Abstract
In mycorrhizal associations, the fungal partner assists its plant host by providing nitrogen (N) in addition to phosphate. Arbuscular mycorrhizal (AM) fungi have access to inorganic or organic forms of N and translocate them via arginine from the extra- to the intraradical mycelium, where the N is transferred to the plant without any carbon skeleton. However, the molecular form in which N is transferred, as well as the involved mechanisms, is still under debate. NH4+ seems to be the preferential transferred molecule, but no plant ammonium transporter (AMT) has been identified so far. Here, we offer evidence of a plant AMT that is involved in N uptake during mycorrhiza symbiosis. The gene LjAMT2;2, which has been shown to be the highest up-regulated gene in a transcriptomic analysis of Lotus japonicus roots upon colonization with Gigaspora margarita, has been characterized as a high-affinity AMT belonging to the AMT2 subfamily. It is exclusively expressed in the mycorrhizal roots, but not in the nodules, and transcripts have preferentially been located in the arbusculated cells. Yeast (Saccharomyces cerevisiae) mutant complementation has confirmed its functionality and revealed its dependency on acidic pH. The transport experiments using Xenopus laevis oocytes indicated that, unlike other plant AMTs, LjAMT2;2 transports NH3 instead of NH4+. Our results suggest that the transporter binds charged ammonium in the apoplastic interfacial compartment and releases the uncharged NH3 into the plant cytoplasm. The implications of such a finding are discussed in the context of AM functioning and plant phosphorus uptake.
Located at the interface between the abiotic and biotic components of terrestrial ecosystems, arbuscular mycorrhizal (AM) fungi are ubiquitous plant symbionts. The mutualistic nature of their interaction with plants is based on nutritional exchanges: as obligate biotrophs, AM fungi depend on a carbon flux from plants, which has been estimated as 5 billion tons per year (Bago et al., 2000). In return, AM fungi supply plants with nutrients directly taken up from the soil thanks to a network of extraradical hyphae that provide an extensive surface area for water and nutrient absorption (Giovannetti et al., 2004).
The uptake of inorganic phosphate (Pi) is considered to be the key physiological process by which AM fungi improve plant growth (Bucher, 2007). AM fungi in fact possess active Pi transporters that acquire Pi from the soil and allow its delivery to the plant (Harrison and van Buuren, 1995). On the other hand, plants are endowed with Pi transporters that are mycorrhizal specific: their role is to acquire the Pi from the interfacial apoplast and to deliver it to the plant cytoplasm. A Medicago truncatula Pi transporter, described as exclusively expressed during AM symbiosis and located in the periarbuscular membrane (Harrison et al., 2002), is not only essential for the acquisition of the Pi delivered by the AM fungus, but also is required to maintain arbuscules and sustain development of the AM fungus (Javot et al., 2007a).
Nitrogen (N) is the other nutrient for which mycorrhizal fungi play a crucial role: plants are in fact completely dependent on N availability in the soil solution for their growth and productivity (Gobert and Plassard, 2008). While the importance of ectomycorrhizal fungi in acquiring N for the plant has convincingly been demonstrated (for review, see Chalot et al., 2006), the role of AM fungi is less clear. N soil sources are either mineral (nitrate and ammonia) or organic (small peptides or amino acids) and AM fungi have been demonstrated through radiotracer experiments to take up inorganic N and translocate it from the extra- to the intraradical hyphae in the form of Arg (Govindarajulu et al., 2005). While it was traditionally believed that amino acids were transferred from the fungus to the plant, current knowledge suggests that N is released in an inorganic form (Govindarajulu et al., 2005). This hypothesis is supported by transcriptional data: transcripts encoding enzymes that assimilate inorganic N have been detected in the extraradical mycelium, whereas those involved in Arg breakdown are expressed in the intraradical fungal structures (Govindarajulu et al., 2005; Lopez-Pedrosa et al., 2006). Furthermore, evidence is now accruing that AM fungi may have access to organic N sources (Hodge et al., 2001; Leigh et al., 2009) thanks to the expression of specific amino acid permeases (Cappellazzo et al., 2008). The way in which N is transferred from the AM fungi to their host is still under debate. Even though the up-regulation of plant nitrate transporters has been observed in mycorrhizal roots of M. truncatula and Lotus japonicus (Hohnjec et al., 2005), NH3/NH4+ is thought to be the preferential form of N released by the fungus (Chalot et al., 2006). So far no plant ammonium transporter (AMT) has been identified as responsible for N uptake at the periarbuscular membrane (Parniske, 2008). Four high-affinity AMTs have been described in L. japonicus, three of which belong to the AMT1 and one to the AMT2 family (Simon-Rosin et al., 2003; D'Apuzzo et al., 2004). However, none of them has been directly associated with the nodule or AM symbiosis. An investigation on the gene expression change of L. japonicus roots upon colonization by Gigaspora margarita has revealed a new AMT, which was the highest up-regulated gene (Guether et al., 2009). Here, we report a detailed description of this novel transporter and include its functional characterization and structural modeling. We demonstrate that it is mycorrhiza specific because it is not up-regulated in the nodules and, further, using laser microdissected cells, we reveal that it is preferentially expressed in arbusculated cells. The implications of such a finding are discussed in the context of AM functioning and plant phosphorus uptake.
RESULTS
Gene Isolation and Phylogenetic Analysis of LjAMT2;2
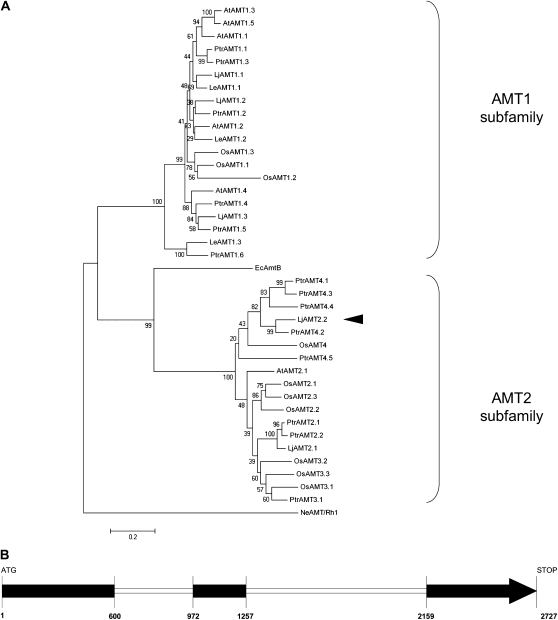
5′- and 3′-RACE PCR was carried out to isolate the full-length cDNA of the LjAMT2;2 transporter. The resulting 1,637-bp cDNA contained an open reading frame encoding a 53-kD polypeptide of 485 amino acid residues (GenBank accession no. FJ668388). The sequence showed that LjAMT2;2 is closely related to LjAMT2;1 described by Simon-Rosin et al. (2003). The phylogenetic analysis confirmed that LjAMT2;2 belongs to subfamily 2 of plant AMTs (Fig. 1A). It clustered into AMT2 subgroup 4 of Oryza sativa and Populus trichocarpa, which are known to present a higher number of AMT members than L. japonicus and Arabidopsis (Arabidopsis thaliana). LjAMT2;2 showed the highest homology to PtrAMT4;2. A recent update of the genomic sequence database of L. japonicus (Sato et al., 2008) has made it possible to identify the genomic DNA (2,730 bp) of LjAMT2;2, which harbors two introns (Fig. 1B).
