Abstract
Objective
Low-level HIV-1 replication may occur during antiretroviral therapy (ART) that suppresses plasma HIV-1 RNA to <50c/mL (suppressive ART). Antiretroviral drugs appear less effective in macrophages and monocytes compared to lymphocytes, both in vitro, and as implied in vivo by greater viral evolution observed during suppressive ART. Our objective was to examine sputum, which is rich in macrophages for evidence of increased HIV-1 replication compared to that in the blood during suppressive ART.
Design
Cross sectional study during suppressive ART. Comparison of HIV-1 DNA sequences derived from induced sputa and peripheral blood mononuclear cells (PBMC).
Methods
Multiple sequences encoding HIV-1 reverse transcriptase, protease, and envelope were generated using single-genome-sequencing. Reverse transcriptase and protease sequences were analyzed for genotypic drug resistance. The evolutionary distances of env sequences from the inferred most recent common ancestor of infection were calculated and CXCR4 co-receptor usage was predicted.
Results
970 bidirectional sequences from 11 individuals were analyzed. HIV-1 env and pol derived from sputa had greater frequency of drug resistance mutations (P = 0.05), evolutionary divergence (P = 0.004) and tendency for CXCR4 usage (P = 0.1) compared to viruses derived from PBMC.
Conclusion
The greater frequency of HIV-1 drug resistance mutations and divergence of HIV-1 env in sputa- compared to PBMC-derived viruses suggests greater HIV-1 replication in the respiratory tract compared to the blood. Characterization of viral evolution over time and by cell-type could identify cells that provide a sanctuary for low-level viral replication in the respiratory tract during suppressive ART.
Keywords: drug resistance, highly active antiretroviral therapy, HIV-1, lung, macrophages, replication, sputum, virus
Introduction
Twenty [1] to forty-five percentage [2] of individuals starting antiretroviral treatment (ART) have been reported to fail to achieve sustained suppression of viral replication. Even during seemingly effective ART, low-level viral replication has been evident in the blood of a subset of individuals[3-6].While many individuals demonstrate little or no HIV-1 replication in the peripheral blood mononuclear cells (PBMC) during ART [7,8], the slow rate of viral decay during ART suggests replenishment of persisting viral reservoirs [9,10]. Several observations suggest that blood may not be the optimal tissue in which to assess low-level replication. First, plasma HIV-1 sequences during and after ART indicate that PBMC are not the principal source of circulating HIV-1[7,11-13]. Second, mucosal associated lymphoid tissue is a primary target of HIV-1 [14], and HIV-1 induces dramatic changes in the mucosal associated lymphoid tissue[15-17] that are not reflected in the peripheral blood. Lastly, two studies of gastrointestinal tissue suggest that low-level replication can occur during ART that suppresses plasma HIV-1 RNA below the limit of detection [17,18].
HIV-1 replication within specific cells may differ during suppressive ART due to the heterogeneity of intracellular antiretroviral (ARV) pharmacokinetics [19]. In vitro studies of macrophages show poor phosphorylation of nucleoside reverse transcriptase inhibitors to the active form [20], and require higher concentrations of protease inhibitors to inhibit viral replication [21]. In addition, increased viral evolution was observed in monocytes compared to lymphocytes separated from cells collected by leukapheresis during suppressive ART [22]. These observations suggest that tissue macrophages may serve as a relative sanctuary during ART without being readily detected in the peripheral blood.
Consistent with this hypothesis, several studies conducted in the pre-ART era[23-26] observed that HIV-1 DNA from alveolar macrophages was compartmentalized and more divergent from the most recent common ancestor (MRCA) of infection when compared to PBMC-derived virus [23,24]; suggesting that alveolar macrophages and/or lymphocytes in the respiratory tract may be a site of increased viral replication. Most of these specimens were obtained during active pulmonary disease, when increased immune activation and viral replication would be expected. High concentrations of HIV-1 in the lung compared to other organs collected during autopsy [25], and different drug-resistance mutations (assessed by consensus sequencing) in bronchoalveolar lavage compared to blood specimens collected during failing ART [26], also suggest that the lung may be a site of increased or differential HIV-1 replication.
