Abstract
Molecular dynamics simulations on the wild-type AmtB protein and its D160A homology model have been performed. Although no significant structural changes due to the mutation of Asp160 were observed, calculations confirmed the critical role of Asp160 for the recognition and binding of NH4+ in AmtB. The carboxyl group of Asp160 is ~8 Å away from NH4+, but their favorable through-space electrostatic interaction is further enhanced by a hydrogen bond chain involving Ala162 (the backbone carbonyl group) and Gly163 (the backbone amide group). This explains the occurrence of the second binding site in AmtB which does not exist in the D160A mutant, as shown in the computed energy profiles. As the initially buried carboxyl group of Asp160 links to the ammonium ion in the periplasmic binding vestibule through a chain of water molecules, a likely deprotonation venue thus is from ammonium to Asp160. Combined QM(PM3)/MM molecular dynamics simulations showed that indeed Asp160 can serve as the proton acceptor and the overall proton transfer process need overcome a barrier of mere 7.7 kcal/mol, which is in good agreement with our previous QM(DFT)/MM optimizations. Significantly, the proton transfer adopts an unconventional mechanism by migrating the negative charge from the carboxyl group of Asp160 to NH4+ via two water molecules, which can be illustrated as –CO2−···H2O···H2O···NH4+ → –COOH···H2O···OH−···NH4+ → –COOH···H2O···H2O···NH3. Apparently, this is also a charge recombination process which is exothermic.
Introduction
Ammonia/ammonium transport channels in Amt/Mep/Rh family membrane proteins 1 are responsible for the acquisition or excretion of ammonium across cellular membranes which either provides a source of nitrogen for amino acid synthesis in bacteria and plants 2 or helps maintain the acid-base balance in animals and human beings.3 Although the extracellular ammonia exists predominantly in the positively charged form under physiological conditions and there have been long-lasting controversies regarding the transport mechanism and the identity of the transported species,4,5 recent X-ray crystallographic structures of both the AmtB protein from Escherichia coli 6–8 and the homologous protein Amt-1 from Archaeoglobus fulgidus 9 show that Amt proteins are homotrimers with one channel at the center of each monomer. Notably, the 20 Å-long channel is narrow and highly hydrophobic and thus conducts ammonia (NH3) rather than ammonium ion (NH4+) which need overcome extremely high desolvation penalty. Most recently, and phenyl Javelle and coworkers investigated the periplasmic recruitment vestibule for NH4+ ring gates of Phe107 and Phe215 outside the E. coli channel using the site-directed mutagenesis technique and approved that the deprotonation of NH4+ is an essential part of the conduction mechanism.10 A similar concept of gas channel has also been proposed for human Rh proteins.5,11 However, we note that this gas channel mechanism is still not universally accepted or simply is only one of several viable transport mechanisms used by Amt proteins.12 For instance, Fong et al. compared the accumulation of methylammonium in the wild-type AmtB and its W148L mutant in response to the electrical potential across the membrane and assumed that AmtB also mediates net transport of the ionic form.13 Moreover, at least for certain plant Amt homologues and Rh proteins, net ammonium ion conductance and thus electrogenic transport has been strongly suggested.14
While it is clear that many structural and mechanistic questions remain to be answered regarding the particular roles of Trp148, Asp160, Phe107, Phe215, His168, His318 etc in the binding and possible deprotonation of NH4+, as well as the gating and selectivity mechanisms of the channel and the subsequent conduction of NH3/NH4+,12,15 the availability of the Amt crystal structures spurred a wave of interests for computational simulations in attempts to get more details at the atomic and dynamical level.16–22 These computational studies unanimously confirmed the binding of NH4+ in the periplasmic recruitment vestibule and the neutral ammonia as the transported species, but differed in the deprotonation mechanism. Of particular interest, however, is the exact role of the highly conserved aspartic acid in the 160th position, which is a helix-capping residue for the fifth helix of AmtB and proximal to the external binding site and the permeation pore, in the ammonium/ammonia transportation.23,24 Site-directed mutagenesis experiments showed that the replacement of Asp160 with alanine completely disables the ammonia/ammonium transport capability, while the introduction of an acidic residue, glutamate, retains most of the activity (71%).23 Whereas these mutagenesis experiments firmly established the central role of Asp160 in the functioning of AmtB, an immediate question is how it functions. In the meanwhile, Marini et al. recently confirmed the importance of a similar aspartate residue among eukaryotic family members as distant as the yeast transporter/sensor Mep2 and the human RhAG and RhCG proteins and assumed that this aspartate residue might play a preserved functional role in Mep/Amt/Rh proteins.24 The static X-ray structure of AmtB shows that Asp160 uses its buried carboxylate ion to form hydrogen bonds with Thr165, Gly164 and Gly163 at the N-terminal end of the fifth helix (M5) and subsequently stabilizes the M4-M5 loop for cation recruitment rather than binds NH4+ directly.6 Indeed, Luzhkov et al.’s computational study showed that the proximal buried Asp160 stabilizes the bound NH4+ at the extracellular binding site by through-space electrostatic interactions and thus is largely responsible for the recruitment and selectivity of the external binding site.18 They also demonstrated that the simulated mutation of Asp160 to Asn causes the loss of binding for NH4+.
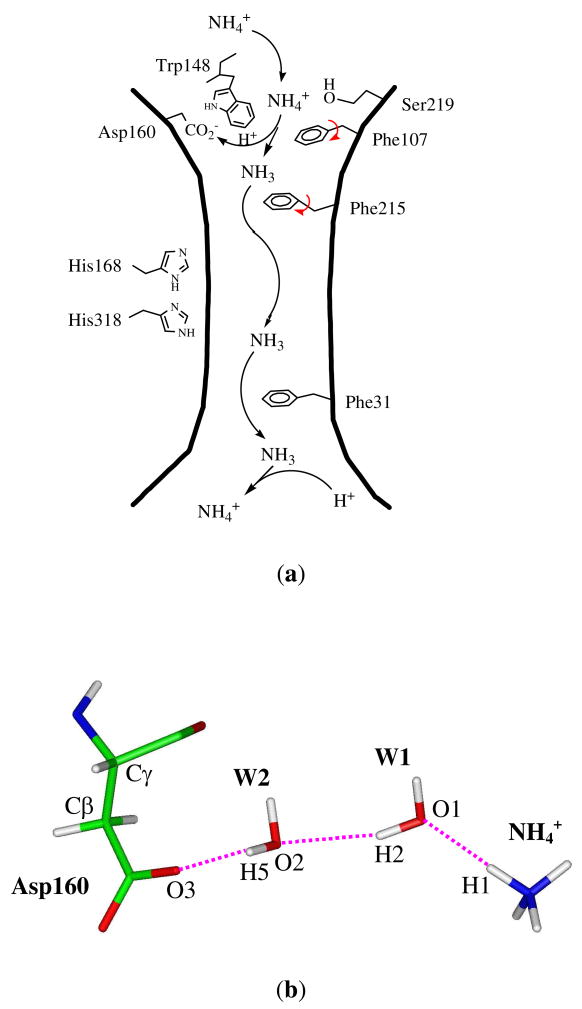
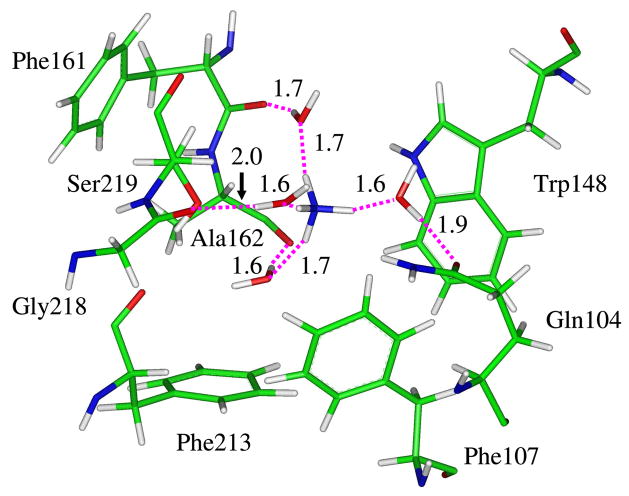
