Abstract
Protein phosphorylation plays an important role in the regulation of centrosome duplication. In budding yeast, numerous lines of evidence suggest a requirement for multiple phosphorylation events on individual components of the centrosome to ensure their proper assembly and function. Here, we report the first example of a single phosphorylation event on a component of the yeast centrosome, or spindle pole body (SPB), that is required for SPB duplication and cell viability. This phosphorylation event is on the essential SPB component Spc29 at a conserved Thr residue, Thr240. Mutation of Thr240 to Ala is lethal at normal gene dosage, but an increased copy number of this mutant allele results in a conditional phenotype. Phosphorylation of Thr240 was found to promote the stability of the protein in vivo and is catalyzed in vitro by the Mps1 kinase. Furthermore, the stability of newly synthesized Spc29 is reduced in a mutant strain with reduced Mps1 kinase activity. These results demonstrate the first evidence for a single phosphorylation event on an SPB component that is absolutely required for SPB duplication and suggest that the Mps1 kinase is responsible for this protein-stabilizing phosphorylation.
Centrosomes are critical for organizing microtubules that make up the mitotic and meiotic spindles that segregate chromosomes during cell division. The duplication of these organelles must be tightly regulated to occur once and only once during each cell cycle to prevent the formation of monopolar or multipolar mitotic spindles that can cause chromosomal instability. The yeast centrosome is called the spindle pole body (SPB)3 and is one of the best characterized microtubule-organizing centers. Although the SPB and the centrosome are morphologically distinct, they share the common function of spindle organization. Many SPB components and regulators of SPB assembly and function are conserved throughout evolution (1). This has made the yeast SPB an excellent model in which to study the regulation of centrosome duplication.
The regulation of centrosome function and duplication by phosphorylation is well documented (2–10). Although several yeast SPB components are phosphoproteins in vivo (11–16), little is known about the specific sites of phosphorylation or the roles these modifications play in the regulation of SPB duplication and function. The yeast cyclin-dependent kinase Cdc28 and the multifunctional Mps1 kinase have both been implicated in the regulation of SPB components by phosphorylation (17–20). Two essential SPB components, Spc42 and Spc110, are phosphorylated by both of these kinases. Prevention of modification by either kinase alone is not detrimental, but the two kinases work in concert with each other to produce a fully functional protein. These examples demonstrate that some SPB components are coordinately regulated by the actions of more than one protein kinase and that an accumulation of hyperphosphorylation, rather than specific individual phosphorylation events, is the predominant mechanism of phosphoregulation of SPB components.
In this study, we demonstrate that a single phosphothreonine, phospho-Thr240, near the C terminus of the SPB component Spc29 is absolutely required for SPB duplication and mitotic progression. The modification promotes the stability of the Spc29 protein and appears to be catalyzed by the Mps1 kinase. These results reveal the first single phosphorylation event known to be essential for SPB duplication and elucidate a mechanism by which cells can achieve tight regulation of centrosome duplication through a cascade of phosphorylation-mediated protein stabilization wherein the yeast cyclin-dependent kinase stabilizes the Mps1 kinase by phosphorylation (19), and the Mps1 kinase in turn stabilizes the Spc29 protein by phosphorylation, ensuring adequate levels of this critical SPB component for the assembly of new spindle poles.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids—All strains used in this study are derivatives of W303 (ade2-1 trp1-1 leu2-3,112 ura3-1 his3-11,15 can1-100, strain W1522) and are described individually in supplemental Table S1.
Microscopy—Indirect immunofluorescence microscopy was performed as described previously (21). Cells were synchronized with mating factor at 25 °C, washed once with water, and released from arrest in yeast extract/peptone/dextrose medium at 36 °C for 2.5 h. For electron microscopic analyses, cells were prepared using the methods described previously (22). The images were collected on a Philips CM10 electron microscope.
Protein Techniques and Kinase Assays—Whole cell lysates were prepared by bead beating in reducing SDS-PAGE sample buffer or by cryolysis and analyzed by immunoblotting on polyvinylidene difluoride membranes. Spc29 protein was detected with an affinity-purified rabbit polyclonal antibody that was diluted 1:500 in blocking buffer (Tris-buffered saline (pH 7.5), 0.05% Tween 20, and 5% nonfat dry milk). Anti-glucose-6-phosphate dehydrogenase antibody (Sigma) was diluted 1:20,000. Anti-green fluorescent protein (GFP) monoclonal antibody (Covance) was diluted 1:5000. Primary antibodies were detected with Alexa Fluor-conjugated goat anti-rabbit or anti-mouse secondary antibodies (Molecular Probes) and visualized on a LI-COR scanner.
