Abstract
Semiconductor quantum dots (QDs) are a new class of fluorescent labels with broad applications in biomedical imaging, disease diagnostics, and molecular and cell biology. In comparison with organic dyes and fluorescent proteins, quantum dots have unique optical and electronic properties such as size-tunable light emission, improved signal brightness, resistance against photobleaching, and simultaneous excitation of multiple fluorescence colors. Recent advances have led to multifunctional nanoparticle probes that are highly bright and stable under complex in vitro and in vivo conditions. New designs involve encapsulating luminescent QDs with amphiphilic block copolymers, and linking the polymer coating to tumor-targeting ligands and drug-delivery functionalities. These improved QDs have opened new possibilities for real-time imaging and tracking of molecular targets in living cells, for multiplexed analysis of biomolecular markers in clinical tissue specimens, and for ultrasensitive imaging of malignant tumors in living animal models. In this article, we briefly discuss recent developments in bioaffinity QD probes and their applications in molecular profiling of individual cancer cells and clinical tissue specimens.
Keywords: nanoparticle, nanocrystal, semiconductor, cancer, tumor, tissue section, live cell
Introduction
The ability to study molecular and cellular events by using fluorescent probes has broadly impacted many areas in biomedical research including cell biology, molecular biology, drug screening, and molecular diagnostics. However, traditional fluorophores such as organic dyes and fluorescent proteins suffer from several intrinsic problems including rapid photobleaching, spectral cross talking, narrow excitation profiles, and limited brightness/signal intensity. In contrast, semiconductor quantum dots (QDs) are fluorescent nanoparticles based on entirely different principles, and exhibit novel optical and electronic properties that are not available from organic dyes and fluorescent proteins. These properties include size- and composition-tunable emission from visible to infrared wavelengths, large absorption coefficients across a wide spectral range, and improved signal brightness and photostability (Alivisatos et al 2005). Due to their broad excitation profiles and narrow/symmetric emission spectra, high-quality QDs are also well suited for multiplexed tagging or encoding, in which multiple colors and intensities are combined to encode thousands of genes, proteins, or small-molecule compounds (Han et al 2001).
These properties have raised new opportunities for analyzing a panel of genes and proteins in cancer cells and clinical tissue specimens. Previous work in cancer molecular profiling has revealed a strong correlation between biomolecular signatures (or biomarkers) and cancer behavior (Liotta and Petricoin 2000). However, clinical tumor specimens (especially human breast and prostate tumors) are highly heterogeneous, containing a mixture of benign, cancerous, and stromal cells. Current technologies for molecular profiling include RT-PCR, gene chips, protein chips, two-dimensional gel electrophoresis, biomolecular mass spectrometry (eg, MALDI-MS, ES-MS, and SELDI-MS), but these technologies are not designed to handle this type of cellular heterogeneities (Liotta and Petricoin 2000). Also, these techniques are “destructive” because they require cells and tissue specimens to be processed into a homogeneous solution, leading to a loss of valuable 3-D cellular and tissue morphological information associated with the original tumor.
The development of nanotechnology, especially bio-conjugated nanoparticles, could provide an essential link by which biomarkers are functionally correlated with disease behavior (Ferrari 2005). In particular, QD probes can be used to quantify a panel of biomarkers on intact cancer cells and tissue specimens, allowing a correlation of traditional histopathology and molecular signatures for the same materials (Gao et al 2003). This integration may be achieved with a spectral imaging microscope that is designed to operate both in the morphological staining mode and the molecular profiling mode. In the following, we discuss the novel optical properties of QDs, recent advances in probe development, and their applications in molecular analysis of intact cancer cells and tissue specimens.
Quantum dots
For use in biology and medicine, QD probes most frequently take the form shown in Figure 1, with an inorganic semiconductor core surrounded by a monolayer of ligands and an amphiphilic polymer coat that is linked to biomolecules. QD cores are most commonly prepared from cadmium selenide (CdSe), a binary semiconductor with size-dependent bandgap energy that can be tuned to emit light of any color throughout the visible spectrum. CdSe QDs have been thoroughly studied and can be produced in large quantities with fluorescence emission efficiencies as high as 90% at room temperature (Qu and Peng 2002; Pan et al 2005; Yin and Alivisatos 2005). These crystalline CdSe cores are synthesized and capped with a protective zinc sulfide (ZnS) shell in a high temperature organic solvent (Murray et al 1993; Hines and Guyot-Sionnest 1996; Dabbousi et al 1997), and are coated with a monolayer of nonpolar coordinating ligands such as trioctylphosphine oxide (TOPO).
