Abstract
Differentiation of bone-resorbing osteoclasts from hematopoietic precursors depends upon expression of the cytokine receptor activator of NFκB ligand (RANKL) by fibroblastic stromal cells, which some evidence suggests are of the osteoblast lineage. We have shown previously that hormonal-responsiveness of the murine RANKL gene is mediated in part by a distal enhancer that binds Runx2, a transcription factor required for commitment to the osteoblast lineage, supporting the idea that osteoclast-supporting stromal cells may be osteoblasts or their progenitors. However, in this study we demonstrate that parathyroid hormone (PTH) stimulation of RANKL in mice is not affected by a significant reduction in the number of osteoblasts. Consistent with this, neither Runx2, nor Cbfb, a binding partner essential for Runx activity, are required for basal or PTH-stimulated RANKL expression in fibroblastic stromal cell models. Nonetheless, RANKL responsiveness to PTH was elevated in cultured calvaria cells expressing high levels of osterix, another transcription factor required for osteoblast differentiation, and this was associated with elevated PTH receptor expression. The responsiveness of RANKL to 1,25-dihydroxyvitamin D3 was not elevated in the osterix-expressing cells. Together, these results suggest that commitment to the osteoblast lineage is not a requirement for RANKL gene transcription in fibroblastic stromal cells but may enhance responsiveness of this gene to specific hormones via control of their receptors.
In the adult skeleton, bone is constantly renewed via the coordinated activity of osteoclasts that resorb bone and osteoblasts that form bone. These cells function within an anatomically distinct structure known as the basic multicellular unit (BMU), in which the osteoclasts are located in the lead and are followed by osteoblasts (1). Because of this organization, bone formation occurs only at sites of prior bone resorption and the recruitment of osteoblasts to such sites is known as coupling. While the mechanisms that underlie coupling are unknown, two different explanations have been proposed. According to the first, release of factors, such as TGFβ, from the bone matrix as a consequence of osteoclast activity recruits osteoblast progenitors and promotes their differentiation (2). Thus, osteoblastogenesis in this “serial” pathway of coupling is a consequence of osteoclastogenesis (3). However, in view of the fact that osteoclastogenesis depends upon support from stromal cells that may be of the osteoblastic lineage, the existence of a parallel pathway has been proposed (4). According to the parallel pathway model, osteoclast and osteoblast differentiation occur simultaneously due in part to the requirement of osteoblast lineage cells for osteoclast differentiation.
The idea that osteoblast lineage cells are required for osteoclast differentiation originated from studies showing that osteoblasts or osteoblast-like cells, not osteoclast precursors, are targets of hormones that stimulate bone resorption (5–7). It has since been demonstrated that these hormones stimulate osteoclast differentiation by acting directly on stromal cells to stimulate expression of receptor activator of NFκB ligand (RANKL),3 suppress expression of the RANKL antagonist osteoprotegerin (OPG), or both (8, 9). However, whether the stromal cells that are the targets of these hormones are in fact osteoblasts or osteoblast precursors is unclear. One reason for this uncertainty is that the calvaria and bone marrow stromal cell cultures commonly used to study osteoblastic cells contain many cell types, including fibroblastic cells that may not be of the osteoblastic lineage (10). In addition, analysis of RANKL expression during osteoblast differentiation in vitro has produced conflicting results with differentiation both promoting (11) and inhibiting (12) RANKL expression. Attempts to identify RANKL-expressing cells in remodeling bone using histologic methods have also produced inconsistent results (8, 13–15). More importantly, conditional ablation of osteoblasts in transgenic mice did not alter osteoclast numbers or bone resorption (16), and many mouse models with increased osteoblast number do not exhibit increased osteoclast number (17–20). Therefore, it remains unclear whether matrix-synthesizing osteoblasts or their precursors are required for the support of osteoclast differentiation. This uncertainty is reflected by the use of deliberately vague terms, such as “stromal” or “stromal/osteoblastic,” when referring to osteoclast support cells.
Whatever their lineage may be, the cells that support osteoclast formation do so by expressing RANKL, which is indispensable for osteoclast formation in vivo (21). To gain insight into the biology of osteoclast support cells, we have sought to understand the mechanisms that control the cell type-specific expression of the murine RANKL gene. We identified a transcriptional enhancer that mediates hormonal control of RANKL in stromal cells (22, 23). This enhancer, designated the RANKL distal control region (DCR), is located 76-kb upstream from the transcription start site. Gel shift and chromatin immunoprecipitation (ChIP) assays revealed that the DCR contains a binding site for runt related transcription factor 2 (Runx2), a transcription factor that is essential for osteoblast differentiation (24, 25). Furthermore, deletion of the Runx2 binding site blunted the hormonal responsiveness conferred by the DCR (22). These results suggested that Runx2 may be a factor linking osteoclast formation to osteoblast formation via cell type-specific control of RANKL expression.
Consistent with the idea that Runx2 is required for RANKL expression in stromal cells, Runx2-deficient mice exhibit a reduced number of osteoclasts (24). Moreover, calvaria cells from Runx2-deficient mice were less capable of supporting osteoclast formation in vitro (26). In contrast, expression of a dominant negative Runx2 protein in a stromal cell line did not alter basal or stimulated RANKL expression (27). In addition, cell lines derived from Runx2-deficient mice or cell lines in which Runx2 was suppressed by RNA interference still expressed RANKL in response to signaling pathways activated by hormones that stimulate bone resorption (28, 29). It has also been recently proposed that Runx2 may exert an inhibitory effect on basal RANKL expression by condensing chromatin and thus reducing transcription (29). Conflicting results were also obtained in two similar transgenic mouse models overexpressing Runx2 in osteoblasts, which showed either an increase in RANKL expression and osteoclast number (30) or no change in these measurements (31). Thus, as with the identity of the cells that produce RANKL, the role of Runx2 in RANKL expression is unclear.
In the present study we investigated the relationship of osteoblasts and their precursors to the cells that support osteoclast differentiation via expression of RANKL. We found that both mature osteoblasts and Runx family proteins are dispensable for RANKL expression. In addition, enrichment of cells committed to the osteoblast lineage was not associated with an increased ability to express RANKL, although it was associated with increased responsiveness to PTH.
EXPERIMENTAL PROCEDURES
Materials—All cells were maintained in α minimal essential medium (Invitrogen, Carlsbad, CA) supplemented with 10% FBS (HyClone, Logan, UT) and 1% each of penicillin, streptomycin, and glutamine (Sigma). Hexadimethrine bromide, puromycin, bovine PTH-(1–34), dibutyryl-cAMP (db-cAMP), and β-mercaptoethanol (BME) were purchased from Sigma. Ganciclovir was purchased from Roche (Nutley, NJ), and osteoprotegerin-Fc (OPG-Fc) was provided by Amgen Inc. (Thousand Oaks, CA). 1,25-Dihydroxyvitamin D3 (1,25(OH)2D3) was purchased from Biomol (Plymouth Meeting, PA) and human PTH-(1–84) was purchased from Bachem California Inc. (Torrance, CA).