Figure 1.
A, An unrooted phylogenetic tree for the amino acid sequences of AMT1/AMT2 plant AMTs. The dendrogram was generated by Mega 4.0 software using ClustalW for the alignment and the neighbor-joining method for the construction of the phylogeny (Tamura et al., 2007). Bootstrap tests were performed using 1,000 replicates. The branch lengths are proportional to the phylogenetic distances. Abbreviations for plant species: At, A. thaliana; Le, Lycopersicon esculentum; Lj, L. japonicus; Os, O. sativa; Ptr, P. trichocarpa; Ec, E. coli; Ne, N. europaea. The mycorrhiza-specific LjAMT2;2 is marked with a black arrowhead. The accession numbers for the AMTs used are as follows: AtAMT1;1 (1703292); AtAMT1;2 (4324714); AtAMT1;3 (5880355); AtAMT1;4 (7450345); AtAMT1;5 (5672513); AtAMT2 (3335376); OsAMT1;1 (AAL05612.1); OsAMT1;2 (AAL05613.1); OsAMT1;3 (AAL05614.1); OsAMT2;1 (BAB87832.1); OsAMT2;2 (NP_915334); OsAMT2;3 (NP_915337.1); OsAMT3;1 (BAC65232.1); OsAMT3;2 (AAO41130); OsAMT3;3 (AK108711); OsAMT4 (AAL58960); LeAMT1;1 (P58905); LeAMT1;2 (O04161); LeAMT1;3 (Q9FVN0); LjAMT1;1 (Q9FSH3); LjAMT1;2 (Q7Y1B9); LjAMT1;3 (Q70KK9); LjAMT2;1 (AAL08212); P. trichocarpa AMTs (Couturier et al., 2007); EcAmtB (NP_286193); and NeRh-1 (NP_840535). B, Intron-exon structure of the LjAMT2;2 gene; exons, black.
AM Symbiosis and Cell Specificity of LjAMT2;2 Expression
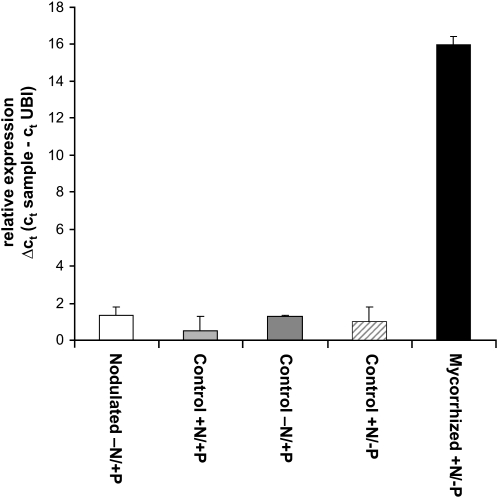
The sequence identified as the LjAMT2;2 transporter originated from an array study performed on L. japonicus roots colonized by Gi. margarita (Guether et al., 2009). The transporter showed a 31,000-fold up-regulation exclusively in mycorrhizal roots. To check the specificity of such an up-regulation, its expression pattern was investigated by comparing it with nodulated roots and the respective controls with high and low N availability: quantitative reverse transcription (RT)-PCR revealed that the LjAMT2;2 transporter expression in nodulated roots remained at control levels (Fig. 2).
Figure 2.
Relative expression of LjAMT2;2 assessed by quantitative RT-PCR in L. japonicus roots after 28 d of mycorrhization by Gi. margarita and 35 d after nodulation by Mesorhizobium loti. The Ct values (threshold cycles) of the samples are corrected against the Ct values of the housekeeping gene ubiquitin (LjUBQ10 [UBI]). Data for each condition are presented as mean + sd and were obtained from three biological and three technical replicates. −N = 10 μm KNO3; +N = 4 mm KNO3; −P = 20 μm PO43−; +P = 500 μm PO43−.
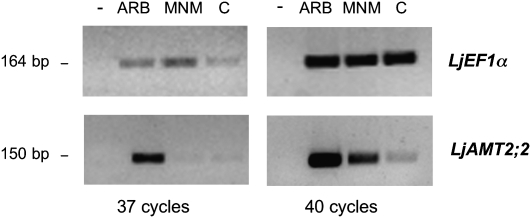
A laser microdissection approach was used to localize the transcript accumulation site. Since Govindarajulu et al. (2005) demonstrated a transfer of N from the AM fungus to the plant during the colonization process, cortical cells were chosen as a first target. Three cell types were collected by a laser microdissection system: arbusculated cells, noncolonized cortical cells from mycorrhizal roots, and cortical cells from nonmycorrhizal roots. RNA extracted from the three cell-type populations was used in semiquantitative RT-PCR experiments. The amount of RNA in the different samples was evaluated by amplifying the Lotus elongation factor (LjEF1α) transcripts with specific primers and conducting 37 and 40 amplification cycles. After 40 cycles, the amplification had reached its plateau in all samples, whereas after 37 cycles it was still in the exponential phase, thus allowing a semiquantitative comparison of transcript abundance. As shown in Figure 3, the LjEF1α transcript abundance in arbusculated cells was comparable with the two other samples.
Figure 3.
RT-PCR analysis of LjAMT2;2 in three different cell-type populations: ARB, arbusculated cells; MNM, noncolonized cortical cells from mycorrhizal roots; and C, cortical cells from nonmycorrhizal roots. LjEF1α amplicons, obtained from parallel control reactions and loaded in the same order as above, were used as internal standards. The sizes of the LjAMT2;2 and LjEF1α amplicons are indicated. Reactions were performed allowing both 37 and 40 cycles of amplifications.
When the specific primers for LjAMT2;2 were used, a fragment of the expected size was present in all the RNA samples after 40 cycles of amplification, with the highest intensity in arbusculated cells, followed by noncolonized cortical cells from mycorrhizal roots and the control (Fig. 3). After 37 cycles of amplification, it became evident that the amount of the amplified product was considerably higher in arbusculated cells than in noncolonized cortical cells from mycorrhizal roots and control cell-type populations, indicating that arbusculated cells are the preferential site for the expression of LjAMT2;2 in a mycorrhizal root. Only faint bands were detected in the two other cortical cell-type populations. This expression pattern was reproducible in two different biological replicates.
