Abstract
Genome-wide scans for recent positive selection in humans have yielded insight into the mechanisms underlying the extensive phenotypic diversity in our species, but have focused on a limited number of populations. Here, we present an analysis of recent selection in a global sample of 53 populations, using genotype data from the Human Genome Diversity-CEPH Panel. We refine the geographic distributions of known selective sweeps, and find extensive overlap between these distributions for populations in the same continental region but limited overlap between populations outside these groupings. We present several examples of previously unrecognized candidate targets of selection, including signals at a number of genes in the NRG–ERBB4 developmental pathway in non-African populations. Analysis of recently identified genes involved in complex diseases suggests that there has been selection on loci involved in susceptibility to type II diabetes. Finally, we search for local adaptation between geographically close populations, and highlight several examples.
The ability to identify the molecular signature of natural selection provides a powerful tool for identifying loci that have contributed to adaptation. Recently, a number of analytical techniques have been developed to identify signals of recent positive selection on a genome-wide scale and applied to polymorphism data from several human populations (Kelley et al. 2006; Voight et al. 2006; Wang et al. 2006; Sabeti et al. 2007; Williamson et al. 2007; Barreiro et al. 2008). Several loci important for human adaptation have been identified or confirmed in these scans: notable examples include a number of genes involved in skin pigmentation (Voight et al. 2006; Sabeti et al. 2007; Williamson et al. 2007); EDAR, involved in hair morphology (Kelley et al. 2006; Fujimoto et al. 2008; Mou et al. 2008); and LCT, at which variants under selection contribute to lactase persistence (Bersaglieri et al. 2004).
The populations studied to date, however, represent a limited sample of human diversity. Most of these studies have relied on either the HapMap (Frazer et al. 2007) or Perlegen (Hinds et al. 2005) datasets, each of which includes samples from only a few populations: one European, one African (or African-American), and one or two East Asian populations. Since selective pressures such as diet, climate, and pathogen load vary greatly across the globe, even on relatively small scales, understanding the genetic response to this environmental variation requires higher geographic resolution in the sampling of human diversity (Prugnolle et al. 2005; Perry et al. 2007; Hancock et al. 2008). In this paper, we present results from a series of genome-wide scans for natural selection using single nuclotide polymorphism (SNP) genotype data from the Human Genome Diversity-CEPH Panel (HGPD), a data set containing 938 individuals from 53 populations typed on the Illumina 650Y platform (Li et al. 2008).
Our goals here were twofold. First, we sought to employ data from the 53 populations of the HGDP to better understand the geographic patterns of selected haplotypes. We find extensive sharing of putative selection signals between genetically similar populations, and limited sharing between genetically distant ones. In particular, Europe, the Middle East, and Central Asia show strikingly similar patterns of putative selection signals.
Second, we sought to identify novel candidate loci that have experienced recent positive selection and relate these signals to phenotypic variation. We identify several novel strong candidates for selection, including C21orf34, a gene of unknown function, and several genes in the NRG–ERBB4 developmental pathway. Interpretation of previous scans for selection has been limited by the relative paucity of information about the genetics of natural variation in humans. Recent genome-wide association studies, however, are beginning to fill this void, and many loci have been identified at which variation influences phenotypes (McCarthy et al. 2008). We have used this information as a guide in the interpretation of our scans for selection. In general, we find limited overlap between the results of genome-wide association studies and our scan for selection, with some notable exceptions, particularly in pigmentation and type II diabetes.
Results
After quality control and removal of related individuals, the HGDP data consist of 657,143 SNPs typed on 938 individuals in 53 populations. For some analyses, each population was treated individually, but for others we found it more powerful to group populations together and increase sample sizes. For these latter analyses, we divided the individuals into eight groups, most of which represent broad geographic regions: Bantu-speaking populations, Biaka Pygmies, Europeans, Middle Easterners, South Asians, East Asians, Oceanians, and Native Americans. These groups were chosen to provide reasonably homogenous sets of populations for analysis, as judged by clustering at randomly chosen loci (Rosenberg et al. 2002; Li et al. 2008). The Mbuti Pygmies and San were dropped from these groups because their large divergence from other African populations means that we might lose power by grouping them with the other Africans, and their small sample sizes indicate that we would have low power in treating them on their own.
Our analyses focus primarily on two haplotype-based tests: iHS (Voight et al. 2006) and XP-EHH (Sabeti et al. 2007). These tests were chosen because previous power analyses suggest they are largely complementary—iHS has good power to detect selective sweeps at moderate frequency (~50%–80%), but low power to detect sweeps that have reached high frequency (>80%) or fixation; in contrast, XP-EHH is most powerful for selective sweeps above 80% frequency (Voight et al. 2006; Sabeti et al. 2007). Some analyses presented here also use FST, a measure of population differentiation which has power to detect selection on standing variation as well as on new selected sites (Innan and Kim 2008), or the CLR test of the allele frequency spectrum (Nielsen et al. 2005; Williamson et al. 2007), an alternative to XP-EHH for detecting high-frequency selective sweeps. Throughout this paper, the “P-values” presented will be empirical P-values; that is, a low P-value indicates that a locus is an outlier with respect to the rest of the genome (Teshima et al. 2006). We find this approach useful because P-values based on an explicit demographic model are unreliable when there is uncertainty in the demographic parameters (as is the case for humans). However, we note that loci detected as being under selection using this approach may be an unrepresentative sample of all truly selected loci; in particular, selection on standing variation and recessive loci are likely to be underrepresented (Teshima et al. 2006).
Assessment of power
The HGDP data present a number of challenges for the detection of selection. First, the data consist largely of tag SNPs selected to maximize coverage of the HapMap populations (Eberle et al. 2007). The allele frequencies and linkage disequilibrium patterns at these SNPs differ from the genome as a whole. Second, the populations of the HGDP have different demographic histories and sample sizes, which may affect power to detect selection.
The selection of tag SNPs from the HapMap is expected to reduce coverage in regions of the genome that show strong evidence of selective sweeps (and thus contain extensive LD) in the HapMap populations. We find this is indeed the case: of the genomic regions with the strongest iHS signals in the HapMap data, about 25% of 200-kb regions contain <20 SNPs on the Illumina chip. This is significantly less than the genome-wide average of about 40 SNPs per 200 kb overall on the Illumina chip (P = 8 × 10−4, P = 9 × 10−3, and P = 2 × 10−6 for regions identified as under selection in the HapMap European, Asian, and Bantu samples, respectively; one-sided t-test) and far fewer than the average 180 SNPs/200 kb in the HapMap. This indicates that power may be reduced in the HGDP for confirming selective sweeps already identified in the HapMap, although it should not affect power to detect novel selection signals in other populations.
To further explore the power to detect selection in this panel, we performed simulations under a simple, three-population model of human demography based on the HapMap (Schaffner et al. 2005) and we approximated the Illumina SNP ascertainment scheme (see Methods; Supplemental Fig. 1). These simulations were designed to guide intuition about the impact of a few chosen parameters on power, rather than to represent a formal null model. One important feature of the demographic model used here is the presence of two population bottlenecks in the non-African populations, with the second bottleneck being stronger in the East Asian population. This demographic model provides a good fit to several aspects of the data for the HapMap populations (data not shown). We use this model here because it is likely to be a good approximation to the demographies of many of the HGDP populations, and because fitting a demographic model to the 53 populations of the HGDP presents significant challenges and no such model is currently available.
As previously reported (Voight et al. 2006; Sabeti et al. 2007), we find that the fraction of extreme iHS scores in a genomic region is a more powerful statistic than the maximum score, while the reverse is true of XP-EHH (data not shown). As noted above, iHS has moderate power to detect a selective sweep that has reached intermediate frequency and little power to detect a sweep near fixation, while XP-EHH is more powerful to detect selective sweeps at or near fixation. Neither test has appreciable power to detect a selective sweep that has not yet reached a frequency >30%. We saw an important effect of demography in these simulations. The power to detect selection is highest in the “African” demography, intermediate in the “European” demography, and lowest in the “East Asian” demography (Supplemental Fig. 2). Although not explicitly included in the simulations, this suggests that power is low for both these tests in Oceania and America, which have experienced more recent and severe bottlenecks (Conrad et al. 2006). This is consistent with the observation that nonequilibrium demographies can inflate haplotype-based test statistics (Macpherson et al. 2008).