Recombinant Mps1 kinase was purified from Escherichia coli BL21 pLysS cells as a glutathione S-transferase fusion using pGEX-6p1 (GE Healthcare). Spc29 was expressed as a maltose-binding protein fusion and purified on amylose affinity resin (New England Biolabs) according to the manufacturer's recommendations. Cdc28-Clb2 complexes purified from yeast using epitope-tagged Clb2 were a kind gift from Sue Jaspersen. Kinase assays were performed as described (18) and visualized by Coomassie Blue staining and autoradiography.
Mass Spectrometry—Purified SPBs were subjected to a triple enzyme digestion as described (23). To enrich for phosphopeptides, a TiO2 method (24) was used on peptide mixtures prior to pressure loading onto a reversed-phase column, and peptides were eluted with a linear gradient of acetonitrile (5–80%). Eluted peptides were analyzed by tandem mass spectrometry (MS/MS) as described (25). Data-dependent MS/MS analysis was performed in an LTQ Orbitrap high resolution mass spectrometer (Thermo Electron, San Jose, CA). Tandem mass spectra were initially analyzed by the software algorithm 2TO3 to identify spectra containing a prominent -H3PO4 or -HPO3 neutral loss from the precursor ion. The resulting tandem mass spectra were searched using SEQUEST against the protein data base to identify the peptide. The resulting SEQUEST output files were filtered by DTASelect for modifications.
Mass spectrometric mapping of in vitro phosphorylation sites was
performed after tryptic digestion of kinase reactions, and samples were
analyzed using a 4000 Q TRAP system (Applied Biosystems, Foster City, CA)
interfaced with a NanoLC-2D instrument (Eksigent, Dublin, CA) for nanoflow
chromatography using an LC Packings PepMap C18 analytical
nanocolumn (75 μm × 15 cm, 3-μm bead size; 100-Å poresize;
Dionex, Sunnyvale, CA) for peptide separation. Precursor ion scanning was used
for phosphopeptide detection and sequencing of the phosphopeptides in the same
mass spectrometric run. First, a negative mode precursor ion scan is acquired,
monitoring for the marker ion 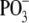 at
m/z -79 over a mass range of 500–1800
m/z. When the signal intensity of the precursor ion scan is
above a threshold of 1000 counts/s, polarity is switched to positive mode, and
a high resolution scan is acquired for charge determination and accurate mass
measurement of the three most intense ions, followed by positive mode MS/MS
sequencing. Tandem mass spectra were searched with MASCOT (Version 2.0, Matrix
Science, London, UK). All MS/MS identifications were manually validated for
quality and phosphorylation site determination.
at
m/z -79 over a mass range of 500–1800
m/z. When the signal intensity of the precursor ion scan is
above a threshold of 1000 counts/s, polarity is switched to positive mode, and
a high resolution scan is acquired for charge determination and accurate mass
measurement of the three most intense ions, followed by positive mode MS/MS
sequencing. Tandem mass spectra were searched with MASCOT (Version 2.0, Matrix
Science, London, UK). All MS/MS identifications were manually validated for
quality and phosphorylation site determination.
