Abstract
The recent development of DNA microarray technology allows us to measure simultaneously the expression levels of thousands of genes and to identify truly correlated genes with anticancer drug response (differentially expressed genes) from many candidate genes. Significance Analysis of Microarray (SAM) is often used to estimate the false discovery rate (FDR), which is an index for optimizing the identifiability of differentially expressed genes, while the accuracy of the estimated FDR by SAM is not necessarily confirmed. We propose a new method for estimating the FDR assuming a mixed normal distribution on the test statistic and examine the performance of the proposed method and SAM using simulated data. The simulation results indicate that the accuracy of the estimated FDR by the proposed method and SAM, varied depending on the experimental conditions. We applied both methods to actual data comprised of expression levels of 12,625 genes of 10 responders and 14 non-responders to docetaxel for breast cancer. The proposed method identified 280 differentially expressed genes correlated with docetaxel response using a cut-off value for achieving FDR <0.01 to prevent false-positive genes, although 92 genes were previously thought to be correlated with docetaxel response ones.
Keywords: differentially expressed genes, false discovery rate, microarray, mixed normal distribution, significance analysis of microarray
Introduction
Genetic markers are promising for our ability to predict the anticancer drug response in individual patients. The recent development of DNA microarray technology allows us to measure simultaneously the expression levels of thousands of genes and to identify truly correlated genes with the anticancer drug response, called differentially expressed genes, from many candidate genes by comparing the gene expression levels between cells or tissues under different conditions. However, since a typical microarray experiment measures the expression levels of thousands of genes with a small sample-size simultaneously, identifying differentially expressed genes poses complex multiple testing problems, and it is difficult to precisely identify differentially expressed genes using traditional statistical methods. The traditional methods such as the two-sample t-test have been used to identify differentially expressed genes [15]. However, such tests often provide unreliable and inaccurate results due to strong parametric assumptions and multiple testing problems. In contrast, Bonferroni correction [5] controlling the family-wise error rate (FWER) is often too conservative, failing to identify differentially expressed genes. In order to solve this problem, the false discovery rate (FDR) is increasingly used. The FDR is defined as the expected proportion of false-positive genes among total identified genes as an index for optimizing the identifiability of differentially expressed genes [4]. Many statistical methods have been proposed for estimating the FDR, i.e. empirical Bayes (EB) method [12], Significance Analysis of Microarray (SAM) [34], and mixture model method (MMM) [24]. Among them, SAM is most widely used for cancer outcome by its attractive advantages in microarray data analysis [10]. Actually, the difficulty of multiplicity problems in simultaneous testing of a large number of genes with a small sample-size data is relieved by SAM through estimating the number of false-positive genes based on a permutation procedure without strict parametric assumptions and replacing the usual t-test statistic with a SAM-statistic [34] or t-type score [24]. Available computer software specific for SAM, also help biological researchers for managing SAM [9]. The precision of estimated FDR in SAM have been examined by many researchers [14, 22, 23, 36]. Among them, Xie et al. (2005) pointed out that the permutation-based methods for FDR estimation such as SAM might overestimate FDR in a certain condition. This suggests the importance of the examination of factors such as target FDR, sample-size, and proportion of differentially expressed genes which may affect the bias and variance of estimated FDR in SAM. If the bias and variance of estimated FDR differ, depending on the experimental condition, we have to choose a suitable method for the experimental condition in a confronted case. We therefore, conducted a simulation study to examine the bias and variance of estimated FDR in SAM.
In this paper, we also propose a new method for estimating the FDR. The proposed method assumes a mixed normal distribution on t-type score, estimating the FDR for a cut-off value based on the numerical integration of probability distribution. Here, the t-type score is a test statistic with a correction term added to the denominator of the Welch type t-statistic in order to stabilize the variation of the denominator [24]. We compared both bias and variance of the estimated FDR between the proposed method and SAM through the simulation study. Additionally, both methods are applied to actual data comprised of the expression levels of 12,625 genes of 10 responders and 14 non-responders to docetaxel for breast cancer (Accession No: GDS360) [20]. Although 92 correlated genes with the docetaxel response were previously identified using a two-sample t-test with the significance level 0.001 [7], there are many false-negative genes among unidentified genes because the adopted significance level is too low to get reasonable result. We, therefore, examined the FDR in this actual data using the proposed method.