Cell Cultures—The Hepa cell line (CRL-2026) was obtained from the American Type Culture Collection. UAMS-32P cells have been described previously (32). Mouse embryonic fibroblasts (MEFs) were obtained from embryonic day (E)-16.5 embryos from WT and Runx2–/– mice, as previously described (33). Primary calvaria cells were harvested from 5-day-old mice as previously described (34). Calvaria cells from adult mice were obtained as described by Tozum et al. (35). Calvarial cell cultures were performed in medium containing 50 μg/ml ascorbic acid (Sigma).
Animal Studies—To investigate the consequences of suppressing bone remodeling on RANKL expression, 6-month-old female Swiss Webster mice were injected intraperitoneally with vehicle or OPG-Fc (10 μg/g body weight) on days 0, 7, and 14. On day 14, osmotic pumps (Model 1003D, Alzet, Cupertino, CA) delivering vehicle or 500 ng/hr hPTH (1–84) in 0.9% NaCl, 1 mg/ml bovine serum albumin, and 1 mm acetic acid were implanted subcutaneously and tissues were harvested 24-h later.
The generation of 3.6Col-tk mice has been described elsewhere (36). For osteoblast ablation, 2-month-old female 3.6Col-tk transgenic mice (n = 18) were injected intraperitoneally daily with vehicle or ganciclovir (8 μg/g body weight) for 2 weeks and then injected intraperitoneally with vehicle or 100 ng/g hPTH-(1–84) 1 h prior to sacrifice.
Generation of osterix-Cre::green fluorescent protein (GFP) mice has been described previously (37). All osterix-Cre::GFP mice utilized for the studies described herein were hemizygous for the transgene.
All mice were fed a standard rodent diet (Harlan-Teklad no. 7004) with water ad libitum and all studies involving mice were approved by the Institutional Animal Care and Use Committees of the University of Arkansas for Medical Sciences and the Central Arkansas Veterans Healthcare System.
Gene Silencing—A set of five lentiviral clones expressing short hairpin RNAs (shRNAs) directed against core-binding factor β (Cbfb) mRNA were obtained from the RNAi Consortium (made available by Sigma). One clone that suppressed Cbfb expression by more than 80% was selected for use in these studies. The sequence of the Cbfb shRNA construct selected was GCTCGAAGAAGAACTCGAGAACTCGAGTTCTCGAGTTCTTCTTCGAGCTTTTTG (only the top strand is shown). The following scrambled shRNA sequence was used as negative control: CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTT. Transduction with lentiviral particles was accomplished by seeding cells in 12-well plates at a density of 25,000 cells/well. Hexadimethrine bromide was added to the culture to a final concentration of 8 μg/ml and the cells were then transduced with lentiviral particles added at a multiplicity of infection of 20. The cells were then incubated for 48 h and placed in medium containing puromycin (10 μg/ml) for 3 days. For gene expression assays, the cells were plated in 12-well plates at a density of 150,000 cells/well.
Immunoblotting—Immunoblots of extracts from cells were performed as previously described (38). Antibodies against Cbfb (sc-56751, Santa Cruz Biotechnology, Santa Cruz, CA) and β-actin (A5316, Sigma) were used at a dilution of 1:200 and 1:1000, respectively.
RNA Analysis—Total RNA was purified from cell cultures using Ultraspec reagent (Biotecx Laboratories, Houston, TX), according to the manufacturer's directions. Taqman quantitative RT-PCR was performed as previously described (22) using the following primer probe sets from Applied Biosystems (Foster City, CA): RANKL, Mm0041908-m1; OPG, Mm00435451_m1; Cbfb, Mm00491551_m1; Cathepsin K (CatK), Mm01255862-g1; A Kinase Anchor Protein 11 (AKAP11), Mm01313936-m1; PTH receptor (PTHR1), Mm00441046_m1; osterix1 (Osx1), Mm00504574_m1; bone sialoprotein (BSP), Mm00492555_m1; Runx1, Mm00486762_m1; Runx2, Mm00501580_m1; Runx3, Mm00490666_m1; GAPDH, Mm99999915_ g1; osteocalcin, forward, 5′-GCTGCGCTCTGTCTCTCTGA-3′, reverse, 5′-TGCTTGGACATGAAGGCTTTG-3′, probe, 5′-FAM-AAGCCCAGCGGCC-NFQ-3′; and ribosomal protein S2, forward, 5′-CCCAGGATGGCGACGAT-3′, reverse, 5′-CCGAATGCTGTAATGGCGTAT-3′, probe, FAM-5′-TCCAGAGCAGGATCC-3′-NFQ.
Electrophoretic Mobility Shift Assay (EMSA)—Nuclear extract preparation and gel mobility shift assays were performed as previously described (39). The following double-stranded oligonucleotides were used in this study (only the top strands are shown): CNS1-OSE2 (5′-AGAATATCACCACATCAAACAC-3). Anti-Runx1 (sc-28679), anti-Runx2 (sc-10758), anti-Runx3 (sc-23576), and non-immune rabbit IgG antibodies were used for supershift assays and were obtained from Santa Cruz Biotechnology.
Cell Sorting—Calvaria cells were cultured to confluence, trypsinized, filtered through a cell strainer and resuspended in 1 ml of Sorting Buffer (25 mm HEPES pH 7.0, 1% heat-inactivated fetal bovine serum, and 1 mm EDTA in Hank's balanced salt solution) at a density of 107 cells/ml. Cells were sorted using a FACSaria (BD, Franklin Lakes, NJ). To minimize false-negative results, we limited sorting of GFP-negative cells to the more negative fraction. Immediately after sorting, cells were resuspended in complete medium and plated in 96-well plates at a density of 104 cells/well. PTH-(1–34) (10–7 m), db-cAMP (1.5 mm), or 1,25(OH2)3 (10–8 m) were added after 6 h, and RNA was extracted after 18 h of incubation using a TaqMan Gene Expression Cells-to-Ct Kit (Ambion, Austin, TX).
Statistics—Data were analyzed using SigmaStat (SPSS Science, Chicago, IL). All values are reported as the mean ± S.D. Differences between group means were evaluated with Student's t test or two-way analysis of variance.