We also investigated the impact of sample size on power. For iHS, the loss of power incurred by decreasing sample size is modest until a threshold of ~40 chromosomes, while XP-EHH maintains power with as few as 20 chromosomes, as long as the reference population is of a fixed sample size (Supplemental Fig. 3). Since many HGDP populations contain around 10 individuals, power may be gained for iHS by grouping together genetically similar populations.
Overview of genomic regions with selection signals
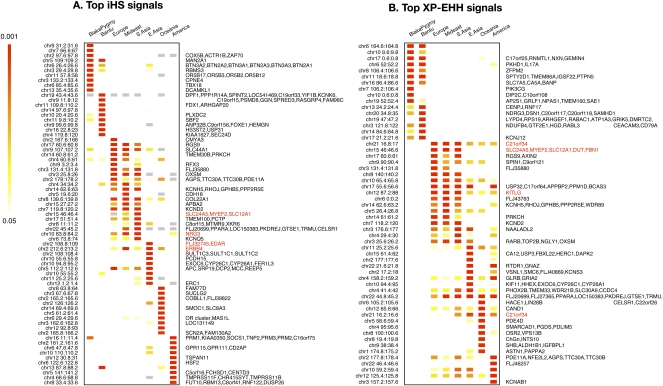
To identify genomic regions that may have been targets of recent selection, we calculated XP-EHH and iHS on each broad population grouping mentioned above and on each individual population. To facilitate comparisons of genomic regions across populations, we then split the genome into nonoverlapping segments of 200 kb and computed, in each segment, the maximum XP-EHH score and the fraction of extreme (|iHS| > 2) iHS scores. The choice of 200 kb as a window size was motivated by the desire to have a sufficient number of SNPs in a window while maintaining a size on the scale of the signal generated by selective sweeps (~0.3–0.5 cM) (Voight et al. 2006). Other window sizes, and the use of a sliding window, gave qualitatively similar results (Supplemental Figs. 6, 7). In each window, we converted the test statistic to an empirical P-value, taking into account the number of SNPs in the window (see Methods). In Figure 1, we show the 10 most extreme windows of the genome from each geographic region for these statistics. The complete lists of regions with empirical P < 0.01 for each geographic region are in Supplemental Figures 13–28.
Figure 1.
Top 10 iHS (A) and XP-EHH (B) signals by population cluster. Each row is a 200-kb genomic window, each column is a geographic region, and each cell is colored according to the position of the window in the empirical distribution of scores for that region. Plotted are the most extreme 10 windows for each geographic region. Gray cells in A are windows that have fewer than 20 SNPs for which iHS was calculated (see Methods). To the right of each row is a list of genes that fall in the window. Windows where the genes are in red are discussed in the text. Note that interpretation of the overlap in XP-EHH signals is complicated by the need for a reference population; see the main text.
A number of interesting patterns emerge from Figure 1. First, there is extensive sharing of extreme iHS and XP-EHH signals between Europe, the Middle East, and Central Asia, while overlap between other regions is much more limited. In fact, 44% of the genomic segments in the 1% tail of iHS in Europe fall in the 5% tail for both the Middle East and Central Asia (89% are shared between Europe and at least one of these two), while only 12% of European signals are present in East Asia by the same criterion. Second, XP-EHH signals seem to be shared on a larger geographic scale than iHS signals. However, the fact that XP-EHH needs a reference population makes overlap hard to interpret; in particular, the lack of overlap between African and non-African groups could be a consequence of use of a reference. To address this, we compared the overlaps in selection signals as judged by the CLR test, which, like XP-EHH, has power to detect high frequency and fixed selective sweeps, but does not rely on a reference population (Williamson et al. 2007). XP-EHH and the CLR test tend to identify the same regions as putative targets of selection and, as with iHS and XP-EHH, signals from the CLR test tend to be shared between Europe, the Middle East, and Central Asia, while sharing between African and non-African populations is very limited (Supplemental Fig. 8). Another concern is that these patterns of overlap may be influenced by the way populations were grouped together for analysis. However, this is not the case; the patterns of overlap hold as well when analysis is performed in each population individually, and both iHS and XP-EHH signals are generally shared between populations in a geographic region (Supplemental Figs. 4, 5).
We also asked how our scans in these pooled populations related to those from the HapMap. We calculated both iHS and XP-EHH on the Phase II HapMap samples and performed the same procedure as above. We found considerable overlap: For iHS, 51% of the windows in the 1% tail in Europe fall in the 5% tail in HapMap Europeans, 63% of signals in East Asia overlap those from the HapMap Asian population by the same criteria, and 38% of signals in the Bantu overlap those identified in the HapMap Yoruba. For XP-EHH, the corresponding figures are 69%, 89%, and 41%. While this is extensive overlap, it is far from complete. One important reason for the incomplete overlap is the vastly different SNP density between the HapMap (which contains 3.1 million SNPs) and the HGDP, especially in regions with strong selection signals in the HapMap, as noted above. Other reasons for incomplete overlap include the presence of some population-specific partial sweeps and incomplete power to detect selection.
Genes and phenotypes under selection
The data in the HGDP permit the use of multiple types of population genetic evidence in evaluating the case for selection on a particular locus. These types of evidence include haplotype structure (Hudson et al. 1994; Sabeti et al. 2002), linkage disequilibrium (Kim and Nielsen 2004; Jensen et al. 2007), the allele frequency spectrum (Smith and Haigh 1974; Tajima 1989), and population differentiation (Lewontin and Krakauer 1973). As an example of how these data allow for the identification of novel selection candidates, we present in Figure 2 the case for selection at C21orf34, a locus of unknown function on chromosome 21.
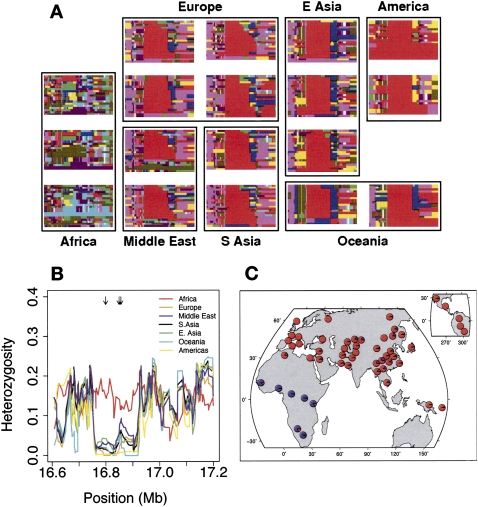
Figure 2.
Evidence for selection in a region containing part of the gene C21orf34. (A) Haplotype plots in a 500-kb region on chromosome 21 surrounding the locus. Each row represents a haplotype, and each column a SNP. Rows are colored the same if and only if the underlying sequence is identical (some low-frequency SNPs are excluded). For full details on the generation of these plots, see Conrad et al. (2006). (B) Heterozygosity in the same region. Lines show heterozygosity calculated in a sliding window of three SNPs across the region in different populations. Black arrows at the top of the plot represent the positions of SNPs with FST > 0.6 (i.e., in the 0.01% tail of worldwide FST). (C). A pie chart of the worldwide distribution of a SNP that tags the red haplotype in A (rs2823850). (Red) The derived allele frequency; (blue) the ancestral allele frequency.
This locus contains some of the most extreme XP-EHH scores in the genome in non-African populations, including the most extreme score in Europe (Fig. 1B). Visualization of the haplotypes in the region (Fig. 2A) revealed a striking lack of diversity in an ~200-kb region in non-African populations; this was confirmed by using a sliding window of heterozygosity (Fig. 2B). If this reduction in diversity and strong haplotype structure had been driven by positive selection in non-African populations, the region should contain SNPs with large population differentiation between African and non-African populations (Slatkin and Wiehe 1998). This is indeed the case: There are a number of SNPs in the region with extreme FST, including the SNP with the highest FST in the HGDP data set, which almost perfectly differentiates African from non-African individuals (Fig. 2C). In sum, these observations suggest that a haplotype in this region swept to near fixation at some point since the out-of-Africa migration. This region of reduced diversity in non-Africans includes the terminal three exons of C21orf34, a gene that is expressed in many tissues (Gardiner et al. 2002), as well as three microRNA genes (mir-99a, let-7c, and mir-125-b2). Currently, there are no known SNPs in any of these potential functional targets.