Materials and Methods
t-type score
For each gene i, i = 1, 2, …, g, the expression level is Xi1, …, Xim from m samples collected from cells or tissues under Condition 1, and Yi1, …, Yin from n samples collected from cells or tissues under Condition 2. A traditional method for testing for a difference in the means between two conditions assuming a normal distribution is the two-sample t-test. However, since thousands of genes are evaluated simultaneously; when some of them have a very small sum of squares under two conditions, their absolute t-statistic becomes very large even though their mean difference is not large. This disadvantage is exacerbated due to the small sample-size. In the case where two-sample t-test is used, therefore, many non-differentially expressed genes are identified as differentially expressed genes. In order to avoid this problem, a new statistic with a correction term added to the denominator of the Welch type t-statistic in order to stabilize the variation of its denominator, called t-type score, has been proposed [24]. We use the t-type score as a test statistic for identifying the differentially expressed genes. Let zi denote the t-type score for gene i,
| (1) |
where and are the sample means for gene i under two conditions respectively, and and are the sample variances for gene i, a0 is the 90th percentile of .
Significance analysis of microarray (SAM)
SAM is often used to estimate the FDR for identifying the differentially expressed genes for cancer outcome [10]. The FDR is estimated through the replications of permutation among all samples for a total of B times. For the bth permutated data, the t-type score is calculated and denoted by i = 1, …, g. When FDRsam denotes the two-sided FDR estimator, FDRsam can be written as
| (2) |
where c1 (>0) and c2 (<0) are the cut-off values, respectively. We can identify over- and under-expressed genes simultaneously using the FDRsam. On the other hand, we formulate the one-sided FDR estimator for each cut-off value (c1, c2) in order to correspond to the FDR estimator of the proposed method. When FDRsam(c1) and FDRsam(c2) denote the one-sided FDR estimator for c1 and c2 respectively, FDRsam(c1) and FDRsam(c2) can be written as
| (3) |
and
| (4) |
respectively.
The FDRsam(c1) is used in order to identify the differentially expressed genes that the gene expression levels under Condition 1 over-express more than under Condition 2. On the other hand, the FDRsam(c2) is used in order to identify the differentially expressed genes that the gene expression levels under Condition 1 under-express more so than under Condition 2.
Proposed FDR estimation method
We propose estimating the FDR assuming a K-component mixed normal distribution on t-type score zi, i = 1, ..., g. The probability density function of K-component mixed normal distribution is
| (5) |
where fk (z; Δk, Vk) denotes the density function of a normal distribution Normal (Δk, Vk) with mean Δk, and variance Vk, and mixed proportion pk. θ represents all unknown parameters {pk, Δk, Vk : k = 1, ..., K} in a K-component mixed normal model. To estimate the all unknown parameters, given z1, …, zg, the following log-likelihood function is maximized.
| (6) |
To obtain the maximum likelihood estimat θ̂, the Newton-Raphson method is used. The one-sided FDR for each cut-off value (c1, c2) is estimated using the parameter estimates θ̂. When PTP1 and PTP2 denote the proportion of total identified positive genes for each cut-off value (c1, c2) respectively, PTP1 and PTP2 can be written as
| (7) |
and
| (8) |
respectively.
Let PFP1 and PFP2 denote the proportion of false-positive genes for each cut-off value (c1, c2) respectively, PFP1 and PFP2 can be written as
| (9) |
| (10) |
Note that f0(z; Δ0, V0) denotes the normal distribution with the smallest absolute mean among f1(z; Δ1, V1), …, fK(z; ΔK, VK), Δ0 = min (|Δ1|, …, |ΔK|). When FDRp(c1) and FDRp(c2) denote the one-sided FDR estimator for each cut-off value (c1, c2) respectively, FDRp(c1) and FDRp(c2) can be written as
| (11) |
and
| (12) |
respectively.
We can determine the cut-off value for the target one-sided FDR by changing c1 and c2 sequentially using Formula (11) and Formula (12).
Simulation study to examine the performance of the proposed method and SAM
In usual microarray experiments, we evaluate the gene expression levels of thousands of genes simultaneously under various experimental conditions. Specifically, target FDR for determining the cut-off value, the sample-size, and the proportion of differentially expressed genes are varied depending on the experimental conditions. We therefore, examined the bias and variance of estimated FDR in both the proposed method and SAM under various experimental conditions through a simulation study. Although we conducted simulation experiments using a three-component model with over-expressed genes and under-expressed genes as well as a two-component model, this paper discusses the result obtained using the two-component model because the results of them were similar.