RESULTS
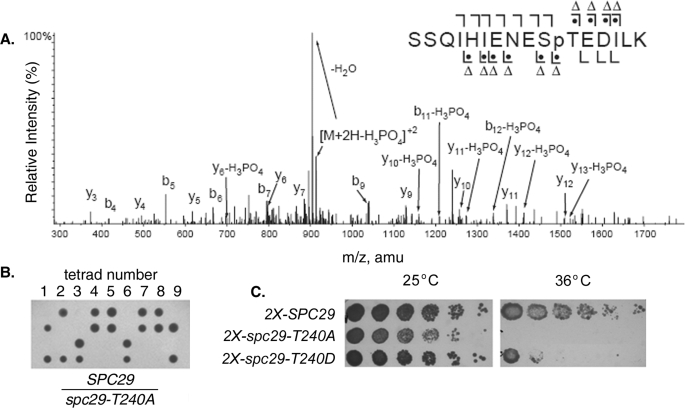
In Vivo Phosphorylation of Spc29 at Thr240 Is Required for Cell Viability—The Spc29 protein is an essential structural component of the central plaque of the SPB and is also one of four known components of the satellite, an early intermediate in the assembly of the SPB (26). Using a mass spectrometry-based analysis of in vivo phosphorylation sites on purified intact SPBs, we identified eight phosphorylation sites on Spc29. A tandem mass fragmentation spectrum for the peptide comprising residues 230–245 of Spc29 (SSQIHIENESTEDILK) from proteolytic digests of purified SPBs (Fig. 1A) demonstrates phosphate modification of Thr240, which was detected in five independent preparations of SPBs that were affinity-purified from Saccharomyces cerevisiae cells using an Mlp2-protein A fusion according to published protocols (27). Thr240 was first chosen for further analysis because it was the only site found that is conserved both throughout Saccharomyces and in the more distantly related fungi Kluyveromyces lactis and Ashbya gosypii.
FIGURE 1.
Phosphorylation of Spc29 at Thr240 is required for SPB
duplication and cell viability. A, tandem mass spectrum of the
Spc29 peptide containing Thr240 from purified intact SPBs. The
b (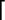 ) and y (
) and y (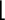 ) ions are
indicated above and below the sequence, respectively. A dot indicates
the presence of phosphate on the fragment ion, and a triangle indicates that
the observed ion has a mass consistent with neutral loss of
H3PO4 (98 Da). B, tetrad dissections from a
heterozygous SPC29/spc29-T240A diploid strain. C, growth of
haploid strains containing two copies of the indicated SPC29 allele
at 25 and 36 °C. Each strain is null for endogenous SPC29.
) ions are
indicated above and below the sequence, respectively. A dot indicates
the presence of phosphate on the fragment ion, and a triangle indicates that
the observed ion has a mass consistent with neutral loss of
H3PO4 (98 Da). B, tetrad dissections from a
heterozygous SPC29/spc29-T240A diploid strain. C, growth of
haploid strains containing two copies of the indicated SPC29 allele
at 25 and 36 °C. Each strain is null for endogenous SPC29.
An allele of the spc29 gene in which the Thr240 codon was mutated to an alanine codon was integrated into the genome in place of wild-type SPC29 to determine the importance of this phosphorylation event. To create this strain, a single copy of SPC29 was first replaced with the Geneticin-resistance marker KanMX in a wild-type diploid strain. The spc29-T240A allele was then integrated directly into the KanMX cassette to direct the mutant allele to the endogenous SPC29 locus. Sporulation and tetrad dissection of this heterozygous SPC29/spc29-T240A diploid strain yielded only two viable spores from each fourspore tetrad (Fig. 1B), and genotypic analysis confirmed that the viable spores harbored a wild-type copy of SPC29 (data not shown). This result demonstrates that cells containing a single copy of spc29-T240A instead of wild-type SPC29 at the endogenous SPC29 locus were inviable. Interestingly, integration of two copies of spc29-T240A at the HIS3 locus conferred a temperature-sensitive-for-growth phenotype, with cells inviable at 36 °C (Fig. 1C). Two copies of a second spc29 allele in which Thr240 was mutated to the phosphomimetic residue aspartate (T240D) were also integrated at the HIS3 locus, conferring a partial temperature-sensitive-for-growth phenotype that was less severe than that observed for the T240A allele (Fig. 1C), suggesting that it is the phosphorylation of this residue that is required for normal protein function.