Identifying sites of low-level replication during apparently effective ART may help develop strategies to improve the success of long-term ART [27]. This pilot study examined induced sputa, rich in alveolar macrophages, to gauge the relative amount of HIV-1 replication compared to blood. Sputa and blood were collected from individuals without respiratory symptoms, and viral replication evaluated by analyses of single viral genome-derived sequences of HIV-1 pol, encoding reverse transcriptase (RT) and protease (PR), and envelope (env), to predict drug resistance and gauge viral divergence.
Methods
Participants and specimens
A cross-sectional study of HIV-1 DNA from blood and sputa was conducted at Seattle Children’s Hospital, among children >6 years old, with no respiratory symptoms, and receiving ART with plasma HIV-1 RNA <50 copies/mL. Informed consent/assent was obtained in accordance with the Institutional Review Board. Alveolar sputa induced by ultrasonic nebulization (Ultra-Neb99, DeVilbiss, Somerset, PA) of hypertonic (3%) saline was expectorated after four minutes of inhalation, and repeated five times [28]. Sputum was treated with 10% dithiothreitol (Calbiochem, San Diego, CA) for 1 2 h, resuspended in PBS, and quantified (Z1 Coulter Counter, Beckman, Brea, CA). Differential cell counts were performed after Wright-Giemsa staining. PBMC (Accuspin tubes, Sigma-Aldrich, St. Louis, MO) and sputa cell DNA was extracted using Gentra DNA Purification System (Minneapolis, MN).
HIV-1 DNA amplification and quantification
Limiting dilution PCR was performed as previously described [6,29]. Multiplexed first round PCR of pol and env used three sets of primers (RTA/RT1, PRA/PR2, and ED31/BH2), followed by separate second round reactions with RTB/RT4, PRB/PR4, and ES7/ES8 to amplify the RT and PR regions of pol, and the C2-V5 region of env. Alternative primers were occasionally used to optimize amplification, including for the first round: RT2, RTC, Polfo (5′-TCAGAGCAGACCAGAGCCAAC-3′; HXB2 coordinates: 1347 1367), PR5, ED5, ED12, ED3 (5′-TTAGGCATCTCCTATGGCAGGAAGAAGCGG-3′; 5957 5986) and ED14 (5′-TCTTGCCTGGAGCTGTTTGATGCCCCAGAC-3′; 7961 7932); and for the second round: PRC, UHGR, RT3, and Polfi (5′-CAACAGCCCCACCAGAAGAGA-3′; 2153 2173). Cycling conditions and other primers have been previously published [6].
Sequencing
Bidirectional sequencing of PCR products was performed as previously described [6] and submitted to GenBank, accession numbers: FJ446721-FJ447017 and FJ447018-FJ447337.
Sequence analysis
Sequences were assembled, edited, aligned, and screened for hypermutation as described [6]. Sequences with ambiguous bases, indicating the possibility of multiple templates, were discarded. Maximum likelihood phylogenetic trees were constructed using the DiverAnalysis web tool (http://indra.mullins.microbiol.washington.edu/cgi-bin/DIVER/diver.cgi). For each individual, reference sequences from the same subtype were used as an out-group. The inferred sequence at the basal node of each participant’s tree was used as an estimate of the MRCA of his/her infection, from which divergence of that participant’s extant viral sequences was calculated. Major drug resistance mutations in RT were defined as per the IAS-USA (http://www.iasusa.org). HIV-1 co-receptor usage was predicted using a X4R5 position-specific scoring matrix (PSSM) [30]. Nonsubtype B sequences were also predicted based on the charge at amino acid positions 11 and 25 [30].