Our recent molecular dynamics (MD) simulations at the molecular mechanical (MM) level,17 however, showed that at the end of the extracellular binding site (labeled as Am16), via two water molecules, NH4+ can effectively form a hydrogen-bonded chain with the negatively charged carboxyl group of Asp160 which otherwise is shielded from the solvent by the indole ring of Trp148 (see Fig. 1). Thus, we proposed that Asp160 is probably the transient proton acceptor from NH4+ or at least the driving force for the deprotonation of NH4+ as water is the ultimate proton acceptor. This is quite analogous to the 8 Å long-range proton transfer between a zinc-bound water and a histidine residue through 2–4 intervening water molecules in carbonic anhydrase II (CAII).25–27 Subsequent quantum mechanical (QM) and combined QM/MM computations on simplified models supported our claim that Asp160 plays a functional role in the deprotonation of NH4+.21 Alternatively, Nygaard et al. suggested a proton transfer path from NH4+ to Asp160 through the backbone carbonyl and amide groups of Ala162 but at the site of Am2 instead of Am1.19 Based on the binding free energies of different species at the external cavities of AmtB and its D160N mutant, Luzhkov et al. proposed that the structural effects of the Asp160 mutation are less important than the replacement of the negative charge.18 Ishikita and Knapp similarly recognized the stabilizing role of Asp160 for NH4+ at the periplasmic binding site and found that Ser219 may act both as a H-bond donor for NH4+ and as a H-bond acceptor for NH3 and the switch of the role played by Ser219 can be crucial in the catalysis of the deprotonation of NH4+.22 Based on the dynamical simulations and pKa analyses at the MM level, Bostick and Brooks20 demonstrated a clear correlation between the protonated form of ammonia/ammonium and the number of available hydrogen bonds, and concluded that the periplasmic end is the deprotonation site, which is in agreement with ours. But they claimed that water is the only plausible proton acceptor for NH4+ and the proton most likely escapes to the periplasm in the form of hydronium.
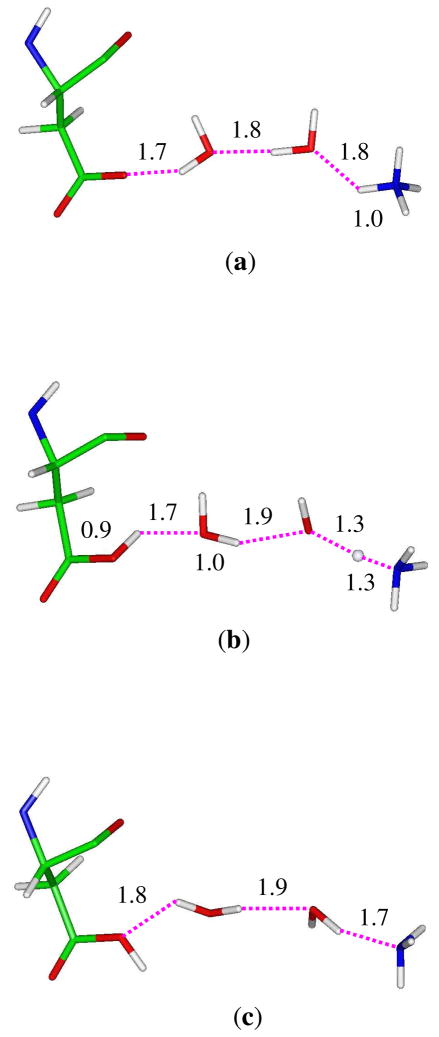
Figure 1.
(a) The modified ammonium/ammonia transport mechanism in AmtB based on the molecular dynamics simulations.17 (b) The proposed deprotonation pathway via a hydrogen bond wire between ammonium and the carboxyl group of Asp160 through two water molecules.
In this work, we further investigated one of the most likely deprotonation pathways derived from our previous MD simulations at the MM level,17 namely a proton transfer from NH4+ to the carboxylate group of Asp160 via water molecules by performing extensive combined QM/MM molecular dynamics simulations and deriving the free energy profile along the proton transfer pathway. To evaluate the probability that water takes the proton,20 we chose a reaction coordinate corresponding to a proton transfer from NH4+ to its adjacent water molecule. Based on the structural changes in the process, a deprotonation mechanism was illustrated. In addition, we studied the homology structure of the D160A mutant and examined the difference of the transport capability between the native AmtB and the D160A variant. All these studies endorse the critical functional role of Asp160, which is not only decisive for the ligand recognition and binding in both the periplasmic and intracellular binding sites, but also the most probable proton acceptor from the ammonium ion via water molecules.
Computational Methods
Detailed description of the computational model for molecular dynamics simulations can be found in our previous work.17 In brief, our complete cubic model consists of a AmtB monomer (PDB entry 1U7G)6 embedded in a phospholipid bilayer of DMPC with water slabs of 30 Å thickness added to the top and bottom of the bilayer. Except aspartatic and glutamic acids with negative charges and lysine residues with positive charges, all rest residues are set neutral. The protonation states of histidine residues, however, are determined based on their individual niches, for instance, both His168 and His318 located within the channel are found to be neutral, but the proton is attached to ND1 in the imidazole ring for His168 whereas the proton is on NE2 for His318. Hydrogen positions are incorporated using the HBUILD facility in CHARMM.28 For the D160A mutant, Asp160 is substituted by an alanine using Accelery’s InsightII package. The simulated box, electrically neutralized by adding 1–3 chloride ions, is further extended with periodic boundary condition (PBC). Since the channel is located in the middle of the protein and external ions except ammonium are far away from the channel, we have not attempted to add NH4+ and Cl− to make up the physiological ionic concentration. The electrostatic interactions are computed with no truncation using the particle mesh Ewald (PME) algorithm.29 The trajectories were generated by performing a number of dynamics simulations which were separated by 0.2~0.4 Å in reaction coordinate with a time step of 1 fs at constant pressure and temperature (300 K). On each window, the first 50~100 ps simulation brought the system to an equilibrium state, followed by the second 100 ps simulation which generated dynamics data for further analyses. Calculations were carried out with CHARMM 28 using the all-atom empirical potential energy function for proteins 30 and phospholipids 31. For the water molecules, the TIP3P potential was used.32 Umbrella sampling technique was adopted to generate the free energy profile or potential of mean force (PMF) along either the transduction trajectory or reaction coordinates and a biasing harmonic potential with a force constant of 15~30 kcal/mol was imposed in simulations. In the simulations, all of the bonds with hydrogen atoms in AmtB except the QM part will be constrained with the SHAKE algorithm (with a tolerance of 10−6).33
In the exploration of the NH4+ deprotonation process with a combined QM/MM energy potential, the QM part consists of the ammonium ion, two water molecules and the carboxyl group of Asp160 where the QM/MM boundary atom Cγ (see Fig. 1) was treated with the generalized hybrid orbital (GHO) method.34 As such, there are 17 QM atoms in total. To make the calculations efficient, we used the semi-empirical PM335 method to obtain the quantum mechanical energy in the combined QM/MM molecular dynamics simulations.