As the framework of simulation, we set the following simulation conditions.
Simulation condition 1
The simulation study was designed to have g (i = 1, …, g) genes in total, with s differentially expressed and g-s non-differentially expressed. Each condition had an equal sample-size N (N = m = n). We generated, for j = 1, …, N,
and
respectively.
Since each population mean of differentially expressed genes was different respectively, we assumed a random effect model, that is, μi ~ Normal (1.0, 0.12), i = 1, …, s.
Simulation condition 2
The total number of replication of permutation (B) was 400 times in SAM.
Simulation condition 3
The proposed method assumes a two-component mixed normal distribution on the t-type score, estimating the parameters (θ̂) by the Newton-Raphson method.
The procedure for conducting the simulation study was as follows:
Step 1. Generate Xij and Yij (i = 1, …, g, j = 1, …, N) according to Simulation Condition 1, calculating the t-type score (zi) of g genes including the s differentially expressed genes and g-s non-differentially expressed genes.
Step 2. Determine a cut-off value (c1) for target FDR (tFDR) by changing the cut-off value sequentially.
Step 3. In SAM, calculate the t-type score ( , i = 1, …, g, b = 1, …, 400) using 400 permutated data according to Simulation Condition 2. In the proposed method, estimate the parameters (θ) of two-component mixed normal distribution according to Simulation Condition 3.
Step 4. Estimate the FDR using Formula (3) in SAM and Formula (11) in the proposed method for a cut-off value (c1).
Step 5. Repeat Steps 1 – 4 1,000 times, calculating the average of the bias of the estimated FDR and the variance of the estimated FDR in both methods.
The three situations of the simulation study were as follows:
Simulation situation 1
Each value is set as g = 3,000, s = 150, and N = 20, calculating the bias and variance of the estimated FDR in both methods when target FDR is set as tFDR = 0.01, 0.05, 0.1, 0.2, and 0.5 respectively.
Simulation situation 2
Each value is set as tFDR = 0.1, g = 3,000, and s = 150, calculating the bias and variance of the estimated FDR in both methods when sample-size is set as N = 5, 10, 20, 40, and 80 respectively.
Simulation situation 3
Each value was set as tFDR = 0.1, g = 3,000, and N = 20, calculating the bias and variance of the estimated FDR in both methods when the number of differentially expressed genes of the total genes is set as s = 30, 75, 150, 300, and 600 respectively.
Results
Results of simulation study
The bias and variance of the estimated FDR by both methods under each simulation situation are shown in Table 1, Table 2, and Table 3 respectively. Table 1 suggests that the bias and variance increase as target FDR becomes high in SAM, whereas the bias and variance were almost constant regardless of the target FDR in the proposed method. Table 2 suggests that the bias increases as the sample-size becomes large in SAM, whereas the bias decreased in the proposed method. In both methods, the variance was almost constant regardless of the sample-size. Table 3 suggests that the absolute bias increases as the number of the differentially expressed genes becomes large in SAM, whereas the bias decreases in the proposed method. In both methods, the variance decreases as the number of differentially expressed genes becomes large. Additionally, when tFDR = 0.5 or s = 600 in SAM and N = 5 or 10 in the proposed method, the absolute bias is larger than 0.01. The variance is smaller than that of SAM under all situations in the proposed method, except for N = 5.
Table 1.
Results of simulation situation 1.
| Target FDR (tFDR) |
Proposed method |
SAM |
||
|---|---|---|---|---|
| Bias | Variance | Bias | Variance | |
| 0.01 | −0.0012 | 0.0057 | 0.0005 | 0.0071 |
| 0.05 | −0.0044 | 0.0163 | 0.0019 | 0.0184 |
| 0.10 | −0.0045 | 0.0214 | 0.0027 | 0.0247 |
| 0.20 | −0.0055 | 0.0239 | 0.0035 | 0.0321 |
| 0.50 | −0.0035 | 0.0154 | 0.0142 | 0.0397 |
Table 2.
