Abstract
Apolipoprotein L1 (APOL1) is a human protein that confers immunity to Trypanosoma brucei infections but can be countered by a trypanosome-encoded antagonist SRA. APOL1 belongs to a family of programmed cell death genes whose proteins can initiate host apoptosis or autophagic death. We report here that all six members of the APOL gene family (APOL1-6) present in humans have rapidly evolved in simian primates. APOL6, furthermore, shows evidence of an adaptive sweep during recent human evolution. In each APOL gene tested, we found rapidly evolving codons in or adjacent to the SRA-interacting protein domain (SID), which is the domain of APOL1 that interacts with SRA. In APOL6, we also found a rapidly changing 13-amino-acid cluster in the membrane-addressing domain (MAD), which putatively functions as a pH sensor and regulator of cell death. We predict that APOL genes are antagonized by pathogens by at least two distinct mechanisms: SID antagonists, which include SRA, that interact with the SID of various APOL proteins, and MAD antagonists that interact with the MAD hinge base of APOL6. These antagonists either block or prematurely cause APOL-mediated programmed cell death of host cells to benefit the infecting pathogen. These putative interactions must occur inside host cells, in contrast to secreted APOL1 that trafficks to the trypanosome lysosome. Hence, the dynamic APOL gene family appears to be an important link between programmed cell death of host cells and immunity to pathogens.
The apolipoprotein L (APOL) gene family is composed of six genes in humans, which are grouped within 619 kb on human chromosome 22 (see Fig. 1; Page et al. 2001). By far, the best-studied family member is APOL1, which encodes a trypanolytic factor in humans and gorillas, lysing pathogenic Trypanosoma brucei subspecies during bloodstream infections (Vanhamme et al. 2003). Most other primates lack APOL1 and do not have trypanolytic activity, except for certain species of baboons, mandrills, and mangabeys (Seed et al. 1990; Lugli et al. 2004). APOL1 is unique among APOL proteins because it can be secreted outside the cell, presumably due to its N-terminal signal peptide. Other APOL proteins do not have this signal peptide and have predicted localizations inside the cell (Page et al. 2001).
Figure 1.
Genome organization and coding exons of the human APOL gene family. (A) The genome organization of the six human APOL genes is shown. (B) Coding exons for human APOL genes and isoforms are shown. Differences between isoforms in noncoding regions are not shown.
In addition to killing parasites, APOL proteins can initiate programmed cell death (PCD) of host cells. Each of the APOL genes putatively contains a BH3 protein domain, which is characteristic of the BH3-only family of pro-apoptosis genes (Liu et al. 2005; Vanhollebeke and Pays 2006). BH3-only proteins function as upstream activators of PCD, responding to stimuli such as cell detachment, cytokine withdrawal, or DNA damage before initiating cell death (Strasser 2005). The BH3 domain itself is a short peptide that binds to a groove on other PCD proteins, such as pro-survival Bcl-2 family members, which can bind BH3-only proteins under normal conditions to prevent cell death. Consistent with other BH3-only proteins, APOL6 was found to cause apoptosis when overexpressed in cancer cells (Liu et al. 2005).
APOL1 was also recently shown to have an intracellular function, causing autophagic death of human cells (Wan et al. 2008). Autophagy is used by cells to “self-eat” during starvation and to recycle cellular contents. Cytosolic contents are gathered in an autophagosome, and trafficked via endosome fusion to the lysosome for degradation (Mizushima 2007). Although autophagy is generally considered to be a pro-survival process, autophagy proteins can cause PCD by a mechanism that is distinct from apoptosis (Degterev and Yuan 2008). The seemingly disparate roles of APOL1 in host autophagic death and trypanolysis are remarkably consistent in that they both utilize endosome trafficking to the lysosome.
Further clues to APOL protein functions come from expression studies, genetic screens, and disease association studies. APOL genes have been implicated in schizophrenia (Mimmack et al. 2002), breast cancer (Dombkowski et al. 2006), cervical cancer (Ahn et al. 2004), and osteoarthritis (Okabe et al. 2007). However, the specific roles of APOL proteins in these diseases are not known. APOL genes are up-regulated by multiple pro-inflammatory signaling molecules, including interferon (IFN)-alpha (Hayashi et al. 2005), IFN-beta (Stojdl et al. 2003), IFN-gamma (Sana et al. 2005), and tumor necrosis factor alpha (TNF-alpha) (Monajemi et al. 2002). These regulations suggest that APOL proteins participate in the immune system, in addition to the role of secreted APOL1 in trypanosome immunity.
Studies of how APOL1 kills trypanosomes have revealed the functions of individual protein domains. These functions may be shared with intracellular APOL proteins by virtue of shared domain architecture. APOL1 associates with a fraction of high-density lipoprotein (HDL) particles in the human bloodstream (it is this association for which the APOL genes were named) (Duchateau et al. 1997). Once introduced into hosts via insect vectors, T. brucei subspecies endocytose HDL particles, likely as a means to obtain both lipids and iron (Green et al. 2003; Vanhollebeke et al. 2008). As the HDL particles are trafficked from endocytic particles to the lysosome, the pH change from pH ∼7 to ∼5 induces a conformational change in the APOL1 membrane addressing domain (MAD). This causes a salt-bridge-linked hinge to open, which releases APOL1 from the HDL particle to insert in the lysosomal membrane (Perez-Morga et al. 2005). After insertion into the lysosomal membrane, APOL1 employs its pore-forming protein domain to create an anion-specific pore, which leads to lysosome swelling and ultimately pathogen lysis. The MAD and pore-forming domain are both required for killing (Perez-Morga et al. 2005).
Two trypanosome subspecies have evolved a means to resist lysis by APOL1, causing the disease “sleeping sickness” in humans. The resistance mechanism of Trypanosoma brucei gambiense is unknown, but the mechanism of Trypanosoma brucei rhodesiense has been discovered (Xong et al. 1998). Trypanosoma brucei rhodesiense encodes the protein SRA that directly binds to APOL1 in the trypanosome lysosome to prevent lysis. SRA interacts with APOL1 using a coiled–coiled interaction at the APOL1 SRA-interacting domain (SID). The SID protein domain was shown to not be required for killing (Vanhamme et al. 2003), except in a recently reported mouse model (Molina-Portela et al. 2008). The role of the SID domain in trypanolysis thus remains somewhat ambiguous.