We next turned our attention from the top signals to specific candidate genes. We took two approaches to identifying selection signals of interest. First, we assembled lists of polymorphisms associated with potentially evolutionarily-relevant traits, and tested whether regions surrounding these polymorphisms show more population differentiation than random regions of the genome (see below, Methods). Second, we considered all genomic windows in the 1% tail of iHS or XP-EHH to be candidates for containing a selective sweep, and all genes within 50 kb of a window as candidates for the target of selection. We then manually examined these regions for genes of interest. In the following sections, we present the results from this analysis.
Pigmentation
Several genes involved in pigmentation have been targets of recent positive selection in non-African populations, including SLC24A5 (Lamason et al. 2005), KITLG (Miller et al. 2007), and SLC45A2 (Norton et al. 2007). Indeed, two of these (KITLG and SLC24A5) appear in the list of the most extreme haplotype patterns in the genome in Figure 1 and Supplemental Figure 8: SLC24A5 has one of the most extreme iHS and XP-EHH signals in Europe, the Middle East and Central Asia (referred to hereafter as West Eurasia) and KITLG has one of the most extreme XP-EHH scores in all non-African populations.
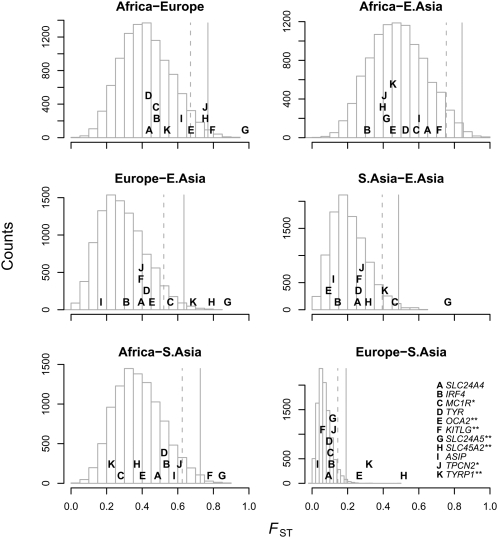
To assess more comprehensively the extent of natural selection in pigmentation genes, we compiled a list of genes currently known to contribute to natural variation in humans from recent GWA studies (Stokowski et al. 2007; Han et al. 2008; Sulem et al. 2007, 2008). Around each pigmentation-associated SNP, we defined a window of 100 kb, and took the maximum FST across SNPs for pairwise comparisons of all continental regions. We used this approach rather than taking FST directly at the association signals because the Illumina chip does not contain most of the SNPs with association signals and, even where it does, the associated SNP may simply be tagging the true causal variant. For comparison, we randomly sampled 10,000 SNPs from the Illumina chip and performed the same procedure. The results for six pairwise comparisons are presented in Figure 3.
Figure 3.
FST around loci involved in natural variation in pigmentation. For each SNP found to be associated with pigmentation in a genome-wide scan, we plot the maximum pairwise FST between geographic regions in a 100-kb window surrounding the SNP in the HGDP data, as well as a histogram of the null distribution calculated by finding the maximum FST in 100-kb windows surrounding each of 10,000 random SNPs. The dotted lines shows the position beyond which 5% of the random SNPs fall, and the solid lines the position beyond which 1% of the random SNPs fall. Gene names that are starred fall in the 5% tail of at least one comparison, and those with two stars fall in the 1% tail of at least one comparison. Letters are positioned along the y-axis to improve readability. The key in the bottom right panel applies to all panels.
Overall, regions of the genome associated with pigmentation tend to have higher FST between Africa and Europe, and between Europe and East Asia, than random regions of the genome (Africa-Europe P = 3 × 10−4, Europe-East Asia P = 1 × 10−4; one-sided Mann-Whitney test). These regions do not show unusually high differentiation between East Asia and Africa (P = 0.51) despite the fact that East Asians have evolved lighter skin since the out-of-Africa expansion. This reflects the fact that most pigmentation genes have been discovered in European or African-American samples and that the evolution of light pigmentation in East Asia seems to have occurred largely via separate genes (Norton et al. 2007). Turning to individual genes, all three of the previously well-supported targets of selection fall well into the 1% tail of at least one comparison. With equally extreme differentiation are OCA2 and TYRP1, previously reported to be candidates for selection based on haplotype structure alone (Voight et al. 2006). These two loci strongly differentiate Europe from Central Asia, in contrast to the overall trend of West Eurasia showing similar patterns of selection (Fig. 1).
Although not identified to date by genome-wide scans for pigmentation variation, we also noticed that other candidate pigmentation loci fall among the top haplotype-based signals of selection. In particular, MLPH shows a strong XP-EHH signal in non-African populations, and RGS19 shows a strong iHS and XP-EHH signal in Bantu populations. MLPH is known to influence pigmentation in mouse (Matesic et al. 2001), dog (Drogemuller et al. 2007), cat (Ishida et al. 2006), and chicken (Vaez et al. 2008), and RGS19 was recently shown to influence pigmentation in mouse (McGowan et al. 2008). These loci have not been identified in genome-wide association studies to date, but we note that there have been no genome-wide admixture mapping studies of pigmentation; this type of study will be necessary to confirm the role of these genes, if any, in between-population variation in pigmentation.
The above results confirm that using the maximum FST in a window around an associated SNP is a relatively sensitive measure for detecting selection, even when the causal SNP may not be present in the data. With this tool in hand, we turn to other phenotypes where the role of selection is unknown.
Disease susceptibility and other quantitative phenotypes
It has been hypothesized that alleles involved in common disease could often be targets of selection (Neel 1962; Di Rienzo and Hudson 2005; Nielsen et al. 2007; Hancock et al. 2008). However, studies of SNPs associated with complex disease have found no evidence that they are significantly more differentiated among populations than random SNPs in the genome (Lohmueller et al. 2006; Myles et al. 2008). Recent genome-wide association studies, along with the genome-wide SNP data from the HGDP, permit a more comprehensive test of this hypothesis. We compiled lists of SNPs associated with several common diseases and quantitative traits for which many associated loci are known (Crohn's disease, type I and II diabetes, height, and lipid levels) from published genome-wide association studies (Scott et al. 2007; Todd et al. 2007; Gudbjartsson et al. 2008; Lettre et al. 2008; Unoki et al. 2008; Weedon et al. 2008; Willer et al. 2008; Yasuda et al. 2008; Zeggini et al. 2008). We applied the above method used for pigmentation to test whether loci associated with any of these other phenotypes are enriched for SNPs with high FST.
Loci involved in lipid levels (Supplemental Fig. 9), susceptibility to Crohn's disease (Supplemental Fig. 10), height (Supplemental Fig. 11), and susceptibility to type I diabetes (Fig. 4) show little evidence of being subject to selection. However, there are a few notable exceptions to this. One such exception is a nonsynonymous SNP (rs3184504) in SH2B3, identified as a risk factor for both type I diabetes and celiac disease (Todd et al. 2007; Hunt et al. 2008). The region appears in the 1% tail of iHS in Europe, and the iHS score on the individual SNP is −2.02 (empirical P = 0.02). This region is also an outlier in the FST comparison between Europe and East Asia (Fig. 4). Interestingly, the risk allele appears on the sweeping haplotype, suggesting that risk for autoimmune disease may have increased as a byproduct of natural selection in some populations.
Figure 4.
FST around loci involved in natural variation in diabetes susceptibility. For each SNP associated with either type I or type II diabetes we plot the maximum pairwise FST between geographic regions in a 100-kb window surrounding the SNP in the HGDP data, as well as a histogram of the null distribution calculated by finding the maximum FST in 100-kb windows surrounding each of 10,000 random SNPs. The dotted lines shows the position beyond which 5% of the random SNPs fall, and the solid lines the position beyond which 1% of the random SNPs fall. Gene names that are starred fall in the 5% tail of at least one comparison, and those with two stars fall in the 1% tail of at least one comparison. Letters are positioned along the y-axis to improve readability. The key in the bottom panel of each column applies to the entire column.