RESULTS
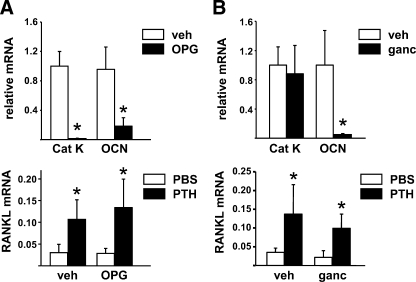
Depletion of Mature Osteoblasts Does Not Alter PTH Stimulation of RANKL—To investigate whether mature osteoblasts are a significant source of RANKL under basal conditions or in response to an osteoclastogenic hormone, we utilized two different approaches to decrease osteoblasts and then measured RANKL mRNA levels in bone in response to exogenous PTH. In the first, adult mice were pretreated with OPG for 2 weeks, a maneuver, which has been shown previously to dramatically reduce both osteoclast and osteoblast number (40). Consistent with this, the expression of cathepsin K, an osteoclast-specific mRNA, and osteocalcin, an osteoblast-specific mRNA, was strikingly reduced in the bone of OPG-treated mice (Fig. 1A). However, despite this reduction in osteoblast formation, infusion of PTH-(1–84) for 24-h stimulated RANKL mRNA to a similar extent in both vehicle- and OPG-treated mice (Fig. 1A).
FIGURE 1.
PTH stimulates RANKL in osteoblast-depleted mice. A, quantitative RT-PCR for cathepsin K (Cat K), OCN, and RANKL mRNA from L5 vertebra of mice pretreated with vehicle or OPG for 2 weeks and then infused with vehicle or PTH for 24 h before RNA preparation. The values represent the mean ± S.D. of 8 to 10 mice per group. *, p < 0.05 versus vehicle or PBS. B, quantitative RT-PCR for the same mRNAs as in A from calvaria of 3.6Col-tk transgenic mice pretreated with vehicle or ganciclovir for 2 weeks and then injected with PBS or PTH and sacrificed after 1 h. All values were normalized to GAPDH mRNA levels. The values represent the mean ± S.D. of 4–5 mice per group. *, p < 0.05 versus vehicle or phosphate-buffered saline.
To confirm the above findings by an alternative approach, we used mice harboring a thymidine kinase transgene under the control of the 3.6 kb Col1a1 promoter (3.6Col-tk mice) (36). In these mice, administration of ganciclovir for 2 weeks killed replicating osteoblast precursors which led to reduced osteoblast number as indicated by reduced osteocalcin mRNA in bone (Fig. 1B). However, a single injection of PTH stimulated RANKL mRNA to a similar extent in both vehicle- and ganciclovir-treated mice (Fig. 1B). Moreover, consistent with the maintenance of basal RANKL expression, osteoclast gene expression was not affected by loss of osteoblasts in this model (Fig. 1B). These results are in agreement with the maintenance of bone resorption in a previous study of osteoblast ablation (16) and suggest that mature osteoblasts are not the main source of RANKL mRNA in bone under basal conditions or after stimulation by PTH.
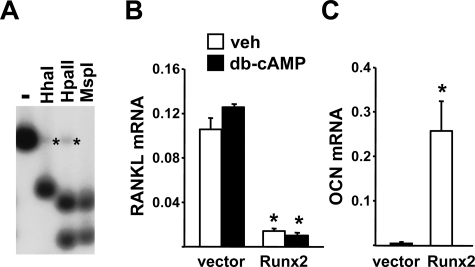
Runx2 Is Not Sufficient for RANKL Expression in Cells Which Normally Do Not Express RANKL—The results of the studies in mice with decreased osteoblast number do not exclude the possibility that committed osteoblast progenitors may be important contributors to basal or hormone-stimulated RANKL levels. Indeed, the presence of a conserved Runx2 binding site in the RANKL DCR suggests that the same factor required for commitment to the osteoblast lineage may play an important role in RANKL transcription. We have shown previously that overexpression of Runx2 in a liver cell line (Hepa) was sufficient to activate the osteocalcin gene but not the RANKL gene (22). However, in this earlier study we found that the CpG island located at the beginning of the endogenous RANKL gene was hypermethylated in Hepa cells leading to suppression of transcription and undetectable levels of RANKL mRNA. Thus it is possible that even if Runx2 is capable of stimulating RANKL transcription in these cells, hypermethylation of the RANKL promoter precluded this from occurring.
To address this possibility, we introduced a bacterial artificial chromosome (BAC) containing the entire murine RANKL gene into Hepa cells by stable transfection. This BAC clone contains regulatory regions, including the DCR, that allow it to be appropriately stimulated by cytokines and hormones in stromal cell lines (22). We verified that the introduced BAC clone was not hypermethylated using Southern blot analysis with methylation-sensitive restriction enzymes (Fig. 2A). RANKL mRNA produced from the integrated BAC DNA was detectable in the transfected cells but was not stimulated by db-cAMP (Fig. 2B). Db-cAMP, rather than PTH, was used here because Hepa cells do not express the PTH receptor (data not shown), and we have shown previously that PTH utilizes the cAMP-protein kinase A (PKA) pathway to stimulate RANKL (32). Importantly, introduction of Runx2 into these cells did not stimulate, but rather suppressed, RANKL expression (Fig. 2B). In contrast, osteocalcin, a known Runx2 target gene, was potently stimulated by Runx2 in the same cells (Fig. 2C). These results confirm that Runx2 is not sufficient for stimulation of RANKL expression.
FIGURE 2.
Runx2 overexpression activates the osteocalcin, but not the RANKL, gene in liver cells. A, genomic DNA from Hepa cells stably transfected with a BAC containing the entire murine RANKL gene (Hepa-BAC1), was digested with PvuI and HindIII followed by digestion with HhaI, HpaII, or MspI (HhaI and HpaII are sensitive to CpG methylation in their recognition sequences). Digests were analyzed by Southern blot with a probe corresponding to the PvuII/HindIII fragment containing RANKL exon 1. Asterisks indicate weak bands resulting from hypermethylated sites in the endogenous RANKL gene in Hepa cells. B and C, quantitative RT-PCR of RANKL (B) or osteocalcin (C) mRNA from Hepa-BAC1 cells transduced with empty vector or Runx2. The RANKL mRNA is derived solely from the BAC1 transgene in the Hepa cells (22). All values were normalized to ribosomal protein S2 mRNA levels. All treatments were performed in triplicate. *, p < 0.05 versus vector-transduced cells.
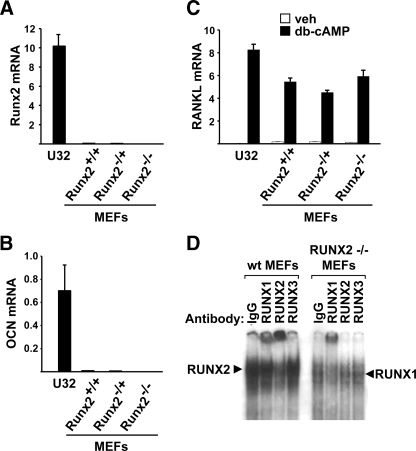
Runx2 Is Not Required for Expression of RANKL in MEFs—Previous studies have shown that calvaria cell cultures from Runx2-deficient mice expressed reduced levels of RANKL mRNA (26). In contrast, calvaria cell lines derived from Runx2-deficient mice displayed normal RANKL expression (28). Therefore, we sought to compare the requirement of Runx2 for RANKL expression in another primary cell model, MEFs. MEFs from wild-type mice and mice lacking one copy of the Runx2 gene expressed very low levels of Runx2 compared with a stromal/osteoblastic cell line, UAMS-32 (Fig. 3A). As expected, Runx2 mRNA was undetectable in MEFs lacking both copies of Runx2. Consistent with the low levels of Runx2, MEFs from all three genotypes expressed low or undetectable osteocalcin mRNA (Fig. 3B), indicating that MEFs do not contain significant numbers of cells committed to the osteoblast lineage. Nonetheless, db-cAMP stimulated RANKL expression to comparable levels in wild type MEFs and UAMS-32 cells (Fig. 3C). It is possible that the low levels of Runx2 present in wild-type MEFs were still required for stimulation of RANKL expression. However, deletion of Runx2 did not alter the stimulation of RANKL by db-cAMP (Fig. 3C).