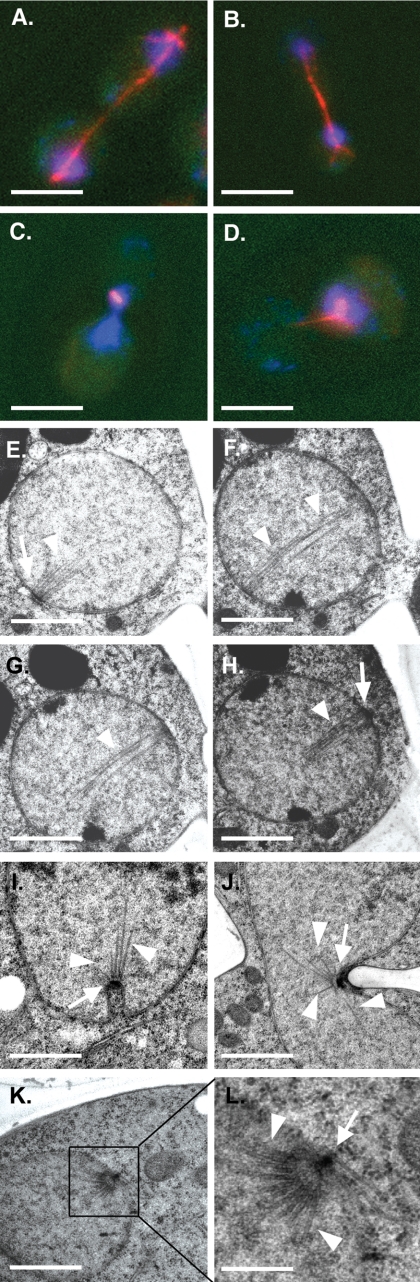
spc29-T240A Cells Fail to Duplicate SPBs—Because of the role played by the SPB in microtubule formation and organization, the mitotic spindles in temperature-sensitive spc29-T240A cells shifted to restrictive temperature were visualized by immunofluorescence and electron microscopy to determine whether the mutation affects SPB function. Wild-type and spc29-T240A cells were synchronized in G1 phase with mating factor at the permissive temperature and then released from arrest at the restrictive temperature. At 2.5 h after release, most wild-type cells had reached late anaphase (judged by morphological analysis and the presence of 2C DNA content by flow cytometric analysis) (data not shown), and samples were prepared for microtubule immunofluorescence. 96% of large budded wild-type cells displayed normal anaphase spindles, characterized by elongated microtubules with two separate DNA masses (n > 100) (Fig. 2, A and B). At this same time point, only 13.5% of spc29-T240A cells (n > 100) displayed normal anaphase spindles, whereas 86.5% of mutant cells had either completely disorganized arrays of microtubules or short microtubule arrays characteristic of monopolar spindles (Fig. 2, C and D). All of the mutant cells with abnormal microtubule organization showed only one DNA mass by 4′,6-diamidino-2-phenylindole staining, indicating that they had failed to segregate their DNA. These results indicate a failure of spc29-T240A cells to complete mitosis and suggest a defect in SPB duplication. This phenotype is similar to that observed in previously described temperature-sensitive spc29 strains (28).
FIGURE 2.
SPB assembly defects in spc29-T240A cells. A–D, immunofluorescence images showing microtubules (red) and DNA (blue) in SPC29 (A and B) and spc29-T240A (C and D) cells after a 2.5-h incubation at 36 °C. Scale bars = 5 μm. E–H, transmission electron microscopic images of four consecutive serial sections of a mitotic spindle in a wild-type SPC29 cell after a 2.5-h incubation at 36 °C. Two duplicated SPBs (arrows) connected by an array of parallel microtubules (arrowheads) are shown. Scale bars = 0.8 μm. I–L, transmission electron microscopic images of spc29-T240A cells after a 2.5-h incubation at 36 °C. Unduplicated SPBs are indicated with arrows, and splayed microtubules, characteristic of monopolar spindles, are indicated with arrowheads. A higher magnification of the monopolar spindle pictured in K is shown in L. Scale bars = 0.8 μm(I–K) and 0.2 μm(L).
Ultrastructural analysis of these cells by electron microscopy confirmed the presence of monopolar spindles. Wild-type and mutant cells from the experiment described above were harvested and processed for electron microscopy. Examination of serial thin sections of these cells demonstrated that whereas all wild-type cells (n = 12) had mitotic spindles with normal arrays of parallel microtubules organized by two SPBs (Fig. 2, E–H), all spc29-T240A cells examined (n = 12) had only one detectable SPB forming a monopolar spindle (Fig. 2, I–L; and supplemental Fig. S1). The microtubules from the single SPBs in these cells were tracked to their ends through consecutive serial sections and were not attached to another SPB. To confirm the absence of a second SPB in these mutant cells, serial sections comprising the entire nucleus of 10 additional spc29-T240A cells were observed to contain only a single SPB. These analyses confirm that prevention of phosphorylation of Spc29 at Thr240 results in a lack of SPB duplication after release from G1 arrest and subsequent mitotic failure.