Results of simulation situation 2.
| Sample-size (N) |
Proposed method |
SAM |
||
|---|---|---|---|---|
| Bias | Variance | Bias | Variance | |
| 5 | −0.0308 | 0.0361 | 0.0005 | 0.0340 |
| 10 | −0.0122 | 0.0257 | 0.0013 | 0.0259 |
| 20 | −0.0045 | 0.0214 | 0.0027 | 0.0247 |
| 40 | −0.0034 | 0.0198 | 0.0042 | 0.0260 |
| 80 | −0.0032 | 0.0205 | 0.0085 | 0.0258 |
Table 3.
Results of simulation situation 3
| Number of differentially expressed genes (s) |
Proposed method |
SAM |
||
|---|---|---|---|---|
| Bias | Variance | Bias | Variance | |
| 30 | −0.0094 | 0.0456 | −0.0004 | 0.0549 |
| 75 | −0.0072 | 0.0290 | 0.0025 | 0.0346 |
| 150 | −0.0045 | 0.0214 | 0.0027 | 0.0247 |
| 300 | −0.0032 | 0.0138 | −0.0025 | 0.0176 |
| 600 | −0.0022 | 0.0087 | −0.0102 | 0.0129 |
Application to actual data
We applied the proposed method and SAM to actual data comprised of the expression levels of 12,625 genes of 10 responders and 14 non-responders to docetaxel for breast cancer (Accession No: GDS360) [20]. This actual data was measured and analyzed in order to identify the correlated genes with the docetaxel response for predicting anti-tumor activity of individual patients [7]. Although 92 correlated genes with the docetaxel response were previously identified using a two-sample t-test (significance level 0.001), it was expected that there would be many false-negative genes among the genes that were not identified because a very strict significance level was used. We identified the correlated genes with docetaxel response based on the FDR using the proposed method and SAM.
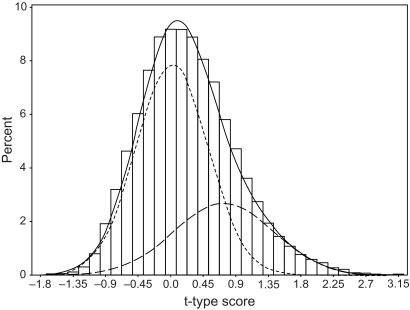
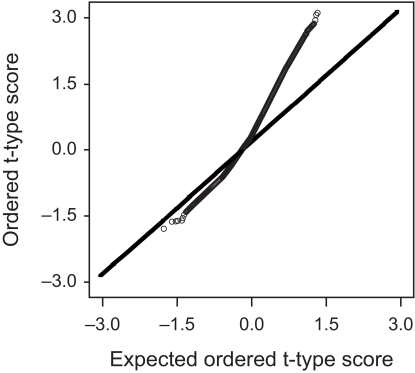
In the proposed method, we assumed five mixed normal distributions on the t-type score with K = 2, …, 5, comparing their fitness by using Akaike Information Criterion (AIC) [1]. AIC is the most well-known criterion for determining the number of components in the model. As a result, we selected a two-component mixed normal distribution from the viewpoint of simplicity of interpretation, although AIC of the two-component model is almost equal to that of a three-component model. The density function of the two-component mixed normal distribution is f(z) = 0.319 f1(z; 0.659, 0.476) + 0.681 f0(z; − 0.057, 0.251). Figure 1 shows a histogram of the t-type score of 12,625 genes and the density function of a two-component mixed normal distribution. As shown in Figure 1, the two-component mixed normal distribution fits the t-type score well. We also calculated the order statistics of zis from raw data, and the expected order statistics of s from 1,000 permutated data. Figure 2 shows the scatter plot of the ordered t-type score versus the expected ordered t-type score in SAM. As shown in Figure 2, it is indicated that there are many differentially expressed genes.
Figure 1.
Histogram of the t-type score and the density function of a two-component mixed normal distribution. The solid line is f, the dotted line is f0, and the broken line is f1 in a two-component mixed normal distribution.
Figure 2.
Scatter plot of the ordered t-type score versus the expected ordered t-type score in SAM.