Functional Expression of LjAMT2;2 in a Yeast Mutant Defective in High-Affinity NH4+ Uptake
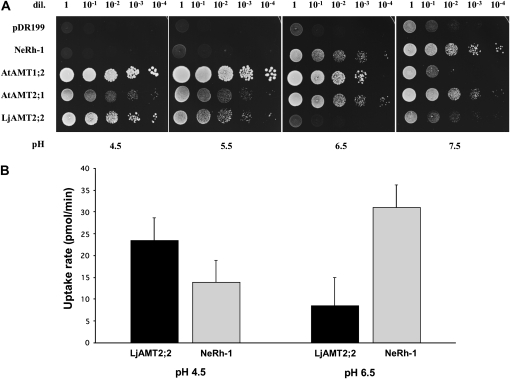
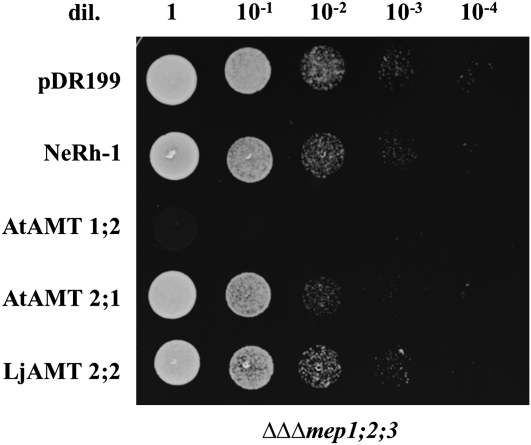
To prove the functionality of LjAMT2;2, a complementation experiment was carried out. The yeast (Saccharomyces cerevisiae) strain 31019b, which is defective in all three endogenous NH4+ transporters (Mep1, Mep2, and Mep3) and is thus unable to grow on a medium containing <5 mm NH4+ as the sole N source (Marini et al., 1997), was used. This strain was transformed with the yeast vector pDR199 that expressed LjAMT2;2 under the control of the constitutive yeast PMA1 promoter. To compare the LjAMT2;2 with already described plant and bacterial NH3/NH4+ transporters/channels, the yeast strain was also transformed with pDR199 expression vectors containing the following coding sequences: the NH3 channel NeRh-1 of Nitrosomonas europaea (Weidinger et al., 2007), the net NH4+ transporter AtAMT1;2 (Neuhäuser et al., 2007), and AtAMT2 of Arabidopsis (Sohlenkamp et al., 2002). Considering the acidic conditions of the periarbuscular space (Guttenberger, 2000; Smith et al., 2001), the yeast growth was analyzed on a yeast nitrogen base (YNB) medium at different pH. As seen in Figure 4A, the yeast growth was restored by the LjAMT2;2 on the minimal medium containing 3 mm NH4+ as the sole N source. The transporter showed a pH dependency and growth promotion was best at an acidic pH of 4.5. At this pH, the amount of NH3 is only approximately 0.002% of the total NH3/NH4+ [pKa(NH4+) = 9.25]. Unlike NH4+, the uncharged NH3 is thought to cross the hydrophobic plasma membrane by simple diffusion, but, at this low pH in the medium, NH3 is apparently almost absent. This indicates that LjAMT2;2 recruits the charged species, NH4+, for transport. The pH dependence of LjAM2;2 has been confirmed using 14C-labeled methylammonium uptake experiments of transformed yeast strains. The yeast expressing the N. europaea Rh-1 channel, which facilitates the bidirectional diffusion of the uncharged NH3 across the membrane, was used as a control.
Figure 4.
A, Growth of NH4+ uptake-deficient yeast (31019b; ΔΔΔmep1;2;3) transformed with NeRh-1, AtAMT1;2, AtAMT2;2, LjAMT2;2, and the empty control plasmid pDR199 on 3 mm ammonium as sole N source at different pH. Shown are serial dilutions (dil.) of cell suspensions ranging from 1 to 1 × 10−4. B, [14C]MeA uptake rates of LjAMT2;2 and NeRh-1 at pH 4.5 and 6.5. Data for each condition are presented as mean + SEM and were obtained from four technical replicates.
Methylammonium (= CH3NH3+ = MeA+) is a widely used transport analog of NH4+ because it has similar chemical characteristics [pKa(MeA+) = 10.64] and is transported by the same uptake systems as NH4+ (Kosola and Bloom, 1994). Its toxicity can be overcome by a simultaneous application of NH4+ (Arst and Cove, 1969) because the transport systems often have a 10-fold greater affinity for NH4+ than for MeA+ (Kosola and Bloom, 1994). The experiment showed a significantly higher 14C-MeA uptake rate for LjAMT2;2 at pH 4.5 than at pH 6.5, whereas better transport rates were obtained for NeRh-1 at pH 6.5 (Fig. 4B). These data confirm the results of the yeast growth experiment. Taken together, the experiments showed an opposite pattern of the pH dependency of N uptake by LjAMT2;2 and NeRh-1 transformed yeast.
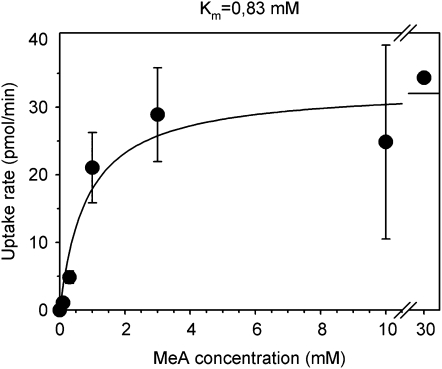
The AMT-deficient strain 31019b carrying the previously mentioned pDR199 constructs was spotted on the YNB medium supplemented with 80 mm MeA and Arg as the metabolizable N source. Figure 5 shows that only AtAMT1;2 efficiently imported the toxic MeA, although LjAMT2;2 transported some 14C-MeA. The capacity of LjAMT2;2 to enhance the permeability to MeA in yeast was further tested at several concentrations. The results indicated that LjAMT2;2 is a high-affinity transporter that was saturable and transported with half-maximal capacity at 0.83 mm (Fig. 6).
Figure 5.
Growth of yeast AMT-deficient strain 31019b on YNB medium supplemented with 80 mm MeA+ and Arg as the sole metabolizable N source. The strain was transformed with NeRh-1, AtAMT1;2, AtAMT2;2, LjAMT2;2, and control plasmid pDR199.
Figure 6.
Concentration-dependent kinetics of [14C]MeA uptake by yeast strain 31019b transformed with the pDR199 LjAMT2;2. Yeast strain 31019b transformed with insert-free pDR199 were used to measure background transport activity, and results are reported as net transport.
Transport Characteristics of LjAMT2;2 in Oocytes
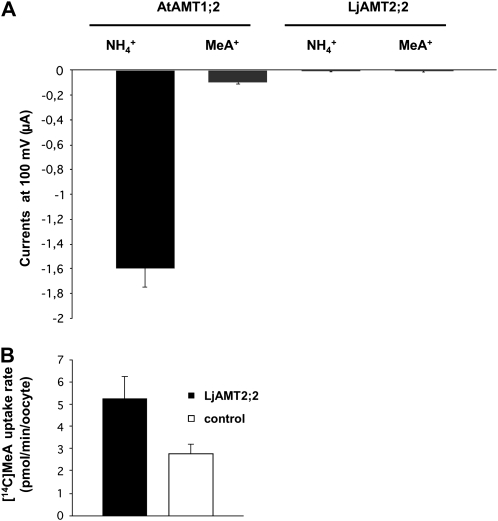
As previously described (Neuhäuser et al., 2007), AtAMT1;2 expressed in Xenopus oocytes caused large ammonium-dependent inward currents. The currents elicited by 3 mm NH4+ were much stronger than those elicited by the equivalent amount of MeA (Fig. 7A). When LjAMT2;2-expressing oocytes were used, surprisingly, NH4+ did not cause any significant change in the inward currents. To exclude that the absence of signals was caused by a nonfunctional protein, the same oocytes were subjected to a [14C]MeA uptake. The measured uptake was significantly higher than in the control oocytes. Oocytes expressing LjAMT2;2 took up the radiotracer by a rate of 2.46 ± 1.45 pmol min−1 per oocyte (Fig. 7B). These results clearly demonstrate that LjAMT2;2 is functional in Xenopus oocytes, but ammonium and MeA transport are not associated with ionic currents.
