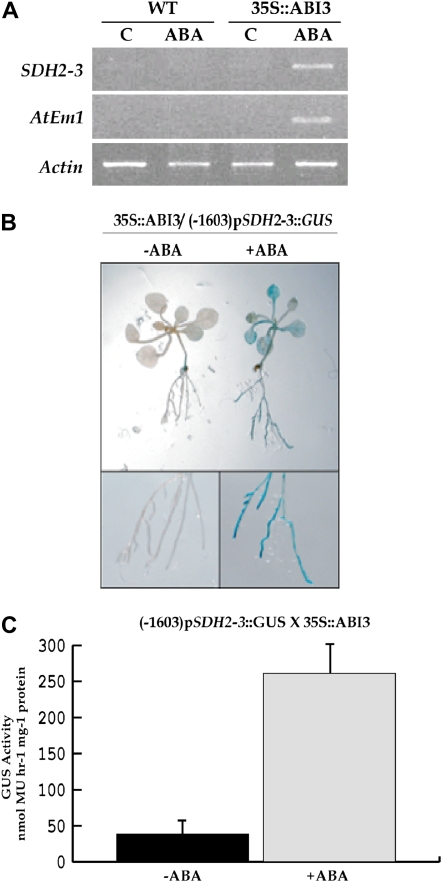
Ectopic expression of ABI3 confers the ability to accumulate seed-specific transcripts in response to ABA in vegetative tissues (Parcy et al., 1994). Therefore, wild-type seedlings and seedlings carrying the ABI3 cDNA fused to the cauliflower mosaic virus 35S promoter were transferred onto plates with 50 μm ABA or without hormone. After 48 h, SDH2-3 transcripts were only observed in ABA-treated 35S∷ABI3 plants, as were the control AtEm1 transcripts (Fig. 3A). To determine if this SDH2-3 mRNA increase is due to promoter activation, 35S∷ABI3 plants were crossed to SDH2-3 promoter (0.4 kb)∷GUS plants. Two-week-old 35S∷ABI3/pSDH2-3∷GUS seedlings were transferred onto plates containing 50 μm ABA, incubated for 48 h, and then stained for GUS. GUS staining was clearly detected in leaves and roots, while plants not treated with ABA showed no GUS expression (Fig. 3B). Furthermore, GUS activity was quantified in protein extracts from leaves of 2-month-old plants. As shown in Figure 3C, ABA strongly induced GUS expression.
Figure 3.
SDH2-3 induction by ABA and ABI3 in vegetative tissue. A, ABA induction of SDH2-3 in 35S∷ABI3 plantlets. RT-PCR analyses of SDH2-3 and AtEm1 transcripts were performed on RNA prepared from 2-week-old wild-type (WT) and transgenic 35S∷ABI3 seedlings incubated for 48 h with or without 50 μm ABA. B, ABA induction of SDH2-3 promoter∷GUS in seedlings ectopically expressing ABI3. Two-week-old (−1,603) pSDH2-3∷GUS/35S∷ABI3 seedlings were incubated for 48 h with or without 50 μm ABA and then stained for GUS activity. C, GUS activity in protein extracts from leaves of 60-d-old plants infiltrated and then incubated for 40 h with or without 10 μm ABA. Four biological replicates were performed. MU, 4-Methylumbelliferone.
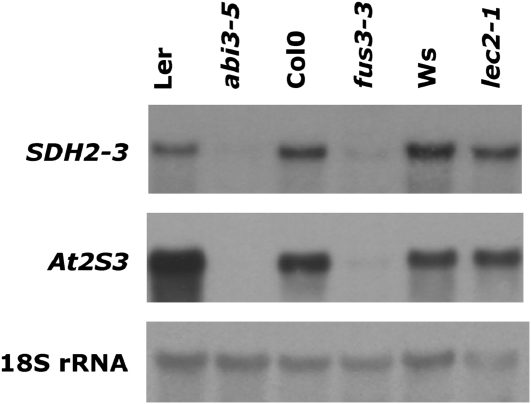
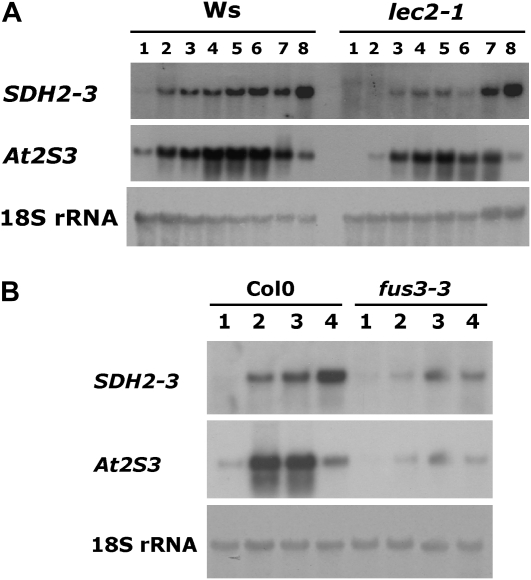
To evaluate the role of FUS3 and LEC2 in SDH2-3 expression, northern-blot analysis was performed using dry seed RNA from fus3-3 and lec2-1 mutants (Fig. 4). SDH2-3 expression was reduced to a similar extent in both fus3-3 and control abi3-5 seeds, as was the At2S3 storage protein gene (Fig. 4). In contrast, lec2-1 had a slight effect, if any, on SDH2-3 and At2S3 transcript levels in dry seeds. We decided to analyze SDH2-3 expression during maturation of lec2-1 seeds, since LEC2 expression decreases toward the end of seed maturation and LEC2 transcript levels become undetectable in dry seeds (Kroj et al., 2003). RNA was extracted at different developmental stages from early maturation to desiccation, and expression was examined by northern blot (Fig. 5A). In wild-type seeds, SDH2-3 transcripts accumulate during the maturation phase of seed development and remain high during the desiccation phase (lane 7) and in dry seeds (lane 8), as reported previously (Elorza et al., 2006). Expression of the At2S3 control albumin gene is similarly induced during maturation, but transcript levels decrease during desiccation. Interestingly, SDH2-3 expression is clearly reduced in lec2-1 seeds before desiccation (lanes 1–6) but not in dry seeds (lane 8) or seeds that have begun to desiccate (lane 7). For comparison, we analyzed fus3-3 developing seeds, since FUS3 mRNA is present until the dry seed stage: expression of SDH2-3 (and At2S3) was drastically reduced in fus3-3 seeds from the beginning of maturation to the dry seed stage (Fig. 5B).
Figure 4.
SDH2-3 expression in fus3-3, lec2-1, and abi3-5 mutant dry seeds. Northern blot analysis of SDH2-3 and At2S3 transcripts was performed on 10 μg of total RNA from mutant dry seeds and their respective wild-type controls. Ws, Wassilewskija.