Discussion
While numerous research has been undertaken related to the bias of the estimated FDR by SAM [14, 22, 23, 36], little is known about the variance of the estimated FDR by SAM. Jung and Jang (2006) [14] noted that SAM can accurately estimate FDR when the target FDR is smaller than 0.1, which is an appropriate value in usual microarray data analysis. Pan (2002 Pan (2003) [22, 23] and Xie et al. (2005) [36] indicated the permutation-based methods for FDR estimation caused overestimation of FDR. In this paper, we examined both bias and variance of the estimated FDR by SAM under various experimental conditions through the simulation study in order to clarify the features of SAM. As a result of the simulation study, we uncovered some problems related to the SAM method. Estimating the distribution of non-differentially expressed genes using permutated data may not lead to precise estimation of FDR. Such a distribution based on the permutation is more dispersed than the true distribution of non-differentially expressed genes, resulting in overestimation of the number of false-positive genes. In particular, this disadvantage was influenced by the target FDR and the sample-size. In contrast, the proposed method estimates directly the distribution of non-differentially expressed genes assuming the mixed normal distribution on the t-type score. Although the estimated FDR by the proposed method was underestimated, the degree of bias of the estimated FDR in both the proposed method and SAM were almost same and the variance of the estimated FDR by the proposed method was smaller than that of SAM under all simulation situations, except for N = 5. The distribution based on the mixed normal distribution might be not more dispersed than the distribution based on the permutation. From the viewpoint of over-dispersion, therefore, the proposed method might precisely estimate the FDR than SAM.
In the simulation study, FDR tended to be underestimated in the proposed method and overestimated in SAM. Although the underestimation was not so large, this may cause the increase of false-positive genes. For instance, when 100 genes are identified as differentially expressed genes with the target FDR 0.1, truly false-positive genes are only 10 with the unbiased FDR, whereas more than 10 false-positive genes may be included in 100 by the underestimation of the FDR. To the contrary, the overestimation may cause the decrease of true-positive genes.
Our simulation study also made clear the different strength of the proposed method and SAM. When the sample-size was as small as 10, the absolute bias in SAM was smaller than that in the proposed method, while the variance was almost the same between them. This strength of SAM may be attractive because microarray experiments are often conducted with small sample-sizes. When the number of differentially expressed genes was as small as 10% of the total genes, FDR were more accurately estimated in SAM than the proposed method. An additional simulation experiment with no differentially expressed genes, i.e. s = 0, revealed that the bias and variance of estimated FDR in SAM were slightly smaller than that in the proposed method. When the sample-size or the number of differentially expressed genes was large, however, both the bias and variance in the proposed method were smaller than those in SAM, probably because SAM could not accurately estimate the distribution of non-differentially expressed genes. The proposed method has an advantage over SAM when the sample-size is greater than 20 or the number of differentially expressed genes is greater than 10% of the total genes. Thus, the proposed method outperforms SAM when the sample-size of each group is more than 20 or the proportion of differentially expressed genes is more than 10% irrespective of the target FDR. Otherwise, SAM outperforms the proposed method.
There would be many over-expressed genes in responder group relative to non-responder group based on both Figures 1–2 in the actual data, whereas under-expressed genes would be few. Table 4 shows the estimated FDR and the number of identified genes in both methods when the cutoff value is changed from 0.1 to 2.0 by 0.1. The number of identified genes was equal between the two methods, because the same t-type score and cut-off value was used. According to the result of simulation study, FDR by the proposed method may be slightly underestimated since the sample-size of the responder group and non-responder group were 10 and 14, respectively, in the actual data. However, the degree of underestimation would not be so large that its influence might be cancelled by taking a slightly smaller value of estimated FDR than the target FDR. For instance, the estimated FDR by the proposed method is 0.007, which corresponded to the cut-off value 1.7 in Table 4, may be appropriate for the target FDR 0.01. If so, 280 genes were identified as the differentially expressed genes correlated with the docetaxel response.
Table 4.
Results of application of the proposed method and SAM. The estimated FDR in both methods, and the number of identified genes for each cut-off value.