Figure 7.
A, Expression of LjAMT2;2 and AtAMT1;2 in oocytes. Shown are inward currents by 3 mm NH4Cl at −100 mV from oocytes injected with equal amounts of cRNA. Data are from three to six oocytes. Similar data were obtained in three independent experiments. Data for each condition are presented as mean + sd and were obtained from more than eight biological replicates. B, [14C]MeA uptake rate of the same oocytes injected with LjAMT2;2 cRNA and of water-injected controls. Data for each condition are presented as mean + sd and were obtained from eight biological replicates for AtAMT1;2 and 20 biological replicates for LjAMT2;2.
Structural Modeling and Probable Transport Mechanism of LjAMT2;2
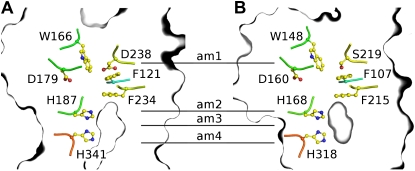
Given that the periarbuscular space is considered an acidic compartment (Guttenberger, 2000), NH4+ is expected to be the dominant species available, whereas NH3 is at vanishing low amounts. Indeed, the growth complementation tests at various pHs indicate that NH4+ is the form recruited by LjAMT2;2. However, the transport assays in oocytes demonstrate that LjAMT2;2 translocates NH3. To resolve this discrepancy, a LjAMT2;2 homology model was set up using the MODELLER software package and was compared with the crystal structure of the bacterial homolog AmtB from Escherichia coli. The ammonium transport mechanism of EcAmtB and the crucially involved amino acids have been deduced from the structure (Khademi et al., 2004; Zheng et al., 2004). In EcAmtB, NH4+ is recruited by the periplasmatic NH4+ binding site (am1), is subsequently deprotonated (between am1 and am2), and is thought to be further translocated as neutral NH3 (am2–am4; Khademi et al., 2004). The amino acids recruiting NH4+ from the periplasm are conserved (Fig. 8). These include the aromatic residues W148 and F107 in EcAmtB, which putatively stabilize the cation by π interactions. The same residues are present in the corresponding positions in LjAMT2;2. S219 in EcAmtB, which forms a hydrogen bond with NH4+ via a hydroxyl oxygen, is replaced by an Asp in LjAMT2;2. As in EcAmtB, two Phe form a gate before the highly hydrophobic pore begins, which contains the two well-conserved pore lining His H187 (EcAmtB: H168) and H341 (EcAmtB: H318; Fig. 8). D179 in LjAMT2;2 corresponds to the functionally important and conserved D160 in EcAmtB (Khademi et al., 2004; Zheng et al., 2004), which stabilizes NH4+ at am1 (Luzhkov et al., 2006). The deprotonation region, as well as the hydrophobic pore of LjAMT2;2, is very similar to those of EcAmtB, suggesting a similar NH4+ recruiting/NH3 translocation mechanism for LjAMT2;2. Such a mechanism is fully supported by our experimental findings on LjAMT2;2 functionality.
Figure 8.
Homology model of LjAMT2;2 (A) and illustration of important residues in the crystal structure of EcAmtB (B). Key residues that are thought to be important for NH4+ binding, subsequent deprotonation, and NH3 translocation in EcAmtB are also found in LjAMT2;2.
DISCUSSION
In mycorrhizal associations, the fungal partner assists its host plant by providing N, in addition to phosphate (Chalot et al., 2006; Smith and Read, 2008). AM fungi have access to inorganic or organic forms of N (Lopez-Pedrosa et al., 2006; Cappellazzo et al., 2008) and translocate them via Arg from the extra- to the intraradical mycelium, where N is transferred to the plant in inorganic form (Govindarajulu et al., 2005; Jin et al., 2005). However, the molecular form in which N is transferred from AM fungi to their hosts is still under debate: while Govindarajulu et al. (2005) considered NH4+, Chalot et al. (2006) added NH3 as a potential candidate. In addition, no plant AMT has been identified so far as being responsible for NH3/NH4+ uptake in AMs (Parniske, 2008). Here, we provide evidence of a plant AMT (LjAMT2;2) involved in N uptake during mycorrhiza symbiosis. In addition to being mycorrhiza specific and preferentially activated in arbusculated cells, the LjAMT2;2 protein is a functional transporter that recruits NH4+ and releases the uncharged NH3 into the plant cytoplasm in a pH-dependent manner.
LjAMT2;2: A Newly Discovered Transporter That Belongs to AMT Family 2
Unlike AMT subfamily 1, members of subfamily 2 are known to harbor some introns in their gene sequences (Couturier et al., 2007). From this point of view, LjAMT2;2, with its two introns, fits well into the AMT2 subfamily. This picture is also confirmed by the phylogenetic analysis that placed LjAMT2;2 in the AMT2 subgroup 4 (Suenaga et al., 2003) of O. sativa and P. trichocarpa (Couturier et al., 2007). Interestingly, transcripts from this subgroup are rare or not detected at all in several species (Suenaga et al., 2003; Couturier et al., 2007). EST sequences have so far only been reported for two members: PtrAMT4;2 in floral buds and PtrAMT4;5 in poplar cultured cells (Couturier et al., 2007). This would seem to suggest that these subgroup members, and perhaps the whole AMT2 subfamily, may have functions that could be tissue specific or activated under specific conditions. Support for this comes from gene knockouts and RNAi-mediated silencing of the AtAMT2 in Arabidopsis, which had no altered phenotype in soil-grown plants (Sohlenkamp et al., 2002).
Expression of LjAMT2;2 Is Mycorrhiza Specific and the Functional Protein Is More Active in an Acidic Environment
In an investigation on the global transcriptome of L. japonicus upon Gi. margarita colonization (Guether et al., 2009), LjAMT2;2 was the highest up-regulated gene in mycorrhizal roots 28 d postinoculation (dpi; 31,000-fold). The gene instead was not differentially regulated in nodules (35 dpi), similarly to the closely related LjAMT2;1 transporter that is expressed constitutively in nodulated and non-nodulated Lotus plants (Simon-Rosin et al., 2003).
Like PT4 transporters, LjAMT2;2 therefore seems to be one of the few mycorrhiza-specific genes that have been detected so far, and its high expression values suggest a switch-on mechanism rather than an up-regulation. This hypothesis will also have to be validated in other nonlegume plants using an AM fungus that does not belong to Gigasporaceae.
The collection of microdissected cells from mycorrhizal and control roots has allowed us to demonstrate that LjAMT2;2 transcripts are mainly localized in arbusculated cells, as already demonstrated for other transporters (Harrison et al., 2002; Balestrini et al., 2007). The functional characterization of LjAMT2;2 by yeast complementation demonstrates its pH dependency, with the highest uptake rates at acidic pH. This is in line with its localization in arbuscule-containing cells, according to the laser microdissection experiments, and particularly in the periarbuscular membrane: the interface compartment that surrounds the arbuscules is, in fact, considered acidic (Guttenberger, 2000; Smith et al., 2001). These data would suggest a similarity with the MtPT4 phosphate transporter of M. truncatula, which is localized in the periarbuscular membrane and shows the highest transport activity under acidic conditions (Harrison et al., 2002).