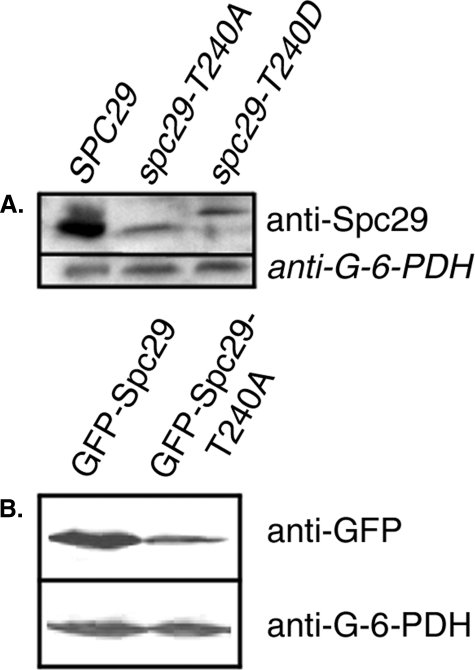
Phosphorylation of Thr240 Stabilizes Spc29—Steady-state levels of wild-type and mutant Spc29 proteins were assayed by Western blotting using an anti-Spc29 polyclonal antibody to determine whether Thr240 phosphorylation affects the stability of Spc29. Prevention of phosphorylation of Thr240 by mutation to alanine greatly reduced the levels of Spc29 protein in cells growing asynchronously at the permissive temperature (Fig. 3A) and also increased the electrophoretic mobility of the entire pool of Spc29. This result suggests that the phosphorylation of Thr240 is required for maintaining levels of Spc29 protein that are sufficient to support SPB duplication and progression through mitosis. Phosphomimetic mutation of Thr240 to aspartate resulted in a slightly more abundant protein relative to Spc29-T240A, coincident with the weak partial rescue of temperature sensitivity observed in spc29-T240D cells (Fig. 1C).
FIGURE 3.
Immunoblots of wild-type and mutant Spc29 proteins. A, levels of the indicated Spc29 protein when under the control of the endogenous SPC29 promoter detected with a rabbit anti-Spc29 polyclonal antibody; B, overexpressed GFP-Spc29 or GFP-Spc29-T240A in a wild-type strain background detected with an anti-GFP monoclonal antibody. G-6-PDH, anti-glucose-6-phosphate dehydrogenase (loading control).
The effect of phosphorylation of Thr240 on Spc29 stability was analyzed further in overexpression studies using strains in which wild-type or non-phosphorylatable alleles of SPC29 under the control of the inducible GAL4 promoter were integrated in the presence of endogenous wild-type SPC29. As shown in Fig. 3B, overexpressed Spc29-T240A protein accumulated at much lower levels than overexpressed wild-type Spc29. Densitometric analysis confirmed that only 46.2% as much Spc29-T240A protein was present in the lysate compared with wild-type Spc29. Southern blot analysis of genomic DNA isolated from these strains showed that only a single copy of each overexpression construct was integrated in each strain (data not shown).