We wondered whether the APOL1–SRA conflict was just one instance of similar, recurrent conflicts that APOL genes have encountered during primate evolution. Since proteins that directly interact with pathogen components often evolve under positive selection, especially at interaction interfaces, we sought to test the APOL gene family for evidence of rapid evolution. We present a detailed evolutionary analysis of the APOL gene family in primates and find evidence that indeed positive selection has occurred—in the entire gene family. We highlight particular protein regions that have evolved under positive selection, and provide strong evidence for additional sites of host–pathogen interaction. Overall, we show that the APOL gene family has been very dynamic during primate evolution, which likely reflects important roles for the family in immunity to pathogens.
Results
The APOL gene family shows evidence of gene gain/loss in primates
We first assembled a complete collection of APOL genes from primate genome sequences. We found that of the seven primate species whose complete genome sequences were available, each had a different complement of APOL genes (Table 1). There was clearly a pattern of gene loss by pseudogenization or outright deletion. For example, the chimpanzee genome, very similar to the human genome, was completely missing both APOL1 and APOL4. Rhesus macaque was completely missing APOL3 and had pseudogenes of APOL1 and APOL5. In contrast to these gene loss events, the macaque, baboon, and marmoset genomes had gained additional APOL genes compared with the human APOL gene cluster.
Table 1.
The APOL gene family is highly polymorphic between primate species
aGene coordinates are given for the following isoforms: APOL1 isoform a, APOL3 isoform 1, APOL4 isoform 1. Gene coordinates for APOL2.1 and APOL7 are given relative to APOL2, and coordinates for marmoset APOL3/L4 are given relative to APOL3.
b(+) Indicates a gene with an intact coding region; − indicates that the gene is not present in the genome; and ps indicates a pseudogene with the genotype shown in parentheses.
cThe ancestries of marmoset APOL1 and APOL2, and APOL3 and APOL4 are too similar to differentiate.
dOnly partial gene sequences were available for baboon APOL1 and marmoset APOL5.
Given the high degree of gene gain/loss in the APOL gene cluster, we determined the phylogenetic relationships between APOL genes in primates. We sequenced APOL genes from additional species and took advantage of a useful feature of APOL genes—they each share a large exon that contains ∼70%–85% of the total coding sequence (Fig. 1). The remaining coding sequences, except for a small C-terminal exon of APOL5, are encoded by one to four small N-terminal exons, which differ between genes and between isoforms of a given gene.
We constructed a phylogenetic tree of all available APOL coding sequences and found patterns of both duplication and pseudogenization (Fig. 2). For each APOL gene, the tree topology matched the established primate phylogeny (Purvis 1995), and major branch points had strong statistical support. APOL5 and APOL6 were each monophyletic and showed no evidence of duplication. And while many of the APOL5 coding sequences were pseudogenes, all 20 APOL6 sequences that we obtained had intact coding regions.
Figure 2.
Phylogenetic relationships in the APOL gene family in primates. A phylogenetic tree was created from the large-exon sequences of each APOL gene, which includes ∼70%–85% of each coding sequence. Pseudogenes are shown in parentheses, and probabilities of individual clades are shown at central topological junctions. Within individual APOL gene clades, sequences cluster according to the established primate phylogeny (Purvis 1995). The tree shows numerous duplications of APOL genes: APOL2.1 is the result of a duplication that occurred after the divergence of APOL1 and APOL2; APOL7 is the result of a duplication that occurred after the divergence of APOL1/L2 and APOL3/L4; and, depending on the location of the tree root, which is unknown, the APOL1/L2 and APOL3/L4 clades may themselves be products of a duplication that occurred early in primate evolution.
APOL1–4 had a dynamic evolutionary history in primates with multiple pseudogenizations and duplications. APOL1 and APOL2, and APOL3 and APOL4 are products of gene duplications that occurred near the divergence of Platyrrhinii (New World monkeys [NWMs]) from Catarrhinii (Old World monkeys [OWMs] and Hominoids), ∼33 Myr ago. We also found APOL gene lineages that are not represented in the human genome. For instance, a paralogous APOL7 lineage arose prior to divergence of APOL1 and APOL2 but is only found as a pseudogene in rhesus macaque and baboon. A more recent duplication of the APOL2 lineage led to the APOL2.1 lineage, which is still represented by presumed intact genes in rhesus macaque and island gibbon. We also found two APOL genes in the prosimians galago and lemur that may be ancestral to simian APOL1–4.
The evolutionary history of APOL1 is of particular interest due to its established role in trypanolysis. We found intact APOL1 in only human and gorilla, which is consistent with the serum resistance of these species to trypanosomes (Seed et al. 1990; Lugli et al. 2004; Poelvoorde et al. 2004). The only other primate species that have shown serum resistance are certain baboons, mangabeys, and mandrills. We did not obtain APOL1 sequences in these species, except in the baboon Papio hamadryas, where we obtained a partial gene sequence. We found pseudogenes of APOL1 in orangutan (nonsense mutation, C862T) and rhesus macaque (frameshift mutation, T700-); both mutations are located in the APOL1 MAD.
Since APOL1 is the only APOL protein that is known to be secreted, we also tried to determine the evolutionary origin of the APOL1 signal peptide. The long and short isoforms of APOL1 share an N-terminal signal peptide for secretion. The first two exons of the long isoform, which encode the signal peptide, have 92 bp of coding sequence not found in the transcripts of other APOL genes. We found these two exons present in the genome sequence of both rhesus macaque APOL1 (pseudogenized) and human APOL2. If we assume that primates do not transcribe these exons in APOL2, as is currently accepted, then APOL1 most likely acquired the exons by a gain-of-function mutation that created a new splice form. This event occurred after the divergence of APOL1 and APOL2 during primate evolution.
APOL1/L2/L2.1, APOL3/L4, and APOL6 show evidence of positive selection during primate evolution
Next, we compared APOL genes, in whole-gene comparisons, for evidence of rapid evolution. For initial analysis, we chose a sampling of species that had whole-genome sequences available and were from the major lineages of simian primates: human and chimpanzee, hominoids; rhesus macaque, OWMs; and white-tufted-ear marmoset, NWMs. These species shared a common ancestor ∼33 Myr ago (Glazko and Nei 2003). We used pairwise alignment tools to identify all APOL genes or gene fragments in these genomes and compared all of the intact, homologous genes by whole-gene dN/dS analysis. In each of the six APOL genes that are present in humans, except APOL1, there was at least one comparison where the dN/dS exceeded one, suggestive of neutral evolution (dN/dS = 1) or positive selection (dN/dS > 1) across the gene family (Table 2). For reference, primate genes have an average whole-gene dN/dS of ∼0.25 (Gibbs et al. 2007).