Figure 5.
SDH2-3 expression is reduced during maturation of lec2-1 seeds. A, Northern-blot analysis of SDH2-3 and At2S3 transcripts during lec2-1 and wild-type (Wassilewskija [Ws]) seed development. Total RNA was extracted from maturing green siliques (thin, approximately 8 mm long [lane 1] to thick, approximately 14 mm long [lane 6]), yellowing siliques (lane 7), and yellow siliques (lane 8). B, Analysis of SDH2-3 and At2S3 transcripts during fus3-3 and wild-type (Col-0) seed development. Total RNA was extracted from maturing green siliques (thin, approximately 8–10 mm long [lane 1] and thick, 12–14 mm long [lane 2]), yellowing siliques (lane 3), and yellow siliques (lane 4).
Therefore, our results reveal that ABI3, FUS3, and LEC2 have a profound effect on SDH2-3 expression and that LEC2 is only necessary before desiccation.
FUS3 and ABI3 Bind the SDH2-3 Promoter
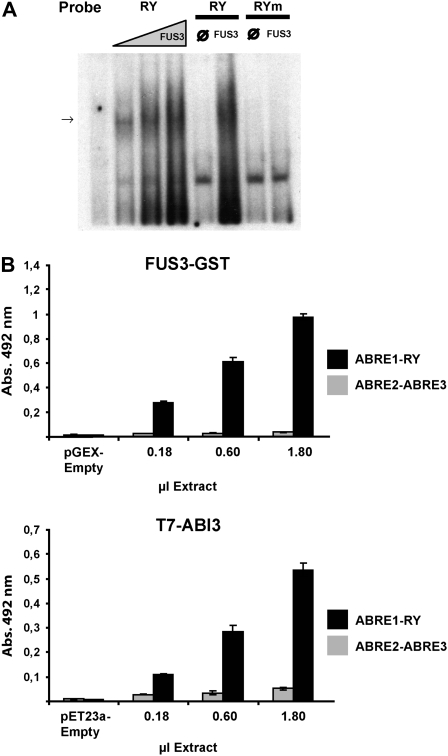
RY motifs are putative targets for B3 domain transcription factors (Suzuki et al., 1997; Reidt et al., 2000; Kroj et al., 2003; Mönke et al., 2004). To investigate whether these factors could recognize in vitro the SDH2-3 RY promoter element, ABI3 and FUS3 proteins expressed in Escherichia coli were tested for their ability to bind to an oligonucleotide containing this element in an electrophoretic mobility shift assay (EMSA). No retardation was observed with the ABI3 protein (data not shown). However, FUS3 was able to bind to the RY probe but not to the RYm element, indicating that RY sequence integrity is required for proper recognition by the transcription factor (Fig. 6A).
Figure 6.
FUS3 and ABI3 bind to probes containing the RY element present in the SDH2-3 promoter. A, EMSA of the native RY probe for increasing concentrations (1:3–1) of a protein extract containing FUS3. Controls were performed with extracts from bacterial cells transformed with the empty pET23a vector (lanes ∅) or the mutated RYm element as probe (lanes RYm). The arrow shows the specific band of the interaction. B, Binding of FUS3 and ABI3 to the ABRE1-RY DNA. Increasing quantities (1:10–1) of extracts from bacteria expressing FUS3-GST or T7 tag-ABI3 were assayed for binding to DNA containing either ABRE1 and RY elements or ABRE2 and ABRE3 elements. Binding was measured by ELISA, with an antibody against GST or T7 tag, conjugated with HRP. Control binding reactions were performed with an extract from bacterial cells transformed with the empty vectors.
Mönke et al. (2004) described a sensitive ELISA-type test to analyze protein-DNA interactions using biotinylated DNA fragments fixed to the solid phase and soluble proteins. Binding of recombinant ABI3 containing the T7 tag at the N-terminal end, and of recombinant FUS3-glutathione S-transferase (GST) to fixed ABRE1-RY and ABRE2-ABRE3 probes, was detected with anti-T7 tag or anti-GST antibodies. FUS3 recognized only the RY-containing element, confirming the results obtained by EMSA (Fig. 6B). In contrast to the EMSA results, binding of ABI3 to the same probe was demonstrated using this system. Both proteins did not bind or bound poorly to the ABRE2-ABRE3 sequence.
Basic Leu Zipper Transcription Factors Bind the SDH2-3 Promoter
ABRE elements (also called G-boxes) are targets for basic Leu zipper (bZIP) transcription factors. Thus, we analyzed the binding properties of bZIP factors to the SDH2-3 promoter. We chose two members of group C of bZIPs (Jakoby et al., 2002), bZIP10 and bZIP25, functionally related to maize Opaque 2 and reported to induce SSP expression synergistically with ABI3 (Lara et al., 2003). The group S1 bZIP53 is a dimerizing partner of bZIP10 and bZIP25 (Ehlert et al., 2006; Weltmeier et al., 2006) and is the only S1 member with an expression pattern in seed development matching that of SDH2-3.
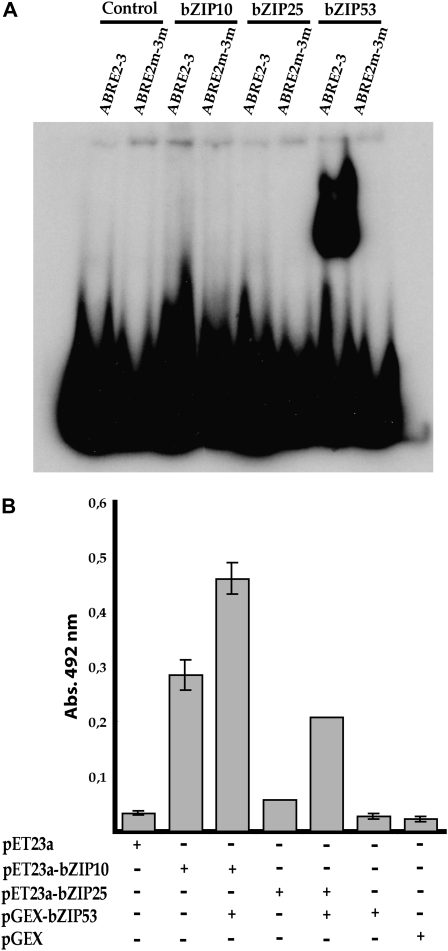
Recombinant proteins were expressed in E. coli and tested for their ability to bind to a probe containing ABRE2 and ABRE3 in an EMSA assay. In this system, bZIP53, but not bZIP10 or bZIP25, was able to bind to the ABRE2-3 probe (Fig. 7A). bZIP53 binding occurs specifically through the ABRE sequences, since mutations at these sequences abolished binding. However, when the ELISA binding test was used to analyze the effect of bZIP53 on bZIP10 and bZIP25 binding to a fixed ABRE2-ABRE3 probe, binding of bZIP10 and, to a lesser extent, of bZIP25 could be detected. More importantly, their binding was enhanced in the presence of bZIP53 (Fig. 7B). Interactions between bZIP53 and either bZIP10 or bZIP25 were also observed in EMSA assays (Supplemental Fig. S1; data not shown).