FIGURE 3.
Runx2 is not required for RANKL expression in MEFs. A–C, quantitative RT-PCR of Runx2 (A), OCN (B), and RANKL (C) mRNA from UAMS-32 cells (U32) or MEFs from wild type (+/+), Runx2 haplo-insufficient (–/+), and Runx2-deficient (–/–) mice treated with vehicle or db-cAMP for 24 h. All values were normalized to ribosomal protein S2 mRNA levels. All treatments were performed in triplicate. D, gel mobility shift assay using nuclear extracts from wild type or Runx2(–/–) MEFs and the Runx2 binding site from the murine RANKL DCR. Control IgG or anti-Runx antibodies were included as indicated.
Although deletion of Runx2 did not alter RANKL expression, it is possible that other members of the Runx family compensated for the lack of Runx2. Indeed, both Runx1 and Runx3 are expressed in MEFs (supplemental Fig. S1). EMSAs using nuclear extracts from Runx2-deficient MEFs revealed that Runx1, but not Runx3, was able to bind the Runx2 binding site from the RANKL DCR (Fig. 3D). Therefore, Runx1 may compensate for the lack of Runx2 in MEFs. Such compensation could also explain the maintenance of RANKL expression in previous Runx2-deletion studies (28, 29).
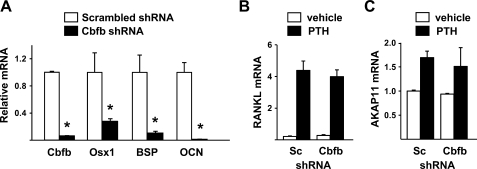
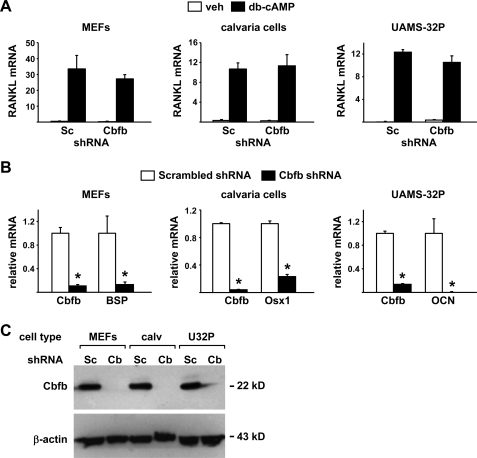
Suppression of Cbfb Does Not Alter Hormonal Control of RANKL—Cbfb is essential for the function of all Runx family members (41, 42), in part by limiting their ubiquitin-mediated degradation (43, 44) and by increasing the affinity of Runx proteins for DNA (45). Therefore, to determine whether any member of the Runx family is required for the expression of RANKL, we silenced Cbfb using shRNAs introduced via lentiviral transduction. A panel of five different shRNAs was screened for the ability to suppress Cbfb expression in UAMS-32 cells and MEFs and one that reduced Cbfb mRNA by more than 80% in both cell models was selected for use (supplemental Fig. S2). Silencing of Cbfb in UAMS-32 cells significantly reduced expression of osterix, bone sialoprotein, and osteocalcin, which are known Runx2 target genes and markers of commitment to the osteoblast lineage (Fig. 4A). However, stimulation of RANKL mRNA by PTH was not affected by Cbfb silencing (Fig. 4B). Similarly, the expression of a control gene, AKAP11, which is located immediately upstream of RANKL on chromosome 14, was not affected by silencing of Cbfb (Fig. 4C).
FIGURE 4.
Suppression of Cbfb does not alter PTH control of RANKL in stromal/osteoblastic cells. Quantitative RT-PCR of mRNA for (A) Cbfb, Osx1, BSP, and OCN or (B) RANKL or (C) AKAP11 from UAMS-32 cells transduced with a lentivirus expressing Scrambled (Sc) or Cbfb shRNA and treated with vehicle or PTH for 4 h. All values were normalized to ribosomal protein S2 mRNA levels. All treatments were performed in triplicate. *, p < 0.05 versus scrambled shRNA.
To confirm the results obtained in a cell line in primary cell models, we silenced Cbfb in MEFs and neonatal mouse calvaria cells (Fig. 5, A–C). Cbfb mRNA and protein levels were reduced by the Cbfb shRNA in all the cell types examined (Fig. 5, B and C) and known Runx target genes expressed in each cell type were also suppressed (Fig. 5B). Nonetheless, silencing of Cbfb did not alter db-cAMP stimulation of RANKL expression in any of these cell types (Fig. 5A). These results show that reduction of Cbfb expression to the point that known Runx2 target genes were potently suppressed did not affect stimulation of RANKL by PKA activation in different cell models. These findings thus demonstrate that Runx proteins are dispensable for RANKL expression in fibroblastic cells.
FIGURE 5.
Suppression of Cbfb does not affect stimulation of RANKL by db-cAMP in different cell types. A, quantitative RT-PCR of RANKL mRNA from MEFs (left), calvaria cells (center), and UAMS-32P cells (right) transduced with a scrambled (Sc) or Cbfb shRNA producing lentivirus and stimulated with vehicle or db-cAMP for 24 h. B, quantitative RT-PCR analysis of Cbfb mRNA in the same cells as in A. In addition, mRNAs for BSP, Osx1, and OCN, were quantified in MEFs, calvaria cells, and UAMS-32P cells, respectively. All values in A and B were normalized to ribosomal protein S2 mRNA levels, and all treatments were performed in triplicate. *, p < 0.05 versus scrambled shRNA. C, anti-Cbfb or anti-β-actin antibodies were used to probe an immunoblot of protein from MEFs, calvaria cells (calv), and UAMS-32P cells (U32P) transduced with scrambled (Sc) or Cbfb (Cb) shRNA producing lentivirus.