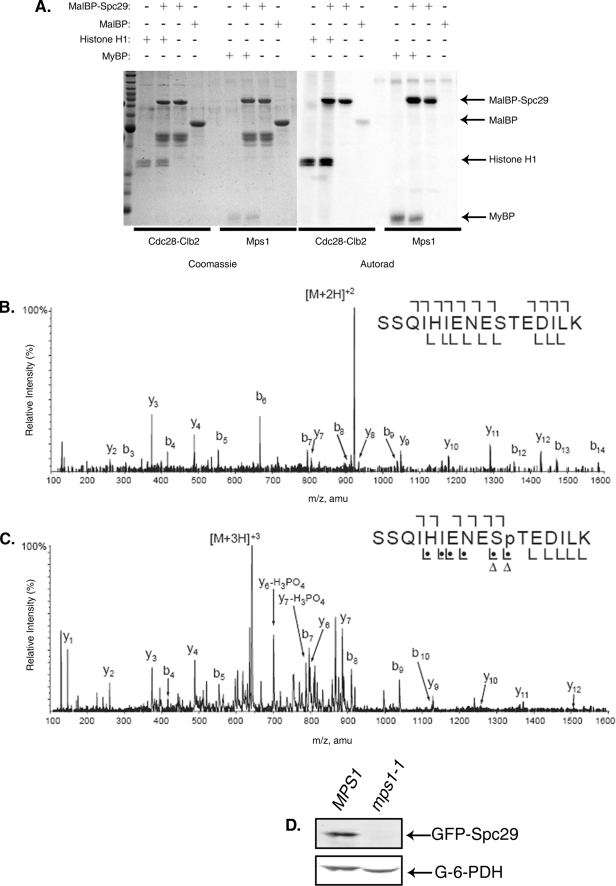
Mps1 Phosphorylates Thr240 of Spc29 in Vitro—We next sought to determine the kinase responsible for regulating the levels of the Spc29 protein via Thr240 phosphorylation. To answer this question, we performed in vitro kinase assays using two kinases known to be involved in the regulation of SPB duplication. The yeast cyclin-dependent kinase Cdc28 and the multifunctional Mps1 kinase were both able to phosphorylate Spc29 to approximately the same extent (Fig. 4A). To determine the specific sites of phosphorylation catalyzed by these two kinases, the phosphorylated proteins were analyzed by mass spectrometry-based phosphorylation site mapping. Phosphorylation of Spc29 by Mps1 was detected at 12 sites, and phosphorylation by Cdc28-Clb2 was detected at five sites (data not shown). Interestingly, although the tryptic peptide containing Thr240 of Spc29 was detected in both Cdc28-Clb2 and Mps1 kinase reactions, it was only to be phosphorylated found in the Mps1 kinase reaction. Annotated tandem mass fragmentation spectra for this peptide from both the Cdc28-Clb2 and Mps1 kinase reactions are shown in Fig. 4, demonstrating the lack of modification of Thr240 in the Cdc28-Clb2 reaction (panel B) and the presence of a phosphate at Thr240 in the Mps1 reaction (panel C). This result confirms that Mps1, but not Cdc28, phosphorylates Thr240 of Spc29 in vitro.
FIGURE 4.
Mps1, but not Cdc28, phosphorylates Thr240in vitro and is required for stabilization of overexpressed Spc29 in vivo. A, in vitro kinase assay showing phosphorylation of maltose-binding protein (MalBP)-tagged Spc29 by both Cdc28-Clb2 and Mps1. Histone H1 and myelin basic protein (MyBP) are positive control substrates for Cdc28 and Mps1, respectively. B and C, tandem mass spectra of the Spc29 peptide containing Thr240 from in vitro kinase reactions in which Cdc28-Clb2 (B) or Mps1 (C) was the kinase. See the legend to Fig. 1A for explanation of symbols. Thr240 is phosphorylated only by Mps1 in vitro. amu, atomic mass units. D, immunoblots of overexpressed GFP-Spc29 in wild-type (MPS1) or mps1-1 cells. G-6-PDH, anti-glucose-6-phosphate dehydrogenase (loading control).
Further support for the hypothesis that Mps1-mediated phosphorylation stabilizes Spc29 in vivo was provided by Western blotting of overexpressed Spc29 in cells harboring the conditional mps1-1 mutation, which is defective in both SPB duplication and the spindle assembly checkpoint due to drastically reduced kinase activity (17, 29). MPS1 or mps1-1 cells with an integrated GAL-GFP-SPC29 overexpression construct were synchronized in G1 phase with mating factor and released from arrest into inducing medium for 2 h. The cells were synchronized in G1 to facilitate the analysis of Spc29 stability at a time when all cells in the culture were actively assembling new SPBs, and the entire experiment was performed at the permissive temperature for the mps1-1 mutation. Fig. 4D shows that overexpressed Spc29 did not accumulate to detectable levels in this mutant strain, whereas Spc29 was readily detectable upon overexpression in an MPS1 strain. This result suggests that Mps1 is the kinase that is responsible for maintaining proper levels of Spc29 protein. The disparity between the levels of overexpressed Spc29-T240A in a wild-type strain background and the levels of overexpressed wild-type Spc29 in an mps1-1 mutant background may reflect additional roles for Mps1 beyond Thr240 phosphorylation in stabilizing the Spc29 protein.