Statistical analysis
The average viral divergence and frequency of sequences with drug resistance mutations were calculated for each PBMC and sputum specimen. Because sequences from individuals on suppressive therapy have been shown to regress toward the MRCA [6,31,32], we analyzed average divergence on the assumption that it will be more sensitive to the minority of sequences which had undergone replication. Differences were compared with a Wilcoxon Two-sided Ranked-Sign Test, using the mean PBMC and sputum values from each individual participant as a paired set.
Results
Fourteen children, 7 to 17 years of age, who had been prescribed ART for a median of 6.1 years, were enrolled. Three participants were excluded from analyses; two had sputa with <2 million cells/specimen; and in the third PCR failed to amplify HIV-1. The remaining 11 participants included in the analysis were 55% (6/11) female, 64% (7/11) African American, 18% (2/11) Hispanic, and 9% (1/11) Caucasian. Clinical characteristics of these participants and their sputum specimens are shown in Table 1. Once ART suppressed the plasma HIV-1 RNA to <50 c/mL, low-level viremias were detected on a median of 3% (range 0 – 17) of determinations, at a median of 97 c/mL (range 51 308), and none of 283 HIV-1 RNA determinations was greater than 400 c/mL. Sputa specimens had a median of 16 (range 2 52) × 106 cells, with a median macrophage to lymphocyte ratio of 7.7 (range 3.2 – >100), which did not correlate with concomitant peripheral blood monocyte to lymphocyte ratio (Spearman’s rank correlation statistic: ρ = 0.17, P = 0.622). Eighteen to 58 (median 28) single-genome sequences/gene were generated from each participant’s specimens with a total of 970 bidirectional sequences analyzed.
Table 1.
Characteristics of participants, sputum, and the number of sequences analyzed.
| [0, 2–6]Clinical Characteristics and Blood Values | [0, 7–9]Sputum Analyses* | |||||||
|---|---|---|---|---|---|---|---|---|
| [0, 4–5]Plasma HIV-1 RNA determinations during ART |
HIV-1 DNA copies per 106 CD4+ in PBMC | # of Sputum Cells (× 106) Collected | Macrophage to Lymphocyte Ratio in Sputum, (Bloodc) | HIV-1 DNA copies per 106 Macrophages and Lymphocytes | ||||
| Participant | ART Regimena | Years on ART | # >50/>400 copies/mL | Total # | ||||
| B1 | A | 7.1 | 0/0 | 40 | 194 | 52 | 3.5 (0.17) | 156 |
| B2 | A | 6.2 | 3/0 | 31 | 593 | 21 | 3.2 (0.26) | 165 |
| C1 | A | 7.0 | 0/0 | 22 | 126 | 11 | 5.2 (0.16) | 342 |
| F1 | A | 3.8 | 5/0 | 30 | 1163 | 4 | 7.7 (0.34) | 27 |
| I2 | B | 1.5 | 0/0 | 8 | 614 | 38 | 5.0 (0.19) | 18 |
| J1 | B | 4.2 | 2/0 | 23 | 451 | 19 | 21.3 (0.20) | 13.3 |
| L1 | A | 7.1 | 2/0 | 37 | 262 | 5 | 4.7 (0.29) | 230 |
| R1 | B | 6.6 | 1/0 | 25 | 228 | 16 | >100:1 (0.42) | 90 |
| S3 | A | 1.6 | 0/0 | 15 | 116 | 3 | >100:1 (0.21) | 20 |
| S4 | A | 0.8 | 3/0 | 21 | 764 | 2 | 20.0 (0.23) | 5 |
| V1 | A | 5.8 | 1/0 | 31 | 2314 | 38 | 15.4 (0.16) | 79 |
| Median | 5.8 | 1/0 | 25 | 451 | 16 | 7.7 (0.21) | 79 | |
| Total | 17/0 | 283 | ||||||
ART, antiretroviral therapy; PBMC, peripheral blood mononuclear cells.