Risk of type II diabetes has been hypothesized to be a target of natural selection in humans due to the effect of the disease on metabolism and energy production (Neel 1962). Indeed, the locus with the strongest impact on disease susceptibility in Europeans, TCF7L2, shows impressive differences in allele frequencies between Africa and East Asia (Fig. 4; Helgason et al. 2007). Overall, we show in Figure 4 that regions of the genome harboring SNPs associated with type II diabetes significantly differentiate Europeans and East Asians from Africans (Europe-Africa P = 0.006, East Asia-Africa P = 0.02, one-sided Mann-Whitney test). There are a number of regions that contain strong outliers in at least one comparison, including TCF7L2, TSPAN8, JAZF1, and ADAMTS9. Other associated regions also show XP-EHH signals well into the 1% tail: these are THADA in East Asia (maximum XP-EHH at rs12474030 of 3.7, empirical P = 1 × 10−4) and an intergenic region on chromosome 11 in Europeans (maximum XP-EHH at rs16936071 of 3.6, empirical P = 2 × 10−4). We note, however, that though these type II diabetes-associated regions are more differentiated than random regions of the genome, the associated SNPs themselves often are not (as in Myles et al. [2008], at a subset of the SNPs considered here). We return briefly to this point in the Discussion.
NRG–ERBB4 pathway
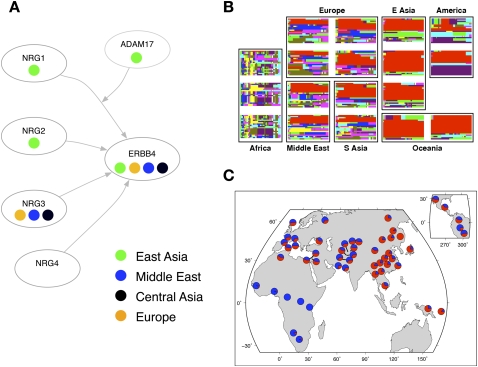
Among the top selection candidates shown in Figure 1, we noticed that two—ERBB4 and NRG3—are, in fact, binding partners (Zhang et al. 1997). Although these two genes are large, and thus contain a number of tested windows, they both are outliers with respect to the rest of the genome even after a conservative Bonferroni correction for the number of windows (empirical P = 0.001 and P = 0.006 in the Middle East for ERBB4 and NRG3, respectively). Further inspection of genes in the NRG–ERBB4 pathway (Kanehisa et al. 2008) revealed a striking alignment of selection signals (Fig. 5A). ERBB4 shows extreme iHS signals in all non-African populations (Fig. 5B,C), NRG3 shows extreme iHS signals in West Eurasian populations, and two other binding partners of ERBB4—NRG1 and NRG2—fall well into the 1% tail of iHS scores in East Asia (Fig. 5A). Further, ADAM17, the gene encoding the enzyme that converts NRG1 to its active form (Mei and Xiong 2008), falls in a region that contains some of the most extreme XP-EHH scores in East Asia (maximum value of XP-EHH in the region of 4.2 at rs2709591, empirical P = 2 × 10−5).
Figure 5.
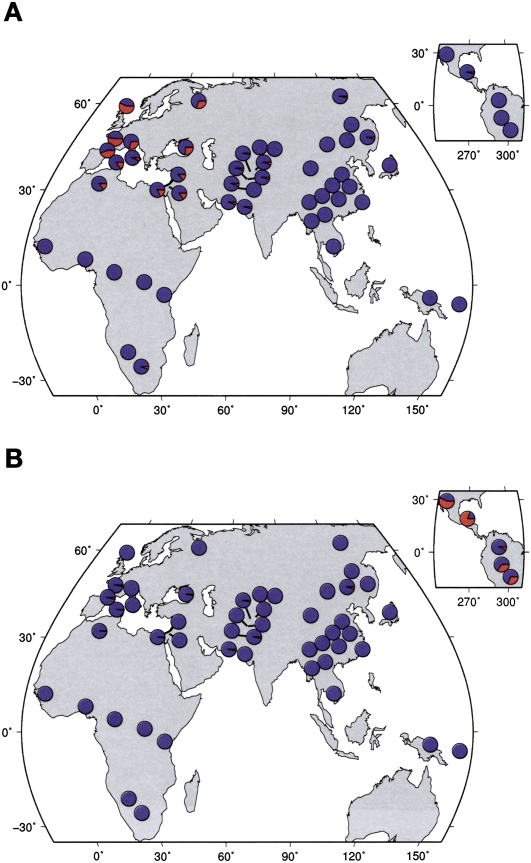
Selection signals in the NRG–ERBB4 pathway. (A) A schematic of the NRG–ERBB4 pathway, drawn from interactions reported in KEGG (Kanehisa et al. 2008) and Mei and Xiong (2008). Each oval represents a gene, and the colored circles denote the geographic regions that have significant selection signals (empirical scores in the top 5% of the distribution). We excluded Oceania and the Americas from this plot since selection scans are expected to have low power in these regions. For ADAM17, the selection statistic is XP-EHH; for the others it is iHS. (B) Haplotype plots at the putative selected region in ERBB4. (C) Worldwide allele frequencies of a SNP that tags the red haplotype in B (rs1505353). (Red) The derived allele; (blue) the ancestral allele.
The NRG–ERBB4 signaling pathway is well-studied and known to be involved in the development of a number of tissues, including heart, neural, and mammary tissue (Gassmann et al. 1995; Tidcombe et al. 2003). Variants in genes in this pathway have been associated with risk of schizophrenia and various psychiatric phenotypes (Stefansson et al. 2002; Hall et al. 2006; Mei and Xiong 2008). We suggest that an unidentified phenotype affected by this pathway has experienced strong recent selection in non-African populations.
Local adaptation
A significant advantage of these data over previous scans for selection is that they allow for the detection of selection on much smaller geographic scales. To identify differential selection between closely related populations, we chose to use FST rather than haplotype-based methods, as haplotype-based signals are largely shared between geographically close populations (Supplemental Figs. 4, 5) and we speculated that selection on a very local scale may lead to only modest allele frequency changes. We manually examined the 100 SNPs that most extremely differentiate in select pairs of populations for evidence of local adaptation (such that all the SNPs mentioned in the following paragraphs fall in the 0.05% tail of the comparison being discussed). We note that though alleles underlying local adaptation may often be population-specific and thus not included on the Illumina chip, a selective sweep should often affect differentiation at nearby tag SNPs (though the magnitude of this effect depends on the levels of migration and the selection coefficient, among other factors; Santiago and Caballero 2005).
Within Africa, we compared the Yoruba to each of the Pygmy populations, and the two Pygmy populations (Mbuti and Biaka) to each other, hypothesizing that the loci involved in reduced stature in Pygmies should be specific to that group (and be detectable by differentiation at nearby tag SNPs). Notably, genes involved in variation in height in Europeans are not enriched for SNPs that strongly differentiate Pygmy from Bantu populations, suggesting that variation in these genes is not responsible for the divergence in phenotype between these two groups (Supplemental Fig. 12). However, among the 100 most differentiated SNPs between the Yoruba and Biaka are two SNPs in genes in the insulin growth factor signaling system: one is a SNP (rs6917747) in an intron of IGF2R, the receptor for the well-studied growth factor IGF2 (FST = 0.54, empirical P = 1 × 10−4). Knockouts of this gene in the mouse lead to fetal overgrowth (Lau et al. 1994). A second SNP (rs9429187) in the gene PIK3R3, which acts downstream of IGF1R (Dey et al. 1998) also differentiates the Biaka and Yoruba (FST = 0.6, empirical P = 1 × 10−4). Considering the defects in the responsiveness of Pygmy cells to IGF1 (Hattori et al. 1996; Jain et al. 1998) without any known causal polymorphism (Bowcock and Sartorelli,1990), we consider both of these genes to be strong candidates for harboring a polymorphism that leads to decreased body size in some Pygmy populations (although we note that the IGF2R polymorphism appears to be specific to Biaka and absent from Mbuti).