Even though weakly, LjAMT2;2 transcripts were also detected in noncolonized cortical cells of mycorrhizal roots. This could be due to a release of NH3/NH4+ from Arg by the intercellular hyphae. However, an activation of gene expression in neighboring cells prior to arbuscule development cannot be excluded. It has in fact been demonstrated that prepenetration responses, as new membrane assembly, precede the colonization of AM fungi in the root cortex (Genre et al., 2008).
The high expression of LjAMT2;2 in arbusculated cells facilitates ammonia flux into the cytosol, but prevents NH4+ accumulation. Due to the negative membrane potential of plant root cells, a massive accumulation of ammonium could be expected if the charged species is transported. Interestingly, all other plant AMTs that have been investigated in detail act as net NH4+ transporters (Mayer et al., 2006; Ludewig et al., 2007). It is known that the plant growth-promoting effect of NH3/NH4+ is reversed at high concentrations (Britto et al., 2001; von Wiren and Merrick, 2004). To prevent severe damage of the membranes by high ammonia concentrations, plants must have evolved protective mechanisms, such as ammonia incorporation into amino acids. In this context, transcriptomic investigations on AMs have so far never detected any activation of plant NH3/NH4+ assimilatory enzymes, thus supporting the hypothesis that the transcriptional activation of protective mechanisms is not required in arbusculated cells. In the presence of an acidic pH in the periarbuscular space, the recognition of NH4+, but transport of NH3, provides an elegant solution that will not lead to the accumulation of NH3/NH4+ to potentially toxic levels.
Ammonium and Phosphate Uptake at the Interface: A Speculative New Model
The structural features of the transporter and the transport experiments suggest that the high-affinity transporter LjAMT2;2 binds NH4+, which makes up ≥99.998% of the total NH3/NH4+ in the periarbuscular space, which has been demonstrated to have a pH of approximately 4.25 (Guttenberger, 2000; Smith et al., 2001).
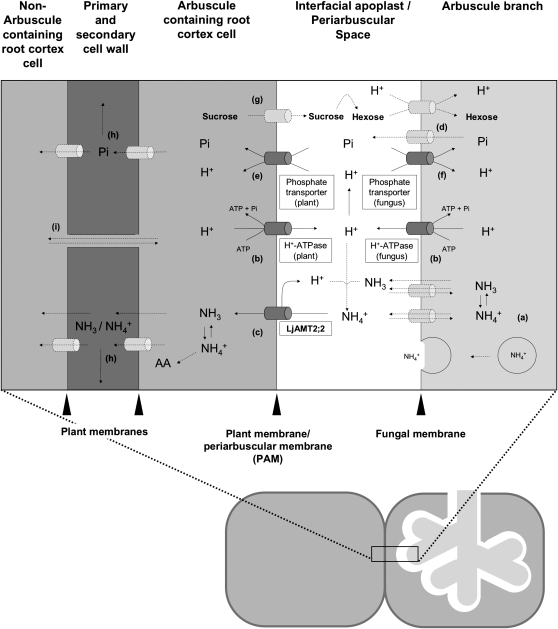
Considering the electroneutral transport shown by the electrophysiological measurements and the MeA assay, the NH4+ ion must be deprotonated prior to its transport across the membrane and then released in its uncharged NH3 form into the plant cytoplasm. We suggest that, together with the protons, which are actively transported into the interfacial apoplast by the plant and also by fungal H+-ATPases (Murphy et al., 1997; Gianinazzi-Pearson et al., 2000; Krajinski et al., 2002; Requena et al., 2003; Balestrini et al., 2007), the protons coming from the NH4+ deprotonation process remain in the interface space and maintain or even reinforce the gradient for H+-dependent transport processes (Fig. 9). This indirect coupling of proton-dependent and proton-independent transport processes could reduce the transport costs that are paid by the plant and the fungus.
Figure 9.
The scheme illustrates N, phosphorus, and carbohydrate exchanges at the mycorrhizal interface according to previous works and the present results. a, NH3/NH4+ is released in the arbuscules from Arg, which is transported from the extra- to the intraradical fungal structures (Govindarajulu et al., 2005). NH3/NH4+ is then translocated by so far unknown mechanisms (transporter, diffusion, or vesicle mediated) into the periarbuscular space, where, due to the acidic environment, its ratio shifts toward NH4+ (>99.99%). b, The acidity of the interfacial apoplast is established by plant and fungal H+-ATPases (Hause and Fester, 2005; Balestrini et al., 2007), thus providing the energy for H+-dependent transport processes. c, The NH4+ ion is deprotonated prior to its transport across the plant membrane via the LjAMT2;2 protein and released in its uncharged NH3 form into the plant cytoplasm. The NH3/NH4+ acquired by the plant is either transported into adjacent cells or immediately incorporated into amino acids. d, Phosphate is released by so far unknown transporters into the interfacial apoplast. e, The uptake of phosphate on the plant side then is mediated by mycorrhiza-specific Pi transporters (Javot et al., 2007b; Guether et al., 2009). f, AM fungi might control the net Pi release by their own Pi transporters, which may reacquire phosphate from the periarbuscular space (Balestrini et al., 2007). g, Plant-derived carbon is released into the periarbuscular space probably as Suc and then cleaved into hexoses by Suc synthases (Hohnjec et al., 2003) or invertases (Schaarschmidt et al., 2006). AM fungi then acquire hexoses (Shachar-Hill et al., 1995; Solaiman and Saito, 1997) and transport them over their membrane by so far unknown hexose transporters. It is likely that these transporters are proton cotransporters as the GpMST1 described for the glomeromycotan fungus Geosiphon pyriformis (Schuessler et al., 2006). Exchange of nutrients between arbusculated cells and noncolonized cortical cells can occur by apoplastic (h) or symplastic (i) ways.
Considering the electroneutral transport of NH3 by LjAMT2;2, it is likely that the plant needs less energy for the uptake of N using this transporter instead of the AMT1 family members, which are based on an electrogenic mechanism. However, it remains to be clarified whether a part of the saved energy has to be paid off to the fungus that, on its hand, has to concentrate ammonium and/or release it into the interfacial apoplast.
CONCLUSION
Ammonium, as ammonium sulfate and ammonium nitrate, is the main form of N supplied to crops when fertilizers are applied. Such N fertilizers are expensive in terms of energy for their synthesis and dangerous for the ecosystem balance because they lead to eutrophication and depletion of soil organic carbon (Tilman et al., 2001; Khan et al., 2007). For these reasons, N fertilizer use surely needs to be reduced in the future. As biofertilizers, AM fungi are an emerging issue in many projects focused on more sustainable agriculture practices (Smith and Read, 2008). In this context, the finding of a plant mycorrhiza-dependent AMT opens new speculation: mycorrhizal fungi could optimize the uptake of N from fertilizers dispersed on agricultural soils and release it as ammonium to the plant.