Results and Discussion
Homology model of the D160A mutant
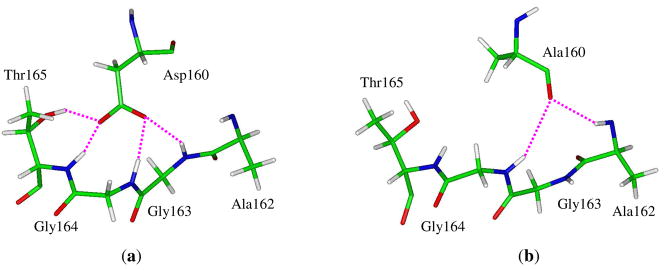
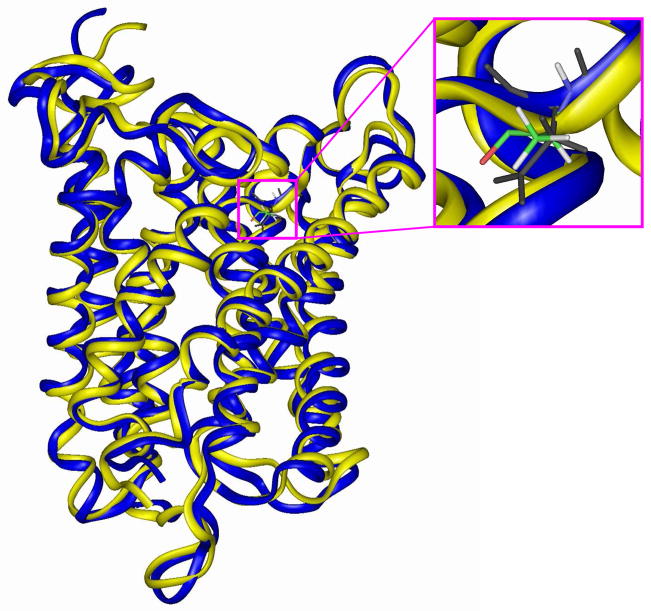
In our previous study of the NH4+/NH3 conductance through the E. coli AmtB,17 we derived the potential energy profile along the reaction coordinate (RC) which is defined as the distance of NH4+/NH3 to the last position (Am4) inside the internal channel pore identified in the AmtB crystal structure,6 and the computations were initiated from the AmtB-NH4+ complex where NH4+ is fully solvated with four water molecules in the first hydration shell (RC = −20.7 Å).17 By mutating Asp160 with alanine at this site, we obtained the D160A variant complexed with a NH4+ just outside the extracellular binding vestibule. After performing 100 ps simulations for equilibrium, we superimposed the final mutant configuration with the wild-type protein and the RMSD is 1.16 Å for the backbone atoms between the two structures, which suggests the high similarity of both structures. But our focal point is the niche of the 160th residue. In AmtB, Asp160 is a helix-capping residue for the fifth helix and its negatively charged carboxyl group is inaccessible to the solvent and used to fix the backbone of the helical structure by forming hydrogen bonds with backbone amide groups of residues Ala162, Gly163, Gly164 and Thr165 (see Fig. 2a).6,7,17 Its main chain carbonyl group, however, directs to the solvent and forms hydrogen bonds with water molecules. With the replacement of the carboxyl group with a hydrogen atom, the main chain carbonyl group of Ala160 in the D160A mutant flips to the position originally taken by the carboxyl group of Asp160 to form hydrogen bonds with the main-chain amide groups of Ala162 and Gly164 to sustain the framework of the fifth helix (see Fig. 2b). As a consequence, no significant structural difference between AmtB and its D160A variant is observed and the mutation of Asp160 neither changes the pore structure nor blocks the entrance of the binding vestibule. Fig. 3 shows the overall surroundings of Asp160/Ala160 in the superimposed wild-type AmtB and its D160A mutant.
Figure 2.
Important hydrogen-bonding interactions (highlighted with magenta dashed lines) between the fifth helix and Asp160 in the wild-type AmtB (a) or Ala160 in the D160A mutant (b).
Figure 3.
The orientations of Asp160 in AmtB (in black) and Ala160 in the D160A mutant (in color) after superimposing these two protein structures (AmtB in blue and D160A mutant in yellow).
Conductance capability of the D160A mutant
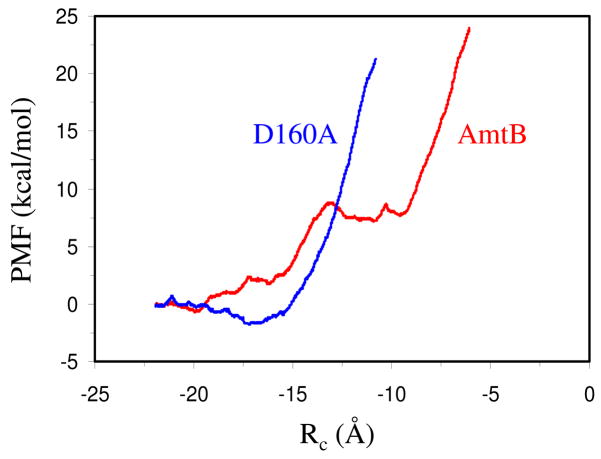
To probe the functional role of Asp160 versus Ala160 in quantitative details, we simulated the binding and transduction of an ammonium ion from outside the binding site to the periplasmic binding vestibule and further into the channel. Fig. 4 compares the free energy (or potential of mean force, PMF) profiles along the NH4+ transduction trajectory through the native AmtB and its D160A mutant channels, where the energies at the starting configurations were referenced as zero. By means of the free energy perturbation and molecular dynamics simulations, Luzhkov et al. found that the mutation of Asp160 to Asn destabilizes the bound NH4+ by 10 kcal/mol.18 Albeit a significant loss of binding stability and heightened barrier, we found that similar to the wild-type AmtB, there is still a shallow periplasmic binding vestibule (RC = −17.5 ~ −15.5 Å) in the D160A mutant which thus can recruit and retain NH4+, as demonstrated in Fig. 4. Fig. 5 presents a snapshot at RC = −17.0 Å showing NH4+ in the extracellular ion-binding vestibule of the D160A mutant. At this snapshot, NH4+ is still hydrated by four water molecules, but all these four water molecules form hydrogen bonds with the backbone carbonyl oxygen atoms of Gln104, Phe161, Ala162, and Gly218. With its further advance in the binding vestibule, NH4+ will lose one water molecule in its first hydration shell, but the energetic loss is largely compensated by the hydrogen bond with the backbone carbonyl oxygen of Phe161 (or Ala162 when NH4+ moves away a little bit) as well as the π-cation interaction with Trp148 and Phe107. This picture is essentially identical to the one in the native AmtB,17 except that there is no carboxylate ion at the distance of around 8 Å to stabilize the bound NH4+ in the D160A mutant via through-space electrostatic attraction.18 Similar to the wild type, however, we also observed the rotation of the phenyl ring of Phe107 at this stage to allow the passage of the ammonium ion. Since the phenyl ring of Phe107 rotates with the approaching of ammonium ion essentially without any additional energetic requirement, Phe107 must play a trivial role in the ammonium/ammonia transportation. The mutagenesis experiments recently conducted by Javelle et al. confirmed that the removal of this phenylalanine gate has little impact on the AmtB activity.10
Figure 4.