DISCUSSION
The results of this study demonstrate that the phosphorylation of a single residue, Thr240, on the essential SPB component Spc29 is required for duplication of the yeast centrosome. This is the first known example of a single phosphorylation site that is required for this critical mitotic event in budding yeast. We have further demonstrated that this phosphorylation event serves to regulate the levels of the Spc29 protein and that prevention of Thr240 modification by mutation to a non-phosphorylatable residue greatly reduces Spc29 stability. Cells with the unstable Spc29-T240A mutant protein are unable to complete mitosis due to SPB duplication failure and spindle assembly defects. Results from in vitro kinase assays as well as in vivo overexpression studies show that Mps1 is likely the kinase responsible for the phosphorylation of Thr240 and stabilization of Spc29 in vivo.
Western analysis indicated that prevention of Thr240 phosphorylation changed the migration of Spc29 in an SDS-polyacrylamide gel. As shown in Fig. 3, wild-type Spc29 appeared as two bands, with the majority of the protein in the faster migrating band, whereas Spc29-T240A appeared as a single band that co-migrated with the faster migrating wild-type band. It is possible that stabilization of Spc29 by phosphorylation of Thr240 permits the accumulation of further phosphate modification and the appearance of a slower migrating hyperphosphorylated form. This hypothesis can be tested through further mutational analysis of the other seven in vivo phosphorylation sites found on Spc29. Regardless, it is clear that the levels of Spc29-T240A are greatly reduced relative to those of wild-type Spc29, which presumably leads to the inability of spc29-T240A cells to construct a second SPB in preparation for mitotic spindle assembly.
Regulation of protein stability has been demonstrated as an important mechanism of control of centrosomal proteins in yeast and mammalian cells (19, 30). Cyclin-dependent kinase-mediated phosphorylation of the Mps1 kinase, which is required for centrosome duplication, prevents its degradation in both of these systems. Subsequent stabilization of Spc29 by Mps1-mediated phosphorylation presents a paradigm by which very tight temporal regulation of the stability of SPB components can be achieved. Elevated cyclin-dependent kinase activity enhances the stability of Mps1, which can in turn promote the stability of Spc29 by phosphorylation of Thr240 at the appropriate point in the cell cycle when the accumulation of SPB components is required to support SPB duplication. Whether or not the proteasome is involved in the apparent lack of stability of Spc29-T240A protein remains to be determined, but it is possible that this modification prevents recognition by a ubiquitin ligase and subsequent proteolysis. Regardless of the mechanism of phosphorylation-mediated stabilization, this “stabilization cascade” appears to be an important means by which cells can regulate the timing of centrosome duplication.
Supplementary Material
Acknowledgments
We thank Janet Meehl for preparation of cells for electron microscopy and Sue Jaspersen for anti-Spc29 antiserum and Cdc28-Clb2 complexes. We also thank Shelly Jones and Chad Pearson for critical analysis of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM51312 (to M. W.) and Grant CA126240-01 from NCI (to Katheryn A. Resing).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
Footnotes
The abbreviations used are: SPB, spindle pole body; GFP, green fluorescent protein; MS/MS, tandem mass spectrometry.