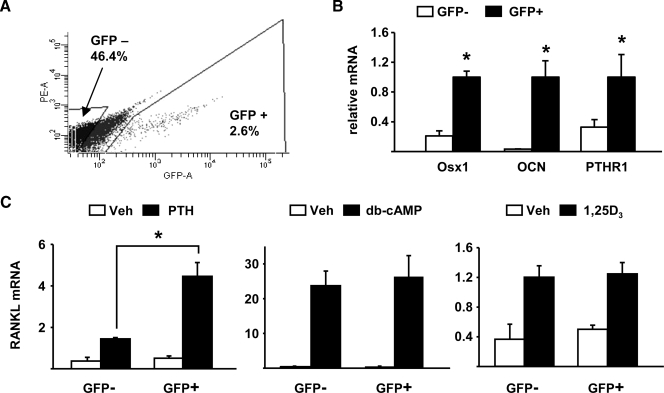
Commitment to the Osteoblast Lineage and RANKL Expression—Finally, to directly compare the ability of cells to express RANKL with their commitment to the osteoblast lineage, we examined RANKL expression in cells from osterix-Cre::GFP transgenic mice, which express a Cre::GFP fusion protein under the control of osterix gene regulatory elements (37). In these mice, cells that express osterix, and are thus committed to the osteoblastic lineage, express GFP. Therefore, we isolated calvaria cells from these mice, sorted them based on GFP expression, and quantified basal and stimulated RANKL mRNA in GFP-positive and GFP-negative cell populations. GFP-positive cells accounted for ∼2.6% of the total sorted cells (Fig. 6A). The GFP-positive population was enriched for cells expressing osterix, osteocalcin, and PTH receptor 1 (PTHR1) (Fig. 6B). PTH stimulation of RANKL was significantly higher in GFP-positive cells (Fig. 6C). This increased responsiveness to PTH was most likely due to higher expression of PTHR1 in GFP-positive cells since db-cAMP, which activates the same PKA-CREB pathway used by PTH to stimulate RANKL (32), stimulated RANKL to the same extent in both GFP-positive and GFP-negative cells (Fig. 6C). Moreover, 1,25(OH)2D3 stimulated RANKL to the same extent in both cell populations (Fig. 6C). Therefore, these results suggest that commitment to the osteoblast lineage, although not required directly for RANKL transcription, may enhance PTH stimulation of RANKL by increasing PTHR1 expression.
FIGURE 6.
Enrichment of osterix-expressing cells is associated with increased PTHR1 expression and increased responsiveness to PTH. A, flow cytometry analysis of calvaria cells from Osx1-Cre::GFP mice. Only a fraction of the GFP-negative cells was collected to avoid including cells expressing low levels of GFP. The GFP-positive cells accounted for 2.6% of the total cell population. B, quantitative RT-PCR for osterix, osteocalcin, and PTHR1 mRNA expression in GFP+ and GFP- calvaria cells from Osx1-Cre::GFP mice. C, quantitative RT-PCR for RANKL mRNA expression in GFP-positive and GFP-negative Osx1-Cre::GFP calvaria cells in response to vehicle, PTH, db-cAMP, and 1,25(OH)2D3 for 24 h. All values were normalized to ribosomal protein S2 mRNA levels, and all treatments were performed in triplicate. *, p < 0.05 versus GFP-negative cells.
DISCUSSION
It is a commonly held view that osteoblasts, or cells of the osteoblast lineage, are important sources of RANKL in bone (14, 46–48). In the present report we show that dramatic reduction of mature osteoblast number by two independent manipulations did not alter either basal or PTH-stimulated RANKL expression in vivo. Likewise, the transcription factor Runx2, which is an essential requirement for osteoblast differentiation, is not required for RANKL expression. Consistent with this, we found that MEFs, a source of fibroblastic cells harboring few committed osteoblasts, robustly expressed RANKL in response to db-cAMP, a surrogate of PTH stimulation of osteoblast-lineage cells. RANKL expression in cells lacking Runx2 was not due to compensation by other members of the Runx family of transcription factors. Lastly, we found that enrichment for cells committed to the osteoblast lineage, as defined by high levels of osterix expression, was not associated with an increased ability to express RANKL, although it was associated with an increased response of RANKL to PTH due to increased PTHR1 expression. Taken together, these results exclude mature osteoblasts as a significant source of RANKL in bone and suggest that if osteoblast precursors are involved in the support of osteoclasts, it is not due to control of RANKL gene expression by osteoblast-specific transcription factors.
Eriksen et al. (49) have suggested that matrix-synthesizing osteoblasts are unlikely to be involved in the support of osteoclastogenesis because osteoclastic bone resorption precedes bone formation thereby preventing osteoblasts from being in the appropriate location for cell-to-cell contact with osteoclast precursors. It is also worth noting that origination of the BMU requires differentiation of osteoclasts at the site to be resorbed prior to the arrival of any osteoblasts (1). The present study, together with previous studies (16), support these ideas because reduction in the number of matrix-synthesizing osteoblasts did not alter RANKL expression or osteoclast differentiation. Therefore, if the coupling of bone formation to bone resorption involves RANKL expression in an osteoblast-lineage cell, such cells must be osteoblast precursors.
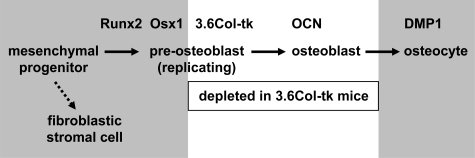
Previous work by us (23) and others (50, 51) clearly demonstrates that PTH stimulation of RANKL is a major contributor to osteoclast formation and the rate of bone remodeling under basal conditions. Therefore, PTH-responsive cells must be a significant source of RANKL in bone, and we have focused our previous studies on the control of the RANKL gene by the PTH-cAMP-PKA-CREB pathway (22, 23, 32). We have also sought to understand the relationship of RANKL-expressing cells to cells of the osteoblast lineage. The results of our studies suggest at least three possible sources of RANKL in bone that are not mutually exclusive: osteoblast precursors, osteocytes, and fibroblastic stromal cells outside the osteoblast lineage (Fig. 7).
FIGURE 7.
RANKL expression in mesenchymal cells. Gene names at the top of the diagram indicate the approximate stage in the osteoblast lineage at which they become active during osteoblast differentiation. Ganciclovir treatment of 3.6Col-tk mice kills replicating pre-osteoblasts and, as a consequence, mature osteoblasts eventually disappear as they die or become osteocytes (as indicated by the white box). The gray boxes indicate cell types that are potentially significant sources of RANKL in bone. Chondrocytes, which may be a significant source of RANKL in growing bone, but not in adult remodeling bone, are not depicted in the diagram. DMP1, dentin matrix protein 1, which is expressed exclusively in osteocytes in bone.
