Abstract
Iron sulfur (Fe-S) clusters are versatile biological cofactors that require biosynthetic systems in vivo to be assembled. In Escherichia coli the Isc (iscRSUA-hscBA-fdx-iscX) and the Suf (sufABCDSE) pathways fulfill this function. Despite extensive biochemical and genetic analysis of both pathways, the physiological function of the A-type proteins of each pathway (IscA and SufA) is still unclear. Studies conducted in vitro suggest two possible functions for A-type proteins, as Fe-S scaffold/transfer proteins or as iron donors during cluster assembly. To resolve this issue, SufA was co-expressed in vivo with its cognate partner proteins from the suf operon, SufBCDSE. Native SufA purified anaerobically using this approach was unambiguously demonstrated to be a [2Fe-2S] protein by biochemical analysis and UV-Visible, Mössbauer, resonance Raman, and EPR spectroscopy. Furthermore, native [2Fe-2S] SufA can transfer its Fe-S cluster to both [2Fe-2S] and [4Fe-4S] apoproteins. These results clearly show that A-type proteins form Fe-S clusters in vivo and are competent to function as Fe-S transfer proteins as purified. This study resolves the contradictory results from previous in vitro studies and demonstrates the critical importance of providing in vivo partner proteins during protein over-expression to allow correct biochemical maturation of metalloproteins.
Keywords: Iron-sulfur, Suf, Biosynthesis, Mösbbauer, A-type protein, Scaffold, Transfer, Ferredoxin, Aconitase
INTRODUCTION
Iron sulfur (Fe-S) clusters are versatile cofactors utilized for diverse biological functions including electron transfer, substrate activation and regulation of gene expression1. A large variety of Different types of Fe-S are found in nature, but the simplest and most abundant forms are [2Fe-2S] and [4Fe-4S] clusters. Assembly of such clusters and maturation of iron-sulfur cluster proteins does not occur spontaneously but instead requires complex biosynthetic machineries. In Escherichia coli for example this process is achieved by the Isc (iscRSUA-hscBA-fdx-iscX) and the Suf (sufABCDSE) pathways 2,3 The inorganic sulfur is supplied by a cysteine desulfurase (IscS/SufS) which catalyzes the PLP-dependent removal of sulfur from L-cysteine and donates that sulfur through sulfur shuttle proteins to a specialized scaffold protein in which the nascent Fe-S cluster is pre-assembled2. The in vivo source of iron is still unclear.
IscU homodimer is a prototype for Fe-S scaffold proteins, endowed with the ability to assemble either [2Fe-2S]2+ clusters or a single [4Fe-4S]2+ cluster4,5 that can be directly transferred to appropriate apoproteins6. IscU has been extensively studied using biochemical, enzymatic and spectroscopic methods. At the present time its role as a scaffold protein is well established. Each pathway also contains an A-type protein (IscA/SufA) that can also assemble Fe-S clusters in vitro and transfer those clusters to apoproteins7-9. E. coli IscA was also reported to bind ferric iron tightly (Kass, 3 × 1019 M-1 and provide iron to IscU during in vitro cluster assembly10. These somewhat contradictory studies led to two proposed functions for the A-type proteins: (1) as alternate Fe-S scaffolds or Fe-S transporters; (2) as Fe donors for cluster assembly on other scaffold proteins. The ambiguity was due, in particular, to the fact that A-type proteins were usually isolated in the apo form and that binding of iron or clusters achieved under in vitro conditions might not be physiologically relevant.
In order to produce SufA in a more physiological context for characterization study, we developed an in vivo expression system that simultaneously expresses all six suf genes products, mimicking the native situation where sufABCDSE are transcribed on a polycistronic mRNA. Using this expression system we structurally and functionally characterized the native SufA by biochemical and spectroscopic approaches. We here show for the first time that SufA exists within cells in the form of an Fe-S protein, containing a [2Fe-2S] cluster that can be transferred to apoprotein targets.