Figure 7.
bZIP53 transcription factor binds the ABRE elements present in the SDH2-3 promoter. A, EMSA of the native (ABRE2-3) and mutated (ABRE2m-3m) probes with protein extracts containing bZIP10, bZIP25, or bZIP53. Controls were performed with extracts from bacterial cells transformed with the empty pET23a vector. Equal concentrations of bZIP proteins in extracts were determined by western blot with appropriate antibodies. B, Binding of bZIP10 and bZIP25 to the ABRE2-ABRE3 DNA. Extracts from bacteria expressing T7 tag-bZIP10 or T7 tag-bZIP25 were assayed for binding to DNA containing ABRE2 and ABRE3 elements in the presence or absence of bZIP53-GST. Binding was measured by ELISA, with an antibody against the T7 tag (bZIP53-GST was not detected). Control binding reactions were performed with extracts from bacterial cells transformed with the empty vectors. Abs., Absorbance.
Altogether, these results are consistent with the hypothesis that the SDH2-3 promoter is a target of bZIP transcription factors and that ABRE boxes are involved in promoter recognition by these factors.
SDH2-3 Is the Main Iron-Sulfur Subunit of Complex II from Mature Seeds
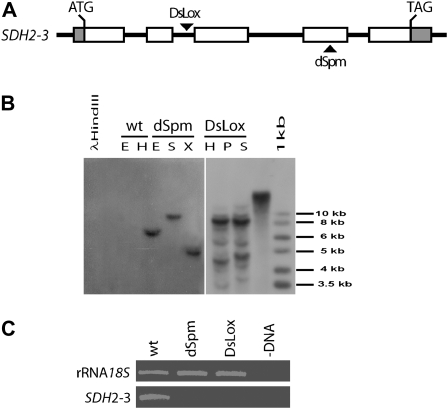
To evaluate the role of SDH2-3 in complex II biogenesis, two sdh2-3 mutant lines were identified and characterized. Insertion/SDH2-3 gene junctions were sequenced, demonstrating that no major deletions or chromosomal rearrangements took place during the insertional events. In the dSpm line, the transposon was confirmed to be in the fourth of five exons, interrupting codon 221, and in the DsLox line, the T-DNA interrupted intron 2 (Fig. 8A). Segregation of Basta resistance and Southern-blot analysis of homozygous sdh2-3 dSpm and DsLox mutant plants (Fig. 8B) were consistent with one dSpm and two DsLox insertions. Northern-blot (data not shown) and reverse transcription (RT)-PCR (Fig. 8C) analyses showed that no SDH2-3 mRNA was detected in mutant plants. Altogether, these results indicate that both mutant lines possess knockout alleles of SDH2-3 and that any phenotypic alteration observed in the dSpm line could be linked to the sdh2-3 mutated allele.
Figure 8.
Identification of knockout mutant sdh2-3 plants. A, Genomic organization of the SDH2-3 gene. Exons are presented as boxes, and insertion sites are indicated by arrowheads. B, Southern-blot analysis was performed with total DNA (6 μg) from wild-type (wt) or homozygous sdh2-3 mutant plants (dSpm and DsLox). DNA was digested with EcoRV (E), HindIII (H), SpeI (S), XbaI (X), or PstI (P) and hybridized with a probe directed to the Basta resistance gene present in the T-DNA (DsLox) and transposon (dSpm). The probe identified one DNA fragment in dSpm mutant plants and at least two DNA fragments in DsLox mutant plants. The DNA fragment in the dSpm mutant and one of the two main DNA fragments in the DsLox mutant have the expected sizes. Phage λ DNA digested with HindIII and a 1-kb ladder were used as size markers. C, RT-PCR analysis of SDH2-3 transcripts in wild-type and mutant plants. Thirty-five amplification cycles were used for SDH2-3 and 15 cycles were used for the 18S rRNA load control. Lane –DNA corresponds to a PCR control without template.
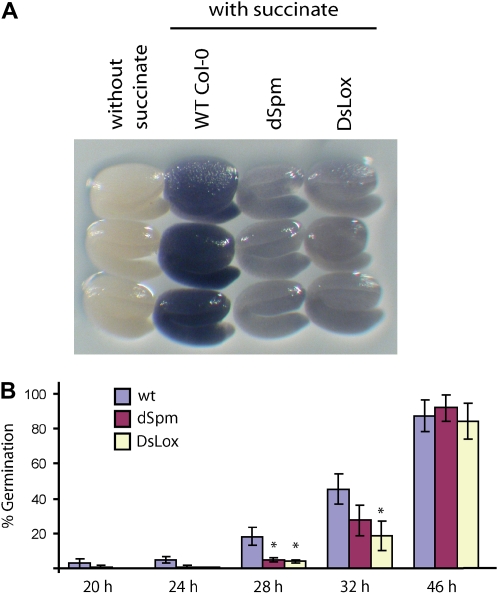
Mature embryos from wild-type, dSpm, and DsLox seeds were assayed for in situ SDH activity as described by Baud and Graham (2006). SDH was clearly detected in wild-type embryos, being homogenous throughout the embryo, and there was negligible background activity in the absence of succinate (Fig. 9A). Interestingly, this activity was greatly reduced in both dSpm and DsLox homozygous sdh2-3 knockout mutants (Fig. 9A), clearly indicating that SDH2-3 codes for most of the iron-sulfur protein of embryo complex II. Nevertheless, SDH activity was detected in the mutant plants, likely resulting from basal expression of SDH2-1 and/or SDH2-2.
Figure 9.
Effects of the sdh2-3 mutations on seed SDH activity and germination. A, SDH activity in mature embryos of wild-type (WT) and sdh2-3 dSpm and DsLox mutant plants. B, Germination of wild-type and sdh2-3 mutant seeds. Values are means ± se of three biological replicates carried out on batches of approximately 100 seeds. Asterisks indicate significant differences according to Student's t test (P < 0.02).