Table 2.
Whole-gene dN/dS comparisons reveal that APOL genes have rapidly evolved in primates of diverse ancestry
aComparisons marked n.a. (not available) were excluded for the following reasons: The gene does not exist in the current genome assembly (chimpanzee APOL1 and APOL4, macaque APOL3, marmoset APOL1 and APOL4), the gene is a putative pseudogene (macaque APOL1 and APOL5, marmoset APOL2), or the gene is not completely sequenced in the current genome assembly (marmoset APOL5).
bNumbers in parentheses indicate the numbers of nonsynonymous and synonymous changes, respectively, between orthologs.
cThe orthology of marmoset APOL3/L4 is unclear. Since it is nearly equally related to human APOL3 and APOL4, we arbitrarily chose to compare it to APOL3.
Next, we tested APOL genes for positive selection with sequences we had obtained during our phylogenetic analysis (see Fig. 2). We combined the analysis of APOL1, APOL2, and APOL2.1 for two reasons. First, phylogenetic analysis showed that these genes shared a very recent common ancestor, some time during primate evolution. Second, we had a relatively small number of sequences for each gene, and additional sequences provided more power to detect positive selection. We combined the analysis of APOL3 and APOL4 for the same reasons. A potential cost to combining the analysis of multiple genes is that gene-specific adaptations will be mixed and possibly be overlooked. We did not test APOL5 because our data consisted mostly of pseudogenes.
We applied multiple maximum likelihood-based tests for evidence of positive selection. With the PAML software package, we found evidence for positive selection in each of our three data sets: APOL1/L2/L2.1, APOL3/L4, and APOL6 (Table 3). With the exception of the N-terminal region of APOL1/L2/L2.1, each gene region tested was also significant. We also tested for positive selection using the more conservative tool PARRIS, which takes into account synonymous rate variation, after accounting for gene conversion with a genetic algorithm for recombination detection (GARD). Using this test, we found evidence for positive selection using PARRIS in APOL3/L4 (P = 0.015) and APOL6 (P = 3.9 × 10−5), but not in APOL1/L2/L2.1.
Table 3.
Evidence for positive selection in primate APOL1/L2/L2.1, APOL3/L4, and APOL6
aWe used maximum-likelihood analysis to estimate whether positive selection had acted on APOL1, APOL2, and APOL2.1 (combined), APOL3 and APOL4 (combined), and APOL6. For APOL1/L2/L2.1 and APOL3/L4, we analyzed the large exon of each gene, representing ∼70%–85% of the coding sequence of each gene (e.g., the APOL2 large exon is 877 bp of the total 1014 bp). For APOL6, we analyzed the entire gene except a repetitive region in the final 19 codons. The individual genes and species analyzed are shown in Figure 2 (APOL1/L2/L2.1 and APOL3/L4) and Figure 3 (APOL6), excluding pseudogenes. Since putative recombination breakpoints were found in APOL1/L2/L2.1 (two breakpoints) and APOL3/L4 (one breakpoint), we analyzed the resulting gene segments in separate PAML analyses. We also performed domain-specific analyses of APOL6.
bCoordinates are given for human APOL1 isoform a (total length, 399 amino acids), human APOL3 isoform 1 (403 amino acids), and human APOL6 (344 amino acids). P is the proportion of codons predicted to evolve with rate dN/dS (for instance, for APOL6 under the comparison of M1 vs. M2 models, 20% of sites are predicted to evolve with an average dN/dS of 3.61); 2 ln lambda is twice the log likelihood difference between nested models.
cWe show results from the comparisons of nested PAML models M1 vs. M2 and M7 vs. M8. In both comparisons, there are two degrees of freedom between models. Bolded P-values are <0.05.
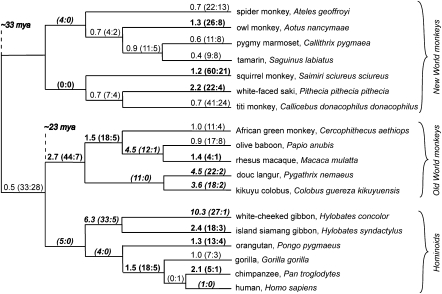
The strongest signature of positive selection was in APOL6, with highly significant findings in PAML analyses of both whole-gene sequences and individual protein domains (see Table 3). To test whether positive selection might be confined to particular primate lineages, we tested NWMs separately from OWMs and Hominoids. In both groupings, we again found evidence for positive selection (using PAML; data not shown). To expand on these findings, we estimated dN/dS on each branch of the primate phylogeny. We found lineages with dN/dS greater than one in each of Hominoids, OWMs, and NWMs (Fig. 3). Clearly, APOL6 experienced strong positive selection throughout simian primate evolution.
Figure 3.
Lineage-specific evidence for positive selection in primate APOL6. A cladogram is shown with maximum-likelihood estimates of lineage-specific dN/dS during primate evolution. Numbers in parentheses are the estimated number of nonsynonymous and synonymous changes, respectively, for each branch. Values of dN/dS are also colored according to the class of dN/dS to which they are predicted to belong (by GABranch analysis): Bolded values belong to class dN/dS = 1.49; bolded, italicized values belong to class dN/dS = 5.92; and nonbolded values belong to class dN/dS = 0.63. Branch lengths in the figure are not proportional to time.
We also asked whether APOL6 showed evidence of positive selection during recent human evolution. To address this question, we sequenced the large exon of APOL6 and neighboring APOL5 in a panel of 24 African Americans. For each gene, we calculated the summary statistic Tajima's D (Table 4; Supplemental Fig. S1). While both genes showed a negative value for Tajima's D (indicating an excess of rare alleles) only APOL6 had a significantly negative D (D = −1.82, P < 0.05). This finding suggested that positive selection indeed acted on APOL6 during recent human evolution. However, since a negative D can also be explained by an expansion in the size of the human population following a recent bottleneck event, we compared the polymorphism and divergence (with chimpanzee orthologs) of the APOL5 and APOL6 genes, using the Hudson–Kreitman–Aguadé (HKA) test (Hudson etal. 1987). We found a significant departure from the neutral mutation hypothesis (see Supplemental Table S2), indicating that the excess of rare polymorphisms in APOL6 was likely due to positive selection, and not demography.