Figure 1.
(A) Schematic representation of bioaffinity quantum dot probes for biological imaging. A CdSe core is surrounded by a ZnS shell that is passivated by hydrophobic TOPO ligands and encapsulated in an amphiphilic polymer. Conjugated PEG molecules serve to reduce nonspecific adsorption, and streptavidin provides a high-affinity linker for conjugation to biotinylated proteins, nucleic acids, or other molecules. (B) Transmission electron micrograph (TEM) of water-soluble quantum dots showing an electron-dense QD core (dark) and a polymer coating as a lighter shade surrounding each QD. (C) Fluorescence micrograph showing that single QDs can be readily observed when the dots are spread on a glass cover slip.
After synthesis, the nonpolar, hydrophobic QDs are transferred to an aqueous phase, a nontrivial process that is crucial to the generation of high-quality QDs. Amphiphilic polymers, such as polyethylene glycol (PEG)-modified lipids and octylamine-modified polyacrylic acid (Dubertret et al 2002; Wu et al 2003), have been used to produce water-soluble QDs that maintain the optical properties and colloidal stability of the original nanoparticles. Previous phase transfer methods involved an exchange of the TOPO ligands for hydrophilic ligands, resulting in QDs with reduced quantum yields and a tendency to aggregate. After transfer to water, QDs may be cross-linked to a variety of biologically active molecules such as antibodies, enzymes, nucleic acids, small molecule inhibitors, or biologically inert PEG. Many coupling schemes have been used to generate cross-links, such as electrostatic adsorption (Mattoussi et al 2000), covalent bond formation (Chan and Nie 1998; Wu et al 2003), and biomolecular bridging via streptavidin-biotin binding (Goldman et al 2002; Wu et al 2003). These methods give rise to QD probes that are highly stable and have excellent affinity toward their targets, although it is difficult to control the number and orientation of biomolecules attached to a single QD.
In comparison with organic dyes and fluorescent protein, QDs have several advantages and unique applications. First, QDs have large molar extinction coefficients on the order of 0.5–5 × 106 M−1cm−1 (Leatherdale et al 2002), about 10–50 times larger than that (5–10 × 104 M−1cm−1) of organic dyes. Thus, QDs are 10–50 times more efficient in adsorbing photons than organic dyes at the same excitation photon flux, leading a significant improvement in probe brightness. Second, QDs are several thousand times more stable against photobleaching than dye molecules, allowing extended imaging and quantitative biomarker studies of cells and tissue specimens. Third, QDs have size- and composition-tunable fluorescence emission from visible to infrared wavelengths, and one light source can be used to excite multiple colors of fluorescence emission. This leads to very large Stokes spectral shifts (measured by the distance between the excitation and emission peaks) that can be used to further improve detection sensitivity. This factor becomes especially important for clinical tissue studies and in-vivo animal imaging due to the high autofluorescence background often seen in complex biomedical specimens. Indeed, the Stokes shifts of semiconductor QDs can be hundreds of nanometers, depending on the wavelength of the excitation light. Organic dye signals are often buried by strong tissue autofluorescence, whereas QD signals can be readily separated from the background by wavelength-resolved or spectral imaging (Gao et al 2004).