Within Western Eurasia, we compared the French, Palestinian, and Balochi populations, as they have the largest sample sizes in their regions. The most extreme SNP (rs4833103) identified in the French–Balochi and French–Palestinian comparisons (French–Balochi FST = 0.69, French–Palestinian FST = 0.6) falls in a cluster of Toll-like receptor genes. A nonsynonymous SNP in TLR6 (rs5743810), a gene involved in the recognition of bacterial pathogens (Ozinsky et al. 2000), is among the highly differentiated SNPs in this cluster (Fig. 6A). This region was previously identified by Todd et al. (2007) as containing SNPs that strongly differentiate populations within Europe; those investigators also noted that the region shows no haplotype-based signature of selection. Also among the most differentiated loci in these comparisons are SNPs in SLC45A2, mentioned above as a locus involved in pigmentation, and a cluster of SNPs in SLC25A13, the gene responsible for type II citrullinemia, a Mendelian disorder of the urea cycle (Kobayashi et al. 1999).
Figure 6.
Worldwide allele frequencies of two nonsynonymous SNPs showing evidence of local adaptation. (A) Frequencies of rs5743810 in TLR6; (B) frequencies of rs12421620 in DPP3. (Red) The frequency of the derived allele; (blue) the frequency of the ancestral allele.
Within East Asia, we chose populations in an attempt to identify SNPs that, like TLR6 and LCT in Europe, show clinal variation in allele frequencies. We examined the tails of the FST distribution between the Dai (a southern Chinese population), Oroqen (a northern Chinese population), and the Han. No SNP showed as striking a pattern as TLR6 in Europe. However, these comparisons identified a number of SNPs in genes related to immunity—a cluster of SNPs in the HLA region (maximum FST of 0.69 at rs1737078) differentiate the Oroqen from the Dai, and SNPs in a cluster of interleukin receptors (maximum FST of 0.68 at rs279545) differentiate the Oroqen from the Han.
Finally, we attempted to use this FST approach to identify putative targets of selection in Oceania and America, the populations that have been most affected by genetic drift. The reduced diversity in these populations, presumably due to bottlenecks in their founding (Conrad et al. 2006; Li et al. 2008), means that power to detect selection in the populations by all current tests is low. Nevertheless, we compared the Yakut (as a Siberian population perhaps most like the ancestors of modern Americans [Li et al. 2008]) and the Maya to identify large allele frequency changes since the founding of the Americas, and the Cambodians and Papuans to identify large allele frequency changes since the founding of Oceania. Among the most differentiated SNPs between the Yakut and Maya is a nonsynonymous SNP (rs12421620) in DPP3, a gene highly expressed in lymphoblasts, the derived allele of which is essentially specific to the Americas (Fig. 6B). Also identified in this comparison is a cluster of interleukin receptors different than the one that differentiates the Oroqen and Han (maximum FST of 0.73 at rs11123915). The comparison between the Cambodian and Papuan populations yielded few immediate selection candidates; the most extreme SNPs fall in or near KCNN3, KCNIP1, and FLJ20366. We emphasize that the interpretation of the forces influencing outlying SNPs in these populations is not straightforward, given the magnitude of genetic drift they have experienced and the lack of a validated demographic model for their histories.
Discussion
We have presented a series of genome-wide scans for natural selection in a worldwide sample of human populations in an effort to refine the geographic patterns of putatively selected haplotypes and identify novel candidate targets of selection.
Geographic patterns of selection
We find that putatively selected haplotypes tend to be shared among geographically close populations. In principle, this could be due to issues of statistical power: broad geographical groupings share a demographic history and thus have similar power profiles. However, strongly selected loci are expected to show geographical patterns largely independent of demography—depending on the relevant selection pressures, they can be highly geographically restricted despite moderate levels of migration, or spread rapidly throughout a species even in the presence of little migration (Nagylaki 1975; Morjan and Rieseberg 2004). Further exploration of the geographic patterns in these data and their implications is warranted, but from the point of view of identifying candidate loci for functional verification, the fact that putatively selected loci often conform to the geographic patterns characteristic of neutral loci is somewhat worrying. This suggests that distinguishing true cases of selection from the tails of the neutral distribution may be more difficult than sometimes assumed, and raises the possibility that many loci identified as being under selection in genome scans of this kind may be false positives. Reports of ubiquitous strong (s = 1 − 5%) positive selection in the human genome (Hawks et al. 2007) may be considerably overstated.
Novel selection candidates
For these reasons, in this paper we have focused on signals of selection in genomic regions for which there is additional evidence for function. One such source of evidence is the alignment of selection signals in a known biological pathway, as seen for the NRG–ERBB4 pathway presented above. False positives arise without regard to genomic annotation, with perhaps some exceptions: for example, recombination rate influences power to detect selection (Nielsen et al. 2007) and varies among functional classes of genes (Frazer et al. 2007), as does gene size (Wang et al. 2003), so signals of selection surrounding multiple genes in a pathway make a compelling suggestion that the pathway has been a target of selection. This principle was exploited by Sabeti et al. (2007) in identifying EDAR and ED2A as targets of selection; this is one of only a few examples where a signal for selection has been successfully linked to a phenotype.
Additional evidence for function comes from association studies. Again, false positives arise largely without regard to function, so the alignment of association and selection signals provides additional evidence that a selection signal is a true positive (unless the association signal itself is a false positive driven by population stratification and a high-FST SNP) (Campbell et al. 2005). This is exemplified by the alignment of selection signals on pigmentation loci in the HGDP data. Following this principle, we have presented two pieces of evidence suggesting that regions associated with risk for type II diabetes have been under selection. First, the distribution of maximum FST is shifted upward in regions showing association with the disease, and a number of these regions are extreme outliers compared with random regions (Fig. 4). Second, several associated regions show extreme XP-EHH scores. Together, these observations suggest that these regions have experienced recent positive selection. However, the fact that the SNPs associated with type II diabetes themselves often do not show high FST makes it plausible that they have not been the actual targets of selection. Instead, selection may be on a related phenotype and result in selection on nearby linked polymorphisms. Interestingly, this seems to be the case for at least one selected locus involved in pigmentation: alleles in KITLG have been associated with blond hair (Sulem et al. 2007), but the presumed target of selection is a nearby polymorphism associated with skin pigmentation (Miller et al. 2007).
In general, we find the evidence for selection on disease risk is not as conclusive as that for selection on pigmentation traits. One parsimonious explanation for this is that selection on disease risk, assuming disease risk is under selection at all, is much weaker than selection on pigmentation. However, the role of the genetic architecture of a trait (the number of loci underlying a trait and their effect sizes and frequencies) in how it responds to selection remains largely unexplored. Since the genetic architecture of pigmentation is relatively simple (compared with other complex traits), perhaps a selection signal on this trait is more readily detected because it is spread across fewer loci. On the other hand, this explanation may confuse cause and effect. Perhaps skin pigmentation has a simpler genetic architecture than other complex traits because it has been subject to recent strong selection—the first moves to a new phenotypic optimum are predicted to be on mutations of large fitness effect (Orr 2002). So assuming a positive correlation between the effects of an allele on fitness and on a trait, it is also plausible that the relatively simple genetic architecture of skin pigmentation is actually a consequence of the strong selection that has acted on this phenotype. Further work on the interplay between genetic architecture and natural selection is needed to clarify these issues.
Conclusions
We have presented here an analysis of a number of putative targets of selection. We expect that our results may be of interest, as the genetics of additional traits are mapped; thus, we have made our data publicly available at http://hgdp.uchicago.edu, as a resource for use in mapping evolutionarily relevant traits. As more genome-wide genotype data become available on more populations, the limiting factors in understanding the loci important in recent human evolution will become the localization and functional verification of precise targets of selection. A full catalog of common sequence variation, such as that envisaged by the 1000 Genomes Project (www.1000genomes.org), will aid the former, and ongoing association studies and functional characterization of the genome (through, for example, the ENCODE project; The ENCODE Project Consortium 2007) will enable the latter. Further work is needed to fully explore how these resources will aid in understanding the mechanisms that underlie phenotypic diversity in our species.