Table 4.
Test statistics for natural selection from human variation in the large exons of APOL5 and APOL6 from 24 African Americans
aThe coding sequence of each large exon was analyzed. Bolded values in the table indicate statistical significance with P < 0.05.
bS indicates segregating sites. In each gene tested, S is also equal to the number of mutations.
chs indicates the number of singleton mutations.
dPi indicates the average number of nucleotide differences between pairs of sequences per site.
eθ indicates the effective population size multiplied by the mutation rate per generation per site.
The APOL6 MAD has a group of rapidly evolving codons that tightly cluster on one face of an alpha helix
We analyzed the particular codons of APOL genes that were predicted to be under positive selection. In APOL1/L2/L2.1, seven of the eight rapidly changing codons were in the SID or just N-terminal to it, which is the region where the trypanosome antagonist SRA interacts with APOL1 (Fig. 4; Supplemental Table S1). In APOL3/L4, one rapidly changing codon was in the MAD, and three were in the region just N-terminal to the SID. And in APOL6, there were rapidly evolving codons throughout the gene. We tested if positive selection acted differently on different APOL6 domains. The MAD, relative to the pore domain and SID, appears to have a higher proportion of codons under positive selection and has experienced stronger overall positive selection (Supplemental Fig. S2). Also in the APOL6 MAD, we found a cluster of five codons that had strong statistical support for positive selection. These five residues appear to be significantly clustered in analyses of either the whole gene (P = 0.004) or the MAD region alone (P = 0.003).
Figure 4.
Codons under positive selection in primate APOL genes, and a cluster of rapidly evolving codons on an APOL6 MAD alpha helix. (A) We used maximum-likelihood-based tests to estimate positive selection in APOL1, APOL2, and APOL2.1 (combined); APOL3 and APOL4 (combined); and APOL6. We did not analyze APOL5 due to lack of available sequences. The genes are shown to scale and are aligned with one another. The dotted lines show two codons that are under positive selection in both APOL1/L2/L2.1 and APOL6. Gene coordinates and amino acids are shown for human genes, and the isoforms shown include APOL1 isoform a and APOL3 isoform1. APOL2.1 is not shown in the figure. Asterisks indicate stop codons. (B) A group of rapidly evolving codons in the APOL6 MAD tightly cluster on one face of an alpha helix, the predicted structure for the region. Rapidly evolving codons are shown in bold. Sites are also shaded according to hydrophobicity, and the helix has 3.6 amino acids per turn. The rapidly evolving sites of the MAD are located at the base of a pH-sensitive hinge structure. The helix appears to be amphipathic, and rapidly evolving sites appear to be at the hydrophobic/hydrophilic interface of the helix. MAD: membrane-addressing domain; SID: SRA-interacting domain.
We mapped the cluster of rapidly evolving codons of the APOL6 MAD onto a predicted protein structure. These codons (codons 165–177) were predicted by prior studies to adopt an alpha helix (Vanhamme et al. 2003; Perez-Morga et al. 2005). We confirmed this prediction with multiple secondary-structure prediction programs (data not shown). We found that the rapidly evolving codons of the APOL6 MAD were tightly clustered on one face of the alpha helix structure (Fig. 4; Supplemental Fig. S3). At first glance, the distributions of rapidly evolving codons appeared spaced apart (see Fig. 4). However, once the region was folded in silico, the rapidly evolving sites were tightly clustered along one side of the helix. The MAD helix was also clearly amphipathic, with the rapidly evolving codons at the interface of the hydrophobic and hydrophilic faces. The locations of these codons within the MAD were also conspicuous—at the very base of the MAD “hinge” structure, which in APOL1 functions as a pH sensor (for model, see Discussion). In the absence of additional structural information, it is unclear whether the codons are oriented on the inner fold of the closed hinge or exposed on the outside of the hinge. We also found a similar group of rapidly evolving codons in the APOL6 SID and adjacent N-terminal region (codons 236–251), but these codons did not meet the criteria for significant clustering (Supplemental Figs. S4, S5).
Discussion
Our study of APOL gene evolution was motivated by the observation that APOL1 is a trypanosome immunity factor that can be antagonized by trypanosome SRA. We hypothesized that this host–pathogen interaction was just one instance of similar conflicts that affected the evolution of APOL genes throughout primate evolution. Indeed, we found that APOL1, and surprisingly other APOL genes that encode exclusively intracellular proteins, have rapidly evolved, in many cases in the same protein region that SRA binds APOL1. However, whereas APOL1 may have rapidly evolved as a result of interactions with antagonists like SRA, the cause of positive selection on other APOL genes is less clear. We favor the hypothesis that other APOL genes also rapidly evolved as a result of interactions with antagonists. The primary difference between secreted APOL1 and other APOL proteins, however, is that other APOL proteins would interact with antagonists inside of host cells.
Antagonists like SRA are a common strategy that pathogens use to enhance infections. Antagonists of host PCD proteins are especially common in viruses, which presumably benefit from preventing, or even causing, cell death (Roulston et al. 1999; Hay and Kannourakis 2002). For example, some viruses encode proteins that are structural mimics of primate BCL2, a pro-survival protein. Viruses use vBcl-2 proteins to inhibit cell death, in part by inhibiting pro-death BH3-only proteins like the APOL genes, to prolong the infection of a cell (Cuconati and White 2002). Other viruses encode antagonists to cause PCD, which allows them to spread to other cells in post-PCD membrane-bound vesicles (Roulston et al. 1999). Pathogenic bacteria and parasites also encode cell-death antagonists, notably bacteria of the genus Chlamydia that broadly antagonize BH3-only proteins (Fischer et al. 2004).