Our finding of increased PTH responsiveness in osterix-expressing cells in vitro suggests that commitment to the osteoblast lineage may enhance the response to PTH via control of PTHR1 expression. Yet, the results of the cell ablation in the 3.6Col-tk transgenic mice demonstrate that replicating 3.6Col-tk-expressing osteoblast precursors do not contribute significantly to basal or PTH-stimulated RANKL expression in vivo. This suggests that if osteoblast precursors are an important source of RANKL, such cells must be at a stage of differentiation that is earlier than the stage at which the 3.6Col-tk transgene becomes active (Fig. 7).
Alternatively, osteocytes may be a significant source of RANKL since they do not replicate and are long-lived, and thus were not killed or depleted in the osteoblast-depletion models (Fig. 7). Consistent with this idea, MLO-Y4 osteocytic cells have been shown to produce RANKL (52, 53). However, it is important to note that osteocytes are not in a location that allows physical interaction with osteoclast precursors to stimulate their differentiation via membrane-bound RANKL, which appears to be the form most important for stimulating osteoclast differentiation (54). Moreover, even if osteocytes do express significant levels of RANKL, this would not constitute a mechanism of coupling since osteocytes are formed at the end of bone formation, while coupling links the end of bone resorption to the beginning of bone formation.
In the present report, we found that commitment to the osteoblast lineage is not required for the ability of cells to express RANKL, suggesting that fibroblastic cells not of the osteoblast lineage, even though they may express lower levels of PTHR1, express RANKL in response to PTH and act as stromal cells for the support of osteoclast differentiation (Fig. 7). This finding is consistent with a previous study that demonstrated stimulation of RANKL by 1,25(OH)2D3 in a variety of fibroblastic cells derived from extra-skeletal tissues such as skin (55). In that previous study, RANKL expression in extraskeletal fibroblasts required co-treatment with dexamethasone whereas RANKL expression in MEFs in the current study did not. However, the requirement for dexamethasone was not due to induction of osteoblast differentiation because dexamethasone did not induce alkaline phosphatase expression in the extraskeletal fibroblasts (55).
Together, these in vitro studies demonstrate that fibroblastic cells from various tissues have the ability to express RANKL in response to the appropriate stimulus. Nonetheless, we have shown previously that activation of the cAMP-PKA-CREB pathway in vivo preferentially stimulated RANKL expression in bone (22). Whether this was due to larger numbers of fibroblastic cells in bone compared with other tissues, such as liver and kidney, is unclear. Thus, it remains possible that, at least in vivo, the RANKL gene is more responsive in fibroblastic cells present in the bone marrow microenvironment than in extraskeletal tissues. However, even if this is the case, the results of the present study suggest that the ability to express RANKL is not due to Runx2 expression and thus is not due to commitment to the osteoblast lineage. Whether the fibroblastic cells in bone that express RANKL represent a multipotential precursor such as the so-called mesenchymal stromal cell (56), or whether they represent a lineage dedicated to the support of osteoclast differentiation, remains to be determined.
Endothelial cells have also been shown to express RANKL and support osteoclastogenesis in vitro in response to cytokines and growth factors (57, 58). Moreover, abundant evidence indicates that activated lymphocytes express RANKL and can support osteoclast formation (59, 60). Whether these cell types contribute significantly to osteoclastogenesis during physiologic bone remodeling remains unclear. To further clarify the identity of osteoclast support cells, it will be important in future studies to determine the relative contribution of RANKL expression by different cell types in vivo via deletion of the RANKL gene in specific genetically defined cell populations.
Our finding that RANKL expression is unaffected in Runx2-deficient cells is consistent with our previous observations that mutation of the Runx2 binding sites in the proximal RANKL promoter did not alter the activity of transcriptional reporter constructs, and that a dominant-negative version of Runx2 had no effect on endogenous RANKL expression in stromal/osteoblastic cells (27). Nonetheless, the evolutionary conservation of the Runx2 binding sites in the DCR and the proximal RANKL promoter suggests that these sites play an important role in RANKL expression. Consistent with this, we have shown previously that Runx2 binds to the DCR enhancer in ChIP assays (22). A potential explanation for these apparently contradictory observations is that Runx2 is required for RANKL expression in cells types other than those examined in our studies. Indeed, a recent report suggests that the Runx2-binding sites in the proximal RANKL promoter are involved in BMP2 stimulated expression of RANKL in chondrocytes (61). Another possibility is that Runx2 is required for expression of RANKL in response to stimuli that were not addressed in our study. However, the present work, together with previous studies examining the requirement of Runx2 for stimulation of RANKL by 1,25(OH)2D3 (28), demonstrates that Runx2 is not required for control of RANKL by two of the major pathways known to control this gene in bone.
In conclusion, we have shown that commitment to the osteoblast lineage is not a prerequisite for the ability of fibroblastic cell types to express RANKL and that mature osteoblasts are not a significant source of RANKL in bone. Together, these findings clarify the stage at which osteoblast progenitors may contribute to osteoclast support but raise the possibility that the coupling of bone formation to bone resorption does not involve expression of RANKL in osteoblast-lineage cells.
Supplementary Material
Acknowledgments
We thank H. M. Kronenberg for providing the Osx1-Cre::GFP transgenic mice and P. E. Cazer and A. DeLoose for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AR049794 (to C. A. O.) and P01 AG13918 (to S. C. M.). This work was also supported by an institutional award from the University of Parma (to C. G.), UAMS Tobacco Settlement Funds, and by Veterans Affairs Merit Reviews (to S. C. M. and R. L. J.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: RANKL, receptor activator of NFκB ligand; BAC, bacterial artificial chromosome; Cbfb, core-binding factor β; db-cAMP, dibutyryl-cAMP; DCR, distal control region; GFP, green fluorescent protein; MEFs, mouse embryonic fibroblasts; PKA, protein kinase A; PTH, parathyroid hormone; OPG, osteoprotegerin; shRNA, short hairpin RNA; EMSA, electrophoretic mobility shift assay; OCN, osteocalcin; Osx1, osterix; BSP, bone sialoprotein.