EXPERIMENTAL SECTION
Bacterial growth and purification of [2Fe-2S] SufA
The strains used to purify SufA were Top10 pGSO164 and Top10 pGSO164 (SufDH128A), containing the entire suf operon under the control of an arabinose-inducible promoter. The strains were grown in 25 ml of LB + Ampicillin (100 μg/μl) overnight. 10 ml of this was used to inoculate 1 liter of LB + Ampicillin media. The cells were allowed to grow at 37°C until mid-log phase. 10 ml of 20% sterile L-arabinose was added per liter of media to induce expression of the sufABCDSE operon. The cells were induced for 3 hrs. After 3 hrs, cells were collected by centrifugation and frozen at -80°C. A modified freeze-thaw procedure was used to purify SufA anaerobically. Briefly, the cell pellet was thawed on ice, and resuspended in the Buffer A containing 25 mM Tris, pH 7.5 with protease inhibitor cocktail (Pierce) and 5 mM ßME. The pellet was refrozen at -80°C for 1 hour. The freeze thaw cycle was repeated at least twice. All steps for the freeze-thaw procedure were carried out in a Coy anaerobic chamber containing a 95% nitrogen/ 5% hydrogen gas mix. The freeze-thaw extract was centrifuged at 20,000 × g for 25 minutes at 4°C in an anaerobically sealed tube. The supernatant was then loaded on a Q-Sepharose anion exchange column, equilibrated with Buffer A, and eluted with a linear gradient of 1 M NaCl in Buffer A. These steps were carried out using an FPLC outside of the Coy chamber but all buffers were extensively nitrogen-sparged to reduce dissolved oxygen prior to use. The fractions containing SufA (as determined by SDS-PAGE) were red. SufA fractions were collected and concentrated in a YM10 centricon (Amicon) according to manufacturer's protocol.
Preparation of 57Fe labeled SufA for Mössbauer Studies
The Top10 pGSO164 (SufDH128A) strain was used to purify 57Fe labeled SufA protein. The cells were grown in a similar manner as described above except that the LB was enriched with 57Fe. 57Fe (Cambridge Isotopes) was resuspended in dilute HCl and added to a final concentration of 10 μM twice: at culture inoculation and at the time of induction with 0.2% L-arabinose. The protein purification was performed as described above. After concentration, holoSufA was frozen in a Mössbauer cup using liquid nitrogen and stored at 77 K.
Fe-S cluster transfer to ferredoxin and Spore photoproduct lyase
E. coli apo-Fdx and B. subtilis apo-SPL were expressed and purified in our laboratory as already described11,12. Apo-Fdx (180 μM) or apo-SPL (80 μM) were incubated anaerobically in 200 μL 0.1 M Tris-HCl pH: 8; 50 mM KCl with native SufA (0.4 iron and sulfur atom/monomer) in order to provide 2 iron and sulfur atoms/ferredoxin and 4 iron and sulfur atoms/SPL. After 1 hour incubation under anaerobic conditions, SufA was separated from SPL on a Ni-NTA column on which SPL is retained since it contains a His-tag at its N-terminus. SufA was recovered in the run through fraction during extensive washing with 0.1 M Tris-HCl pH: 8; 50 mM KCl buffer whereas SPL was collected in the 400 mM imidazole fraction. Fe-S transfer to SPL was monitored by assaying the corresponding imidazole fraction for iron and sulfur content and for its UV-vis. spectrum. In the case of ferredoxin, Fe-S transfer was followed by monitoring the UV-vis. absorption in the 300-600 nm region and by EPR analysis of the SufA-Fdx protein mixture after reduction with 2 mM dithionite.
Activation of Aconitase (AcnA) using native SufA
All the following procedures were performed anaerobically in the glove box at 18 °C. Aconitase A (0.2 nmol), in its apoform, was incubated in 50 mM Tris-HCl pH: 7.6 containing 5 mM DTT with a 10-fold molar excess of the native SufA (0.4 iron and sulfur atom/monomer) in order to provide 4 iron and 4 sulfur atoms/AcnA. After 15 min incubation, Aconitase activity was assayed as described by Gardner, P.R., and Fridovitch, I.13 by monitoring the formation of NADPH by the increase in absorbance at 340 nm. For that, AcnA-SufA proteins were added to 0.6 mM MnCl2, 25 mM citrate, 0.5 U isocitric dehydrogenase, 0.25 mM NADP+, 50 mM Tris-HCl, pH 7.6, in a 100-μL final volume. The 100 % activity corresponds to the activity of the chemically reconstituted AcnA (recAcnA) prepared by incubating apo-AcnA with 4 molar excess of ferrous iron and sulfur for 30 min. in the presence of 5 mM DTT (5 μmol/min/mg). The values reported for the specific activity of AcnA (SufA-recAcnA, recAcnA and apoAcnA) correspond to the average of three independent experiments.