Potential of mean force (PMF) profiles for the passage of an ammonium ion across the AmtB (in red) and the D160A mutant (in blue) proteins. The free energies at the starting configurations are referenced as zero.
Figure 5.
Snapshot showing the surrounding for the ammonium ion in the extracellular binding site in the D160A mutant (RC = −17.0 Å).
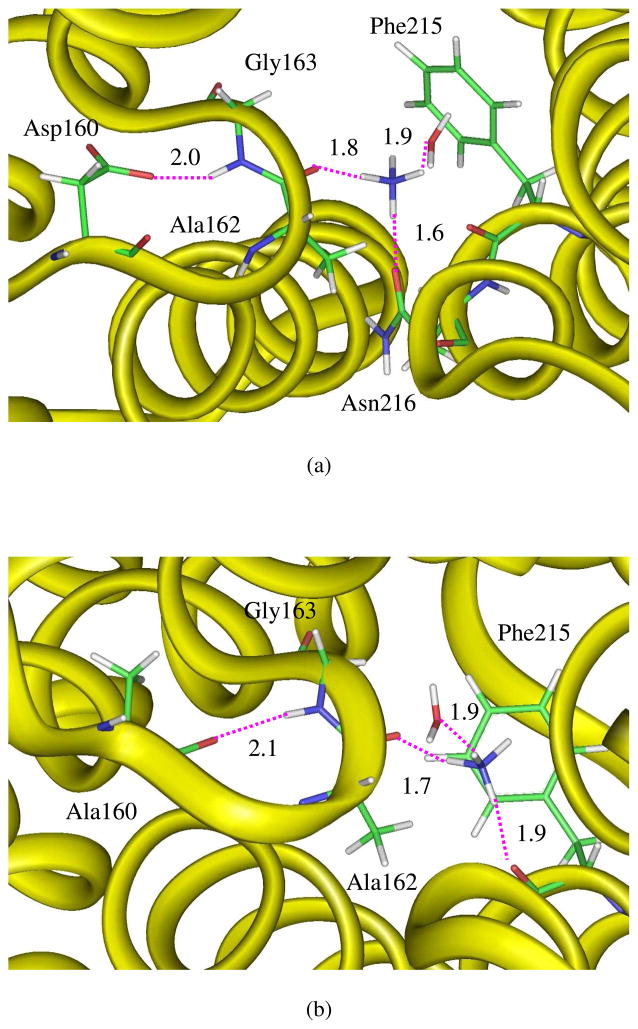
Interestingly, significant disparity occurs with the continuing advance of NH4+ into the channel. Due to the highly hydrophobic and narrow nature of the channel, NH4+ will lose its most if not all hydration water molecules and this desolvation process is extremely endothermic, as evidenced by the high solvation free energy of ammonium ion in water (~80 kcal/mol).36 Thus, a steep increase of energy is observed for the transport of NH4+ in the D160A mutant after the extracellular binding site (see the blue curve in Fig. 4). In sharp contrast, in the native AmtB, an intracellular binding site in the range of −12.2 ~ −9.2 Å is identified. Structural analyses indicated that at this site the second gate, namely the phenyl ring of Phe215, opens only slightly and NH4+ moves around of the phenyl ring to form hydrogen bonds with either Asn216 or His168 which has been speculated as a possible proton acceptor.7 However, it seems that the stabilization of NH4+ in the second binding site in the native AmtB comes from the negatively charged carboxyl group of Asp160 which is only ~8 Å away and forms a hydrogen-bonded chain with NH4+ via the backbones of Ala162 and Gly163 (Fig. 6a). When NH4+ moves deeper, the only coupled water which can go through the channel together with the ammonium ion joins in the hydrogen-bond wire and makes the ammonium ion remain stable in a ~3 Å long range. With two opposite charges located at the two ends of a hydrogen-bonded wire, the hydrogen bonding interaction or stabilization is expected to be enhanced remarkably. By analyzing the binding energies, Luzhkov et al. pointed out that the electrostatic interactions, notably from the proximal carboxylate ion of Asp160, are the largest stabilizing factor for NH4+ at the periplasmic binding vestibule.18 But we note that there is no directionality for through-space electrostatic interactions. Thus, simple electrostatic model can not explain the appearance of the intracellular binding site in AmtB. With the mutation of Asp160 with alanine, the carbonyl group of Ala160 flips and is similarly able to form hydrogen-bond wire with NH4+ via Ala162 and Gly163 (Fig. 6b). But the magnitude of stabilization must be greatly reduced compared with the AmtB case, and as such, there is no intracellular or second binding site for ammonium ion in the D160A mutant.
Figure 6.
Snapshots illustrating the surroundings for the ammonium ion (a) in the intracellular binding region of the native AmtB (RC = −11.3 Å) and (b) in a similar position in the D160A mutant (RC = −10.8 Å) which is not a binding site.
Role of Asp160 for the intracellular binding site in the wild-type AmtB
While strong electrostatic attraction between the negatively charged carboxyl group of Asp160 and the positively charge ammonium ion has been recognized due to their relatively short distance (~8 Å),18 Fig. 6 suggests that Asp160 may have a secondary stabilizing role by influencing the hydrogen bond between NH4+ and the main-chain carbonyl group of Ala162 and resulting in the second binding site which locates around the phenyl ring of Phe215. Although overwhelming information on hydrogen bonds can be found in the literature and hydrogen-bonding interactions conventionally fall into the category of weak interactions with a strength ranging from 2 to 10 kcal/mol,37 new types of strong hydrogen bonds have been proposed and investigated. These new types include charge assisted hydrogen bonds (CAHBs),38,39 low barrier hydrogen bonds (LBHBs),40 dihydrogen bonds (DHBs)41 and resonance-assisted hydrogen bonds (RAHBs)38,42,43. Among these strong hydrogen bonds, CAHB refers to a case where a positive or negative charge on the proton donating or accepting group remarkably increases the strength of the hydrogen bond while RAHB highlights the cooperativity between π-electron delocalization and hydrogen bonds. The hydrogen bonds between the carboxyl group of Asp160 and the backbone amine group of Gly163 as well as between the ammonium ion and the backbone carbonyl group of Ala162 seem to satisfy the requirements for both CAHB and RAHB (see Fig. 6a). The negatively charged carboxyl group strongly polarizes the hydrogen donor, the amine group of Gly163 which conjugates with the adjacent carbonyl group of Ala162, resulting in an enhanced shifting (polarization) of electron density from the amine group to the carbonyl oxygen. Since the carbonyl oxygen forms hydrogen bond with the positively charged ammonium ion, the electron density accumulation on the oxygen atom further consolidates its position as a hydrogen bond acceptor and strengthens the hydrogen bonding interaction with NH4+ which is dominated by the electrostatic forces.43
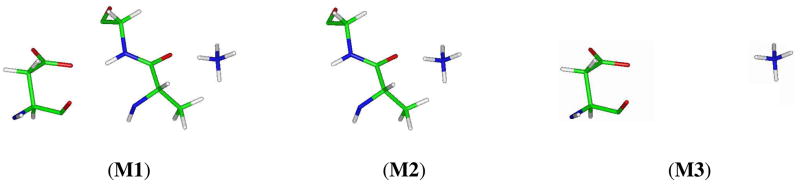
To explore the impact of the negatively charged carboxyl group of Asp160 on the hydrogen bonding between NH4+ and the backbone carbonyl group of Ala162, we performed QM calculations at the MP2/6-31+G(d) level on reduced models including Asp160, Ala162, Gly163 and NH4+ as plotted in Fig. 7 with broken bonds saturated with hydrogen atoms. The structural parameters for the models are taken from the configurations shown in Fig. 6. Since here we focus on the hydrogen bonding interaction of NH4+ with the carbonyl group of Ala162, we take NH4+ as one fragment A and the rest as another fragment B, and define the binding energy ΔE as the energy difference between the complex AB and fragments A and B with the basis set superposition error (BSSE) correction. Among the three simplified models shown in Fig. 7, M1 refers to the interaction of NH4+ with both Ala162 and Asp160, while models M2 and M3 correspond to the interaction of NH4+ with either Ala162 or Asp160 alone. Thus, the change of the binding energy (ΔΔE) in M1 compared with the summation of binding energies in M2 and M3 reflects the coupling effect, or the enhanced hydrogen bonding interaction due to the so-called CAHB and RAHB effects. Table 1 compiles the computation results. While the through-space electrostatic attraction between Asp160 and NH4+ reaches as high as −39.5 kcal/mol although they are separated by ~8 Å, the hydrogen bond between NH4+ and the carbonyl group of Ala162 is far stronger than conventional hydrogen bonds. With the carboxyl group at the other end of the conjugated segment, the polarization induced by the anion shifts electron density along the conjugated bonds to the carbonyl oxygen atom, and eventually enhanced the binding to NH4+ by 9.0 kcal/mol. The mutation of Asp160 to Ala160 reduces the extra coupling energy to only 2.3 kcal/mol. Furthermore, if we consider the bonding between the carboxylate ion and the amine group of Gly163 similarly enhanced by NH4+, the energy gain for the overall AmtB-NH4+ complex would be even much more than 9.0−2.3=6.7 kcal/mol, and eventually results in the second intracellular binding site in addition to the first periplasmic binding site.