Based on our findings, we predict there are at least two types of antagonists of APOL proteins, each with distinct mechanisms: SID antagonists and MAD antagonists (Fig. 5). We predict that there are additional SID antagonists, of which SRA is one example, that interact with the SIDs of various APOL proteins. In each of our comparisons of APOL genes—APOL1/L2/L2.1, APOL3/L4, and APOL6—we found rapidly evolving codons in the SID or in the adjacent N-terminal region. In APOL1, this observation fits our expectation that interactions like APOL1-SRA have over time led to positive selection. But there appear to be more SID antagonists like SRA that are currently unknown that affect APOL proteins on the inside of host cells.
Figure 5.
Cartoon model for how MAD and SID antagonists use distinct mechanisms to cause or prevent APOL6-mediated PCD of host cells. (A, i) In the absence of pathogen antagonists, we predict that the APOL6 MAD functions like the APOL1 MAD during trypanosome infections. That is, at pH 7 in an endosome, the MAD hinge is in closed conformation with salt bridges between adjacent alpha helices. At pH 5 in an endolysosome, the hinge opens and allows APOL1 to dissociate from HDL particles to form a pore in the lysosomal membrane, killing the trypanosome. An important difference between APOL1 and APOL6 is that secreted APOL1 kills trypanosomes, whereas APOL6 is not secreted and induces apoptosis of host cells. (ii) We predict that pathogen antagonists manipulate APOL6 function by interacting at the base of the MAD hinge—a region under strong positive selection (shown in dark gray). By recognizing this critical region, antagonists may be able to manipulate APOL6 in different ways. One possibility is that antagonists promote infections by preventing APOL6-mediated apoptosis. In this case, antagonists interact with the MAD hinge to keep it from opening (shown in figure), prolonging infection of the cell. Alternately, antagonists may further infections by prematurely causing apoptosis. In this case, antagonists could interact with the MAD base to open it, initiating apoptosis prematurely (not shown in figure). (B) The trypanosome antagonist SRA binds APOL1 to prevent APOL1-mediated trypanosome lysis during infections. The same region of APOL1 that SRA binds, the SID, is under positive selection in APOL6 (shown in dark gray) and other APOL genes. Hence, we predict that additional SID antagonists, like SRA but distinct in mechanism from MAD antagonists, interact with the SID to block APOL-mediated cell death of host cells during infections.
SRA and other predicted SID antagonists affect the functions of host proteins differently. SRA prevents pathogen death, while SID antagonists of intracellular APOL proteins would prevent host cell death. Hence, we expect to find these SID antagonists encoded in intracellular pathogens, which would benefit from inhibiting PCD. Our conclusions lend further support to recent findings (Molina-Portela et al. 2008) that the SID domain must carry out an essential role in APOL function, else its antagonism by pathogens would have resulted in simple deletions of the SID domains.
Interestingly, the specific interaction of APOL1 and SRA appears to be relatively “new.” The secreted form of APOL1, critical for trypanolysis, is an evolutionary innovation via novel alternate splicing in primates. In this way, a gene that functioned in host cell death, possibly as an intracellular defense during infections, became co-opted to kill pathogens directly. SRA also appears to be the product of an evolutionary innovation in trypanosomes, in which a VSG gene encoding a cell-surface antigen underwent a 378-bp deletion to become an APOL1 antagonist (De Greef and Hamers 1994; Campillo and Carrington 2003). Although the interaction of APOL1 and SRA is novel in primates, it serves as a model for how predicted SID antagonists have likely interacted with APOL proteins throughout simian primate evolution.
Next, we predict that pathogens encode MAD antagonists to specifically interact with the MAD of APOL6. Unlike other APOL genes, APOL6 was the only APOL gene with multiple rapidly evolving codons in the MAD and was not pseudogenized in any species that we tested. These observations suggest that APOL6 has a function that is necessary and distinct from the other APOL genes and may encounter a novel class of antagonist.
The rapidly evolving codons in the APOL6 MAD tightly cluster on one face an alpha helix. In other immunity genes, similar clusters interact directly with pathogen proteins. For example, in the primate antiviral gene TRIM5 the viral-capsid specificity-determining domain is a tightly clustered region of 13 amino acids under strong positive selection (Sawyer et al. 2005). The rapidly evolving codons in the APOL6 MAD are positioned in a very curious region—at the base of the hinge. In APOL1, the MAD adopts a hinge structure that opens when the surrounding pH changes from 7 to 5, during trafficking from endosome to lysosome. How might MAD antagonists affect APOL6 by binding the hinge base? One possibility is that antagonists bind the hinge base to lock the hinge closed (see Fig. 5). This would likely keep APOL6 from committing its sole known function—initiating apoptosis. Alternately, MAD antagonists may bind the hinge base to open it early, perhaps by cleaving it, causing the cell to initiate apoptosis prematurely. Since the hinge base has rapidly evolved throughout simian primate evolution, we would expect to find MAD antagonists in the pathogens of many primate species.
Other regions of APOL genes, in addition to the MAD and SID, have also changed rapidly. We identified rapidly evolving codons in the APOL6 pore-forming domain and in the region between the MAD and SID in multiple APOL genes (see Fig. 4). APOL proteins may also interact with antagonists in these regions, implying that pathogens use diverse mechanisms to interact with APOL proteins.
The functions of APOL proteins inside host cells are still largely unknown. They are capable of causing PCD by both the intrinsic apoptosis pathway (APOL6) and by autophagic death (APOL1). How these deaths are regulated is not yet known. One possibility is that APOL proteins directly sense elements of infections and trigger PCD as a defense. Alternately, APOL proteins may have a housekeeping role, like other BH3-only proteins, and have simply been co-opted by pathogen antagonists to cause or prevent PCD during infections. For example, pathogens like the bacteria Francisella tularensis may use the APOL proteins to escape lysosomal degradation (Checroun et al. 2006). A final alternative is that APOL proteins kill intracellular pathogens directly, especially those confined to specific intracellular compartments with host cells.
We have shown that the APOL gene family experienced dynamic evolution in simian primates. These events, however, are not confined to the past since modern-day primate populations may harbor important polymorphisms in APOL genes. We found that APOL6 may have experienced adaptive sweeps during recent human evolution. In APOL1, loss of function alleles have already been shown to be clinically important, where homozygous loss conferred susceptibility to Trypanosoma evansi infections (Vanhollebeke et al. 2006). The history of strong positive selection on APOL genes suggests that variants of APOL proteins may be clinically important because they confer resistance, or susceptibility, to modern-day human diseases.