Cluster transfer in the presence of BPS
Apo-Aconitase (0.2 nmol) was incubated anaerobically in 50 mM Tris-HCl, pH 7.6, 5 mM DTT with either [2Fe-2S] SufA (providing 4 equivalents of Fe and S atoms/apoAcnA) or 4-fold molar excess of Fe2+ and S2-. BPS was added at the same time as iron and sulfide (or [2Fe-2S] SufA) in both cases and the mixtures were incubated 15 min. and then aconitase activity was measured as described above.
Iron, sulfide, and protein analysis
The protein concentration was measured using Bradford's assay with 2 mg/ml BSA as standard14. Beinert's assay was used for sulfide estimation and both the Ferrozine assay and ICP-AES were used for iron estimation15,16.
Spectroscopic methods
UV-visible spectra were recorded with a Cary 1 Bio (Varian) spectrophotometer. EPR spectra were recorded on a Bruker EMX (9.5 GHz) EPR spectrometer equipped with an ESR 900 helium flow cryostat (Oxford Instruments). Double integrals of the EPR signals and spin concentration were obtained through the Win-EPR software using the spectrum of a 200 μM Cu(EDTA) standard recorded under non saturating conditions. Mössbauer spectra were recorded at 4.2 K on either a weak-field spectrometer using a permanent magnet of 50 mT or on a strong-field spectrometer furnished with a Janis CNDT/SC SuperVaritemp cryostat equipped with an 8-T superconducting magnet. Both spectrometers operate in a constant acceleration mode in a transmission geometry. The spectra were analyzed by using the program WMOSS (WEB research). The isomer shifts are reported in reference to the centroid of a room temperature spectrum of a metallic Fe foil. Resonance Raman spectra of frozen samples held at 15 K were recorded using a Jobin-Yvon U1000 double additive monochromator equipped with a front-illuminated LN_2 -cooled CCD detector. The excitation wavelength was obtained from the 441.6 nm line of a HeCd laser (Liconix). Spectral resolution was 4 cm-1 in these experimental conditions.
RESULTS
SufA is a [2Fe-2S] protein
Following co-expression of the SufABCDSE proteins in E. coli, SufA was purified anaerobically. The SufA-containing fraction was red and its UV-Visible spectrum displayed absorption bands at λmax=330, 420, and 460 nm (Fig. S1B) as previously reported for reconstituted [2Fe-2S]2+ SufA from E. chrysanthemi9. During a systematic screen to mutate His residues of SufD, we observed that the fraction containing SufA was deeper in red in the SufABCDSE expression system with a SufDH128A mutation than that from the wild-type SufABCDSE expression system. Indeed, SufA purified from the SufABCDSE(SufDH128A) expression system shows more intense light absorption bands and higher iron and sulfur content (0.6 iron and 0.7 sulfur/SufA monomer) (Fig. 1 & Fig. S1B). To firmly define the type of cluster on isolated native SufA this preparation was then used for further biochemical, spectroscopic (Electron Paramagnetic Resonance, Raman Resonance and Mössbauer spectroscopy) and functional characterization. The origin of the effect of the SufDH128A mutation is the subject of a separate study.
Figure 1.
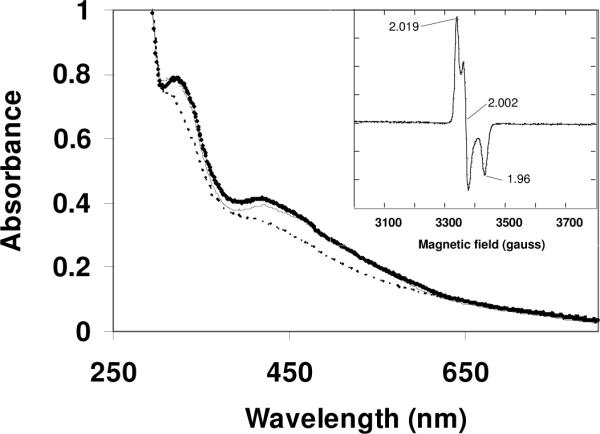
UV-Vis spectra of SufA isolated from anaerobic anion exchange chromatography of freeze-thaw lysates from SufABCDSE(SufDH128A) expression system in 0.1 M Tris-HCl pH: 8; 50 mM KCl, before (bold trace), after incubation anaerobically for 60 min with 5 mM DTT (dashed trace) and re-exposure to air for 1 minute (thin trace). Inset: EPR spectrum of SufA reduced with 5 mM DTT. Microwave power, 0.1 mW; modulation amplitude, 1 mT; Receiver gain: 2×104;
Temperature, 9.5 K.