Methods
Phasing
Extensive testing of the most reliable approach to phasing this type of data was performed by Conrad et al. (2006); we followed their approach closely. Briefly, phasing was done using fastPHASE, with the settings that allow variation in the switch rate between subpopulations. The populations were grouped into subpopulations corresponding to the clusters identified in Rosenberg et al. (2002). Haplotypes from the HapMap YRI and CEU populations were included as known, as they were phased in trios and are highly accurate. HapMap JPT and CHB genotypes were also included to help with the phasing.
Simulations
Simulations were done using a hybrid coalescent/forward-time approach in a three-population demographic model optimized to produce HapMap-like data. We used the “cosi” demographic model with slight modifications (Schaffner et al. 2005). In each simulation, the population was initialized by coalescent simulation with “cosi” until the time point before the out-of-Africa split. From that point on, haplotypes were simulated forward in time using a Wright-Fisher model. Following Hoggart et al. (2007), all parameters were scaled by a factor of five to increase efficiency—that is, the branch lengths and populations sizes were decreased by a factor of five and the mutation, recombination, and mutation rates were all increased by a factor of five. We simulated selected alleles by randomly placing a single selected mutation on each tree during the Wright-Fisher stage of the simulation. This means that the ultimate allele frequency in these simulations is stochastic, and the number in each allele frequency bin varies somewhat. We simulated 2500 regions of 500 kb with a recombination map (including hotspots) generated by cosi using the genome-wide average of 1 cM/Mb as the baseline rate (Schaffner et al. 2005).
In all simulations, we attempted to match the ascertainment to that of the real data as much as possible. This involved a two-step ascertainment process. In the first step, we approximately matched the joint allele frequency spectrum to the HapMap using rejection sampling. We estimated the joint allele frequency spectrum of the simulations, f(x), and that of the HapMap, g(x), on a 12 × 12 × 12 grid, then accepted a simulated allele with probability P = f(x)/Mg(x). To exactly match the distribution, M should be the maximum of the ratio of the two densities, but in this situation there is a tradeoff between precision and speed of the simulations—we found a value of M = 8 to produce satisfactory results (Supplemental Fig. 1). In the second step, we implemented a greedy algorithm for tag SNP selection (Carlson et al. 2004), following an approximation of the tagging strategy used by Illumina in their design of the 650K chip (Eberle et al. 2007). Considered for inclusion were all SNPs with a minor allele frequency over 0.05. We included all such SNPs that tagged at least one other SNP in the “European” population with an r2 threshold of 0.7, or that tagged at least two other SNPs in the “East Asian” population with r2 > 0.8, or that tagged at least two other SNPs in the “African” population with r2 > 0.7. We then randomly selected common SNPs (MAF > 0.05) from each population to increase SNP density by one third. This procedure provided a good match to the Illumina chip (Supplemental Fig. 1).
We calculated iHS and XP-EHH on these simulations as described below in a window of forty SNPs (this is ~200 kb at the Illumina SNP density) surrounding the selected site, treating either the maximum XP-EHH or fraction of SNPs with |iHS| > 2 as the test statistic. Critical values were obtained by 1000 neutral simulations. For XP-EHH, we used the African population as the reference for the simulations of selection in Europe and East Asia, and the European population as the reference for simulations of selection in Africa. The genetic map used was estimated on each population individually using LDHat (Myers et al. 2005), and then averaged across populations.
Calculation of test statistics
iHS was calculated as in Voight et al. (2006), XP-EHH as in Sabeti et al. (2007), and CLR as in Nielsen et al. (2005). iHS was calculated on all SNPs with a minor allele frequency of at least 5%. XP-EHH requires the definition of a reference population—for non-African populations, we used the chromosomes from the Bantu-speaking populations as a reference, and for the Bantu and Biaka Pygmy populations we used the European chromosomes as a reference population. For analyses on the HapMap, we used the CEPH population as a reference for the Yoruba, and the Yoruba as a reference for the CEPH and Asian populations. The CLR test was calculated every 20 kb across each chromosome in each population. In each 200 kb genomic window, the fraction of SNPs with |iHS| > 2 and the maximum XP-EHH and CLR were used as test statistics. The genetic map used was that released by the HapMap Consortium (www.hapmap.org); this map is averaged across all three HapMap populations and thus is unlikely to be influenced by selection in any individual population. We determined the ancestral state of each SNP by comparison to the chimpanzee genome. Due to the different demographic histories of the X chromosome and the autosomes, data from the X chromosome were normalized separately.
To convert the test statistics from each window into an empirical P-value, we binned windows by number of SNPs in increments of 20 SNPs. For iHS, all windows with <20 SNPs were dropped. Within each bin, for each window i, the fraction of windows with a value of the statistic greater than that in i is the empirical P-value. To generate Figure 1, we took the complete 1% tail of each test statistic and collapsed adjacent windows, assigning the minimum P-value to the collapsed window. Plotted are the most extreme 10 such windows from each geographic region.
FST analysis
To generate Figures 3 and 4, we compiled a list of polymorphisms known to associate with the phenotype of interest from genome-wide association studies. For each pair of populations, we calculated FST using the Weir and Cockerham estimator (Cockerham and Weir 1986). For each SNP, we found the SNP closest to it on the Illumina chip, defined a window of 50 kb on either side of that SNP, and took the maximum FST in that window as the test statistic. To generate a null distribution, we performed the same procedure on a random sample of 10,000 SNPs from the Illumina chip.
Available resources
We implemented a database of our results based on the generic genome browser (Stein et al. 2002), available at http://hgdp.uchicago.edu. The software used for simulation and calculation of iHS and XP-EHH, as well as the raw phased data, are available at this same site.
Acknowledgments
We thank Molly Przeworski, members of the Pritchard, Przeworski, and Stephens laboratory groups, and George Perry for discussions; Paul Scheet for suggestions regarding phasing; Melissa Hubisz for the code for the CLR statistic; the anonymous reviewers for helpful comments; and NSF award CNS-0619926 for computer resources. This work was supported by a Packard Foundation grant to J. Pritchard. J. Pickrell was supported by an NIH training grant to the University of Chicago. J.N. was supported by a US National Science Foundation postdoctoral research fellowship in bioinformatics. M.W.F. was supported by NIH grant GM28016. J. Pritchard is an investigator of the Howard Hughes Medical Institute.
Footnotes
[Supplemental material is available online at www.genome.org. The data from this study is publicly available at http://hgdp.uchicago.edu/.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.087577.108.