Methods
Primate DNA sequences
GenBank accession numbers for sequences generated in this study are FJ177967–FJ178000.
We obtained the DNA sequences and annotation of human APOL genes from manually annotated entries in GenBank (Lander et al. 2001; Benson et al. 2008). We used the BLAT tool to obtain sequences from whole-genome assemblies in the UCSC genome browser database (http://genome.ucsc.edu/) (Kent 2002; Karolchik et al. 2008) for the following primate species (assembly dates in parentheses): chimpanzee, Pan troglodytes (March 2006) (Chimpanzee Sequencing and Analysis Consortium 2005); orangutan, Pongo pygmaeus (July 2007) (data not shown); rhesus macaque, Macaca mulatta (Jan 2006) (Gibbs et al. 2007); white-tufted-ear marmoset Callithrix jacchus (June 2007) (data not shown). We also obtained sequences from shotgun-sequenced BAC clones from GenBank for these species: baboon, Papio hamadryas; owl monkey, Aotus nancymaae; and ring-tailed lemur, Lemur catta (data not shown). We obtained sequences from assembling reads from trace archives in GenBank for lowland gorilla, Gorilla gorilla; and baboon, Papio hamadryas (data not shown). Our laboratory also obtained DNA or RNA for sequencing from the Integrated Primate Biomaterials and Information Resource (IPBIR) (Coriell Institute for Medical Research, Camden, NJ) for the following species: island siamang gibbon, Hylobates syndactylus; white-cheeked gibbon, Hylobates concolor; olive baboon, Papio anubis (APOL6 only); patas monkey, Erythrocebus patas; African green monkey, Cercopithecus aethiops; douc langur, Pygathrix nemaeus; kikuyu colobus, Colobus guereza kikuyuensis; purple-faced langur, Trachypithecus vetulus vetulus; black-handed spider monkey, Ateles geoffroyi; Bolivian gray titi monkey, Callicebus donacophilus donacophilus; pygmy marmoset, Callithrix pygmaea; red-chested mustached tamarin, Saguinus labiatus; squirrel monkey, Saimiri sciureus sciureus; and white-faced saki, Pithecia pithecia pithecia.
DNA from 24 African Americans was obtained from a Human Variation Panel from Coriell Institute for Medical Research, catalog no. HD24AA.
Primate gene sequencing and assembly
Genes were amplified by polymerase chain reaction (PCR) using the PCR Super Mix HiFi Kit (Invitrogen). Reverse-transcriptase PCR was also performed for some genes using the SuperScript III RT/Platinum Taq Kit (Invitrogen). We cloned some gene fragments using the TA-cloning vector pCR2.1 (Invitrogen) prior to sequencing. All sequencing was performed by the Sanger method. Base calling was performed with phred, sequence assembly with Phrap, and viewing with Consed (Ewing and Green 1998; Ewing et al. 1998; Gordon 2003). After assembly, we analyzed only those bases that had phred quality scores of at least q20 (<1% chance error). We detected human heterozygotes using PolyPhred version 6.15 (Nickerson et al. 1997) and required at least twice redundant read coverage at each nucleotide position. All polymorphisms were confirmed by visual inspection.
Evolutionary analyses
Whole-gene values for dN/dS were calculated by the method of Comeron (1995) with K-estimator (Comeron 1999). For genes with more than one known isoform, whole-gene dN/dS analysis was performed on the following isoforms: APOL1 isoform a, APOL3 isoform 3, and APOL4 isoform 1. The numbers of nonsynonymous and synonymous changes in primate lineages were estimated using maximum likelihood analysis in PAML (Yang 1997).
Multiple alignments were made with ClustalW (Higgins and Sharp 1988) and checked manually for proper alignment (for alignments, see Supplemental Figs. S6–S9). Phylogenetic trees were estimated by Bayesian inference using Mr Bayes (Huelsenbeck and Ronquist 2001). The program was run for 1 million generations and samples taken every 100 generations. We used the following settings: General Time Reversible (GTR) evolutionary model, gamma-distributed rate variation across sites with a proportion of invariable sites, and default settings for prior distributions. All other settings were defaults. Software programs were run locally or using the website Mobyle (mobyle.pasteur.fr).
Predictions of individual codons that evolved under positive selection were performed by maximum likelihood using nssites model comparisons in PAML. To identify positive selection, we required a significant difference in log likelihoods, by chi-squared testing, between nested PAML models M1 and M2, and M7 and M8. Model M1 allows codons to belong to one of two categories of dN/dS, one with dN/dS < 1 and the other with dN/dS = 1. Model M2 is the same as M1 except that it adds a third category with dN/dS > 1. Model M7 allows codons to have dN/dS values according to a beta distribution (two parameters). Model M8 is the same as M7 except that it adds a discrete category of dN/dS of with dN/dS > 1. Codons predicted to be under positive selection were identified by Bayes-Empirical-Bayes (BEB) analysis. We required that rapidly evolving codons be predicted in both M1 vs. M2 and M7 vs. M8 model comparisons with P-values of greater than 0.95. All P-values are reported from BEB analysis using model M2 because this combination provided the most conservative estimates of codons that experienced strong positive selection. Lineage-specific evidence for positive selection was performed using free-ratio analysis in PAML.
We also used the HyPhy software package (www.datamonkey.org) to estimate positive selection (Pond et al. 2005). First, we used the GARD tool to estimate if gene conversion or recombination had occurred in test sequences (Kosakovsky Pond et al. 2006). Our parameters to GARD included the HKY85 nucleotide substitution bias model, a beta-gamma distribution for rate variation, and three rate classes. If gene conversion was predicted, we tested each gene segment between putative breakpoints for positive selection, using a phylogenetic tree constructed specifically for that segment. To identify particular codons that evolved under positive selection, we used FEL and REL with the HKY85 model. To identify whole-gene evidence for positive selection, we used PARRIS with the HKY85 model. To estimate lineage-specific values of dN/dS we used GABranch (Pond and Frost 2005). We tested for differences in positive selection between APOL6 domains with the HyPhy batch file dNdSDistributionComparisons.bf. We used default values as starting parameters (shown in Supplemental Fig. S2), and obtained similar results using random starting values.
We used DnaSP to calculate test statistics for natural selection from human variation data (Rozas et al. 2003). Tajima's D measures the difference between the number of segregating sites in a sequence alignment and the average number of nucleotide differences between pairs of sequences (Tajima 1989). We also used DnaSP to perform the HKA test (Hudson et al. 1987), in which we tested all substitutions between compared genes.