Gel filtration chromatography revealed that SufA migrates as a single peak at an apparent molecular weight of 30.2 kDa, close to the expected molecular weight of a SufA dimer (26.6 kDa) (not shown). EPR analysis revealed that reduction with 5 mM dithiothreitol (DTT) generates a paramagnetic SufA protein characterized by an S = 1/2 EPR signal (Fig. 1, inset) whose g values as well as temperature dependence and microwave power saturation properties are consistent with a [2Fe-2S]1+ cluster. The EPR signal integrates to only 10% of total iron.
That the cluster is not destroyed during treatment with DTT is supported by: (i) the UV-visible spectrum of SufA after reduction (Fig. 1); (ii) an almost full restoration of the initial absorption bands after short re-exposure to air (Fig. 1); (iii) the conservation of Fe and S after chromatography of the reduced SufA over a desalting column (data not shown).
The resonance Raman spectrum of SufA recorded at 15 K (Fig. S2) shows intense bands at 286 and 402 cm-1 with additional bands at 350, 382, and 393 cm-1. The band at 286 cm-1 is diagnostic of [2Fe-2S]2+ clusters and is not seen for [4Fe-4S]2+ clusters or mononuclear iron sites17. Mössbauer spectrum for 57Fe-labeled SufA recorded at 4.2 K in 50 mT (Fig. 2A) consists of an intense symmetric quadrupole doublet (gray solid line in Fig. 2A) and a weak asymmetric quadrupole doublet (dashed line in Fig. 2A). The Mössbauer parameters determined for the intense doublet (ΔEQ = 0.53 ± 0.03 mm/s and δ = 0.28 ± 0.02 mm/s) are indicative of a [2Fe-2S]2+ cluster with cysteinyl ligands, while that of the weak doublet (ΔEQ = 2.68 mm/s and δ = 1.22 mm/s) suggest non-specifically bound ferrous ions. Moreover, the spectrum recorded in a strong applied field of 6 T (Fig. 2B) shows that the intense doublet (detected in the weak-field spectrum) arises from a diamagnetic system. This observed diamagnetism is consistent with the S = 0 ground state expected for a [2Fe-2S]2+ cluster comprising two antiferromagnetically coupled ferric ions. Detailed analysis of the data yields that the intense doublet accounts for 89% of the total Fe absorption. Thus, the Mössbauer data show clearly that a majority (i.e., 89%) of the Fe atoms in purified SufA is present in the form of a [2Fe-2S]2+ cluster with the remaining Fe atoms (11%) in non-specifically bound ferrous ion form.
Figure 2.
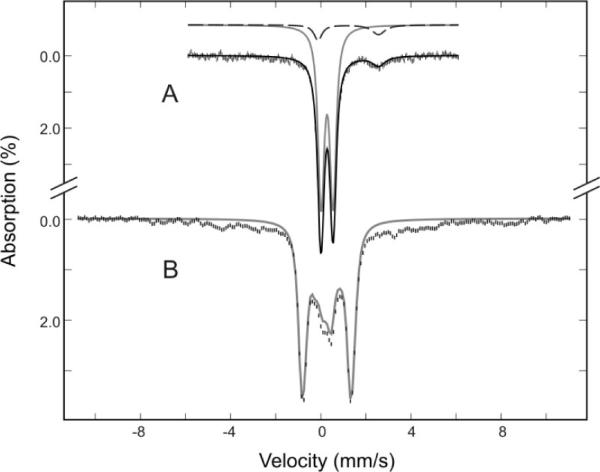
Mössbauer spectra of 57Fe-labeled SufA purified from SufABCDSE(SufDH128A) expression system recorded at 4.2 K in a magnetic field of 50 mT (A) or 6 T (B). The data can be decomposed into a major spectral component (gray solid lines) arising from a diamagnetic [2Fe-2S]2+ cluster and a minor component (dashed line in A) corresponding to non specifically bound FeII.
All spectroscopic data indicate that native SufA, co-expressed with SufBCDSE, is a [2Fe-2S] protein. Taking into consideration the iron and sulfur content, it may be estimated that in our best as-isolated preparations approximately 50% of SufA dimer contains a [2Fe-2S] cluster.