Imaging and tracking of cellular events
Live cell imaging often involves transgenic expression of fluorescently tagged proteins (Miyawaki et al 2003), but this technique is labor-intensive and the resulting fluorescent proteins are not sufficiently bright or stable for single-molecule imaging and tracking. To address this problem, recent work has shown that QDs microinjected into the cytoplasm of a single cell of a frog embryo could maintain stable and bright fluorescence over long periods of excitation and observation, allowing real time imaging of embryonic development and cellular tracking (Dubertret et al 2002). Importantly, injection of a large number of QD particles into an individual cell did not appear to negatively impact embryonic development or cause toxicity. In 2004, Derfus et al (2004) demonstrated that QDs conjugated to organelle-targeted peptides could translocate to either nuclei or mitochondria following microinjection into the cytoplasm. Microinjection is, however, a labor-intensive technique and requires delicate manipulation of one cell at a time. Other techniques for QD delivery across cellular membranes include chemical-mediated transfection (cationic peptides, cationic lipids, transferrin proteins) and electroporation (Chen and Gerion 2004; Derfus et al 2004; Jaiswal et al 2004). Many types of cells are also able to engulf or take up QDs spontaneously via endocytosis, which has led to a QD-based cellular motility assay (Pellegrino et al 2003; Voura et al 2004) as well as imaging of cancer cell extravasation in living animals (Voura et al 2004). However, these delivery methods are all hampered by aggregation of QDs in the cytoplasm or trapping of the QD probes in endosomes, vesicles, and other intracellular organelles. Working with largely aggregated QDs, Nan et al (2005) measured the motions of multi-protein molecular machines such as kinesin and dynein. By tracking the paths of cationic peptide-conjugated QDs inside cells, their data showed directional, non-Brownian motion that was consistent with the involvement of molecular motors. Recent work in our own lab has shown that cellular membrane pores formed by bacterial toxins like streptolysin O (SLO) are large enough to allow the diffusion of single QDs into the cellular cytoplasm, while maintaining cellular viability. A major finding is that the SLO-delivered QD probes are primarily single dots and they maintain their fluorescence and blinking characteristics. Figure 2 shows confocal fluorescence images of single dots and aggregates in the cytoplasm of live fibroblast cells.
Figure 2.
Fluorescence images of single quantum dots and aggregates in the cytoplasm of live fibroblast cells. (A) Single QDs observed in the cytoplasm after delivery via SLO toxin. (B) QD aggregates observed in the cytoplasm after a long period of time following SLO delivery (40 hours). Microtubules were visualized by GFP-tubulin protein expression. Both images were obtained with a spinning disk confocal microscope (Perkin Elmer), with the focal plane near the center of each cell.
It is also significant that QD probes have been used to image and track cell surface proteins with considerable success. In particular, Lidke et al (2004) conjugated epidermal growth factor (EGF) to QDs in order to fluorescently image the binding of EGF to erbB/HER receptors on the membranes of cells. The authors were able to observe individual blinking QDs on the cell surface (corresponding to single receptor molecules), and record the endocytosis and endosomal trafficking of the receptors in real time. The high photostability and brightness of these probes made possible the observation of a previously unreported transport mechanism from cellular filopodia to the cell body (Lidke et al 2005). Similarly, Dahan et al (2003) reported that QDs conjugated to antibody fragments against the GlyR receptor could be used to observe single receptors on the membranes of neurons. The use of QD probes has allowed real-time imaging and tracking of single receptor molecules over an extended period of time.
Moleculr profiling of individual cancer cells
Several groups have used multicolor QD probes for molecular profiling of fixed cancer cells. Wu et al reported the potential of QDs in 2001 with a variety of multicolor labeling experiments on fixed cells, demonstrating significant sensitivity and photostability of QD probes compared to the best available organic dyes (Wu et al 2003). Now that these probes are commercially available, their superiority to conventional dyes has been well established in many fixed cell labeling experiments. With their narrow emission bandwidths, QDs have also been used to simultaneously label up to 5 biomarkers on the same fixed cells, a task that is nearly impossible with conventional dyes due to fluorophore crosstalk and the need for multiple excitation sources. In another development, QDs have been used to simultaneously label fixed cells for mRNA and protein, combining immunocytochemistry and fluorescence in situ hybridization (Matsuno et al 2005). Furthermore, QDs have been used as dual-modality imaging probes, showing bright contrast in fluorescent micrographs of cells and tissues, and their corresponding electron micrographs (Nisman et al 2004; Giepmans et al 2005). It is clear that QD probes have opened new possibilities in multiplexed analysis of cellular biomarkers or antigens. As an example, Figure 3 shows color fluorescence images of human prostate cancer cells that were stained with up to five QD colors, allowing multiplexed analysis of up to five tumor antigens on single intact cancer cells.