References
- 1.Jaspersen, S. L., and Winey, M. (2004) Ann. Rev. Cell Dev. Biol. 20 1-28 [DOI] [PubMed] [Google Scholar]
- 2.Casenghi, M., Barr, F. A., and Nigg, E. A. (2005) J. Cell Sci. 118 5101-5108 [DOI] [PubMed] [Google Scholar]
- 3.Cha, H., Hancock, C., Dangi, S., Maiguel, D., Carrier, F., and Shapiro, P. (2004) Biochem. J. 378 857-865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang, P., Senga, T., and Hamaguchi, M. (2007) Oncogene 26 4357-4371 [DOI] [PubMed] [Google Scholar]
- 5.Kinoshita, K., Noetzel, T. L., Pelletier, L., Mechtler, K., Drechsel, D. N., Schwager, A., Lee, M., Raff, J. W., and Hyman, A. A. (2005) J. Cell Biol. 170 1047-1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fumoto, K., Hoogenraad, C. C., and Kikuchi, A. (2006) EMBO J. 25 5670-5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hergovich, A., Cornils, H., and Hemmings, B. A. (2007) Biochim. Biophys. Acta, 1784, 3-15 [DOI] [PubMed] [Google Scholar]
- 8.Tarapore, P., Okuda, M., and Fukasawa, K. (2002) Cell Cycle 1 75-81 [PubMed] [Google Scholar]
- 9.Chen, Z., Indjeian, V. B., McManus, M., Wang, L., and Dynlacht, B. D. (2002) Dev. Cell 3 339-350 [DOI] [PubMed] [Google Scholar]
- 10.Okuda, M., Horn, H. F., Tarapore, P., Tokuyama, Y., Smulian, A. G., Chan, P. K., Knudsen, E. S., Hofmann, I. A., Snyder, J. D., Bove, K. E., and Fukasawa, K. (2000) Cell 103 127-140 [DOI] [PubMed] [Google Scholar]
- 11.Donaldson, A. D., and Kilmartin, J. V. (1996) J. Cell Biol. 132 887-901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ficarro, S. B., McCleland, M. L., Stukenberg, P. T., Burke, D. J., Ross, M. M., Shabanowitz, J., Hunt, D. F., and White, F. M. (2002) Nat. Biotechnol. 20 301-305 [DOI] [PubMed] [Google Scholar]
- 13.Friedman, D. B., Sundberg, H. A., Huang, E. Y., and Davis, T. N. (1996) J. Cell Biol. 132 903-914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira, G., Knop, M., and Schiebel, E. (1998) Mol. Biol. Cell 9 775-793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogel, J., Drapkin, B., Oomen, J., Beach, D., Bloom, K., and Snyder, M. (2001) Dev. Ccell 1 621-631 [DOI] [PubMed] [Google Scholar]
- 16.Wigge, P. A., Jensen, O. N., Holmes, S., Soues, S., Mann, M., and Kilmartin, J. V. (1998) J. Cell Biol. 141 967-977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castillo, A. R., Meehl, J. B., Morgan, G., Schutz-Geschwender, A., and Winey, M. (2002) J. Cell Biol. 156 453-465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman, D. B., Kern, J. W., Huneycutt, B. J., Vinh, D. B. N., Crawford, D. K., Steiner, E., Scheiltz, D., Yates, J., III, Resing, K. A., Ahn, N. G., Winey, M., and Davis, T. N. (2001) J. Biol. Chem. 276 17958-17967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaspersen, S. L., Huneycutt, B. J., Giddings, T. H., Jr., Resing, K. A., Ahn, N. G., and Winey, M. (2004) Dev. Cell 7 263-274 [DOI] [PubMed] [Google Scholar]
- 20.Huisman, S. M., Smeets, M. F., and Segal, M. (2007) J. Cell Sci. 120 435-446 [DOI] [PubMed] [Google Scholar]
- 21.Jaspersen, S. L., Giddings, T. H., Jr., and Winey, M. (2002) J. Cell Biol. 159 945-956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giddings, T. H., Jr., O'Toole, E. T., Morphew, M., Mastronarde, D. N., McIntosh, J. R., and Winey, M. (2001) Methods Cell Biol. 67 27-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacCoss, M. J., McDonald, W. H., Saraf, A., Sadygov, R., Clark, J. M., Tasto, J. J., Gould, K. L., Wolters, D., Washburn, M., Weiss, A., Clark, J. I., and Yates, J. R., III (2002) Proc. Natl. Acad. Sci. U. S. A. 99 7900-7905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cantin, G. T., Shock, T. R., Park, S. K., Madhani, H. D., and Yates, J. R., III (2007) Anal. Chem. 79 4666-4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venable, J. D., and Yates, J. R., III (2004) Anal. Chem. 76 2928-2937 [DOI] [PubMed] [Google Scholar]
- 26.Adams, I. R., and Kilmartin, J. V. (1999) J. Cell Biol. 145 809-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niepel, M., Strambio-de-Castillia, C., Fasolo, J., Chait, B. T., and Rout, M. P. (2005) J. Cell Biol. 170 225-235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elliott, S., Knop, M., Schlenstedt, G., and Schiebel, E. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 6205-6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winey, M., Goetsch, L., Baum, P., and Byers, B. (1991) J. Cell Biol. 114 745-754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisk, H. A., and Winey, M. (2001) Cell 106 95-104 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.