References
- Barreiro L.B., Laval G., Quach H., Patin E., Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat. Genet. 2008;40:340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- Bersaglieri T., Sabeti P.C., Patterson N., Vanderploeg T., Schaffner S.F., Drake J.A., Rhodes M., Reich D.E., Hirschhorn J.N. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowcock A., Sartorelli V. Polymorphism and mapping of the IGF1 gene, and absence of association with stature among African Pygmies. Hum. Genet. 1990;85:349–354. doi: 10.1007/BF00206760. [DOI] [PubMed] [Google Scholar]
- Campbell C.D., Ogburn E.L., Lunetta K.L., Lyon H.N., Freedman M.L., Groop L.C., Altshuler D., Ardlie K.G., Hirschhorn J.N. Demonstrating stratification in a European American population. Nat. Genet. 2005;37:868–872. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- Carlson C.S., Eberle M.A., Rieder M.J., Yi Q., Kruglyak L., Nickerson D.A. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerham C.C., Weir B.S. Estimation of inbreeding parameters in stratified populations. Ann. Hum. Genet. 1986;50:271–281. doi: 10.1111/j.1469-1809.1986.tb01048.x. [DOI] [PubMed] [Google Scholar]
- Conrad D.F., Jakobsson M., Coop G., Wen X., Wall J.D., Rosenberg N.A., Pritchard J.K. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat. Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- Dey B.R., Furlanetto R.W., Nissley S.P. Cloning of human p55 gamma, a regulatory subunit of phosphatidylinositol 3-kinase, by a yeast two-hybrid library screen with the insulin-like growth factor-I receptor. Gene. 1998;209:175–183. doi: 10.1016/s0378-1119(98)00045-6. [DOI] [PubMed] [Google Scholar]
- Di Rienzo A., Hudson R.R. An evolutionary framework for common diseases: The ancestral-susceptibility model. Trends Genet. 2005;21:596–601. doi: 10.1016/j.tig.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Drogemuller C., Philipp U., Haase B., Gunzel-Apel A.-R., Leeb T. A noncoding melanophilin gene (MLPH) SNP at the splice donor of exon 1 represents a candidate causal mutation for coat color dilution in dogs. J. Hered. 2007;98:468–473. doi: 10.1093/jhered/esm021. [DOI] [PubMed] [Google Scholar]
- Eberle M.A., Ng P.C., Kuhn K., Zhou L., Peiffer D.A., Galver L., Viaud-Martinez K.A., Lawley C.T., Gunderson K.L., Shen R., et al. Power to detect risk alleles using genome-wide tag SNP panels. PLoS Genet. 2007;3:1827–1837. doi: 10.1371/journal.pgen.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto A., Kimura R., Ohashi J., Omi K., Yuliwulandari R., Batubara L., Mustofa M.S., Samakkarn U., Settheetham-Ishida W., Ishida T., et al. A scan for genetic determinants of human hair morphology: EDAR is associated with Asian hair thickness. Hum. Mol. Genet. 2008;17:835–843. doi: 10.1093/hmg/ddm355. [DOI] [PubMed] [Google Scholar]
- Gardiner K., Slavov D., Bechtel L., Davisson M. Annotation of human chromosome 21 for relevance to Down syndrome: Gene structure and expression analysis. Genomics. 2002;79:833–843. doi: 10.1006/geno.2002.6782. [DOI] [PubMed] [Google Scholar]
- Gassmann M., Casagranda F., Orioli D., Simon H., Lai C., Klein R., Lemke G. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature. 1995;378:390–394. doi: 10.1038/378390a0. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson D.F., Walters G.B., Thorleifsson G., Stefansson H., Halldorsson B.V., Zusmanovich P., Sulem P., Thorlacius S., Gylfason A., Steinberg S., et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- Hall J., Whalley H.C., Job D.E., Baig B.J., McIntosh A.M., Evans K.L., Thomson P.A., Porteous D.J., Cunningham-Owens D.G., Johnstone E.C., et al. A neuregulin 1 variant associated with abnormal cortical function and psychotic symptoms. Nat. Neurosci. 2006;9:1477–1478. doi: 10.1038/nn1795. [DOI] [PubMed] [Google Scholar]
- Han J., Kraft P., Nan H., Guo Q., Chen C., Qureshi A., Hankinson S.E., Hu F.B., Duffy D.L., Zhao Z.Z., et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4:e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock A.M., Witonsky D.B., Gordon A.S., Eshel G., Pritchard J.K., Coop G., Di Rienzo A. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet. 2008;4:e32. doi: 10.1371/journal.pgen.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y., Vera J.C., Rivas C.I., Bersch N., Bailey R.C., Geffner M.E., Golde D.W. Decreased insulin-like growth factor I receptor expression and function in immortalized African Pygmy T cells. J. Clin. Endocrinol. Metab. 1996;81:2257–2263. doi: 10.1210/jcem.81.6.8964861. [DOI] [PubMed] [Google Scholar]
- Hawks J., Wang E.T., Cochran G.M., Harpending H.C., Moyzis R.K. Recent acceleration of human adaptive evolution. Proc. Natl. Acad. Sci. 2007;104:20753–20758. doi: 10.1073/pnas.0707650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A., Palsson S., Thorleifsson G., Grant S.F.A., Emilsson V., Gunnarsdottir S., Adeyemo A., Chen Y., Chen G., Reynisdottir I., et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat. Genet. 2007;39:218–225. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- Hinds D.A., Stuve L.L., Nilsen G.B., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- Hoggart C.J., Chadeau-Hyam M., Clark T.G., Lampariello R., Whittaker J.C., De Iorio M., Balding D.J. Sequence-level population simulations over large genomic regions. Genetics. 2007;177:1725–1731. doi: 10.1534/genetics.106.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R.R., Bailey K., Skarecky D., Kwiatowski J., Ayala F.J. Evidence for positive selection in the superoxide dismutase (Sod) region of Drosophila melanogaster. Genetics. 1994;136:1329–1340. doi: 10.1093/genetics/136.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt K.A., Zhernakova A., Turner G., Heap G.A.R., Franke L., Bruinenberg M., Romanos J., Dinesen L.C., Ryan A.W., Panesar D., et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H., Kim Y. Detecting local adaptation using the joint sampling of polymorphism data in the parental and derived populations. Genetics. 2008;179:1713–1720. doi: 10.1534/genetics.108.086835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y., David V.A., Eizirik E., Schaffer A.A., Neelam B.A., Roelke M.E., Hannah S.S., O'Brien S.J, Menotti-Raymond M. A homozygous single-base deletion in MLPH causes the dilute coat color phenotype in the domestic cat. Genomics. 2006;88:698–705. doi: 10.1016/j.ygeno.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Jain S., Golde D.W., Bailey R., Geffner M.E. Insulin-like growth factor-I resistance. Endocr. Rev. 1998;19:625–646. doi: 10.1210/edrv.19.5.0348. [DOI] [PubMed] [Google Scholar]
- Jensen J.D., Thornton K.R., Bustamante C.D., Aquadro C.F. On the utility of linkage disequilibrium as a statistic for identifying targets of positive selection in nonequilibrium populations. Genetics. 2007;176:2371–2379. doi: 10.1534/genetics.106.069450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley J.L., Madeoy J., Calhoun J.C., Swanson W., Akey J.M. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 2006;16:980–989. doi: 10.1101/gr.5157306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Nielsen R. Linkage disequilibrium as a signature of selective sweeps. Genetics. 2004;167:1513–1524. doi: 10.1534/genetics.103.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Sinasac D.S., Iijima M., Boright A.P., Begum L., Lee J.R., Yasuda T., Ikeda S., Hirano R., Terazono H., et al. The gene mutated in adult-onset type II citrullinaemia encodes a putative mitochondrial carrier protein. Nat. Genet. 1999;22:159–163. doi: 10.1038/9667. [DOI] [PubMed] [Google Scholar]
- Lamason R.L., Mohideen M.-A.P.K., Mest J.R., Wong A.C., Norton H.L., Aros M.C., Jurynec M.J., Mao X., Humphreville V.R., Humbert J.E., et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- Lau M.M., Stewart C.E., Liu Z., Bhatt H., Rotwein P., Stewart C.L. Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality. Genes & Dev. 1994;8:2953–2963. doi: 10.1101/gad.8.24.2953. [DOI] [PubMed] [Google Scholar]