A deeper understanding of the fine regulation of fungal and plant AMTs seems to be mandatory.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Inoculation Methods
Gigaspora margarita Becker and Hall (strain deposited in the Bank of European Glomales as BEG 34) was used as fungal inoculum. The mycorrhization method of Lotus japonicus (Regel) K. Larsen (Gifu; wild type) is described in detail (Guether et al., 2009). For the nodulation experiment, Lotus seedlings were grown in vermiculite and a B & D nutrient solution (Broughton and Dilworth, 1971) containing 10 μm or 4 mm KNO3, respectively. A part of the N-starved plants was inoculated with the Mesorhizobium loti strain NZP2235. Bacterial cells from a 2-d-old liquid culture (5 mL) were centrifuged, washed, and suspended in 50 mL of water. Twenty-eight days after fungal inoculation, samples were cut from the mycorrhizal roots after observation under a stereomicroscope. Some segments were stained with 0.1% cotton blue in lactic acid and the infection was quantified, as described by Trouvelot et al. (1986); other fragments were processed for cellular and molecular analysis. Root segments from three independent experiments were analyzed. The average root colonization (61%) was in the same range as for the reported microarray study (Guether et al., 2009). The roots inoculated with M. loti were checked for the presence of pinkish nodules 35 dpi and harvested.
RNA Isolation, cDNA Synthesis, and Real-Time RT-PCR
The RNA isolation, cDNA synthesis, and quantitative RT-PCR methods have already been described in detail (Guether et al., 2009). Prior to quantitative RT-PCR, gene-specific primers for LjAMT2;2 were tested on genomic DNA and cDNA for amplification. Because RNA extracted from mycorrhizal roots contains plant and fungal RNAs, the specificity of the primer pair was also analyzed by PCR amplification of Gi. margarita genomic DNA. No amplification products were obtained on fungal DNA. The oligonucleotide sequences for the LjAMT2;2 were as follows: forward primer, 5′-ACACATGCTTGCACTGCTACC-3′; reverse primer, 5′-CTGCCCATCCTTGAACAACCC-3′.
5′-RACE and 3′-RACE
5′-RACE and 3′-RACE were performed with the aforementioned total RNA extracted from the mycorrhized roots using a SMART RACE cDNA amplification kit (CLONTECH). The gene-specific primer sequences used are as follows: reverse primer, 5′-GTGCTGCCCATCCTTGAACAACC-3′; forward primer, 5′-GAGACCCTTACACAGTGAGCATGGA-3′. PCR was performed according to the CLONTECH protocol using the Advantage 2 PCR enzyme system and 35 cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 2 min, and a final extension at 72°C for 10 min. The RACE products were subjected to electrophoresis, cloned in pCRII (TOPO cloning kit; Invitrogen), and analyzed by DNA sequencing.
Plasmid Constructs
The coding region of LjAMT2;2 (1,455 bp) was amplified from the aforementioned cDNA by PCR using the Advantage 2 PCR enzyme system (CLONTECH) and the following oligonucleotides: forward primer, 5′-CGTACATTAAACATGTCTACTGTT-3′; reverse primer, 5′-TTTAGTTTGAGGTCATCTCGA-3′. The PCR product was cloned in the pCRII vector (TOPO cloning kit; Invitrogen) and verified via full-length sequencing. For oocyte and yeast (Saccharomyces cerevisiae) expression, the open reading frame of LjAMT2;2 was subcloned into the oocyte expression vector pOO2 and the yeast expression vector pDR199. The following AMTs were used as controls for the yeast expression experiment: AtAMT1;2 (Neuhäuser et al., 2007); AtAMT2;1 (B. Neuhäuser and U. Ludewig, unpublished data); NeRh-1 (Weidinger et al., 2007). The GenBank accession numbers for the coding sequences of the control AMTs are AF110771, AF182039, and NC_004757.
Expression in Yeast
The plasmids containing the respective open reading frames were heat shock transfected in the ura− AMT-defective yeast strain 31019b; ΔΔΔmep1;2;3 (Marini et al., 1997). The N-deficient growth medium was YNB without amino acids and ammonium sulfate (Difco), supplemented with 3% Glc and 3 mm NH4Cl as the sole N source. No buffer was added. The growth of the yeast was not affected by the expression of the different constructs under nonselective conditions.
Yeast Uptake
Yeast was grown in a liquid Arg medium until the OD595 reached 0.6–0.8. Cells were harvested, washed, and resuspended in an uptake buffer (50 mm potassium phosphate buffer supplemented with 0.6 m sorbitol, pH 6) to a final OD595 of 5. Before the uptake, cells were energized by adding 10 μL 1 m Glc to 100 μL cells OD595 of 5 and incubated for 7 min at 30°C. The uptake was started by adding 110 μL of uptake buffer containing [14C]MeA (2.11 GBq/mmol; Amersham Bioscience) in different concentrations. Samples (50 μL) samples were taken after 30, 60, 120, and 300 s and washed in 4 mL of an ice-cold washing solution (uptake buffer containing 100 mm of nonlabeled MeA). Immediately after taking the sample, the solution was filtered through glass fiber filters (GF/C; Whatman). The filters containing the yeast were washed three times with 4 mL of ice-cold washing solution, placed in scintillation vials, 4 mL of a scintillation buffer were added, and the activity was analyzed by liquid scintillation counting. The concentration dependence of the uptake was fitted using the following equation: I = Imax/(1 + Km/c), where Imax is the maximal uptake rate at the saturating concentration, Km is the substrate concentration that permits half-maximal uptake, and c is the experimentally used concentration.
Electrophysiological Measurements, Preparation, and Injection of Oocytes
These methods have been described in more detail elsewhere (Mayer et al., 2006). Briefly, oocytes were taken from adult females, dissected by a collagenase treatment (2 mg/mL, 1.5 h), and injected with 50 nL of cRNA (1 ng/nL). The oocytes were kept in ND96 (Kitayama et al., 1996) for 3 d at 16°C and then placed in a small recording chamber. The recording solution was (in mm): 110 choline chloride, 2 CaCl2, 2 MgCl2, 5 MES, pH adjusted with Tris to 4.5 (LjAMT2;2) and 5.5 (AtAMT1;2). Variable ammonium concentrations were added as NH4Cl salt. Currents without added ammonium were subtracted at each voltage. Error bars show se for repetitions n = 20 or n = 8 for LjAMT2;2 and AtAMT1;2, respectively.
Radiotracer Uptake into Xenopus laevis Oocytes
The ammonium analog 14C-methylammonium (Amersham Biosciences; specific activity: 2.11 GBq/mmol) was used as a radiotracer. The previously described choline-based buffer was used for uptake experiments. Batches of >12 oocytes were incubated for 15 min in 500 μL of the respective buffer, which contained a 1/30 dilution (total concentration, 3 mm) of 14C-labeled and nonlabeled methylammonium at room temperature. Then the oocytes were carefully washed five times in 1 mL of ice-cold buffer containing 100 mm of unlabeled methylammonium and aliquoted (four oocytes per scintillation vial). After solubilization with 20 μL 10% SDS, 4 mL of scintillation buffer was added and the activity was analyzed by liquid scintillation counting (Wallac 1414 WinSpectral liquid scintillation counter).