Figure 3.
Color fluorescence images of human prostate cancer cells stained with multicolor QD-antibody conjugates. (a) Staining of membrane antigen E-cadherin with QD655 (emission wavelength = 655 nm); (b) staining of cytoplasmic antigen vimentin with QD525; (c) staining of nuclear antigen HIF1alpha with QD655; and (d) multiplexed staining of tumor antigens with QD525 (labeling RANKL), 565(N-cadherin), 605(EF1aplha), 655(E-cadherin) and 705 (vimentin). The cell nuclei were counterstained blue with DAPI in (a) and (b).
It is also important to note that the use of multiplexed QD probes further allows spatial mapping of tumor antigens on single cells. For example, Figure 4 shows fluorescence images of single breast cancer cells labeled with a cocktail of antibody-QD probes, together with control PBMC cells that do not express the tumor antigens. Remarkably, all three colors were observed from breast tumor cells (a and b), while no QD binding was detected with either the control QDs (c) or the control PBMC cells (d). Due to antigen clustering and colocalization, fluorescence imaging showed overlapping Her-2 and epithelial specific antigen (ESA or EpCam) signals on the surface of BT-474 cells (a). The BT-20 cells were found to have a ring of expressed ESA on the cellular peripherals and a much lower level of Her-2. In contrast to stable QD signals, fluorescence from dye-labeled cells was photobleached in less than 20 seconds (e and f), preventing quantitative imaging or spectroscopic studies at the single-cell level. QDs are considerably brighter and more photostable than organic fluorophores, which should allow sensitive detection of low-abundance cellular targets through signal averaging and background subtraction. For statistical studies of heterogeneous cell populations, we measured more than 100 cancer cells with a fluorescence microscope and a spectrometer. The results reveal that the BT-474 cells are clustered in a “high Her-2” area, and that BT-20 cells are clustered in a “low Her-2” area. In clinical samples, a panel of selected markers could be analyzed to provide information on disease staging and treatment options.
Figure 4.
Multiplexed QD staining and spatial mapping of tumor antigens on single breast cancer cells. (a) Breast tumor BT-474 cell, and (b) breast tumor BT-20 cell; both were stained with cytokeratin-QD (green), Her-2-QD (yellow), epithelial specific antigen-QD (red), and the nuclear dye DAPI (blue). Control data: (c) BT-474 cancer cells treated with QDs conjugated to nonspecific bovine serum albumin, and (d) peripheral blood mononuclear cells (PBMC) treated with the same QD-antibody conjugates used in panels (a) and (b). (e) Fluorescence image of BT-474 cells labeled with FITC-conjugated antibody against cytokeratin, and (f) photobleached image after 20 seconds.
Clinical tissue specimens and correlation with tumor behavior
A major application of quantum dots will be in multiplexed labeling and molecular analysis of pathological tissue specimens. In comparison with single cells, clinical tissue specimens are often highly heterogeneous (containing different cell populations in various microenvironments) and are much more difficult to analyze. Taking advantage of the high photostability of QDs, Tokumasu and Dvorak (2003) were able to collect 40 consecutive optical sections using confocal microscopy and generated a 3-D reconstructed, high-resolution image of the membrane domain band 3 in erythrocytes. Ness et al (2003) developed an immunohisto-chemical protocol that combines conventional enzymatic signal amplification and QD labeling to detect intracellular antigens in rat and mouse brain tissue sections. Their study showed that QD immunofluorescence labeling had greater sensitivity than similar IHC approaches using conventional dyes (Ness et al 2003). Wu et al (2003) developed reliable and specific QD probes to localize the breast cancer cell surface marker Her2, cytoskeleton fibers, and nuclear antigens in fixed cells, live cells, and tissue sections, with a substantial increase in brightness and photostability as compared to organic dyes. Taking advantage of the superior photostability of QDs, Ferrara et al (2006) were able to obtain a 3D visualization of the vascular endothelium from an en face preparation of human coronary artery by taking large z-stacks series. All these studies demonstrate that QDs are excellent probes with improved signal-to-noise ratios, and are well suited for studying complex biological problems (Alivisatos et al 2005).