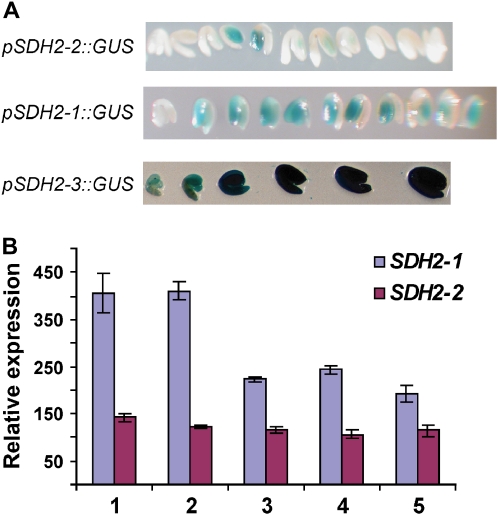
SDH2-1 and SDH2-2 transcripts are low in dry seeds (Elorza et al., 2006). To analyze their expression during embryo development, we used plants transformed with fusions of the SDH2-1 and SDH2-2 promoters to the GUS reporter gene (Elorza et al., 2004). Embryos from the early maturation stage to the desiccation stage were dissected and stained for GUS activity (Fig. 10A). Whereas no or very weak GUS staining was observed for the SDH2-2 promoter, GUS expression was detected for the SDH2-1 promoter. Interestingly, SDH2-1 promoter activity decreased during embryo development (Fig. 10A), but GUS staining was not completely eliminated in dry seeds. Furthermore, data on SDH2-1 and SDH2-2 expression from public expression databases confirm that seed SDH2-1 expression decreases from the torpedo to the green cotyledon stage and that SDH2-2 is expressed at a very low level, if any (Fig. 10B; http://www.bar.utoronto.ca; Schmid et al., 2005).
Figure 10.
Expression of SDH2 genes during embryo maturation. A, Embryos manually dissected from seeds were stained for GUS activity. Plants carrying the promoters of SDH2-1, SDH2-2, and SDH2-3 fused to the GUS gene have been described (Elorza et al., 2004). Walking-stick to dry seed embryos were stained for 60 h (pSDH2-2∷GUS) or 14 h (pSDH2-1∷GUS and pSDH2-3∷GUS). B, Expression of SDH2-1 and SDH2-2 during seed development. Data are from Schmid et al. (2005) and were obtained at www.bar.utoronto.ca. Mean signal intensities were averaged from three replicates. Expression was analyzed in whole seeds with mid to late torpedo embryos (lane 1), late torpedo to early walking-stick embryos (lane 2), walking-stick to early curled cotyledon embryos (lane 3), curled cotyledons to early green cotyledons (lane 4), and green cotyledons (lane 5).
Germination Is Retarded in Seeds Lacking a Functional SDH2-3 Gene
Homozygous sdh2-3 mutant plants showed no obvious phenotypic defects during vegetative or reproductive growth when compared with wild-type plants, at least under the growth conditions used (Supplemental Fig. S2). These results indicate that SDH2-3 is not an essential gene for Arabidopsis growth and development. Given its expression pattern, we decided to investigate the germination of mutant sdh2-3 and wild-type seeds. Germination of sdh2-3 mutant seeds was retarded compared with that in the wild type (Fig. 9B), suggesting an important role of SDH2-3 for seed germination.
DISCUSSION
The specific expression pattern of SDH2-3 during seed maturation raises interesting questions about its regulation and function. A similar pattern has been described only once for a mitochondrial protein, a pea (Pisum sativum) LEA protein. This protein may be involved in protecting the inner mitochondrial membrane during seed desiccation (Grellet et al., 2005; Tolleter et al., 2007); however, no data concerning the regulation of its expression are available. Here, we have performed a detailed characterization of the SDH2-3 promoter and identified key regulatory elements and transcription factors involved in its regulation. Moreover, analysis of sdh2-3 loss-of-function plants revealed its participation in early stages of seed germination.
ABRE and RY Motifs Are Major cis-Elements Regulating SDH2-3 Expression
ABREs (G-boxes) have been implicated in SSP and LEA protein gene regulation and shown to function effectively when two copies are located in tandem or when it is associated with a coupling or enhancer element like the RY motif (Busk and Pagès, 1998; Leung and Giraudat, 1998; Nambara and Marion-Poll, 2003). In the SDH2-3 promoter, ABRE2 and ABRE3 are separated by a short 6-bp sequence and ABRE1 is located eight nucleotides upstream of the RY enhancer element (Fig. 1A). When ABRE and RY motifs were mutated individually, promoter activities were significantly reduced, indicating that they all act synergistically to give high expression in mature seeds (Fig. 1B). Since ABRE2 mutation almost abolishes GUS expression, this element appears to be the most important by itself. Considering that the mutations introduced in ABRE and RY boxes have been shown to abolish the binding of bZIP and B3 domain transcription factors, respectively (Reidt et al., 2000; Bensmihen et al., 2002), these results suggest that transcription factors from these families may be directly involved in SDH2-3 regulation.
B3 Domain Transcription Factors Regulate SDH2-3 Expression in Vivo and Interact with the SDH2-3 Promoter in Vitro
ABI3, FUS3, and LEC2 master regulators exhibit partially overlapping expression patterns and participate in an intricate and not fully understood network of cross-regulations involved in most seed maturation aspects, including storage compound synthesis (To et al., 2006; Santos-Mendoza et al., 2008, and refs. therein). We have used the severe abi3-5, fus3-3, and lec2-1 mutants to address their role in SDH2-3 expression. Here, we have demonstrated that SDH2-3 transcript level is drastically reduced in abi3-5, fus3-3, and maturing lec2-1 seeds (Figs. 2B, 4, and 5). These results obtained with individual mutants show that these three plant-specific transcription factors containing the conserved B3 DNA-binding domain regulate SDH2-3 expression, likely at the level of transcription from the SDH2-3 promoter, as shown for ABI3 (Figs. 2, C and D, and 3).
LEC2, FUS3, and ABI3 expression begins early (from the heart stage) in maturation (Parcy et al., 1994; Kroj et al., 2003). However, LEC2 expression decreases during desiccation, and only ABI3 and FUS3, whose expression is maintained, are required to establish tolerance to desiccation. Interestingly, dependence of SDH2-3 expression on LEC2 correlates with the known LEC2 expression pattern, since once seeds entered desiccation, SDH2-3 expression was not or was only slightly affected by the lec2-1 mutation. Therefore, our results are consistent with a model in which SDH2-3 expression is regulated by the three transcription factors before desiccation, and then LEC2 becomes dispensable.