SufA is an [Fe-S] transport protein
To investigate whether native SufA has the ability to transfer its cluster to target proteins (as was previously demonstrated in vitro with chemically reconstituted SufA protein), apo ferredoxin (Fdx) from E. coli was used as a cluster acceptor protein. The holoprotein of Fdx contains a [2Fe-2S] cluster that exhibits unique spectroscopic properties, allowing easy monitoring of the cluster transfer reaction. ApoFdx was incubated anaerobically with sufficient [2Fe-2S] SufA to provide a 2 fold-molar excess of Fe and S with respect to apoFdx. The UV-Vis spectrum of the SufAFdx mixture after 60 min incubation indicates formation of holoFdx with λmax at 415 and 460 nm11 (Fig. 3). Subsequent reduction of the SufA-Fdx mixture with an excess of dithionite confirmed the formation of holoFdx, as unambiguously indicated by the appearance of an absorption band at 550 nm (indicated by an arrow in Fig. 3), characteristic of reduced [2Fe-2S]+ Fdx8, with concomitant disappearance of the 415 and 460 nm bands (Fig. 3).
Figure 3.
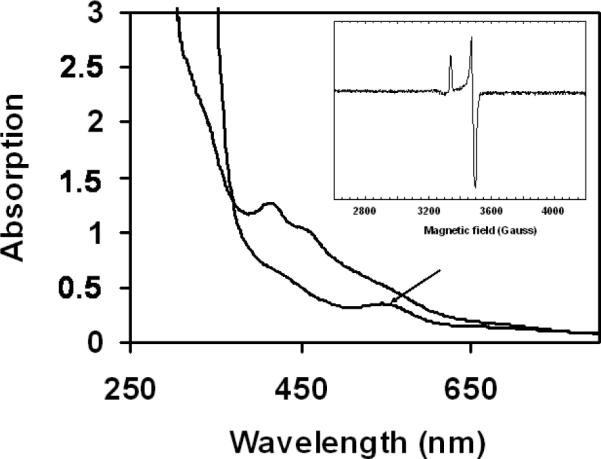
[2Fe-2S] cluster transfer from SufA to apoferredoxin. UV-Vis spectrum of the protein mixture after 1 hour incubation before (upper line) and after (lower line) reduction with 2 mM dithionite. Arrow indicates absorbance at 550 nm. Inset: EPR spectrum of the protein mixture after reduction with dithionite. Temperature: 10 K; microwave power: 0.1 mW; receiver gain: 2×104; modulation amplitude: 1 mT.
An EPR signal from the reduced sample was also observed whose g values were distinct from those of SufA and matched those of holoFdx11 (Fig. 3, inset). Assuming complete reduction, EPR quantification indicates that 80% of the [2Fe-2S] clusters present in SufA were transferred to Fdx. In contrast, after 60 min incubation with a two-fold molar excess of Fe and S in the form of ferrous iron and sulfide salts and in the absence of SufA only 6% of the Fdx protein was converted into holoFdx.
To investigate whether SufA could provide Fe-S clusters also to [4Fe-4S] target enzymes, native SufA was anaerobically incubated with the apoform of B. subtilis spore photoproduct lyase (SPL) or the apoform of E. coli aconitase A (AcnA), two well-characterized [4Fe-4S] proteins12,18. Following incubation, both apoSPL and apoAcnA convert into mature [4Fe-4S] holoproteins, as shown by spectroscopy and activity assays, respectively, with concomitant formation of apoSufA. A reducing agent, DTT, was absolutely required during this maturation process, as expected for a process converting ferric ions present in [2Fe-2S]2+ clusters to a mixture of ferric and ferrous ions in [4Fe-4S]2+ clusters. In addition, no cluster transfer from [2Fe-2S] SufA to apoSPL or apoAcnA could be observed when DTT was omitted from the reaction mixture or when apoSPL or apoAcnA were first pretreated with DTT and then desalted anaerobically over a G-25 column to remove DTT before transfer reaction. The results are presented in Fig. 4 and Fig. 5 for apoSPL and apoAcnA respectively.
Figure 4.
Iron-sulfur cluster transfer from native [2Fe-2S] SufA to apo Spore photoproduct lyase (SPL). ApoSPL (80 μM) was incubated with 10 molar excess of native SufA (0.4 iron and sulfide/monomer) in 0.1 M Tris-HCl pH: 8; 50 mM KCl, in the presence (A) or in the absence (B) of 5 mM DTT. The reaction was monitored by UV-vis. absorption spectroscopy before separation onto Ni-NTA column.
Figure 5.