Most studies on QD fluorescent labeling have been carried out with cells (both live and fixed) (Tokumasu and Dvorak 2003; Wu et al 2003; Lidke et al 2004) or freshly harvested tissues (Ness et al 2003; Ferrara et al 2006). However, most available clinical specimens are archived, formalin fixed, paraffin-embedded (FFPE) tissues that might be several decades old. Since the clinical outcomes of these tissues are already known, it will be great value to use these specimens for examining the relationship between molecular profile and clinical outcome. Compared with cells or animal tissues, archived clinical specimens need special treatment such as antigen retrieval, and their background autofluorescence is generally much stronger than that observed in cells. Our group has developed highly successful procedures for QD staining of archival FFPE tissue specimens. One example is to study the epithelial-mesenchymal transition (EMT) process in the progression of prostate cancer to the bone. EMT is a normal biological mechanism first reported in embryonic development and later found to be involved in cancer metastasis (Huber et al 2005). During EMT, cancer cells undergo phenotypical changes and become more invasive, characterized by changes in the profile change of cellular adhesion molecules, particularly, an increase of N-cadherin and a loss of E-cadherin. Other important markers include the cytoskeleton proteins vimentin, cytokeratin 18 and RANKL. In one example, we have used QD-conjugated secondary antibodies (Qdots Corporation, now Invitrogen) for molecular profiling of two FFPE androgen-repressed prostate cancer (ARCaP) cells lines. These two cell lines were selected because they represent two phenotypes at the two ends of the EMT process during prostate cancer progression. The ARCaPE is more epithelial-like and less invasive, while the ARCaPM has more mesenchymal characteristics and more invasive (Zhau et al 1996). Our QD staining studies have achieved simultaneous staining of 4 different biomarkers with expression profiles consistent with western blot data (Figure 5). Moreover, QD staining also provides spatial localization information (both inter- and intracellular) which is not possible with western blot or any molecular biology techniques. We have also found that staining of FFPE cells requires longer incubation time (overnight at 4°C vs 1 hour at room temperature) and a higher QD-secondary antibody concentration than that required for freshly fixed cells.
Figure 5.
Multiplexed QD profiling of four tumor biomarkers using two FFPE prostate cancer cell lines with distinct bone-metastasis behaviors. The four markers, all associated with epithelial-mesenchymal transition (EMT), are N-cadherin, EF (elongation factor)-1alpha, E-cadherin, and vimentin, and their corresponding QD colors are 565 nm, 605 nm, 655 nm, and 705 nm. The cell nuclei were counterstained blue by DAPI, and the spectra were captured under blue excitation. (a) Color fluorescence image of highly metastatic prostate cancer cells (clone ARCaPm); (b) single-cell fluorescence spectrum obtained from image (a); (c) color fluorescence image of benign prostate cancer cells (clone ARCaPe); (d) single-cell spectrum obtained from image (c). The relative abundance of these markers is consistent with previous western blotting data (not shown). Note that individual cancer cells have heterogeneous expression patterns, that the single-cell data in (b) and (d) are representative of a heterogeneous cell population.
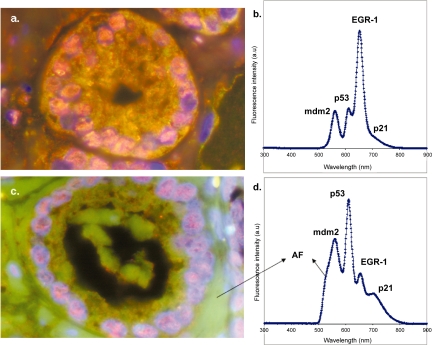
For molecular profiling of clinical FFPE prostate specimens, we also selected four tumor antigens (mdm-2, p53, EGR-1 and p21) as a model system for technology development. These markers are known to be important in prostate cancer diagnosis and are correlated with tumor behavior (Hernandez et al 2003; Mora et al 2005). As shown in Figure 6, we were able to detect all 4-markers in the tissue specimens, but the autofluorescence was higher than that observed in FFPE cells. Compared with FFPE cells, clinical tissue specimens may require harsher antigen retrieval conditions (EDTA buffer vs citrate buffer) and generally have stronger autofluorescence. On the other hand, autofluorescence can be desirable by serving as counterstaining of tissue morphology. Autofluorescence can be separated from the QD signal by intentionally illuminating the sample to bleach it out while leaving the QDs bright enough for imaging and spectral analysis. Of course, spectral unmixing algorithms can be developed or obtained commercially (Mansfield et al 2005) to separate the background fluorescence from the real QD signals. In summary, these results demonstrate the feasibility of using QDs as fluorescent labels for molecular profiling of FFPE clinical specimens. With continuous efforts in optimizing the experimental conditions, we believe that QD probes hold great promise in multiplexed molecular profiling of clinical tissue specimens and correlation of biomarkers with disease behavior.