The described in vivo analysis of mutants in ABI3, FUS3, and LEC2 does not elucidate if these transcription factors directly or indirectly trigger SDH2-3 expression. Recent studies have demonstrated that they directly controlled the induction of SSP gene expression, recognizing the RY motifs present in the promoters of these target genes (Ezcurra et al., 2000; Reidt et al., 2000; Kroj et al., 2003; Mönke et al., 2004; Braybrook et al., 2006). Accordingly, we found that FUS3 is able to specifically bind the RY element of the SDH2-3 promoter in EMSA assays (Fig. 6A), in agreement with observations made on the Vicia faba legumin and the Arabidopsis At2S3 albumin promoters (Reidt et al., 2000; Kroj et al., 2003). Only using a more sensitive ELISA method have we also shown that ABI3 interacts directly with a probe containing the RY element (Fig. 6B). Altogether, our results suggest that ABI3 and FUS3 interact, probably with different affinities or by different mechanisms, with the SDH2-3 promoter and directly regulate its expression. It is worth noting that, to the best of our knowledge, there are no previous reports showing that these B3 domain master regulators may directly regulate a gene involved in primary metabolism. Nevertheless, it has to be pointed out that indirect regulatory effects for B3-type transcription factors have also been reported (Gazzarrini et al., 2004; To et al., 2006) and cannot be ruled out in the case of SDH2-3, especially concerning its regulation by LEC2.
bZIP Transcription Factors Bind the SDH2-3 Promoter
Emerging models of SSP and LEA gene regulation by ABI3 suggest that, in addition to a likely weak direct interaction with RY promoter elements, ABI3 is recruited at the promoter level by bZIP proteins interacting with ABRE elements. For instance, ABI3 interacts with the bZIP transcription factor ABI5, which in turn is able to bind to LEA promoters and regulate their expression through a domain unique to ABI3 and not present in FUS3 or LEC2 (Nakamura et al., 2001). We found that SDH2-3 expression was not reduced in abi5 mutant plants (data not shown), suggesting that this bZIP transcription factor is not implicated in regulating the SDH2-3 promoter. Similarly, ABI3 is also able to interact with bZIP10 and bZIP25 in regulating SSP genes (Lara et al., 2003).
We have found that ABRE elements in the SDH2-3 promoter are strongly bound by bZIP53 (Fig. 7A) but less prominently by bZIP10 and bZIP25 binding (Fig. 7B). Furthermore, band-shift assays performed in the presence of bZIP53 and either bZIP25 or bZIP10 revealed interactions between the bZIP factors and the increased DNA-binding capacity of the heterodimers (Supplemental Fig. S1; data not shown). These data support the idea that the ABRE elements present in the SDH2-3 promoter are recognized by group C bZIP/group S1 bZIP heterodimers.
Based on our results and on previous studies (Ezcurra et al., 2000; Kroj et al., 2003; Lara et al., 2003), we propose a model that predicts that the activation of SDH2-3 would require proteins of the B3 (ABI3, FUS3, and LEC2) and bZIP (bZIP53 and either bZIP10 or bZIP25) classes. According to this model, FUS3 (and probably LEC2) and bZIP53/group C bZIP would recognize RY and ABRE sequences, respectively, whereas ABI3 may interact weakly with the SDH2-3 promoter and also be tethered to the promoter through interactions with the bZIP heterodimers. Accordingly, mutations in RY would prevent FUS3 binding, and consequently, SDH2-3 expression would be reduced. Similarly, mutations in ABRE boxes preclude bZIP heterodimer binding to the promoter and recruitment of ABI3, resulting in a drastic reduction of SDH2-3 promoter transactivation, as observed when ABI3 itself is inactivated.
At present, only a few promoters of mitochondrial protein genes have been analyzed (Zabaleta et al., 1998; Satoh et al., 2002; Welchen et al., 2004; Dojcinovic et al., 2005; Weltmeier et al., 2006; González et al., 2007; Ho et al., 2007). However, to the best of our knowledge, the SDH2-3 promoter is the first embryo-specific promoter characterized for a mitochondrial respiratory protein and the first gene promoter from TCA cycle enzymes to be characterized in plants.
Respiratory Complex II in Mature Seeds and during Germination Mainly Contains SDH2-3
Several processes essential for seed viability and germination occur during the maturation phase in seed development. Although we may speculate that SDH2-3 induction is part of the metabolic adaptations occurring during maturation, and that SDH2-3 is important for SDH activity under the conditions prevailing during maturation and desiccation stages, the lack of any visible phenotype in developing seeds from homozygous sdh2-3 knockout plants indicates that SDH2-3 is not essential for seed set and viability. The fact that SDH2-1 is residually expressed in maturing embryos (Fig. 10) may explain this observation.
The steady-state abundance of SDH2-3 transcripts is high in mature embryos, whereas SDH2-1and SDH2-2 are expressed at very low levels (Elorza et al., 2006; Fig. 10). These observations correlate well with data obtained for SDH activity (Fig. 9A), which is greatly reduced in the two sdh2-3 homozygous mutants. Residual SDH activity detected in the knockout lines is likely due to the presence of SDH2-1 and/or SDH2-2 and is sufficient to sustain germination. We have previously found that mitochondrial complex II is essential for gametophyte development in Arabidopsis using heterozygous mutant plants for the flavoprotein gene (SDH1; León et al., 2007). However, the lack of sdh1/sdh1 homozygous seeds precluded the analysis of the effect of SDH absence on seed development and germination.
The imbibing seed resumes metabolic activity within minutes of water entering its cells, and rapid increases in respiration rate accompany the earliest stages of germination (Bewley and Black, 1994). Logan et al. (2001) showed that succinate-dependent O2 consumption and citric acid cycle activity increased rapidly during imbibition of maize (Zea mays) embryos. We have determined that 2-thenoyltrifluoroacetone, a complex II inhibitor, completely blocks germination (data not shown), strongly supporting an essential role of complex II in this process. An appealing hypothesis is that high SDH2-3 expression in mature embryos allows for a rapid increase in SDH activity upon imbibition, before expression of SDH2-1 and SDH2-2. This assumption is supported by the observation that germination is retarded in the sdh2-3 mutants (Fig. 9B). During germination and postgerminative growth, SDH2-3 transcripts decline, and SDH2-1 and SDH2-2 transcript levels increase (Elorza et al., 2006), suggesting that the embryo-specific SDH2-3-containing complex II is replaced by a complex II containing SDH2-1 or SDH2-2. This turnover may explain why the lack of SDH2-3 has only an effect on germination kinetics. To our knowledge, this is the first study concerning the expression of TCA cycle genes during seed development and the characterization of phenotypic alterations during germination in mutant lines.
MATERIALS AND METHODS
Plant Growth and Transformation
Arabidopsis (Arabidopsis thaliana ecotype Columbia [Col-0], Landsberg erecta [Ler], or C24) seeds were cold treated for 48 h at 4°C in darkness and then germinated and grown hydroponically at 20°C to 24°C under a 16-h-light/8-h-dark cycle (Gibeaut et al., 1997). Agrobacterium tumefaciens-mediated transformation of Arabidopsis plants was accomplished using the floral dip protocol (Clough and Bent, 1998). Seeds of the T1 generation were selected for resistance to hygromycin. At least 12 independent transgenic lines were obtained for each construct, and transgene presence was verified by PCR.