To measure the statistical significance of clusters of rapidly evolving codons, we used a permutation test. For the cluster in the APOL6 MAD, we measured the probability that five rapidly evolving codons out of 12 would cluster within 13 sites from a total of 325 sites. This cluster was also significant if we only analyzed the MAD (five of five codons clustered within 13 sites from a total of 67 sites). These tests assumed that all sites tested were equally likely to have experienced positive selection.
Protein secondary structures were predicted using the software Phyre (Bennett-Lovsey et al. 2008).
Acknowledgments
We thank David Hockenbery for helpful discussions. We thank Sara Sawyer and members of the Malik laboratory for feedback on our manuscript. We thank Janet Young for help with software. This study was funded by a post-doctoral fellowship from the SpinOdyssey Foundation and the American Cancer Society (no. PF-07-260-01-DDC, E.E.S.) and a grant from the President's Circle Foundation of the Fred Hutchinson Cancer Research Center. H.S.M. was supported by a Burroughs Wellcome Fund Investigator Award in Pathogenesis Award and a Searle Scholar Award.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. FJ177967–FJ178000.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.085647.108.
References
- Ahn W.S., Bae S.M., Lee J.M., Namkoong S.E., Han S.J., Cho Y.L., Nam G.H., Seo J.S., Kim C.K., Kim Y.W. Searching for pathogenic gene functions to cervical cancer. Gynecol. Oncol. 2004;93:41–48. doi: 10.1016/j.ygyno.2003.11.031. [DOI] [PubMed] [Google Scholar]
- Bennett-Lovsey R.M., Herbert A.D., Sternberg M.J., Kelley L.A. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campillo N., Carrington M. The origin of the serum resistance associated (SRA) gene and a model of the structure of the SRA polypeptide from Trypanosoma brucei rhodesiense. Mol. Biochem. Parasitol. 2003;127:79–84. doi: 10.1016/s0166-6851(02)00306-7. [DOI] [PubMed] [Google Scholar]
- Checroun C., Wehrly T.D., Fischer E.R., Hayes S.F., Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Comeron J.M. A method for estimating the numbers of synonymous and nonsynonymous substitutions per site. J. Mol. Evol. 1995;41:1152–1159. doi: 10.1007/BF00173196. [DOI] [PubMed] [Google Scholar]
- Comeron J.M. K-Estimator: Calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics. 1999;15:763–764. doi: 10.1093/bioinformatics/15.9.763. [DOI] [PubMed] [Google Scholar]
- Cuconati A., White E. Viral homologs of BCL-2: Role of apoptosis in the regulation of virus infection. Genes & Dev. 2002;16:2465–2478. doi: 10.1101/gad.1012702. [DOI] [PubMed] [Google Scholar]
- De Greef C., Hamers R. The serum resistance-associated (SRA) gene of Trypanosoma brucei rhodesiense encodes a variant surface glycoprotein-like protein. Mol. Biochem. Parasitol. 1994;68:277–284. doi: 10.1016/0166-6851(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Degterev A., Yuan J. Expansion and evolution of cell death programmes. Nat. Rev. Mol. Cell Biol. 2008;9:378–390. doi: 10.1038/nrm2393. [DOI] [PubMed] [Google Scholar]
- Dombkowski A.A., Cukovic D., Novak R.F. Secretome analysis of microarray data reveals extracellular events associated with proliferative potential in a cell line model of breast disease. Cancer Lett. 2006;241:49–58. doi: 10.1016/j.canlet.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Duchateau P.N., Pullinger C.R., Orellana R.E., Kunitake S.T., Naya-Vigne J., O'Connor P.M., Malloy M.J., Kane J.P. Apolipoprotein L, a new human high density lipoprotein apolipoprotein expressed by the pancreas. Identification, cloning, characterization, and plasma distribution of apolipoprotein L. J. Biol. Chem. 1997;272:25576–25582. doi: 10.1074/jbc.272.41.25576. [DOI] [PubMed] [Google Scholar]
- Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M.C., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fischer S.F., Vier J., Kirschnek S., Klos A., Hess S., Ying S., Hacker G. Chlamydia inhibit host cell apoptosis by degradation of proapoptotic BH3-only proteins. J. Exp. Med. 2004;200:905–916. doi: 10.1084/jem.20040402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs R.A., Rogers J., Katze M.G., Bumgarner R., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Glazko G.V., Nei M. Estimation of divergence times for major lineages of primate species. Mol. Biol. Evol. 2003;20:424–434. doi: 10.1093/molbev/msg050. [DOI] [PubMed] [Google Scholar]
- Gordon D. Viewing and editing assembled sequences using Consed. Curr. Protoc. Bioinformatics. 2003;11.2 doi: 10.1002/0471250953.bi1102s02. [DOI] [PubMed] [Google Scholar]
- Green H.P., Del Pilar Molina Portela M., St Jean E.N., Lugli E.B., Raper J. Evidence for a Trypanosoma brucei lipoprotein scavenger receptor. J. Biol. Chem. 2003;278:422–427. doi: 10.1074/jbc.M207215200. [DOI] [PubMed] [Google Scholar]
- Hay S., Kannourakis G. A time to kill: Viral manipulation of the cell death program. J. Gen. Virol. 2002;83:1547–1564. doi: 10.1099/0022-1317-83-7-1547. [DOI] [PubMed] [Google Scholar]
- Hayashi J., Stoyanova R., Seeger C. The transcriptome of HCV replicon expressing cell lines in the presence of alpha interferon. Virology. 2005;335:264–275. doi: 10.1016/j.virol.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Higgins D.G., Sharp P.M. CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Hudson R.R., Kreitman M., Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Karolchik D., Kuhn R.M., Baertsch R., Barber G.P., Clawson H., Diekhans M., Giardine B., Harte R.A., Hinrichs A.S., Hsu F., et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond S.L., Posada D., Gravenor M.B., Woelk C.H., Frost S.D. GARD: A genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Liu Z., Lu H., Jiang Z., Pastuszyn A., Hu C.A. Apolipoprotein l6, a novel proapoptotic Bcl-2 homology 3-only protein, induces mitochondria-mediated apoptosis in cancer cells. Mol. Cancer Res. 2005;3:21–31. [PubMed] [Google Scholar]