| Cut-off value |
Estimated FDR |
Number of identified genes | |
|---|---|---|---|
| Proposed method | SAM | ||
| 0.1 | 0.504 | 0.748 | 6,433 |
| 0.2 | 0.464 | 0.612 | 5,685 |
| 0.3 | 0.420 | 0.487 | 4,935 |
| 0.4 | 0.373 | 0.381 | 4,227 |
| 0.5 | 0.324 | 0.291 | 3,612 |
| 0.6 | 0.274 | 0.225 | 3,008 |
| 0.7 | 0.226 | 0.172 | 2,506 |
| 0.8 | 0.181 | 0.132 | 2,063 |
| 0.9 | 0.141 | 0.101 | 1,680 |
| 1.0 | 0.106 | 0.076 | 1,371 |
| 1.1 | 0.078 | 0.059 | 1,087 |
| 1.2 | 0.056 | 0.044 | 877 |
| 1.3 | 0.039 | 0.033 | 691 |
| 1.4 | 0.026 | 0.024 | 571 |
| 1.5 | 0.017 | 0.018 | 451 |
| 1.6 | 0.011 | 0.014 | 357 |
| 1.7 | 0.007 | 0.011 | 280 |
| 1.8 | 0.004 | 0.008 | 218 |
| 1.9 | 0.003 | 0.006 | 171 |
| 2.0 | 0.002 | 0.006 | 119 |
Acknowledgments
This study was supported by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NiBio) of Japan. Akihiro Hirakawa, Chikuma Hamada, Isao Yoshimura contributed equally to this work.
Footnotes
Copyright in this article, its metadata, and any supplementary data is held by its author or authors. It is published under the Creative Commons Attribution By licence. For further information go to: http://creativecommons.org/licenses/by/3.0/.
References
- [1].Akaike H. Information theory and an extension of the maximum likelihood principle. In: Petrov BN, Csaki F, editors. 2nd international symposium on information theory. Akademiai Kiado; Budapest: 1973. pp. 267–81. [Google Scholar]
- [2].Allison DB, Gadbury GL, Heo M, et al. A mixture approach for the analysis of microarray gene expression data. Computational Statistics and Data Analysis. 2002;39:1–20. [Google Scholar]
- [3].Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene change. Bioinformatics. 2001;17:509–19. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- [4].Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- [5].Bonferroni CE. l1 calcolo delle assicurazioni su gruppi di teste. Studi in Onore del Professore Salvatore Ortu Crboni Rome Italy. 1935:13–60. [Google Scholar]
- [6].Broberg P. Statistical methods for ranking differentially expressed genes. Genome Biology. 2003;4(6):R41. doi: 10.1186/gb-2003-4-6-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chang JC, Wooten EC, Tsimelzon A, et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet. 2003;362:362–9. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- [8].Chen Y, Dougherty ER, Bittner ML. Ratio-based decisions and the quantitative analysis of cDNA microarray images. Journal of Biomedical Optics. 1997;2:364–7. doi: 10.1117/12.281504. [DOI] [PubMed] [Google Scholar]
- [9].Chu G, Narasimhan B, Tibshirani R, et al. 2005. SAM “Significance Analysis of Microarray” Users Guide and Technical Document Accessed 1 September 2007. URL: http://www-stat.stanford.edu/~tibs/SAM/
- [10].Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. Journal of the National Cancer Institute. 2007;99:147–57. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- [11].Efron B, Tibshirani JR. Introduction to the bootstrap. Chapman and Hall; 1993. [Google Scholar]
- [12].Efron B, Tibshirani R, Storey JD, et al. Empirical bayes analysis of microarray experiment. Journal of the American Statistical Association. 2001;456:1151–60. [Google Scholar]
- [13].Jung S. Sample-size for FDR-control in microarray data analysis. Bioinformatics. 2005;21(14):3097–3014. doi: 10.1093/bioinformatics/bti456. [DOI] [PubMed] [Google Scholar]
- [14].Jung S, Jang W. How accurately can we control the FDR in analyzing microarray data? Bioinformatics. 2006;22(14):1730–6. doi: 10.1093/bioinformatics/btl161. [DOI] [PubMed] [Google Scholar]
- [15].Kohane I, Kho A, Butte A. Microarrays for an integrative genomics. The MIT Press; 2002. [Google Scholar]
- [16].Kooperberg C, Aragaki A, Strand AD, et al. Significance testing for small microarray experiments. Statistics in Medicine. 2005;24:2281–98. doi: 10.1002/sim.2109. [DOI] [PubMed] [Google Scholar]
- [17].Lee M, Kuo F, Whitmore G, et al. Importance of replication in microarray gene expression studies: Statistical methods and evidence from repetitive cDNA hybridizations. Proceedings of the National Academy of Sciences of the U.S.A. 2000;97(18):9834–39. doi: 10.1073/pnas.97.18.9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].McLachlan G, Peel D. Finite mixture models. Wiley New York: 2000. [Google Scholar]