Laser Microdissection
Mycorrhizal and nonmycorrhizal root segments were fixed in freshly prepared Methacarn (absolute methanol:chloroform:glacial acetic acid [6:3:1]) at 4°C overnight for paraffin embedding (Balestrini et al., 2007). A Leica AS laser microdissection system (Leica Microsystems) was used to isolate cells from the prepared tissue sections as described in Balestrini et al. (2007) and Guether et al. (2009). After collection, the RNA extraction buffer from the PicoPure kit (Arcturus Engineering) was added and samples were incubated at 42°C for 30 min, centrifuged at 800g for 2 min, and stored at −80°C. Then, for the following RNA extraction steps, about 1,500 cells were pooled for each cell-type population in a single tube with a final volume of 50 μL.
RNA Extraction and RT-PCR on Microdissected Samples
RNA extractions were performed with a slightly modified PicoPure kit protocol (Arcturus Engineering), as described by Balestrini et al. (2007). RNA quantification was obtained using the NanoDrop 1000 spectrophotometer. A one-step RT-PCR kit (Qiagen) was used for the RT-PCR experiments that were conducted on the RNA extracted from the several samples. Reactions were carried out as described in detail in Guether et al. (2009). Amplification reactions with specific primers for LjAMT2;2 and the housekeeping gene LjEF1α (Guether et al., 2009; annealing temperature 65°C and 60°C, respectively) were allowed to proceed for different numbers of cycles (37 and 40) to determine the exponential amplification phase. The RT-PCR experiments were conducted on at least two independent biological and technical replicates.
Homology Modeling
The homology models were based on four high-resolution structures of AmtBs from Escherichia coli (PDB ID: 1U77, 1XQE, 2NS1, 3B9Y) and one high-resolution structure of Amt-1 from Archaeglobus fulgidus (PDB ID: 2B2H). Twenty-one amino acids of the N-terminal moiety and 69 amino acids of the C-terminal moiety of the primary sequence of LjAMT2;2 were truncated. Multiple sequence alignments based on primary sequences and the secondary structures of the templates were performed with the ESPRESSO method from the T-COFFEE program. Afterward, the sequence alignments were inspected visually and manually corrected. Homology modeling of LjAMT2;2 was performed with the MODELLER software package (version 9v5; http://salilab.org/modeller/modeller.html). Graphic representations were prepared using Pymol software (DeLano, 2002).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number FJ668388.
Acknowledgments
We thank Mara Novero for her assistance in the sandwich preparation, Thomas Ott for providing us with the M. loti strain NZP2235, and Jorge Gómez-Ariza for his assistance with Figure 9.
This work was supported by the European Union as part of the INTEGRAL project (Marie Curie Research Training Network; project reference no. 505227 to M.G.); by the INTEGRAL Project, the Italian Ministers of University, Research and Environment (FISR project “Soilsink”), and the Compagnia di San Paolo Foundation (grants to P.B.); and by the German Research Foundation (grants to U.L.).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Paola Bonfante (p.bonfante@ipp.cnr.it).
References
- Arst HN, Cove DJ (1969) Methylammonium resistance in Aspergillus nidulans. J Bacteriol 98 1284–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago B, Pfeffer PE, Shachar-Hill Y (2000) Carbon metabolism and transport in arbuscular mycorrhizas. Plant Physiol 124 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini R, Gomez-Ariza J, Lanfranco L, Bonfante P (2007) Laser microdissection reveals that transcripts for five plant and one fungal phosphate transporter genes are contemporaneously present in arbusculated cells. Mol Plant Microbe Interact 20 1055–1062 [DOI] [PubMed] [Google Scholar]
- Britto DT, Siddiqi MY, Glass AD, Kronzucker HJ (2001) Futile transmembrane NH4(+) cycling: a cellular hypothesis to explain ammonium toxicity in plants. Proc Natl Acad Sci USA 98 4255–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton WJ, Dilworth MJ (1971) Control of leghaemoglobin synthesis in snake beans. Biochem J 125 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher M (2007) Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol 173 11–26 [DOI] [PubMed] [Google Scholar]
- Cappellazzo G, Lanfranco L, Fitz M, Wipf D, Bonfante P (2008) Characterization of an amino acid permease from the endomycorrhizal fungus Glomus mosseae. Plant Physiol 147 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalot M, Blaudez D, Brun A (2006) Ammonia: a candidate for nitrogen transfer at the mycorrhizal interface. Trends Plant Sci 11 263–266 [DOI] [PubMed] [Google Scholar]
- Couturier J, Montanini B, Martin F, Brun A, Blaudez D, Chalot M (2007) The expanded family of ammonium transporters in the perennial poplar plant. New Phytol 174 137–150 [DOI] [PubMed] [Google Scholar]
- D'Apuzzo E, Rogato A, Simon-Rosin U, El Alaoui H, Barbulova A, Betti M, Dimou M, Katinakis P, Marquez A, Marini AM, et al (2004) Characterization of three functional high-affinity ammonium transporters in Lotus japonicus with differential transcriptional regulation and spatial expression. Plant Physiol 134 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002) The Pymol Molecular Graphics System. http://pymol.sourceforge.net (October 20, 2008)
- Genre A, Chabaud M, Faccio A, Barker DG, Bonfante P (2008) Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinazzi-Pearson V, Arnould C, Oufattole M, Arango M, Gianinazzi S (2000) Differential activation of H+-ATPase genes by an arbuscular mycorrhizal fungus in root cells of transgenic tobacco. Planta 211 609–613 [DOI] [PubMed] [Google Scholar]
- Giovannetti M, Sbrana C, Avio L, Strani P (2004) Patterns of below-ground plant interconnections established by means of arbuscular mycorrhizal networks. New Phytol 164 175–181 [DOI] [PubMed] [Google Scholar]
- Gobert A, Plassard C (2008) The beneficial effect of mycorrhizae on N utilization by the host-plant: myth or reality? In A Varma, ed, Mycorrhiza. Springer-Verlag, Berlin, pp 209–240
- Govindarajulu M, Pfeffer PE, Jin H, Abubaker J, Douds DD, Allen JW, Bucking H, Lammers PJ, Shachar-Hill Y (2005) Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 435 819–823 [DOI] [PubMed] [Google Scholar]
- Guether M, Balestrini R, Hannah MA, Udvardi MK, Bonfante P (2009) Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol 182 200–212 [DOI] [PubMed] [Google Scholar]
- Guttenberger M (2000) Arbuscules of vesicular-arbuscular mycorrhizal fungi inhabit an acidic compartment within plant roots. Planta 211 299–304 [DOI] [PubMed] [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ, van Buuren ML (1995) A phosphate transporter from the mycorrhizal fungus Glomus versiforme. Nature 378 626–629 [DOI] [PubMed] [Google Scholar]
- Hause B, Fester T (2005) Molecular and cell biology of arbuscular mycorrhizal symbiosis. Planta 221 184–196 [DOI] [PubMed] [Google Scholar]