- Lettre G., Jackson A.U., Gieger C., Schumacher F.R., Berndt S.I., Sanna S., Eyheramendy S., Voight B.F., Butler J.L., Guiducci C., et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R.C., Krakauer J. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics. 1973;74:175–195. doi: 10.1093/genetics/74.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Lohmueller K.E., Mauney M.M., Reich D., Braverman J.M. Variants associated with common disease are not unusually differentiated in frequency across populations. Am. J. Hum. Genet. 2006;78:130–136. doi: 10.1086/499287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson J.M., Gonzalez J., Witten D.M., Davis J.C., Rosenberg N.A., Hirsh A.E., Petrov D.A. Nonadaptive explanations for signatures of partial selective sweeps in Drosophila. Mol. Biol. Evol. 2008;25:1025–1042. doi: 10.1093/molbev/msn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matesic L.E., Yip R., Reuss A.E., Swing D.A., O'Sullivan T.N., Fletcher C.F., Copeland N.G., Jenkins N.A. Mutations in Mlph, encoding a member of the Rab effector family, cause the melanosome transport defects observed in leaden mice. Proc. Natl. Acad. Sci. 2001;98:10238–10243. doi: 10.1073/pnas.181336698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M.I., Abecasis G.R., Cardon L.R., Goldstein D.B., Little J., Ioannidis J.P.A., Hirschhorn J.N. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- McGowan K.A., Li J.Z., Park C.Y., Beaudry V., Tabor H.K., Sabnis A.J., Zhang W., Fuchs H., de Angelis M.H., Myers R.M., et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat. Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei L., Xiong W.-C. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat. Rev. Neurosci. 2008;9:437–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.T., Beleza S., Pollen A.A., Schluter D., Kittles R.A., Shriver M.D., Kingsley D.M. Cis-regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131:1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morjan C.L., Rieseberg L.H. How species evolve collectively: Implications of gene flow and selection for the spread of advantageous alleles. Mol. Ecol. 2004;13:1341–1356. doi: 10.1111/j.1365-294X.2004.02164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou C., Thomason H., Willan P., Clowes C., Harris W., Drew C., Dixon J., Dixon M., Headon D. Enhanced ectodysplasin-A receptor (EDAR) signaling alters multiple fiber characteristics to produce the East Asian hair form. Hum Mutat. 2008;29:1405–1411. doi: 10.1002/humu.20795. [DOI] [PubMed] [Google Scholar]
- Myers S., Bottolo L., Freeman C., McVean G., Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Myles S., Davison D., Barrett J., Stoneking M., Timpson N. Worldwide population differentiation at disease-associated SNPs. BMC Med. Genomics. 2008;1:22. doi: 10.1186/1755-8794-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagylaki T. Conditions for the existence of clines. Genetics. 1975;80:595–615. doi: 10.1093/genetics/80.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel J.V. Diabetes mellitus: A “thrifty” genotype rendered detrimental by “progress”? Am. J. Hum. Genet. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Williamson S., Kim Y., Hubisz M.J., Clark A.G., Bustamante C. Genomic scans for selective sweeps using SNP data. Genome Res. 2005;15:1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Hellmann I., Hubisz M., Bustamante C., Clark A.G. Recent and ongoing selection in the human genome. Nat. Rev. Genet. 2007;8:857–868. doi: 10.1038/nrg2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton H.L., Kittles R.A., Parra E., McKeigue P., Mao X., Cheng K., Canfield V.A., Bradley D.G., McEvoy B., Shriver M.D., et al. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol. Biol. Evol. 2007;24:710–722. doi: 10.1093/molbev/msl203. [DOI] [PubMed] [Google Scholar]
- Orr H.A. The population genetics of adaptation: The adaptation of DNA sequences. Evolution. 2002;56:1317–1330. doi: 10.1111/j.0014-3820.2002.tb01446.x. [DOI] [PubMed] [Google Scholar]
- Ozinsky A., Underhill D.M., Fontenot J.D., Hajjar A.M., Smith K.D., Wilson C.B., Schroeder L., Aderem A. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc. Natl. Acad. Sci. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G.H., Dominy N.J., Claw K.G., Lee A.S., Fiegler H., Redon R., Werner J., Villanea F.A., Mountain J.L., Misra R., et al. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle F., Manica A., Charpentier M., Guegan J.F., Guernier V., Balloux F. Pathogen-driven selection and worldwide HLA class I diversity. Curr. Biol. 2005;15:1022–1027. doi: 10.1016/j.cub.2005.04.050. [DOI] [PubMed] [Google Scholar]
- Rosenberg N.A., Pritchard J.K., Weber J.L., Cann H.M., Kidd K.K., Zhivotovsky L.A., Feldman M.W. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Sabeti P.C., Reich D.E., Higgins J.M., Levine H.Z.P., Richter D.J., Schaffner S.F., Gabriel S.B., Platko J.V., Patterson N.J., McDonald G.J., et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Sabeti P.C., Varilly P., Fry B., Lohmueller J., Hostetter E., Cotsapas C., Xie X., Byrne E.H., McCarroll S.A., Gaudet R., et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago E., Caballero A. Variation after a selective sweep in a subdivided population. Genetics. 2005;169:475–483. doi: 10.1534/genetics.104.032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner S.F., Foo C., Gabriel S., Reich D., Daly M.J., Altshuler D. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 2005;15:1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L.J., Mohlke K.L., Bonnycastle L.L., Willer C.J., Li Y., Duren W.L., Erdos R., Stringham H.M., Chines P.S., Jackson A.U., et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M., Wiehe T. Genetic hitch-hiking in a subdivided population. Genet. Res. 1998;71:155–160. doi: 10.1017/s001667239800319x. [DOI] [PubMed] [Google Scholar]
- Smith J.M., Haigh J. The hitch-hiking effect of a favourable gene. Genet. Res. 1974;23:23–35. [PubMed] [Google Scholar]
- Stefansson H., Sigurdsson E., Steinthorsdottir V., Bjornsdottir S., Sigmundsson T., Ghosh S., Brynjolfsson J., Gunnarsdottir S., Ivarsson O., Chou T.T., et al. Neuregulin 1 and susceptibility to schizophrenia. Am. J. Hum. Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., et al. The generic genome browser: A building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokowski R.P., Pant P.V.K., Dadd T., Fereday A., Hinds D.A., Jarman C., Filsell W., Ginger R.S., Green M.R., van der Ouderaa F.J., et al. A genomewide association study of skin pigmentation in a South Asian population. Am. J. Hum. Genet. 2007;81:1119–1132. doi: 10.1086/522235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulem P., Gudbjartsson D.F., Stacey S.N., Helgason A., Rafnar T., Magnusson K.P., Manolescu A., Karason A., Palsson A., Thorleifsson G., et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- Sulem P., Gudbjartsson D.F., Stacey S.N., Helgason A., Rafnar T., Jakobsdottir M., Steinberg S., Gudjonsson S.A., Palsson A., Thorleifsson G., et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat. Genet. 2008;40:835–837. doi: 10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima K.M., Coop G., Przeworski M. How reliable are empirical genomic scans for selective sweeps? Genome Res. 2006;16:702–712. doi: 10.1101/gr.5105206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidcombe H., Jackson-Fisher A., Mathers K., Stern D.F., Gassmann M., Golding J.P. Neural and mammary gland defects in ErbB4 knockout mice genetically rescued from embryonic lethality. Proc. Natl. Acad. Sci. 2003;100:8281–8286. doi: 10.1073/pnas.1436402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd J.A., Walker N.M., Cooper J.D., Smyth D.J., Downes K., Plagnol V., Bailey R., Nejentsev S., Field S.F., Payne F., et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoki H., Takahashi A., Kawaguchi T., Hara K., Horikoshi M., Andersen G., Ng D., Holmkvist J., Borch-Johnsen K., Jorgensen T., et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40:1098–1102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- Vaez M., Follett S.A., Bed'hom B., Gourichon D., Tixier-Boichard M., Burke T. A single point-mutation within the melanophilin gene causes the lavender plumage colour dilution phenotype in the chicken. BMC Genet. 2008;9:7. doi: 10.1186/1471-2156-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B.F., Kudaravalli S., Wen X., Pritchard J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journals/pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Li S., Zhang Y., Zheng H., Xu Z., Ye J., Yu J., Wong G.K.-S. Vertebrate gene predictions and the problem of large genes. Nat. Rev. Genet. 2003;4:741–749. doi: 10.1038/nrg1160. [DOI] [PubMed] [Google Scholar]
- Wang E.T., Kodama G., Baldi P., Moyzis R.K. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc. Natl. Acad. Sci. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon M.N., Lango H., Lindgren C.M., Wallace C., Evans D.M., Mangino M., Freathy R.M., Perry J.R.B., Stevens S., Hall A.S., et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer C.J., Sanna S., Jackson A.U., Scuteri A., Bonnycastle L.L., Clarke R., Heath S.C., Timpson N.J., Najjar S.S., Stringham H.M., et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson S.H., Hubisz M.J., Clark A.G., Payseur B.A., Bustamante C.D., Nielsen R. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journals.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K., Miyake K., Horikawa Y., Hara K., Osawa H., Furuta H., Hirota Y., Mori H., Jonsson A., Sato Y., et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat. Genet. 2008;40:1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- Zeggini E., Scott L.J., Saxena R., Voight B.F., Marchini J.L., Hu T., de Bakker P.I.W., Abecasis G.R., Almgren P., Andersen G., et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Sliwkowski M.X., Mark M., Frantz G., Akita R., Sun Y., Hillan K., Crowley C., Brush J., Godowski P.J., et al. Neuregulin-3 (NRG3): A novel neural tissue-enriched protein that binds and activates ErbB4. Proc. Natl. Acad. Sci. 1997;94:9562–9567. doi: 10.1073/pnas.94.18.9562. [DOI] [PMC free article] [PubMed] [Google Scholar]