Figure 6.
Multiplexed QD staining of archived FFPE clinical specimen from human prostate cancer patients, and comparison between two different glands on the same tissue specimen. Four tumor biomarkers (mdm-2, p53, EGR-1 and p21) were labeled with four colors of QDs emitting at 565 nm, 605 nm, 655nm, and 705 nm respectively. (a) Color fluorescence image of QD-stained tissue specimens showing just one gland; (b) representative fluorescence spectrum obtained from single cells in the gland (image a); (c) color fluorescence image of the same QD-stained tissue specimens but showing a different gland; (d) representative fluorescence spectrum obtained from single cells in the second gland (image c). Note that the biomarker profile is remarkably different for different glands. This ability to measure cellular heterogeneity on the same tumor specimen will be crucial for clinical applications. AF stands for autofluorescence.
In conclusion, semiconductor quantum dots have emerged as a new class of fluorescent tags for molecular profiling of single cancer cells and clinical tissue specimens. With only a single light source, multicolor fluorescence imaging allows rapid screening and selection of cancer cells, and wavelength-resolved spectroscopy provides quantitative data on the expression levels of multiple bio-markers. It should be possible to simultaneously determine the expression levels of 8–10 genes or proteins in single cells. With spectroscopic multiplexing, it might even be possible to analyze 50–100 of genes and proteins on morphologically intact cells or tissue specimens. In addition to molecular pathology and in-vitro diagnostics, QD probes have shown promise as contrast agents for in vivo tumor imaging in living animals (Gao and Nie 2004; Voura et al 2004; Stroh et al 2005). Thus, we expect QD-based imaging and diagnostic technologies to have broad applications in linking biomolecular signatures with disease behavior, and will play a significant role in personalized and predictive medicine.
Acknowledgments
We are grateful to Dr Xiaohu Gao for helpful discussions, to Dr Leland Chung, and Dr Haiyan Zhau for providing the ARCaP cells and Dr Milton Datta for providing human prostate cancer samples. The work was supported in part by NIH grants (P20 GM072069, R01 CA108468-01, and U54CA119338) and the Georgia Cancer Coalition Distinguished Cancer Scholars Program (to S.N.). A.M.S. acknowledges the Whitaker Foundation for generous fellowship support.