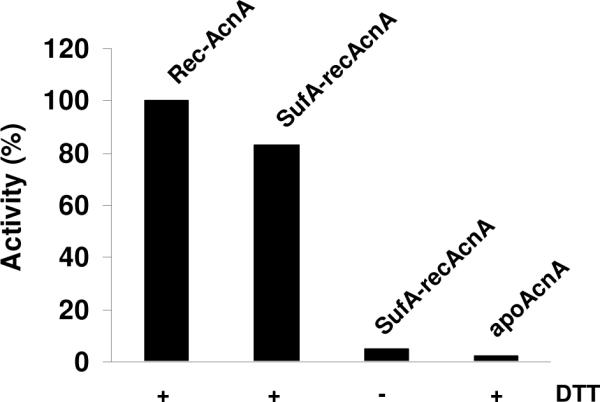
Activation of apo-aconitase by [2Fe-2S] SufA (SufA-recAcnA). Apo-Aconitase (0.2 nmol) was incubated anaerobically with [2Fe-2S] SufA (10 equivalents) in 50 mM Tris-HCl, pH 7.6 for 15 min. and aconitase activity was assayed as described in the experimental part. As controls, 0.2 nmol of either pure chemically reconstituted AcnA (recAcnA) or apoprotein (apoAcnA) were also assayed for aconitase activity. In all cases, proteins (recAcnA, SufA-recAcnA and apoAcnA) were added to 0.6 mM MnCl2, 25 mM citrate, 0.1 mg isocitric dehydrogenase (0.5 U), 0.25 mM NADP+, in the presence (+) or in the absence (-) of 5 mM DTT in a 100 μL final volume and the formation of NADPH monitored by UV-visible absorption spectroscopy.
In the case of apoSPL, incubation with native SufA in the presence of 5 mM DTT led to a change in the UV-visible spectrum of SufA with incubation time, with a decrease of the broad absorption in the 500-700 nm region and disappearance of the bands at 460 and 330 nm. The final UV-vis. spectrum is similar to that of a reconstituted holoSPL containing a [4Fe-4S]2+ cluster with one major broad absorption band at 420 nm (Fig. 4A). Using a His-tagged SPL protein, it was possible to purify it through a Ni-NTA affinity column, and under these conditions SPL was shown to contain around 3.2-3.3 iron and sulfur/monomer. In the absence of DTT or when apoSPL pretreated with DTT was used as a target, no change in the UV-visible spectrum of SufA was observed over the same incubation time and no iron and sulfur could be detected in the His-tagged SPL fraction after separation onto a Ni-NTA column showing that there is no cluster transfer under these conditions (Fig. 4B). In the case of apoAcnA (Fig. 5), an activity assay was used to monitor the assembly of the [4Fe-4S] cluster during incubation with [2Fe-2S] SufA. Activation of aconitase (80% yield) by native SufA was observed after 15 min incubation in the presence of DTT whereas less than 5% activation occurred when DTT was omitted from the reaction mixture, an activity close to that found with apo-AcnA (Fig. 5). All these studies clearly show that native SufA has the ability to transfer its Fe-S cluster to both [2Fe-2S] and [4Fe-4S] apoproteins, a function expected for an Fe-S scaffold protein. However, transfer for [4Fe-4S] apoproteins specifically requires a reducing agent such as DTT.
Intact cluster transfer from SufA
In order to determine whether the [4Fe-4S] clusters assembled in AcnA involves intact cluster transfer from native SufA, or arises through spontaneous self-assembly using Fe and sulfide ions transiently released in solution from the [2Fe-2S] cluster of SufA, we monitored cluster assembly in AcnA by measuring aconitase activity during anaerobic reaction with SufA in the presence of an iron chelator. Bathophenanthroline sulfonate (BPS) proved suitable because it did not inhibit aconitase activity and did not chelate iron out of native SufA at the concentrations used in the experiments (data not shown). BPS is expected to be an inhibitor of the cluster transfer reaction only in the case of a transient disassembly of SufA cluster but not if the mechanism implies a direct transfer. As shown in Fig. 6, BPS indeed efficiently inhibits apoAcnA activation by iron and sulfide. In contrast, when iron and sulfur are provided by SufA almost no inhibition was observed at concentrations up to 400 μM of BPS. These results thus strongly support a direct cluster transfer mechanism during maturation of AcnA by SufA.
Figure 6.

Intact cluster transfer from SufA. Apo-Aconitase (0.2 nmol) was incubated anaerobically in 50 mM Tris-HCl, pH 7.6, 5 mM DTT with either [2Fe-2S] SufA (providing 4 equivalents of Fe and S atoms/apoAcnA) (black bars) or 4-fold molar excess of Fe2+ and S2- (hatched bars). Increased amounts of BPS were added at the same time as iron and sulfide (or [2Fe-2S] SufA) in both cases and the mixtures were incubated 15 min. and then aconitase activity was measured. The values reported for the activity of AcnA correspond to the average of three independent experiments.