Figure 7.
Models for quantum mechanical calculations to evaluate the charge-enhanced hydrogen bonding interaction between the ammonium ion and the carbonyl group of Ala162.
Table 1.
Quantum mechanical calculations of binding energies of ammonium ion with model systems of E. coli AmtB and the D160A mutant at the MP2/6-31+G(d) level (kcal/mol)
| System | ΔE1 (Model M1) | ΔE2 (Model M2) | ΔE3 (Model M3) | ΔΔE1) |
|---|---|---|---|---|
| AmtB | −77.5 | −29.0 | −39.5 | −9.0 |
| D160A | −26.3 | −21.4 | −2.6 | −2.3 |
ΔΔE = ΔE1 − ΔE2 − ΔE3.
Thus, the carboxyl group of Asp160 is responsible for not only the ammonium uptake in the periplasmic binding site as well recognized,18 but also the existence of the second binding site in AmtB which is only about 6 kcal/mol higher in energy than the first binding site. But we note that this second binding site was not identified in the crystal structure,6 and it locates between the positions of Am1 for NH4+ and Am2 for NH3. Our previous MM simulations17 suggested that the deprotonation of NH4+ occurs in the first rather than the second binding site, which will be investigated in the following subsection.
Deprotonation of NH4+ in the periplasmic binding site
As it was proposed that the NH4+ deprotonation most probably occurs in the periplasmic binding vestibule and the highly conserved Asp160 is likely the proton acceptor,17,21 we performed combined QM/MM molecular simulations to explore the possible proton transfer pathways by deriving the free energy profile along reaction coordinates. Whereas localized proton transfers have been well studied,44 long-range proton transfers in biochemical processes are more challenging to quantify as the exact transfer pathways and rate-limiting factors are difficult to identify due to the involvement of many residues and solvents.26,45 One prominent difficulty in the theoretical studies of long-range proton transfers is the proper definition of a reaction coordinate to characterize the progress of long-range proton transfer.27 A promising solution is to define reaction coordinates for long-range proton transfers by considering the movement of the excess charge center.27,46 For our current case where only two water molecules are involved as shown in Fig. 1b, however, a linear combination of donor-proton and acceptor-proton distances might be effective.27 In general, the to Asp160 could occur via a sequential hopping of protons through proton transfer from NH4+ water molecules analogous to the von Grotthuss mechanism which explains the anomalously high proton mobility in water,47 or via a stepwise mechanism in which NH4+ first loses a proton to the water molecule in its first hydration shell, followed by a proton transfer to the next water, and finally to the carboxylate group. In either way, the departure of a proton from NH4+ initiates the whole process. Thus, we focus on the proton H1 as shown in Fig. 1b and choose the distance difference between the bonds N-H1 and O1-H1 as the reaction coordinate in the simulations. To preserve the connectivity of the hydrogen bonded chain, in the simulations the heavy atoms were subject to harmonic constraint with 10 kcal/mol·Å2 force. As such, the calculated value of barrier corresponds to its upper bound.
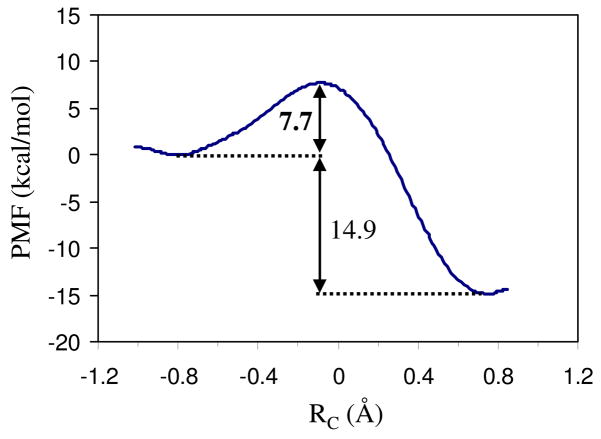
Fig. 8 shows the energy profile where the proton transfer reaction proceeds from left (negative values of the reaction coordinate correspond to the reactant state) to right (positive values of the reaction coordinate correspond to the product state). Our simulations suggest that the reaction occurs fast as the barrier is only 7.7 kcal/mol which is in good agreement with the value of 7.2 kcal/mol derived from our previous QM(DFT)/MM single-point optimizations.21 The combination of the positive and negative charges eventually stabilizes the system by 14.9 kcal/mol. At the reactant state which is located in the range of RC = −0.85 ~ −0.75 Å, the ammonium ion forms a hydrogen bond network with three water molecules as well as the hydroxyl group of Ser219. The carboxyl group of Asp160, however, forms hydrogen bonds with the hydroxyl group of Thr165 and one water molecule (the latter is shown in Fig. 9a). At this state, an ideal hydrogen-bonded chain from NH4+ to the –CO2− of Asp160 is observed as shown in Fig. 9a. The subsequent proton transfer experiences a transition state at the vicinity of −0.09 Å where the transferred proton H1 lingers in the midst of the donor and acceptor. Fig. 9b shows a snapshot of the reactive part in the transition state. Of significant interest, the negatively charged carboxylate ion of Asp160 has already captured a proton from the nearby water molecule (W2), which has simultaneously got a proton from W1 to maintain its neutral structure. As a consequence, the water molecule (W1) in the first hydration shell of NH4+ is deprotonated to generate a hydroxide anion in the transition state. This is, however, consistent with the QM/MM single-point optimizations at the DFT level,21 which also suggested the existence of a hydroxide ion in the intermediate state except that the hydroxide ion is in the W2 position. Thus, apart from the migration of a positive charge in a proton transfer process,47 there is also a possibility that a negative charge migrates until the quench of charges in a proton transfer process. This unconventional alternative mechanism was recently proposed and demonstrated computationally in two solution systems and in CAII by Cui and coworkers.48 Based on the molecular dynamics simulations at the DFT level, Tuckerman et al. also found that hydrated hydroxide anion with a 3-fold oxygen coordination plays a crucial role for the fast transport of OH− in aqueous solution.49 In the present study of the transition state of the deprotonation of NH4+ in AmtB, the hydroxide ion is also stabilized by a hydrogen bond network, including water (W2), NH4+ and the amine group in the indole ring of Trp148. A snapshot showing the niche of the hydroxide anion at the transition state is presented in the Supporting Information.