References
- Alivisatos AP, Gu WW, Larabell C. Quantum dots as cellular probes. Annu Rev Biomed Eng. 2005;7:55–76. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- Chan WCW, Nie SM. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–8. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- Chen F, Gerion D. Fluorescent CdSe/ZnS nanocrystal-peptide conjugates for long-term, nontoxic imaging and nuclear targeting in living cells. Nano Lett. 2004;4:1827–32. [Google Scholar]
- Dabbousi BO, RodriguezViejo J, Mikulec FV, et al. (CdSe) ZnS core-shell quantum dots: Synthesis and characterization of a size series of highly luminescent nanocrystallites. J Phys Chem B. 1997;101:9463–75. [Google Scholar]
- Dahan M, Levi S, Luccardini C, et al. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science. 2003;302:442–5. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- Derfus AM, Chan WCW, Bhatia SN. Intracellular delivery of quantum dots for live cell labeling and organelle tracking. Adv Mater. 2004;16:961–6. [Google Scholar]
- Dubertret B, Skourides P, Norris DJ, et al. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science. 2002;298:1759–62. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- Ferrara DE, Weiss D, Carnell PH, et al. Quantitative 3D fluorescence technique for the analysis of en face preparations of arterial walls using quantum dot nanocrystals and two-photon excitation laser scanning microscopy. Am J Physiol Regul Integr Comp Physiol. 2006;290:R114–R23. doi: 10.1152/ajpregu.00449.2005. [DOI] [PubMed] [Google Scholar]
- Ferrari M. Cancer nanotechnology: Opportunities and challenges. Nat Rev Cancer. 2005;5:161–71. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- Gao XH, Nie SM. Molecular profiling of single cells and tissue specimens with quantum dots. Trends Biotechnol. 2003;21:371–3. doi: 10.1016/S0167-7799(03)00209-9. [DOI] [PubMed] [Google Scholar]
- Gao XH, Cui YY, Levenson RM, et al. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22:969–76. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- Giepmans BNG, Deerinck TJ, Smarr BL, et al. Correlated light and electron microscopic imaging of multiple endogenous proteins using quantum dots. Nat Methods. 2005;2:743–9. doi: 10.1038/nmeth791. [DOI] [PubMed] [Google Scholar]
- Goldman ER, Balighian ED, Mattoussi H, et al. Avidin: A natural bridge for quantum dot-antibody conjugates. J Am Chem Soc. 2002;124:6378–82. doi: 10.1021/ja0125570. [DOI] [PubMed] [Google Scholar]
- Han MY, Gao XH, Su JZ, et al. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat Biotechnol. 2001;19:631–5. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- Hernandez I, Maddison LA, Wei YL, et al. Prostate-specific expression of p53(R172L) differentially regulates p21, Bax, and mdm2 to inhibit prostate cancer progression and prolong survival. Mol Cancer Res. 2003;1:1036–47. [PubMed] [Google Scholar]
- Hines MA, Guyot-Sionnest P. Synthesis and characterization of strongly luminescing ZnS-capped CdSe nanocrystals. J Phys Chem. 1996;100:468–71. [Google Scholar]
- Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–58. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Jaiswal JK, Goldman ER, Mattoussi H, et al. Use of quantum dots for live cell imaging. Nat Methods. 2004;1:73–8. doi: 10.1038/nmeth1004-73. [DOI] [PubMed] [Google Scholar]
- Leatherdale C, Woo W, Mikulec F, et al. On the absorption cross section of CdSe nanocrystal quantum dots. J Phys Chem B. 2002;106:7619–22. [Google Scholar]
- Lidke DS, Lidke KA, Rieger B, et al. Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. J Cell Biol. 2005;170:619–26. doi: 10.1083/jcb.200503140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidke DS, Nagy P, Heintzmann R, et al. Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nat Biotechnol. 2004;22:198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- Liotta L, Petricoin E. Molecular profiling of human cancer. Nat Rev Genet. 2000;1:48–56. doi: 10.1038/35049567. [DOI] [PubMed] [Google Scholar]
- Mansfield JR, Gossage KW, Hoyt CC, et al. Autofluorescence removal, multiplexing, and automated analysis methods for in-vivo fluorescence imaging. J Biomed Opt. 2005;10 doi: 10.1117/1.2032458. [DOI] [PubMed] [Google Scholar]