DISCUSSION
All the results reported here, namely the spectroscopic characterization and the Fe and S analysis, unambiguously demonstrate that SufA as isolated from E. coli cells harbours a [2Fe-2S] cluster. Both Mössbauer and resonance Raman data are in agreement with complete cysteinyl coordination for the cluster. Since there are only 3 cysteine residues per SufA monomer, this suggests one [2Fe-2S] cluster at the dimer interface, consistent with structural studies19,20. The presence of a [2Fe-2S]+1 cluster was also demonstrated by EPR spectroscopy after reduction with DTT. It is interesting to mention that this is the first report of a A-type Fe-S scaffold protein in the one-electron reduced state.
Most of the Fe-S proteins involved in Fe-S biogenesis (such as IscU, IscA, SufA and SufB) are usually purified in the apoform when they are expressed without their cognate partners. Since protein-protein interactions have been shown to carefully regulate the activity of the individual proteins, the maturation of metal centers would likely depend on the presence of correct partner proteins21,22. Furthermore, Fe-S biogenesis pathways are subject to complex in vivo regulation in response to demand for Fe-S cluster protein maturation. Thus, analysis of Fe-S assembly proteins presents a number of technical challenges due to the dynamic aspects of in vivo cluster assembly process. In the present work, detection of a cluster in the isolated SufA was possible only because of co-expression of the full suf operon. These cellular conditions probably lead to saturation of in vivo target proteins with Fe-S clusters thus allowing accumulation of labile intermediates in proteins, like SufA, that are involved in Fe-S cluster biogenesis. The fact that the best preparations here nevertheless are substoichiometric in terms of [2Fe-2S] clusters might also be due to partial loss of the labile clusters during purification.
Our results also demonstrate that SufA can transfer Fe-S clusters to both [2Fe-2S] and [4Fe-4S] apoproteins, as shown in the case of ferredoxin, spore photoproduct lyase (SPL) and aconitase. This supports the notion that SufA has a cluster scaffold or cluster shuttle protein function, with a labile cluster that can be transferred to apoprotein targets. Interestingly, in vitro assembly of [4Fe-4S] clusters in aconitase or SPL only proceeds if DTT, a reducing agent, is present in the reaction mixture, in agreement with the requirement for two electrons during formation of a [4Fe-4S] cluster from 2×[2Fe-2S] clusters. The detailed mechanism of formation of a [4Fe-4S] cluster in aconitase from [2Fe-2S] clusters from SufA is an interesting issue that will be addressed in future studies. Recently, reduced Fdx was shown to donate electrons for the conversion of two [2Fe-2S] clusters to one [4Fe-4S] cluster within the IscU from A. vinelandii 4. Whether such a reductive coupling mediated by DTT occurs on SufA prior to cluster transfer or on the target protein after transfer is an opened question requiring further investigation. What we clearly demonstrate here is that cluster assembly in target proteins does not occur through a transient disassembly of the [2Fe-2S] cluster in native SufA into free Fe and S in solution. Indeed, a strong iron chelator did not inhibit cluster transfer from SufA to aconitase, supporting a direct and intact cluster transfer mechanism, and confirming previous reports using reconstituted holoSufA or holoIscA proteins 9.
All these results now firmly establish that SufA functions as a Fe-S scaffold or Fe-S transporter protein in vivo rather than as a Fe donor for delivery of only Fe during cluster assembly. Furthermore our results emphasize that in vivo Fe-S cluster formation is a dynamic process that involves complex interactions between all proteins encoded by the suf operon.
Supplementary Material
ACKNOWLEDGMENT
This work was supported in part by National Institutes of Health grants GM81706 (to F.W.O.), and GM47295 (to B. H. H.), a Cottrell Scholar Award (to F.W.O.), and from the Centre National de la Recherche Scientifique, the Agence Nationale pour la Recherche (Programmes blancs BIOSUF), the Centre à l'Energie Atomique, and the Université Joseph Fourier (Grenoble) (to S.O.Ch.). We would like to thanks L. Bordes (DSV/iBiTec-S/CEA-Grenoble) for resonance Raman and M. Atta and A. Chandor (iRTSV/LCBM/CEA-Grenoble, France) for the gift of the SPL enzyme.