For germination assays, surface-sterilized seeds were stratified at 4°C for 48 h in the dark, sown on half-concentrated Murashige and Skoog medium solidified with 0.8% (w/v) agar, and incubated at 20°C to 24°C under a 16-h-light/8-h-dark cycle. Germination was scored at different times based on radicle emergence.
Heterozygous abi3-5, fus3-3, and lec2-1 seeds were obtained from the Arabidopsis Biological Resource Center (ABRC; http://www.arabidopsis.org/abrc). Homozygous mature abi3-5 and lec2-1 mutant seeds were identified in siliques of heterozygous plants by their characteristic phenotype and sown to get homozygous mutant plants. Homozygous fus3-3 seeds were selected before desiccation, because of the ability of immature embryos to germinate on 0.5× Murashige and Skoog medium solidified with 0.8% (w/v) agar supplemented with 1% (w/v) Suc. Growing plantlets were transferred and grown hydroponically. Seeds from a homozygous transgenic Arabidopsis line (A19, C24 background) carrying a transcriptional fusion between the double enhanced cauliflower mosaic virus 35S promoter and the ABI3 cDNA (35S∷ABI3) were kindly provided by François Parcy (Commissariat à l'Energie Atomique-INRA-Université Joseph Fourier, Grenoble, France).
Homozygous transgenic plants carrying a SDH2-3 promoter fragment (1,603 or 391 bp) and the SDH2-3 5′ untranslated region (UTR) fused to GUS (Elorza et al., 2004) were crossed with homozygous abi3-5 and 35S∷ABI3 plants and their respective controls (Ler and C24). F1 seeds from the abi3-5 cross were germinated on hygromycin to select for the pSDH2-3∷GUS construct, and homozygous abi3-5 F2 seeds were identified by their green phenotype. F1 seeds from the 35S∷ABI3 cross were germinated on hygromycin (pSDH2-3∷GUS construct) and kanamycin (35S∷ABI3 construct).
Promoter Mutagenesis and GUS Activity Assays
Mutagenesis was performed by PCR, using a construct containing the SDH2-3 promoter and 5′ UTR fused to GUS as a template (construct P5; Elorza et al., 2004). Two PCRs were carried out for each mutant with the same template. For the promoter mutated in the RY element (RYm), one amplification was done with primers SDH2-3F and RYmR and the other was done with primers SDH2-3R and RYmF (Supplemental Table S1). Both amplification products were purified by electrophoresis through agarose gels, and their mixture was used as a template for a third PCR with primers SDH2-3F and SDH2-3R. This PCR product was cloned into pGEM-T plasmid (Promega), and the DNA fragment obtained by digestion with BamHI and NcoI was ligated into pCAMBIA 1381 (http://www.cambia.org).
The same procedure was employed for constructs mutated in ABRE1 (ABRE1m), ABRE2 (ABRE2m), or ABRE3 (ABRE3m), with SDH2-3F, SDH2-3R, and the following primers: for ABRE1m, ABRE1mF and ABRE1mR; for ABRE2m, ABRE2mF and ABRE2mR; and for ABRE3m, ABRE3mF and ABRE3mR (for primer sequences, see Supplemental Table S1). To obtain the construct mutated in the three ABRE elements (3xABREm), the ABRE1 mutant was used as a template in the first step and mutation in ABRE2 was introduced as described. In the second step, mutation in ABRE3 was introduced with primers 3XABREmF and 3XABREmR.
The structures of constructs were verified by DNA sequencing. All constructs contain a promoter fragment (391 bp) and the 5′ UTR up to the first SDH2-3 codon in frame with the GUS reporter gene and were introduced into A. tumefaciens GV3101 by electroporation. Arabidopsis transgenic plants carrying the mutations in the SDH2-3 promoter fused to the GUS reporter gene were obtained, and soluble extracts of plant tissues were assayed for GUS activity by fluorometric measurements using 4-methylumbelliferyl-β-d-glucuronide (Sigma-Aldrich) as a substrate (Jefferson, 1987). Protein concentrations were determined by the Bradford method (Bradford, 1976), and GUS activities were calculated as nanomoles of 4-methylumbelliferone per hour per milligram of protein. Histochemical GUS staining was performed as described (Elorza et al., 2004).
Nucleic Acid Preparation and Analysis
Total DNA was prepared from green leaves according to Ausubel et al. (1994). Southern-blot analysis was performed by standard procedures with a 32P-labeled probe derived from the Basta resistance gene by PCR amplification between primers BastaF and BastaR. Total RNA was extracted from seeds (50 mg) by the method of Vicient and Delseny (1999) and further purified using the RNeasy Plant Mini Kit (Qiagen), according to the manufacturer's protocol. Northern-blot analyses were performed as described (Elorza et al., 2006). Probes were obtained by PCR amplification using the following primer pairs: for SDH2-3, sdh2-58 and sdh2-63; for At2S3, At2S3F and At2S3R; and for 18S rRNA, 18SF and 18SR.
cDNA synthesis was performed on 2 μg of RNA with random hexamers as primers, according to the instructions of the ImProm-II RT system (Promega). PCR amplifications were performed in 50 μL, with one-twentieth of the cDNA and 0.5 units of AmpliTaq Gold (Applied Biosystems). The following primer pairs were used: for SDH2-3, sdh2-57 or sdh2-50 and sdh2-51; for the LEA AtEm1 transcript, Em1F and Em1R; for actin mRNA, actinF and actinR; and for 18S rRNA, 18SF and 18SR.
EMSA
The cDNAs encoding AtbZIP10, AtbZIP25, AtbZIP53, and ABI3 proteins were cloned into the expression vector pET23a (Novagen), and FUS3 cDNA was cloned into pGEX-2T vector (Amersham Biosciences) as a translational fusion to GST. Expression in Escherichia coli, preparation of protein extracts, and EMSAs were as described previously (Lara et al., 2003). To obtain the DNA probes, overlapping oligonucleotides were annealed and end labeled with [32P]dATP by the fill-in reaction (Klenow exo-free DNA polymerase; USB Corporation [http://www.usbweb.com]) and purified by 8% PAGE (39:1 cross-linking). The following oligonucleotides were used: for the wild-type RY probe, RY1 and RY2; for the mutated RYm probe, RYm1 and RYm2; for the wild-type ABRE2-3 probe, ABRE2-3a and ABRE2-3b; and for the mutated ABRE2m-3 m probe, ABRE2m-3mF and ABRE2m-3mR.