- Matsuno A, Itoh J, Takekoshi S, et al. Three-dimensional imaging of the intracellular localization of growth hormone and prolactin and their mRNA using nanocrystal (quantum dot) and confocal laser scanning microscopy techniques. J Histochem Cytochem. 2005;53:833–8. doi: 10.1369/jhc.4A6577.2005. [DOI] [PubMed] [Google Scholar]
- Mattoussi H, Mauro JM, Goldman ER, et al. Self-assembly of CdSe-ZnS quantum dot bioconjugates using an engineered recombinant protein. J Am Chem Soc. 2000;122:12142–50. [Google Scholar]
- Michalet X, Pinaud FF, Bentolila LA, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–44. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millo O, Katz D, Cao YW, et al. Imaging and spectroscopy of artificial-atom states in core/shell nanocrystal quantum dots. Phys Rev Lett. 2001;86:5751–4. doi: 10.1103/PhysRevLett.86.5751. [DOI] [PubMed] [Google Scholar]
- Mitchell GP, Mirkin CA, Letsinger RL. Programmed assembly of DNA functionalized quantum dots. J Am Chem Soc. 1999;121:8122–3. [Google Scholar]
- Miyawaki A, Sawano A, Kogure T. Lighting up cells: labelling proteins with fluorophores. Nat Cell Biol. 2003:S1–S7. [PubMed] [Google Scholar]
- Mora GR, Olivier KR, Mitchell RF, et al. Regulation of expression of the early growth response gene-1 (EGR-1) in malignant and benign cells of the prostate. Prostate. 2005;63:198–207. doi: 10.1002/pros.20153. [DOI] [PubMed] [Google Scholar]
- Murray CB, Norris DJ, Bawendi MG. Synthesis and characterization of nearly monodisperse CdE (E = S, Se, Te) semiconductor nanocrystallites. J Am Chem Soc. 1993;115:8706–15. [Google Scholar]
- Nan XL, Sims PA, Chen P, et al. Observation of individual microtubule motor steps in living cells with endocytosed quantum dots. J Phys Chem B. 2005;109:24220–4. doi: 10.1021/jp056360w. [DOI] [PubMed] [Google Scholar]
- Ness JM, Akhtar RS, Latham CB, et al. Combined tyramide signal amplification and quantum dots for sensitive and photostable immunofluorescence detection. J Histochem Cytochem. 2003;51:981–7. doi: 10.1177/002215540305100801. [DOI] [PubMed] [Google Scholar]
- Nisman R, Dellaire G, Ren Y, et al. Application of quantum dots as probes for correlative fluorescence, conventional, and energy-filtered transmission electron microscopy. J Histochem Cytochem. 2004;52:13–8. doi: 10.1177/002215540405200102. [DOI] [PubMed] [Google Scholar]
- Pan DC, Wang Q, Jiang SC, et al. Synthesis of extremely small CdSe and highly luminescent CdSe/CdS core-shell nanocrystals via a novel two-phase thermal approach. Adv Mater. 2005;17:176–9. [Google Scholar]
- Pellegrino T, Parak W, Boudreau R, et al. Quantum dot-based cell motility assay. Diferentiation. 2003;71:542–8. doi: 10.1111/j.1432-0436.2003.07109006.x. [DOI] [PubMed] [Google Scholar]
- Qu LH, Peng XG. Control of photoluminescence properties of CdSe nanocrystals in growth. J Am Chem Soc. 2002;124:2049–55. doi: 10.1021/ja017002j. [DOI] [PubMed] [Google Scholar]
- Stroh M, Zimmer JP, Duda DG, et al. Quantum dots spectrally distinguish multiple species within the tumor milieu in vivo. Nat Med. 2005;11:678–82. doi: 10.1038/nm1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumasu F, Dvorak J. Development and application of quantum dots for immunocytochemistry of human erythrocytes. Journal of Microscopy-Oxford. 2003;211:256–61. doi: 10.1046/j.1365-2818.2003.01219.x. [DOI] [PubMed] [Google Scholar]
- Voura E, Jaiswal J, Mattoussi H, et al. Tracking metastatic tumor cell extravasation with quantum dot nanocrystals and fluorescence emission-scanning microscopy. Nat Med. 2004;10:993–8. doi: 10.1038/nm1096. [DOI] [PubMed] [Google Scholar]
- Wu XY, Liu HJ, Liu JQ, et al. Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat Biotechnol. 2003;21:41–6. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- Yin Y, Alivisatos AP. Colloidal nanocrystal synthesis and the organic-inorganic interface. Nature. 2005;437:664–70. doi: 10.1038/nature04165. [DOI] [PubMed] [Google Scholar]
- Zhau HYE, Chang SM, Chen BQ, et al. Androgen-repressed phenotype in human prostate cancer. Proc Natl Acad Sci U S A. 1996;93:15152–7. doi: 10.1073/pnas.93.26.15152. [DOI] [PMC free article] [PubMed] [Google Scholar]