Footnotes
Supporting Information Available. Comparison of anion exchange chromatography elution profiles and optical spectra of SufA purified from wild-type SufABCDSE and SufABCDSE (SufDH128A) expression systems, and resonance Raman spectrum of native 57Fe-S SufA. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- (1).Johnson MK. Curr Opin Chem Biol. 1998;2:173–81. doi: 10.1016/s1367-5931(98)80058-6. [DOI] [PubMed] [Google Scholar]
- (2).Fontecave M, Ollagnier-de-Choudens S. Arch Biochem Biophys. 2008;474:226–37. doi: 10.1016/j.abb.2007.12.014. [DOI] [PubMed] [Google Scholar]
- (3).Johnson DC, Dean DR, Smith AD, Johnson MK. Annu Rev Biochem. 2005;74:247–81. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- (4).Chandramouli K, Unciuleac M-C, Naik S, Dean DR, Huynh BH, Johnson MK. Biochemistry. 2007;46:6804–6811. doi: 10.1021/bi6026659. [DOI] [PubMed] [Google Scholar]
- (5).Agar JN, Krebs C, Frazzon J, Huynh BH, Dean DR, Johnson MK. Biochemistry. 2000;39:7856–62. doi: 10.1021/bi000931n. [DOI] [PubMed] [Google Scholar]
- (6).Unciuleac M-C, Chandramouli K, Naik S, Mayer S, Huynh BH, Johnson MK, Dean DR. Biochemistry. 2007;46:6812–21. doi: 10.1021/bi6026665. [DOI] [PubMed] [Google Scholar]
- (7).Krebs C, Agar JN, Smith AD, Frazzon J, Dean DR, Huynh BH, Johnson MK. Biochemistry. 2001;40:14069–80. doi: 10.1021/bi015656z. [DOI] [PubMed] [Google Scholar]
- (8).Ollagnier-de-Choudens S, Mattioli T, Takahashi Y, Fontecave M. J Biol Chem. 2001;276:22604–7. doi: 10.1074/jbc.M102902200. [DOI] [PubMed] [Google Scholar]
- (9).Ollagnier-de Choudens S, Nachin L, Sanakis Y, Loiseau L, Barras F, Fontecave M. J Biol Chem. 2003;278:17993–8001. doi: 10.1074/jbc.M300285200. [DOI] [PubMed] [Google Scholar]
- (10).Ding H, Clark RJ. Biochem J. 2004;379:433–40. doi: 10.1042/BJ20031702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ta DT, Vickery LE. J Biol Chem. 1992;267:11120–5. [PubMed] [Google Scholar]
- (12).Chandor A, Berteau O, Douki T, Gasparutto D, Sanakis Y, Ollagnier-de-Choudens S, Atta M, Fontecave M. J Biol Chem. 2006;281:26922–31. doi: 10.1074/jbc.M602297200. [DOI] [PubMed] [Google Scholar]
- (13).Gardner PR, Fridovich I. J Biol Chem. 1992;267:8757–63. [PubMed] [Google Scholar]
- (14).Bradford MM. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- (15).Beinert H. Methods Enzymol. 1978;54:435–45. doi: 10.1016/s0076-6879(78)54027-5. [DOI] [PubMed] [Google Scholar]
- (16).Beinert H. Anal Biochem. 1983;131:373–8. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- (17).Fu W, Drozdzewski PM, Davies MD, Sligar SG, Johnson MK. J Biol Chem. 1992;267:15502–10. [PubMed] [Google Scholar]
- (18).Kennedy MC, Stout CD. In: Advances in Inorganic Chemistry. Sykes A, editor. Vol. 38. Academic Press Inc.; New York: 1992. pp. 323–339. [Google Scholar]
- (19).Morimoto K, Yamashita E, Kondou Y, Lee SJ, Arisaka F, Tsukihara T, Nakai M. J Mol Biol. 2006;360:117–32. doi: 10.1016/j.jmb.2006.04.067. [DOI] [PubMed] [Google Scholar]
- (20).Wada K, Hasegawa Y, Gong Z, Minami Y, Fukuyama K, Takahashi Y. FEBS Lett. 2005;579:6543–8. doi: 10.1016/j.febslet.2005.10.046. [DOI] [PubMed] [Google Scholar]
- (21).Layer G, Gaddam SA, Ayala-Castro CN, Ollagnier-de Choudens S, Lascoux D, Fontecave M, Outten FW. J Biol Chem. 2007;282:13342–50. doi: 10.1074/jbc.M608555200. [DOI] [PubMed] [Google Scholar]
- (22).Eccleston JF, Petrovic A, Davis CT, Rangachari K, Wilson RJ. J Biol Chem. 2006;281:8371–8. doi: 10.1074/jbc.M513455200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