Figure 8.
Potential of mean force (PMF) profile for the process of proton transfer from the ammonium ion to the nearest water molecule which links to the carboxyl group of Asp160 via another water molecule.
Figure 9.
Snapshots for the structural changes of the reactive part along the deprotonation of NH4+ in the combined QM/MM molecular dynamics simulations: (a) reactant state (RC = −0.8 Å); (b) transition state (RC = −0.1 Å); (c) product state (RC = 0.7 Å).
To probe when this series of covalent bond cleavage/formation in the proton transfers from W1 to W2 and from W2 to Asp160 happen, we traced the distance changes for hydrogen bonds O3-H5 and O2-H2 along the reaction coordinate. While the changes for the bonds O1-H1 and N-H1 are smooth as their difference is used as the reaction coordinate, there are dramatic changes for the bonds O2-H2 and O3-H5 at RC = −0.6 Å which is still quite close to the reactant state rather than to the transition state. When RC is less than −0.6 Å, we observe large fluctuations of these hydrogen bond distances which fall into two groups, one is at around 1.8–2.0 Å and the other varies at 2.3–3.2 Å. These two groups correspond to the two O-H bonds in a water molecule. Once RC moves over −0.6 Å, however, the O2-H2 and O3-H5 bonds fluctuates little at 1.0 Å, implying strong chemical bonds. Thus, the proton transfers along the hydrogen bond wire W1-W2-Asp160 are sequential with very little energetic costs, and the net outcome is the generation of a hydroxide ion as an intermediate. In other words, most of the barrier (7.7 kcal/mol) corresponds to the breaking of the N-H1 bond and the formation of the O1-H1 bond.
Conclusion
The analyses of the D160A computational model revealed that the mutation of the carboxyl group with a hydrogen atom results in the flipping of the backbone carbonyl group which forms hydrogen bonds with the residues in the fifth helix and consequently helps support the scaffold of the protein. Thus, the role of Asp160 should be primarily functional. In the E. coli AmtB, although the negatively charged carboxyl group of Asp160 is buried and about 8 Å away from the transported ammonium ion, there is significant stabilizing through-space electrostatic interaction between them. If NH4+ does not deprotonate in the periplasmic binding site and continues to move into the channel, this favorable electrostatic interaction will be further enhanced by a hydrogen bond chain through Ala162 (the backbone C=O group) and Gly163 (the backbone N-H group). This explains the occurrence of the intracellular binding site in AmtB but none in the D160A mutant, as exhibited in the computed energy profiles. In brief, our computations reinforced the essential role of Asp160 in the uptake of ammonium ions in AmtB via long-range electrostatic attraction from the carboxylate ion, and this conclusion is in consistent with previous findings.
To explore the deprotonation mechanism, we carried out combined QM/MM molecular dynamics simulations at the periplasmic binding site to derive the energy profile along one of the most likely proton transfer pathways, namely from NH4+ to the carboxylate group of Asp160 via two water molecules. Significantly, instead of using the well-known Grotthuss mechanism, AmtB adopts an unconventional mechanism in which a negative charge migrates from the carboxylate ion of Asp160 to NH4+ through a water chain. The proton transfer includes two steps and the first step is the sequential proton transfers from the adjacent water molecule (W2) to Asp160, and from another water molecule (W1) to W2. Since W1 is in the first hydration shell of NH4+, an ion pair OH−···NH4+ is thus formed after the first proton transfer step. The second step concerns the charge annealing after one proton is transferred from NH4+ to OH−. Overall, at the present QM(PM3)/MM level, the reaction barrier is only 7.7 kcal/mol, suggesting a fast process for the deprotonation of the ammonium ion in the periplasmic binding vestibule of AmtB.
Finally, we would like to point out that the recent fine work done by Javelle et al. is not against the hypothesis that the NH4+ deprotonation occurs in the periplasmic binding vestibule.10 Their site-directed mutagenesis experiment showed that the AmtB activity absolutely requires Phe215 but not Phe107. Whereas our simulations confirmed the trivial role of Phe107 in the ammonium transportation and deprotonation, Phe215 plays a structural role by blocking the way for NH4+ and confines the latter in the periplasmic binding site until its deprotonation. Our previous simulations have already demonstrated the neutral ammonia can easily open the second gate and pass Phe215 to enter and eventually go through the channel.17
Supplementary Material
Acknowledgments
This work was supported by the Keck foundation, the National Institute of Health (NIH) and by funds from the Faculty Research and Creative Activities Support Fund, Western Michigan University.