- Lugli E.B., Pouliot M., Portela Mdel P., Loomis M.R., Raper J. Characterization of primate trypanosome lytic factors. Mol. Biochem. Parasitol. 2004;138:9–20. doi: 10.1016/j.molbiopara.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Mikkelsen T.S., Hillier L.W., Eichler E.E., Zody M.C., Jaffe D.B., Yang S., Endard W., Waterston R.H. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Mimmack M.L., Ryan M., Baba H., Navarro-Ruiz J., Iritani S., Faull R.L., McKenna P.J., Jones P.B., Arai H., Starkey M., et al. Gene expression analysis in schizophrenia: Reproducible up-regulation of several members of the apolipoprotein L family located in a high-susceptibility locus for schizophrenia on chromosome 22. Proc. Natl. Acad. Sci. 2002;99:4680–4685. doi: 10.1073/pnas.032069099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: Process and function. Genes & Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Molina-Portela M.P., Samanovic M., Raper J. Distinct roles of apolipoprotein components within the trypanosome lytic factor complex revealed in a novel transgenic mouse model. J Exp Med. 2008;205:1721–1728. doi: 10.1084/jem.20071463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monajemi H., Fontijn R.D., Pannekoek H., Horrevoets A.J. The apolipoprotein L gene cluster has emerged recently in evolution and is expressed in human vascular tissue. Genomics. 2002;79:539–546. doi: 10.1006/geno.2002.6729. [DOI] [PubMed] [Google Scholar]
- Nickerson D.A., Tobe V.O., Taylor S.L. PolyPhred: Automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe T., Ohmori Y., Tanigami A., Hishigaki H., Suzuki Y., Sugano S., Kawaguchi A., Nakaya H., Wakitani S. Detection of gene expression in synovium of patients with osteoarthritis using a random sequencing method. Acta Orthop. 2007;78:687–692. doi: 10.1080/17453670710014400. [DOI] [PubMed] [Google Scholar]
- Page N.M., Butlin D.J., Lomthaisong K., Lowry P.J. The human apolipoprotein L gene cluster: Identification, classification, and sites of distribution. Genomics. 2001;74:71–78. doi: 10.1006/geno.2001.6534. [DOI] [PubMed] [Google Scholar]
- Perez-Morga D., Vanhollebeke B., Paturiaux-Hanocq F., Nolan D.P., Lins L., Homble F., Vanhamme L., Tebabi P., Pays A., Poelvoorde P., et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science. 2005;309:469–472. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- Poelvoorde P., Vanhamme L., Van Den Abbeele J., Switzer W.M., Pays E. Distribution of apolipoprotein L-I and trypanosome lytic activity among primate sera. Mol. Biochem. Parasitol. 2004;134:155–157. doi: 10.1016/j.molbiopara.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Pond S.L., Frost S.D. A genetic algorithm approach to detecting lineage-specific variation in selection pressure. Mol. Biol. Evol. 2005;22:478–485. doi: 10.1093/molbev/msi031. [DOI] [PubMed] [Google Scholar]
- Pond S.L., Frost S.D., Muse S.V. HyPhy: Hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Purvis A. A composite estimate of primate phylogeny. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1995;348:405–421. doi: 10.1098/rstb.1995.0078. [DOI] [PubMed] [Google Scholar]
- Roulston A., Marcellus R.C., Branton P.E. Viruses and apoptosis. Annu. Rev. Microbiol. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- Rozas J., Sanchez-DelBarrio J.C., Messeguer X., Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Sana T.R., Janatpour M.J., Sathe M., McEvoy L.M., McClanahan T.K. Microarray analysis of primary endothelial cells challenged with different inflammatory and immune cytokines. Cytokine. 2005;29:256–269. doi: 10.1016/j.cyto.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Sawyer S.L., Wu L.I., Emerman M., Malik H.S. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed J.R., Sechelski J.B., Loomis M.R. A survey for a trypanocidal factor in primate sera. J. Protozool. 1990;37:393–400. doi: 10.1111/j.1550-7408.1990.tb01163.x. [DOI] [PubMed] [Google Scholar]
- Stojdl D.F., Lichty B.D., tenOever B.R., Paterson J.M., Power A.T., Knowles S., Marius R., Reynard J., Poliquin L., Atkins H., et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 2003;4:263–275. doi: 10.1016/s1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Strasser A. The role of BH3-only proteins in the immune system. Nat. Rev. Immunol. 2005;5:189–200. doi: 10.1038/nri1568. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhamme L., Paturiaux-Hanocq F., Poelvoorde P., Nolan D.P., Lins L., Van Den Abbeele J., Pays A., Tebabi P., Van Xong H., Jacquet A., et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003;422:83–87. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- Vanhollebeke B., Pays E. The function of apolipoproteins L. Cell. Mol. Life Sci. 2006;63:1937–1944. doi: 10.1007/s00018-006-6091-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhollebeke B., Truc P., Poelvoorde P., Pays A., Joshi P.P., Katti R., Jannin J.G., Pays E. Human Trypanosoma evansi infection linked to a lack of apolipoprotein L-I. N. Engl. J. Med. 2006;355:2752–2756. doi: 10.1056/NEJMoa063265. [DOI] [PubMed] [Google Scholar]
- Vanhollebeke B., De Muylder G., Nielsen M.J., Pays A., Tebabi P., Dieu M., Raes M., Moestrup S.K., Pays E. A haptoglobin–hemoglobin receptor conveys innate immunity to Trypanosoma brucei in humans. Science. 2008;320:677–681. doi: 10.1126/science.1156296. [DOI] [PubMed] [Google Scholar]
- Wan G., Zhaorigetu S., Liu Z., Kaini R., Jiang Z., Hu C.A. Apolipoprotein L1, a novel Bcl-2 homology domain 3-only lipid-binding protein, induces autophagic cell death. J. Biol. Chem. 2008;283:21540–21549. doi: 10.1074/jbc.M800214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xong H.V., Vanhamme L., Chamekh M., Chimfwembe C.E., Van Den Abbeele J., Pays A., Van Meirvenne N., Hamers R., De Baetselier P., Pays E. A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell. 1998;95:839–846. doi: 10.1016/s0092-8674(00)81706-7. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]