- [19].McLachlan G, Do K, Ambroise C. Analyzing microarray gene expression data. Wiley New York: 2004. [Google Scholar]
- [20].National Center for Biotechnology Information Gene Expression Omnibus. Breast cancer and docetaxel treatment. Accession No: GDS360. Accessed 9 April 2007. URL: http://www.ncbi.nlm.nih.gov/geo/
- [21].Newton MA, Kendziorski CM, Richmond CS, et al. On differential variability of expression ratios; improving statistical inference about gene expression changes from microarray data. Journal of Computational Biology. 2001;8:37–52. doi: 10.1089/106652701300099074. [DOI] [PubMed] [Google Scholar]
- [22].Pan W. A comparative review of statistical methods for discovering differentially expressed genes in replicated microarray experiments. Bioinformatics. 2002;18(4):546–56. doi: 10.1093/bioinformatics/18.4.546. [DOI] [PubMed] [Google Scholar]
- [23].Pan W. On the use of permutation in and the performance of a class of nonparametric methods to detect differential gene expression. Bioinformatics. 2003;19(11):1333–40. doi: 10.1093/bioinformatics/btg167. [DOI] [PubMed] [Google Scholar]
- [24].Pan W, Lin J, Le C. A mixture model approach to detecting differentially expressed genes with microarray data. Functional and Integrative Genomics. 2003;3:117–24. doi: 10.1007/s10142-003-0085-7. [DOI] [PubMed] [Google Scholar]
- [25].Pawitan Y, Michiels S, Koscielny S, et al. False discovery rate, sensitivity and sample-size for microarray studies. Bioinformatics. 2005;21(13):3017–3024. doi: 10.1093/bioinformatics/bti448. [DOI] [PubMed] [Google Scholar]
- [26].Pawitan Y, Murthy K, Michiels S, et al. Bias in the estimation of false discovery rate in microarray studies. Bioinformatics. 2005;21(20):3865–872. doi: 10.1093/bioinformatics/bti626. [DOI] [PubMed] [Google Scholar]
- [27].Ploner A, Calza S, Gusnanto A, et al. Multidimensional local false discovery rate for microarray data. Bioinformatics. 2006;22(5):556–65. doi: 10.1093/bioinformatics/btk013. [DOI] [PubMed] [Google Scholar]
- [28].Pounds S, Morris WS. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-value. Bioinformatics. 2003;19(10):1236–42. doi: 10.1093/bioinformatics/btg148. [DOI] [PubMed] [Google Scholar]
- [29].Pounds S, Cheng C. Robust estimation of the false discovery rate. Bioinformatics. 2006;22(16):1979–87. doi: 10.1093/bioinformatics/btl328. [DOI] [PubMed] [Google Scholar]
- [30].Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate procedure. Bioinformatics. 2003;19(3):368–75. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- [31].Schena M, Sharon D, Heller R, et al. Parallel genome human analysis: Microarray based-expression monitoring of 1000 genes. Proceedings of the National Academy of Sciences of the U.S.A. 1997;93(20):10614–9. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Storey JD, Tibshirani R. Statistical significance for genome-wide experiments. Proceedings of the National Academy of Sciences of the U.S.A. 2003;100(16):9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Thomas J, Olson J, Tapscotto S, et al. An efficient and robust statistical modeling approach to discover differentially expressed genes using genomic expression profiles. Genome Research. 2001;11:1227–36. doi: 10.1101/gr.165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tusher V, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the U.S.A. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wu B, Guan Z, Zhao H. Parametric and nonparametric FDR estimation revisited. Biometrics. 2006;62:735–44. doi: 10.1111/j.1541-0420.2006.00531.x. [DOI] [PubMed] [Google Scholar]
- [36].Xie Y, Pan W, Khodursky AB. A note on using permutation-based false discovery rate estimates to compare different analysis methods for microarray data. Bioinformatics. 2005;21:4280–8. doi: 10.1093/bioinformatics/bti685. [DOI] [PubMed] [Google Scholar]
- [37].Zhao Y, Pan W. Modified nonparametric approaches to detecting differentially expressed genes in replicated microarray data. Bioinformatics. 2003;19(9):1046–54. doi: 10.1093/bioinformatics/btf879. [DOI] [PubMed] [Google Scholar]