Footnotes
Supporting Information Available: Complete citation for reference 30 and a snapshot showing the interactions of the hydroxide anion with surrounding residues and solvent molecules at the transition state in the deprotonation of ammonium cation. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jayakumar A, Hwang SJ, Fabiny JM, Chinault AC, Barnes EMJ. J Bacteriol. 1989;171(996) doi: 10.1128/jb.171.2.996-1001.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]; Marini AM, Vissers S, Urrestarazu A, Andre B. EMBO J. 1994;13:3456. doi: 10.1002/j.1460-2075.1994.tb06651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ninnemann O, Jauniaux JC, Frommer WB. EMBO J. 1994;13:3464. doi: 10.1002/j.1460-2075.1994.tb06652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Marini AM, Matassi G, Raynal V, Andre B, Cartron JP, Cherif-Zahar B. Nature Genetics. 2000;26:341. doi: 10.1038/81656. [DOI] [PubMed] [Google Scholar]; Westhoff CM, Ferreri-Jacobia M, Mak DOD, Foskett JK. J Biol Chem. 2002;277:12499. doi: 10.1074/jbc.C200060200. [DOI] [PubMed] [Google Scholar]
- 2.von Wirén N, Merrick M. Trends Curr Genet. 2004;9:95. [Google Scholar]
- 3.Knepper MA, Packer R, Good DW. Physiol Rev. 1989;69:179. doi: 10.1152/physrev.1989.69.1.179. [DOI] [PubMed] [Google Scholar]
- 4.Soupene E, Ramirez RM, Kustu S. Mol Cell Biol. 2001;21:5733. doi: 10.1128/MCB.21.17.5733-5741.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Soupene E, Lee H, Kustu S. Proc Natl Acad Sci USA. 2002;99:3926. doi: 10.1073/pnas.062043799. [DOI] [PMC free article] [PubMed] [Google Scholar]; Meier-Wagner J, Nolden L, Jakoby M, Siewe R, Kramer R, Burkovski A. Microbiol. 2001;147:135. doi: 10.1099/00221287-147-1-135. [DOI] [PubMed] [Google Scholar]; Bakouh N, Benjelloun F, Hulin P, Brouillard F, Edelman A, Cherif-Zahar B, Planelles G. J Biol Chem. 2004;279:15975. doi: 10.1074/jbc.M308528200. [DOI] [PubMed] [Google Scholar]
- 5.Ripoche P, Bertrand O, Gane P, Birkenmeier C, Colin Y, Cartron JP. Proc Natl Acad Sci USA. 2004;101:17222. doi: 10.1073/pnas.0403704101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khademi S, O’Connell J, Remis J, Robles-Colmenares Y, Miericke LJW, Stroud RM. Science. 2004;305:1587. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 7.Zheng L, Kostrewa D, Berneche S, Winkler FK, Li XD. Proc Natl Acad Sci USA. 2004;101:17090. doi: 10.1073/pnas.0406475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khademi S, Stroud RM. Physiology. 2006;21:419. doi: 10.1152/physiol.00051.2005. [DOI] [PubMed] [Google Scholar]; Khademi S, Stroud RM. Gas channels for ammonia. In: Grisshammer R, Buchanan SK, editors. Structural Biology of Membrane Proteins. Royal Society of Chemistry; Cambridge, UK: 2006. p. 212. [Google Scholar]
- 9.Andrade SLA, Dickmanns A, Ficner R, Einsle O. Proc Natl Acad Sci USA. 2005;102:14994. doi: 10.1073/pnas.0506254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Javelle A, Lupo D, Ripoche P, Fulford T, Merrick M, Winkler FK. Proc Natl Acad Sci USA. 2008;105:5040. doi: 10.1073/pnas.0711742105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conroy MJ, Bullough PA, Merrick M, Avent ND. Brit J Haematol. 2005;131:543. doi: 10.1111/j.1365-2141.2005.05786.x. [DOI] [PubMed] [Google Scholar]; Lupo D, Li XD, Durand A, Tomizaki T, Cherif-Zahar B, Matassi G, Merrick M, Winkler FK. Proc Natl Acad Sci USA. 2007;104:19303. doi: 10.1073/pnas.0706563104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Javelle A, Lupo D, Li XD, Chami M, Ripoche P, Merrick M, Winkler FK. J Struct Biol. 2007;158:472. doi: 10.1016/j.jsb.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Fong RN, Kim KS, Yoshihara C, Inwood WB, Kustu S. Proc Natl Acad Sci USA. 2007;104:18706. doi: 10.1073/pnas.0709267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludewig U, von Wirén N, Frommer WB. J Biol Chem. 2002;277:13548. doi: 10.1074/jbc.M200739200. [DOI] [PubMed] [Google Scholar]; Mayer M, Dynowski M, Ludewig U. Biochem J. 2006;396:431. doi: 10.1042/BJ20060051. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mayer M, Schaaf G, Mouro I, Lopez C, Colin Y, Neumann P, Cartron JP, Ludewig U. J Gen Physiol. 2006;127:133. doi: 10.1085/jgp.200509369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Severi E, Javelle A, Merrick M. Mol Memb Biol. 2007;24:161. doi: 10.1080/09687860601129420. [DOI] [PubMed] [Google Scholar]
- 16.Liu YM, Hu XH. J Phys Chem A. 2006;110:1375. doi: 10.1021/jp054261c. [DOI] [PubMed] [Google Scholar]; Lamoureux G, Klein ML, Berneche S. Biophys J. 2007;92:L82. doi: 10.1529/biophysj.106.102756. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yang H, Xu Y, Zhu W, Chen K, Jiang H. Biophys J. 2007;92:877. doi: 10.1529/biophysj.106.090191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Y, Cao Z, Mo Y. J Am Chem Soc. 2006;128:10876. doi: 10.1021/ja0631549. [DOI] [PubMed] [Google Scholar]
- 18.Luzhkov VB, Almloef M, Nervall M, Aaqvist J. Biochemistry. 2006;45:10807. doi: 10.1021/bi0610799. [DOI] [PubMed] [Google Scholar]
- 19.Nygaard TP, Rovira C, Peters GH, Jensen MO. Biophys J. 2006;91:4401. doi: 10.1529/biophysj.106.089714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bostick DL, Brooks CL., III PLoS Comput Biol. 2007;3:231. doi: 10.1371/journal.pcbi.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao Z, Mo Y, Thiel W. Angew Chem Int Ed. 2007;46:6811. doi: 10.1002/anie.200701348. [DOI] [PubMed] [Google Scholar]
- 22.Ishikita H, Knapp EW. J Am Chem Soc. 2007;129:1210. doi: 10.1021/ja066208n. [DOI] [PubMed] [Google Scholar]
- 23.Javelle A, Severi E, Thornton J, Merrick M. J Biol Chem. 2004;279:8530. doi: 10.1074/jbc.M312399200. [DOI] [PubMed] [Google Scholar]
- 24.Marini AM, Boeckstaens M, Benjelloun F, Cherif-Zahar B, Andre B. Curr Genet. 2006;49:364. doi: 10.1007/s00294-006-0062-5. [DOI] [PubMed] [Google Scholar]
- 25.Eriksson AE, Jones TA, Liljas A. Proteins: Struct, Funct, Genet. 1988;4:274. doi: 10.1002/prot.340040406. [DOI] [PubMed] [Google Scholar]; Tu C, Silverman DN, Forsman C, Jonsson BH, Lindskog S. Biochemistry. 1989;28:7913. doi: 10.1021/bi00445a054. [DOI] [PubMed] [Google Scholar]; Christianson DW, Fierke CA. Acc Chem Res. 1996;29:331. [Google Scholar]; Lu D, Voth GA. J Am Chem Soc. 1998;120:4006. [Google Scholar]; Cui Q, Karplus M. J Phys Chem B. 2003;107:1071. [Google Scholar]
- 26.Toba S, Colombo G, Merz KM. J Am Chem Soc. 1999;121:2290. [Google Scholar]
- 27.König PH, Ghosh N, Hoffmann M, Elstner M, Tajkhorshid E, Frauenheim T, Cui Q. J Phys Chem A. 2006;110:548. doi: 10.1021/jp052328q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187. [Google Scholar]