- Hodge A, Campbell CD, Fitter AH (2001) An arbuscular mycorrhizal fungus accelerates decomposition and acquires nitrogen directly from organic material. Nature 413 297–299 [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Perlick AM, Puhler A, Kuster H (2003) The Medicago truncatula sucrose synthase gene MtSucS1 is activated both in the infected region of root nodules and in the cortex of roots colonized by arbuscular mycorrhizal fungi. Mol Plant Microbe Interact 16 903–915 [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Vieweg ME, Puhler A, Becker A, Kuster H (2005) Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol 137 1283–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ (2007. a) A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 104 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H, Pumplin N, Harrison MJ (2007. b) Phosphate in the arbuscular mycorrhizal symbiosis: transport properties and regulatory roles. Plant Cell Environ 30 310–322 [DOI] [PubMed] [Google Scholar]
- Jin H, Pfeffer PE, Douds DD, Piotrowski E, Lammers PJ, Shachar-Hill Y (2005) The uptake, metabolism, transport and transfer of nitrogen in an arbuscular mycorrhizal symbiosis. New Phytol 168 687–696 [DOI] [PubMed] [Google Scholar]
- Khademi S, O'Connell J, Remis J, Robles-Colmenares Y, Miericke LJW, Stroud RM (2004) Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 Å. Science 305 1587–1594 [DOI] [PubMed] [Google Scholar]
- Khan SA, Mulvaney RL, Ellsworth TR, Boast CW (2007) The myth of nitrogen fertilization for soil carbon sequestration. J Environ Qual 36 1821–1832 [DOI] [PubMed] [Google Scholar]
- Kitayama S, Morita K, Dohi T (1996) Uptake and release of dopamine through the rat dopamine transporter expressed in Xenopus laevis oocyte: evaluation by voltammetric measurement of intracellular dopamine concentration. Neurosci Lett 211 132–134 [DOI] [PubMed] [Google Scholar]
- Kosola KR, Bloom AJ (1994) Methylammonium as a transport analog for ammonium in tomato (Lycopersicon esculentum L.). Plant Physiol 105 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajinski F, Hause B, Gianinazzi-Pearson V, Franken P (2002) Mtha1, a plasma membrane H+-ATPase gene from Medicago truncatula, shows arbuscule-specific induced expression in mycorrhizal tissue. Plant Biol 4 754–761 [Google Scholar]
- Leigh J, Hodge A, Fitter AH (2009) Arbuscular mycorrhizal fungi can transfer substantial amounts of nitrogen to their host plant from organic material. New Phytol 181 199–207 [DOI] [PubMed] [Google Scholar]
- Lopez-Pedrosa A, Gonzalez-Guerrero M, Valderas A, Azcon-Aguilar C, Ferrol N (2006) GintAMT1 encodes a functional high-affinity ammonium transporter that is expressed in the extraradical mycelium of Glomus intraradices. Fungal Genet Biol 43 102–110 [DOI] [PubMed] [Google Scholar]
- Ludewig U, Neuhauser B, Dynowski M (2007) Molecular mechanisms of ammonium transport and accumulation in plants. FEBS Lett 581 2301–2308 [DOI] [PubMed] [Google Scholar]
- Luzhkov VB, Almlof M, Nervall M, Aqvist J (2006) Computational study of the binding affinity and selectivity of the bacterial ammonium transporter AmtB. Biochemistry 45 10807–10814 [DOI] [PubMed] [Google Scholar]
- Marini AM, Soussi-Boudekou S, Vissers S, Andre B (1997) A family of ammonium transporters in Saccharomyces cerevisiae. Mol Cell Biol 17 4282–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M, Dynowski M, Ludewig U (2006) Ammonium ion transport by the AMT/Rh homologue LeAMT1;1. Biochem J 396 431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy PJ, Langridge P, Smith SE (1997) Cloning plant genes differentially during colonization of roots of Hordeum vulgare by the vesicular-arbuscular mycorrhizal fungus Glomus intraradices. New Phytol 135 291–301 [Google Scholar]
- Neuhäuser B, Dynowski M, Mayer M, Ludewig U (2007) Regulation of NH4+ transport by essential cross talk between AMT monomers through the carboxyl tails. Plant Physiol 143 1651–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske M (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6 763–775 [DOI] [PubMed] [Google Scholar]
- Requena N, Breuninger M, Franken P, Ocon A (2003) Symbiotic status, phosphate, and sucrose regulate the expression of two plasma membrane H+-ATPase genes from the mycorrhizal fungus Glomus mosseae. Plant Physiol 132 1540–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al (2008) Genome structure of the legume Lotus japonicus. DNA Res 15 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaarschmidt S, Roitsch T, Hause B (2006) Arbuscular mycorrhiza induces gene expression of the apoplastic invertase LIN6 in tomato (Lycopersicon esculentum) roots. J Exp Bot 57 4015–4023 [DOI] [PubMed] [Google Scholar]
- Schuessler A, Martin H, Cohen D, Fitz M, Wipf D (2006) Characterization of a carbohydrate transporter from symbiotic glomeromycotan fungi. Nature 444 933–936 [DOI] [PubMed] [Google Scholar]
- Shachar-Hill Y, Pfeffer PE, Douds D, Osman SF, Doner LW, Ratcliffe RG (1995) Partitioning of intermediary carbon metabolism in vesicular-arbuscular mycorrhizal leek. Plant Physiol 108 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Rosin U, Wood C, Udvardi MK (2003) Molecular and cellular characterisation of LjAMT2;1, an ammonium transporter from the model legume Lotus japonicus. Plant Mol Biol 51 99–108 [DOI] [PubMed] [Google Scholar]
- Smith SE, Dickson S, Smith FA (2001) Nutrient transfer in arbuscular mycorrhizas: how are fungal and plant processes integrated? Aust J Plant Physiol 28 683–694 [Google Scholar]
- Smith SE, Read DJ (2008) Mycorrhizal Symbiosis. Academic Press, London
- Sohlenkamp C, Wood CC, Roeb GW, Udvardi MK (2002) Characterization of Arabidopsis AtAMT2, a high-affinity ammonium transporter of the plasma membrane. Plant Physiol 130 1788–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solaiman MDZ, Saito M (1997) Use of sugars by intraradical hyphae of arbuscular mycorrhizal fungi revealed by radiorespirometry. New Phytol 136 533–538 [DOI] [PubMed] [Google Scholar]
- Suenaga A, Moriya K, Sonoda Y, Ikeda A, Von WN, Hayakawa T, Yamaguchi J, Yamaya T (2003) Constitutive expression of a novel-type ammonium transporter OsAMT2 in rice plants. Plant Cell Physiol 44 206–211 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tilman D, Fargione J, Wolff B, D'Antonio C, Dobson A, Howarth R, Schindler D, Schlesinger WH, Simberloff D, Swackhamer D (2001) Forecasting agriculturally driven global environmental change. Science 292 281–284 [DOI] [PubMed] [Google Scholar]
- Trouvelot A, Kough JL, Gianinazzi-Pearson V (1986) Mesure du taux de mycorhization VA d'un système radiculaire. In Proceedings of the 1st ESM. INRA Press, Paris, pp 217–221
- von Wiren N, Merrick M (2004) Regulation and function of ammonium carriers in bacteria, fungi, and plants. In E Boles, R Krämer, eds, Molecular Mechanisms Controlling Transmembrane Transport. Springer, Berlin, pp 95–120
- Weidinger K, Neuhäuser B, Gilch S, Ludewig U, Meyer O, Schmidt I (2007) Functional and physiological evidence for a rhesus-type ammonia transporter in Nitrosomonas europaea. FEMS Microbiol Lett 273 260–267 [DOI] [PubMed] [Google Scholar]
- Zheng L, Kostrewa D, Berneche S, Winkler FK, Li XD (2004) The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc Natl Acad Sci USA 101 17090–17095 [DOI] [PMC free article] [PubMed] [Google Scholar]