DNA-Protein Interaction Analysis by ELISA
The ELISA technique was used to study DNA-protein interaction as described by Mönke et al. (2004). Streptavidin-coated microwell strips (Nunc) were used, and the following double-stranded biotinylated DNA fragments were prepared according to Mönke et al. (2004): for the ABRE2-ABRE3 probe, ABRE2-ABRE3F and ABRE2-ABRE3R; and for the ABRE1-RY probe, ABRE1-RYF and ABRE1-RYR. Antibodies against T7 tag (Novagen) and GST (Amersham Biosciences) conjugated with horseradish peroxidase (HRP) were used to detect bound transcription factors. HRP activity was determined by adding 60 μL of a solution containing 0.015% hydrogen peroxide (Sigma-Aldrich) and o-phenylenediamine dihydrochloride (one tablet from Dako [http://www.dako.com] dissolved in 3 mL). After incubation at 37°C, the reaction was stopped with 60 μL of 2 n HCl, and the A492 was measured with a plate reader (Tecan).
Isolation of Insertional sdh2-3 Mutants
Insertional mutant lines were searched at two Web sites: the Arabidopsis Insertion Database (http://atidb.org/cgi-perl/index) and the T-DNAExpress (http://signal.salk.edu/cgi-bin/tdnaexpress). Two mutant alleles were identified, one in the Wisconsin DsLox T-DNA population (DsLox503G03) and the other in the Sainsbury Laboratory Arabidopsis Transposants dSpm population (line SM_3.623). Arabidopsis seeds from these lines (Col-0 background) were obtained from the Nottingham Arabidopsis Stock Centre (University of Nottingham) and the ABRC, respectively. To isolate homozygous mutant plants, seedlings were genotyped by a PCR-based approach using total DNA extracted from one cotyledon or one small leaf and primers flanking the insertion point for the wild-type allele and a gene-specific and left border-specific primer pair for the mutant alleles. For the dSpm line, primers sdh2-57 and sdh2-51 were used for the wild-type allele and primers dspm3′ and sdh2-51 were used for the mutant one. For the DsLox line, the wild-type allele was amplified with sdh2-50 and sdh2-51 and the mutant allele was amplified with LBW and sdh2-51.
Histochemical SDH Assay
A simplified protocol, based on the procedure described by Baud and Graham (2006), was used. Seeds were imbibed for 12 h at 4°C in the dark, excised embryos were incubated for 1 h at 37°C in 50 mm sodium phosphate (pH 7.6) and 150 mm NaCl, and finally, staining was performed by incubation for 3 h at 30°C in 50 mm sodium phosphate (pH 7.6), 7.5 mm succinate (fresh), and 2.4 mm nitroblue tetrazolium (Calbiochem). As controls, embryos were incubated in parallel in the same buffer without succinate. After staining, embryos were washed with distilled water and viewed using a stereoscopic microscope (Nikon SMZ800), and photographs were taken with a Nikon Coolpix 4500 CCD digital camera.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AB013395 and AJ278912.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Binding of bZIP25 and bZIP53 proteins to the ABRE2-3 sequence of the SDH2-3 promoter.
Supplemental Figure S2. SDH2-3 is not essential for Arabidopsis growth and development.
Supplemental Table S1. List of oligonucleotides.
Supplementary Material
Acknowledgments
We are greatly indebted to Simon Litvak and Laura Tarragó-Litvak for their constant encouragement. We also thank François Parcy for the 35S∷ABI3 line and Rosario Alonso for her help with the EMSA and ELISA assays. The ABRC and the Nottingham Arabidopsis Stock Centre are acknowledged for providing us with the mutant insertion lines.
This work was supported by Fondecyt-Chile (research grant no. 1060485 and Ph.D. grant no. AT–4040013 to H.R.), by the Millennium Nucleus for Plant Functional Genomics, Millennium Scientific Initiative Program, Mideplan, Chile (grant no. P06–009–F), and by AECI-Spain (grant nos. A/012927/07 and B019552/08).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Xavier Jordana (xjordana@bio.puc.cl).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1994) Current Protocols in Molecular Biology. Wiley Interscience, New York
- Baud S, Graham IA (2006) A spatiotemporal analysis of enzymatic activities associated with carbon metabolism in wild-type and mutant embryos of Arabidopsis using in situ histochemistry. Plant J 46 155–169 [DOI] [PubMed] [Google Scholar]
- Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F, Giraudat J, Parcy F (2002) The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD, Black M (1994) Seeds: Physiology of Development and Germination. Plenum Press, New York
- Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci USA 103 3468–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk PK, Pagès M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37 425–435 [DOI] [PubMed] [Google Scholar]
- Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Ehlers Loureiro M, Fernie AR (2003) Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species of tomato. Plant Physiol 133 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Dojcinovic D, Krosting J, Harris AJ, Wagner DJ, Rhoads DM (2005) Identification of a region of the Arabidopsis AtAOX1a promoter necessary for mitochondrial retrograde regulation of expression. Plant Mol Biol 58 159–175 [DOI] [PubMed] [Google Scholar]
- Ehlert A, Weltmeier F, Wang X, Mayer CS, Smeekens S, Vicente-Carbajosa J, Dröge-Laser W (2006) Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J 46 890–900 [DOI] [PubMed] [Google Scholar]
- Elorza A, León G, Gómez I, Mouras A, Holuigue L, Araya A, Jordana X (2004) Nuclear SDH2-1 and SDH2-2 genes, encoding the iron-sulfur subunit of mitochondrial complex II in Arabidopsis, have distinct cell-specific expression patterns and promoter activities. Plant Physiol 136 4072–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elorza A, Roschzttardtz H, Gómez I, Mouras A, Holuigue L, Araya A, Jordana X (2006) A nuclear gene for the iron-sulfur subunit of mitochondrial complex II is specifically expressed during Arabidopsis seed development and germination. Plant Cell Physiol 47 14–21 [DOI] [PubMed] [Google Scholar]
- Ezcurra I, Wycliffe P, Nehlin L, Ellerstrom M, Rask L (2000) Transactivation of the Brassica napus promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J 24 57–66 [DOI] [PubMed] [Google Scholar]
- Figueroa P, León G, Elorza A, Holuigue L, Araya A, Jordana X (2002) The four subunits of mitochondrial respiratory complex II are encoded by multiple nuclear genes and targeted to mitochondria in Arabidopsis thaliana. Plant Mol Biol 50 725–734 [DOI] [PubMed] [Google Scholar]
- Figueroa P, León G, Elorza A, Holuigue L, Jordana X (2001) Three different genes encode the iron-sulfur subunit of succinate dehydrogenase in Arabidopsis thaliana. Plant Mol Biol 46 241–250 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 7 373–385 [DOI] [PubMed] [Google Scholar]