References
- 1.Parfitt, A. M. (2002) Bone 30 5–7 [DOI] [PubMed] [Google Scholar]
- 2.Bonewald, L. F., and Mundy, G. R. (1990) Clin. Orthop. Relat Res. 250 261–276 [PubMed] [Google Scholar]
- 3.Manolagas, S. C., Jilka, R. L., Bellido, T., O'Brien, C. A., and Parfitt, A. M. (1996) in Principles of Bone Biology (Bilezikian, J. P., Raisz, L. G., and Rodan, G. A., eds), Academic Press, New York
- 4.Manolagas, S. C. (2000) Endocr. Rev. 21 115–137 [DOI] [PubMed] [Google Scholar]
- 5.Rodan, G. A., and Martin, T. J. (1982) Calcif. Tissue Int. 34 311. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi, N., Akatsu, T., Udagawa, N., Sasaki, T., Yamaguchi, A., Moseley, J. M., Martin, T. J., and Suda, T. (1988) Endocrinology 123 2600–2602 [DOI] [PubMed] [Google Scholar]
- 7.Udagawa, N., Takahashi, N., Akatsu, T., Sasaki, T., Yamaguchi, A., Kodama, H., Martin, T. J., and Suda, T. (1989) Endocrinology 125 1805–1813 [DOI] [PubMed] [Google Scholar]
- 8.Lacey, D. L., Timms, E., Tan, H. L., Kelley, M. J., Dunstan, C. R., Burgess, T., Elliott, R., Colombero, A., Elliott, G., Scully, S., Hsu, H., Sullivan, J., Hawkins, N., Davy, E., Capparelli, C., Eli, A., Qian, Y. X., Kaufman, S., Sarosi, I., Shalhoub, V., Senaldi, Guo, J., Delaney, J., and Boyle, W. J. (1998) Cell 93 165–176 [DOI] [PubMed] [Google Scholar]
- 9.Yasuda, H., Shima, N., Nakagawa, N., Yamaguchi, K., Kinosaki, M., Mochizuki, S., Tomoyasu, A., Yano, K., Goto, M., Murakami, A., Tsuda, E., Morinaga, T., Higashio, Udagawa, N., Takahashi, N., and Suda, T. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bilic-Curcic, I., Kronenberg, M., Jiang, X., Bellizzi, J., Mina, M., Marijanovic, I., Gardiner, E. M., and Rowe, D. W. (2005) Genesis. 43 87–98 [DOI] [PubMed] [Google Scholar]
- 11.Huang, J. C., Sakata, T., Pfleger, L. L., Bencsik, M., Halloran, B. P., Bikle, D. D., and Nissenson, R. A. (2004) J. Bone Miner. Res. 19 235–244 [DOI] [PubMed] [Google Scholar]
- 12.Gori, F., Hofbauer, L. C., Dunstan, C. R., Spelsberg, T. C., Khosla, S., and Riggs, B. L. (2000) Endocrinology 141 4768–4776 [DOI] [PubMed] [Google Scholar]
- 13.Kartsogiannis, V., Zhou, H., Horwood, N. J., Thomas, R. J., Hards, D. K., Quinn, J. M. W., Niforas, P., Ng, K. W., Martin, T. J., and Gillespie, M. T. (1999) Bone 25 525–534 [DOI] [PubMed] [Google Scholar]
- 14.Silvestrini, G., Ballanti, P., Patacchioli, F., Leopizzi, M., Gualtieri, N., Monnazzi, P., Tremante, E., Sardella, D., and Bonucci, E. (2005) J. Mol. Histol. 36 59–67 [DOI] [PubMed] [Google Scholar]
- 15.Ikeda, T., Utsuyama, M., and Hirokawa, K. (2001) J. Bone Miner. Res. 16 1416–1425 [DOI] [PubMed] [Google Scholar]
- 16.Corral, D. A., Amling, M., Priemel, M., Loyer, E., Fuchs, S., Ducy, P., Baron, R., and Karsenty, G. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 13835–13840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabatakos, G., Sims, N. A., Chen, J., Aoki, K., Kelz, M. B., Amling, M., Bouali, Y., Mukhopadhyay, K., Ford, K., Nestler, E. J., and Baron, R. (2000) Nat. Med. 6 985–990 [DOI] [PubMed] [Google Scholar]
- 18.Jochum, W., David, J. P., Elliott, C., Wutz, A., Plenk, H. J., Matsuo, K., and Wagner, E. F. (2000) Nat. Med. 6 980–984 [DOI] [PubMed] [Google Scholar]
- 19.Li, X., Ominsky, M. S., Niu, Q. T., Sun, N., Daugherty, B., D'Agostin, D., Kurahara, C., Gao, Y., Cao, J., Gong, J., Asuncion, F., Barrero, M., Warmington, K., Dwyer, D., Stolina, M., Morony, S., Sarosi, I., Kostenuik, P. J., Lacey, D. L., Simonet, W. S., Ke, H. Z., and Paszty, C. (2008) J. Bone Miner. Res. 23 860–869 [DOI] [PubMed] [Google Scholar]
- 20.Akune, T., Ohba, S., Kamekura, S., Yamaguchi, M., Chung, U. I., Kubota, N., Terauchi, Y., Harada, Y., Azuma, Y., Nakamura, K., Kadowaki, T., and Kawaguchi, H. (2004) J. Clin. Investig. 113 846–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong, Y. Y., Yoshida, H., Sarosi, I., Tan, H. L., Timms, E., Capparelli, C., Morony, S., Oliveira, d. S. A., Van, G., Itie, A., Khoo, W., Wakeham, A., Dunstan, C. R., Lacey, D. L., Mak, T. W., Boyle, W. J., and Penninger, J. M. (1999) Nature 397 315–323 [DOI] [PubMed] [Google Scholar]
- 22.Fu, Q., Manolagas, S. C., and O'Brien, C. A. (2006) Mol. Cell. Biol. 26 6453–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galli, C., Zella, L. A., Fretz, J. A., Fu, Q., Pike, J. W., Weinstein, R. S., Manolagas, S. C., and O'Brien, C. A. (2008) Endocrinology 149 146–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komori, T., Yagi, H., Nomura, S., Yamaguchi, A., Sasaki, K., Deguchi, K., Shimizu, Y., Bronson, R. T., Gao, Y. H., Inada, M., Sato, M., Okamoto, R., Kitamura, Y., Yoshiki, S., and Kishimoto, T. (1997) Cell 89 755–764 [DOI] [PubMed] [Google Scholar]
- 25.Otto, F., Thornell, A. P., Crompton, T., Denzel, A., Gilmour, K. C., Rosewell, I. R., Stamp, G. W., Beddington, R. S., Mundlos, S., Olsen, B. R., Selby, P. B., and Owen, M. J. (1997) Cell 89 765–771 [DOI] [PubMed] [Google Scholar]
- 26.Gao, Y. H., Shinki, T., Yuasa, T., Kataoka-Enomoto, H., Komori, T., Suda, T., and Yamaguchi, A. (1998) Biochem. Biophys. Res. Commun. 252 697–702 [DOI] [PubMed] [Google Scholar]
- 27.O'Brien, C. A., Kern, B., Gubrij, I., Karsenty, G., and Manolagas, S. C. (2002) Bone 30 453–462 [DOI] [PubMed] [Google Scholar]
- 28.Notoya, M., Otsuka, E., Yamaguchi, A., and Hagiwara, H. (2004) Biochem. Biophys. Res. Commun. 324 655–660 [DOI] [PubMed] [Google Scholar]