- 29.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J Chem Phys. 1995;103:8577. [Google Scholar]
- 30.MacKerell AD, et al. J Phys Chem B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 31.Schlenkrich M, Brickmann J, MacKerell AD, Jr, Karplus M. An empirical potential energy function for phopholipids: Criteria for parameter optimization and applications. In: Merz KM Jr, Roux B, editors. Biological Membranes: A Molecular Perspective from Computation and Experiment. Birkhäuser; Boston: 1996. p. 31. [Google Scholar]
- 32.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926. [Google Scholar]
- 33.van Gunsteren WF, Berendsen HJC. Mol Phys. 1977;34:1311. [Google Scholar]
- 34.Gao JL, Amara P, Alhambra C, Field MJ. J Phys Chem A. 1998;102:4714. [Google Scholar]
- 35.Stewart JJP. J Comput Chem. 1989;10:209. [Google Scholar]
- 36.Pearson RGJ. Am Chem Soc. 1986;108:6109. [Google Scholar]; Cao Z, Lin M, Zhang Q, Mo Y. J Phys Chem A. 2004;108:4277. [Google Scholar]
- 37.Scheiner S. Ann Rev Phys Chem. 1994;45:23. doi: 10.1146/annurev.pc.45.100194.000323. [DOI] [PubMed] [Google Scholar]; Scheiner S. Hydrogen Bonding: A Theoretical Perspective. Oxford University Press; New York: 1997. [Google Scholar]; Jeffrey GA. An Introduction to Hydrogen Bonding. Oxford University Press; New York: 1997. [Google Scholar]; Desiraju GR, Steiner T. The Weak Hydrogen Bond In Structural Chemistry and Biology. Oxford University Press; New York: 2001. [Google Scholar]
- 38.Gilli P, Bertolasi V, Pretto L, Ferretti V, Gilli G. J Am Chem Soc. 2004;126:3845. doi: 10.1021/ja030213z. [DOI] [PubMed] [Google Scholar]
- 39.Gora RW, Grabowski SJ, Leszczynski J. J Phys Chem A. 2005;109:6397. doi: 10.1021/jp044072e. [DOI] [PubMed] [Google Scholar]
- 40.Cleland WW, Frey PA, Gerlt JA. J Biol Chem. 1998;273:25529. doi: 10.1074/jbc.273.40.25529. [DOI] [PubMed] [Google Scholar]; Cleland WW, Kreevoy MM. Science. 1994;264:1887. doi: 10.1126/science.8009219. [DOI] [PubMed] [Google Scholar]; Frey PA, Whitt SA, Tobin JB. Science. 1994;264:1927. doi: 10.1126/science.7661899. [DOI] [PubMed] [Google Scholar]; Warshel A, Papazyan A, Kollman PA. Science. 1995;269:102. doi: 10.1126/science.7661987. [DOI] [PubMed] [Google Scholar]
- 41.Custelcean R, Jackson JE. Chem Rev. 2001;101:1963. doi: 10.1021/cr000021b. [DOI] [PubMed] [Google Scholar]; Grabowski SJ, Sokalski WA, Leszczynski J. J Phys Chem A. 2005;109:4331. doi: 10.1021/jp0444215. [DOI] [PubMed] [Google Scholar]; Lee JC, Jr, Peris E, Rheingold AL, Crabtree RH. J Am Chem Soc. 1994;116:11014. [Google Scholar]; Lough AJ, Park S, Ramachandran R, Morris RH. J Am Chem Soc. 1994;116:8356. [Google Scholar]; Richardson TB, de Gala S, Crabtree RH, Siegbahn PEM. J Am Chem Soc. 1995;117:12875. [Google Scholar]
- 42.Gilli G, Bellucci F, Ferretti V, Bertolasi V. J Am Chem Soc. 1989;111:1023. doi: 10.1021/ja00269a047. [DOI] [PubMed] [Google Scholar]; Gilli P, Bertolasi V, Ferretti V, Gilli G. J Am Chem Soc. 1994;116:909. [Google Scholar]; Gilli P, Bertolasi V, Ferretti V, Gilli G. J Am Chem Soc. 2000;122:10405. [Google Scholar]; Gilli P, Bertolasi V, Pretto L, Ly3ka A, Gilli G. J Am Chem Soc. 2002;124:13554. doi: 10.1021/ja020589x. [DOI] [PubMed] [Google Scholar]; Bertolasi V, Gilli P, Ferretti V, Gilli GJ. Am Chem Soc. 1991;113:4917. [Google Scholar]; Beck JF, Mo Y. J Comput Chem. 2007;28:455. doi: 10.1002/jcc.20523. [DOI] [PubMed] [Google Scholar]
- 43.Gilli G, Gilli P. J Mol Struct. 2000;552:1. [Google Scholar]
- 44.Hwang JK, Warshel A. J Am Chem Soc. 1996;118:11745. [Google Scholar]; Gao JL, Truhlar DG. Ann Rev Phys Chem. 2002;53:467. doi: 10.1146/annurev.physchem.53.091301.150114. [DOI] [PubMed] [Google Scholar]; Hammes-Schiffer S. Biochemistry. 2002;41:13335. doi: 10.1021/bi0267137. [DOI] [PubMed] [Google Scholar]; Cui Q, Karplus M. J Am Chem Soc. 2002;124:3093. doi: 10.1021/ja0118439. [DOI] [PubMed] [Google Scholar]; Kiefer PM, Hynes JT. J Phys Chem A. 2003;107:9022. [Google Scholar]; Benkovic SJ, Hammes-Schiffer S. Science. 2003;301:1196. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]; Liang Z, Klinman JP. Curr Opin Struct Biol. 2004;14:648. doi: 10.1016/j.sbi.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 45.Nagle JF, Morowitz HJ. Proc Natl Acad Sci USA. 1978;75:298. doi: 10.1073/pnas.75.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nagle JF, Mille M. J Chem Phys. 1981;74:1367. [Google Scholar]; Lu D, Voth GA. Proteins: Struct, Funct, Genet. 1998;33:119. [PubMed] [Google Scholar]; Sham YY, Muegge I, Warshel A. Proteins: Struct, Funct, Genet. 1999;36:484. [PubMed] [Google Scholar]; Pomès R, Roux B. Biophys J. 2002;82:2304. doi: 10.1016/S0006-3495(02)75576-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wu Y, Voth GA. Biophys J. 2003;85:864. doi: 10.1016/S0006-3495(03)74526-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Popovic DM, Stuchebrukhov AA. J Am Chem Soc. 2004;126:1858. doi: 10.1021/ja038267w. [DOI] [PubMed] [Google Scholar]; Bondar AN, Fischer S, Smith JC, Elstner M, Suhai S. J Am Chem Soc. 2004;126:14668. doi: 10.1021/ja047982i. [DOI] [PubMed] [Google Scholar]; Schutz CN, Warshel A. J Phys Chem B. 2004;108:2066. [Google Scholar]; Frank RAW, Titman CM, Pratap JV, Luisi BF, Perham RN. Science. 2004;306:872. doi: 10.1126/science.1101030. [DOI] [PubMed] [Google Scholar]; Fisher Z, Prada JAH, Tu C, Duda D, Yoshioka C, An H, Govindasamy L, Silverman DN, McKenna R. Biochemistry. 2005;44:1097. doi: 10.1021/bi0480279. [DOI] [PubMed] [Google Scholar]; Braun-Sand S, Burykin A, Chu ZT, Warshel A. J Phys Chem B. 2005;109:583. doi: 10.1021/jp0465783. [DOI] [PubMed] [Google Scholar]; Riccardi D, König P, Guo H, Cui Q. Biochemistry. 2008;47:2369. doi: 10.1021/bi701950j. [DOI] [PubMed] [Google Scholar]
- 46.Pomès R, Roux B. Biophys J. 1998;75:33. doi: 10.1016/S0006-3495(98)77492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chakrabarti N, Tajkhorshid E, Roux B, Pomès R. Structure. 2004;12 doi: 10.1016/j.str.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 47.de Grotthuss CJT. Ann Chim Phys. 1806;58:54. [Google Scholar]
- 48.Riccardi D, König P, Prat-Resina X, Yu HB, Elstner M, Frauenheim T, Cui Q. J Am Chem Soc. 2006;128:16302. doi: 10.1021/ja065451j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tuckerman ME, Chandra A, Marx D. Acc Chem Res. 2006;39:151. doi: 10.1021/ar040207n. [DOI] [PubMed] [Google Scholar]; Tuckerman ME, Marx D, Parrinello M. Nature. 2002;417:925. doi: 10.1038/nature00797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.