- Gibeaut M, Hulett J, Cramer G, Seemann J (1997) Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol 115 317–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge B, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis AB13 gene by positional cloning. Plant Cell 4 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González DH, Welchen E, Attallah CV, Comelli RN, Mufarrege EF (2007) Transcriptional coordination of the biogenesis of the oxidative phosphorylation machinery in plants. Plant J 51 105–116 [DOI] [PubMed] [Google Scholar]
- Grellet J, Benamar A, Teyssier E, Avelange-Macherel MH, Grunwald D, Macherel D (2005) Identification in pea seed mitochondria of Late Embryogenesis Abundant protein able to protect enzymes from drying. Plant Physiol 137 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signalling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho LHM, Giraud E, Lister R, Thirkettle-Watts D, Low J, Clifton R, Howell KA, Carrie C, Donald T, Whelan J (2007) Characterization of the regulatory and expression context of an Alternative Oxidase gene provides insights into cyanide-insensitive respiration during growth and development. Plant Physiol 143 1519–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7 106–111 [DOI] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5 387–405 [Google Scholar]
- Kroj T, Savino G, Valon C, Giraudat J, Parcy F (2003) Regulation of storage protein gene expression in Arabidopsis. Development 130 6065–6073 [DOI] [PubMed] [Google Scholar]
- Landschütze V, Willmitzer L, Müller-Röber B (1995) Inhibition of flower formation by antisense repression of mitochondrial citrate synthase in transgenic potato plants leads to a specific disintegration of the ovary tissues of flower. EMBO J 14 660–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara P, Oñate-Sánchez L, Abraham Z, Ferrándiz C, Díaz I, Carbonero P, Vicente-Carbajosa J (2003) Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J Biol Chem 278 21003–21011 [DOI] [PubMed] [Google Scholar]
- Lemaitre T, Urbanczyk-Wochniak E, Flesch V, Bismuth E, Fernie AR, Hodges M (2007) NAD-dependent isocitrate dehydrogenase mutants of Arabidopsis suggest the enzyme is not limiting for nitrogen assimilation. Plant Physiol 144 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire B, Oyedotun K (2002) The Saccharomyces cerevisiae mitochondrial succinate:ubiquinone oxidoreductase. Biochim Biophys Acta 1553 102–116 [DOI] [PubMed] [Google Scholar]
- León G, Holuigue L, Jordana X (2007) Mitochondrial complex II is essential for gametophyte development in Arabidopsis. Plant Physiol 143 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49 199–222 [DOI] [PubMed] [Google Scholar]
- Logan DC, Millar AH, Sweetlove LJ, Hill SA, Leaver CJ (2001) Mitochondrial biogenesis during germination in maize embryos. Plant Physiol 125 662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luerssen H, Kirik V, Herrmann P, Misera S (1998) FUSCA3 encodes a protein with a conserved VP1/ABI3-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J 15 755–764 [DOI] [PubMed] [Google Scholar]
- Millar A, Eubel H, Jänsch L, Kruft V, Heazlewood J, Braun H (2004) Mitchondrial cytochrome c oxidase and succinate dehydrogenase complexes contain plant specific subunits. Plant Mol Biol 56 77–90 [DOI] [PubMed] [Google Scholar]
- Mönke G, Altschmied L, Tewes A, Reidt W, Mock HP, Bäumlein H, Conrad U (2004) Seed-specific transcription factors ABI3 and FUS3: molecular interaction with DNA. Planta 219 158–166 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26 627–635 [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2003) ABA action and interaction in seeds. Trends Plant Sci 8 213–217 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR (2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50 1093–1106 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AM, Loureiro ME, Ratcliffe RG, Sweetlove LJ, Fernie AR (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms JJJ, Léon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM (1993) Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana: a comparative study using abscisic acid-insensitive abi3 mutants. Plant Physiol 102 1185–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J (1994) Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6 1567–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidt W, Wohlfarth T, Ellerstrom M, Czihal A, Tewes A, Ezcurra I, Rask L, Bäumlein H (2000) Gene regulation during late embryogenesis: the RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J 21 401–408 [DOI] [PubMed] [Google Scholar]
- Roos MH, Tielens AGM (1994) Differential expression of two succinate dehydrogenase subunit-B genes and a transition in energy metabolism during the development of the parasitic nematode Haemonchus contortus. Mol Biochem Parasitol 66 273–281 [DOI] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54 608–620 [DOI] [PubMed] [Google Scholar]
- Satoh R, Nakashima K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2002) ACTCAT, a novel cis-acting element for proline- and hypoosmolarity-responsive expression of the ProDH gene encoding proline dehydrogenase in Arabidopsis. Plant Physiol 130 709–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studart-Guimarães C, Fait A, Nunes-Nesi A, Carrari F, Usadel B, Fernie AR (2007) Reduced expression of succinyl CoA ligase can be compensated for by an upregulation of the γ-amino-butyrate (GABA) shunt in illuminated tomato leaves. Plant Physiol 145 626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR (1997) The conserved B3 domain of VIVIPAROUS I has a cooperative DNA binding activity. Plant Cell 9 799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolleter D, Jaquinod M, Mangavel C, Passirani C, Saulnier P, Manon S, Teyssier E, Payet N, Avelange-Macherel MH, Macherel D (2007) Structure and function of a mitochondrial Late Embryogenesis Abundant protein are revealed by desiccation. Plant Cell 19 1580–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient C, Delseny M (1999) Isolation of total RNA from Arabidopsis thaliana seeds. Anal Biochem 268 412–413 [DOI] [PubMed] [Google Scholar]
- Welchen E, Chan RL, González DH (2004) The promoter of the Arabidopsis nuclear gene COX5b-1, encoding subunit 5b of the mitochondrial cytochrome c oxidase, directs tissue-specific expression by a combination of positive and negative regulatory elements. J Exp Bot 55 1997–2004 [DOI] [PubMed] [Google Scholar]
- Weltmeier F, Ehlert A, Mayer CS, Dietrich K, Wang X, Schütze K, Alonso R, Harter K, Vicente-Carbajosa J, Dröge-Laser W (2006) Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J 25 3133–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankovskaya V, Horsefield R, Törnroth S, Luna-Chavez C, Miyoshi H, Léger C, Byrne B, Cecchini G, Iwata S (2003) Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299 700–704 [DOI] [PubMed] [Google Scholar]
- Zabaleta E, Heiser V, Grohmann L, Brennicke A (1998) Promoters of nuclear-encoded respiratory chain complex I genes from Arabidopsis thaliana contain a region essential for anther/pollen-specific expression. Plant J 15 49–59 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.