- 29.Mori, K., Kitazawa, R., Kondo, T., Maeda, S., Yamaguchi, A., and Kitazawa, S. (2006) J. Cell. Biochem. 98 1629–1644 [DOI] [PubMed] [Google Scholar]
- 30.Geoffroy, V., Kneissel, M., Fournier, B., Boyde, A., and Matthias, P. (2002) Mol. Cell. Biol. 22 6222–6233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, W. G., Toyosawa, S., Furuichi, T., Kanatani, N., Yoshida, C., Liu, Y., Himeno, Narai, S., Yamaguchi, A., and Komori, T. (2001) J. Cell Biol. 155 157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu, Q., Jilka, R. L., Manolagas, S. C., and O'Brien, C. A. (2002) J. Biol. Chem. 277 48868–48875 [DOI] [PubMed] [Google Scholar]
- 33.Nagy, A., Gertsenstein, M., Vintersten, K., and Behringer, R. R. (2003) Manipulating the Mouse Embryo: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 34.Bellido, T., Ali, A. A., Plotkin, L. I., Fu, Q., Gubrij, I., Roberson, P. K., Weinstein, R. S., O'Brien, C. A., Manolagas, S. C., and Jilka, R. L. (2003) J. Biol. Chem. 278 50259–50272 [DOI] [PubMed] [Google Scholar]
- 35.Tozum, T. F., Oppenlander, M. E., Koh-Paige, A. J., Robins, D. M., and McCauley, L. K. (2004) Calcif. Tissue Int. 75 60–70 [DOI] [PubMed] [Google Scholar]
- 36.Jilka, R. L., O'Brien, C. A., Ali, A. A., Roberson, P. K., Weinstein, R. S., and Manolagas, S. C. (2009) Bone 44 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodda, S. J., and McMahon, A. P. (2006) Development 133 3231–3244 [DOI] [PubMed] [Google Scholar]
- 38.Lin, S. C., Yamate, T., Taguchi, Y., Borba, V. Z., Girasole, G., O'Brien, C. A., Bellido, T., Abe, E., and Manolagas, S. C. (1997) J. Clin. Investig. 100 1980–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Brien, C. A., and Manolagas, S. C. (1997) J. Biol. Chem. 272 15003–15010 [DOI] [PubMed] [Google Scholar]
- 40.Ominsky, M. S., Li, X., Asuncion, F. J., Barrero, M., Warmington, K. S., Dwyer, D., Stolina, M., Geng, Z., Grisanti, M., Tan, H. L., Corbin, T., McCabe, J., Simonet, W. S., Ke, H. Z., and Kostenuik, P. J. (2008) J. Bone Miner. Res. 23 672–682 [DOI] [PubMed] [Google Scholar]
- 41.Miller, J., Horner, A., Stacy, T., Lowrey, C., Lian, J. B., Stein, G., Nuckolls, G. H., and Speck, N. A. (2002) Nat. Genet. 32 645–649 [DOI] [PubMed] [Google Scholar]
- 42.Kundu, M., Javed, A., Jeon, J. P., Horner, A., Shum, L., Eckhaus, M., Muenke, M., Lian, J. B., Yang, Y., Nuckolls, G. H., Stein, G. S., and Liu, P. P. (2002) Nat. Genet. 32 639–644 [DOI] [PubMed] [Google Scholar]
- 43.Huang, G., Shigesada, K., Ito, K., Wee, H. J., Yokomizo, T., and Ito, Y. (2001) EMBO J. 20 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lien, C. Y., Lee, O. K., and Su, Y. (2007) Stem Cells (Dayt) 25 1462–1468 [DOI] [PubMed] [Google Scholar]
- 45.Tang, Y. Y., Crute, B. E., Kelley, J. J., Huang, X., Yan, J., Shi, J., Hartman, K. L., Laue, T. M., Speck, N. A., and Bushweller, J. H. (2000) FEBS Lett. 470 167–172 [DOI] [PubMed] [Google Scholar]
- 46.Tat, S. K., Pelletier, J. P., Lajeunesse, D., Fahmi, H., Duval, N., and Martel-Pelletier, J. (2008) Bone 43 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kearns, A. E., Khosla, S., and Kostenuik, P. J. (2008) Endocr. Rev. 29 155–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mak, K. K., Bi, Y., Wan, C., Chuang, P. T., Clemens, T., Young, M., and Yang, Y. (2008) Dev. Cell 14 674–688 [DOI] [PubMed] [Google Scholar]
- 49.Eriksen, E. F., Eghbali-Fatourechi, G. Z., and Khosla, S. (2007) J. Bone Miner. Res. 22 1–6 [DOI] [PubMed] [Google Scholar]
- 50.Miao, D. S., He, B., Lanske, B., Bai, X. Y., Tong, X. K., Hendy, G. N., Goltzman, D., and Karaplis, A. C. (2004) Endocrinology 145 2046–2053 [DOI] [PubMed] [Google Scholar]
- 51.Ueno, Y., Shinki, T., Nagai, Y., Murayama, H., Fujii, K., and Suda, T. (2003) J. Cell. Biochem. 90 267–277 [DOI] [PubMed] [Google Scholar]
- 52.Zhao, S., Kato, Y., Zhang, Y., Harris, S., Ahuja, S. S., and Bonewald, L. F. (2002) J. Bone Miner. Res. 17 2068–2079 [DOI] [PubMed] [Google Scholar]
- 53.You, L., Temiyasathit, S., Lee, P., Kim, C. H., Tummala, P., Yao, W., Kingery, W., Malone, A. M., Kwon, R. Y., and Jacobs, C. R. (2008) Bone 42 172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hikita, A., Yana, I., Wakeyama, H., Nakamura, M., Kadono, Y., Oshima, Y., Nakamura, K., Seiki, M., and Tanaka, S. (2006) J. Biol. Chem. 281 36846–36855 [DOI] [PubMed] [Google Scholar]
- 55.Quinn, J. M., Horwood, N. J., Elliott, J., Gillespie, M. T., and Martin, T. J. (2000) J. Bone Miner. Res. 15 1459–1466 [DOI] [PubMed] [Google Scholar]
- 56.Keating, A. (2006) Curr. Opin. Hematol. 13 419–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collin-Osdoby, P., Rothe, L., Anderson, F., Nelson, M., Maloney, W., and Osdoby, P. (2001) J. Biol. Chem. 276 20659–20672 [DOI] [PubMed] [Google Scholar]
- 58.Ishida, A., Fujita, N., Kitazawa, R., and Tsuruo, T. (2002) J. Biol. Chem. 277 26217–26224 [DOI] [PubMed] [Google Scholar]
- 59.Eghbali-Fatourechi, G., Khosla, S., Sanyal, A., Boyle, W. J., Lacey, D. L., and Riggs, B. L. (2003) J. Clin. Investig. 111 1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walsh, M. C., Kim, N., Kadono, Y., Rho, J., Lee, S. Y., Lorenzo, J., and Choi, Y. (2006) Annu. Rev. Immunol. 24 33–63 [DOI] [PubMed] [Google Scholar]
- 61.Usui, M., Xing, L., Drissi, H., Zuscik, M., O'Keefe, R., Chen, D., and Boyce, B. F. (2008) J. Bone Miner. Res. 23 314–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.