Included a median of 34.5% (range 5 – 70.5%) squamous epithelial cells and 29% (range 3 – 82%) neutrophils.
Antiretoviral Regimens: A = three class combination with nucleoside reverse transcriptase inhibitors, nonnucleoside reverse transcriptase inhibitor, and protease inhibitor; B = two class combination with nucleoside reverse transcriptase inhibitors and protease inhibitors (lopinavir with ritonavir).
estimated by QUALITY program [29] applied to limiting dilution PCR of env.
ratio of monocytes to lymphocyes in the concomitant peripheral blood.
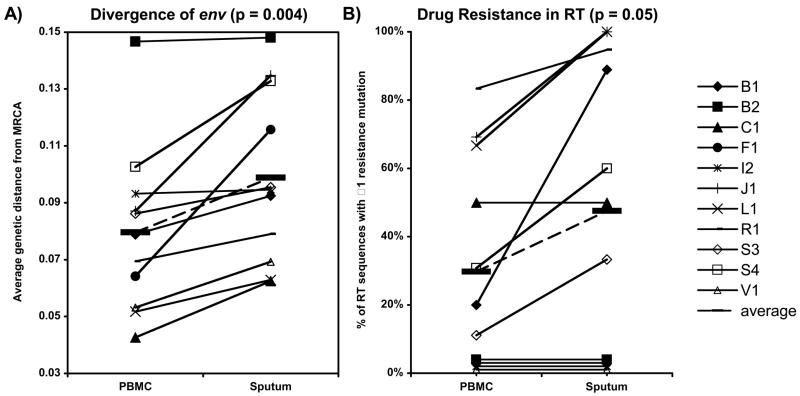
The mean divergence of the sputum-derived HIV-1 env sequences from the MRCA in all 11 participants was greater than PBMC-derived sequences (P = 0.004) (Fig. 1a). The mean divergence of all sputa-derived sequences was 9.9% versus 8.0% for all PBMC-derived sequences. This corresponds to a mean of >11 additional nucleotide changes in sputum- compared to PBMC-derived env sequences over the approximately 625 base pair region.
Fig. 1.
HIV-1 sequences derived from induced sputa compared to those from peripheral blood mononuclear cells (PBMC).
The mean value for each participant’s sequences is represented by a symbol (see key to right). Plots show (a) HIV-1 mean env divergence from the ancestor of infection, and (b) frequency of RT sequences with ≥1 drug resistance mutation. Values from each participant’s sputum and PBMC are connected with a line. The mean of all participants’ values is shown for each parameter (bold hashes connected by dashed line). P values refer to the differences between sputum and PBMC values using Wilcoxon Two-sided Ranked-Sign Test to compare the paired mean values from each individual. Values corresponding to 0% on panel B are splayed slightly for better visualization.
Major protease inhibitor-associated mutations were only found in one participant (J1), whereas mutations associated with resistance to nucleoside analog reverse transcriptase inhibitors (NRTI) were detected in 7 of 11 and two of these also had resistance to nonnucleoside reverse transcriptase inhibitors (NNRTI) (Fig. 1b). All seven of these participants had treatment with mono- or dual-NRTI prior to highly active ART. In 6 of these 7 participants the percentage of sequences with drug resistance mutations was greater in sputum- compared to PBMC-derived sequences and in one (C1) it was equal (P = 0.05). Averaging all participants, the mean percentage of sequences with drug resistant HIV-1 was 52% from sputa versus 36% from PBMC.
Env codons associated with the use of the CXCR4 co-receptor (X4 sequences) were found in 6 of 11 participants. In five of the six, the percentage of X4 sequences was greater in sputa- compared to PBMC-derived sequences (P = 0.1), with overall means of (41%) versus (31%). The PSSM algorithm is less well validated in nonsubtype B virus. Scoring of basic amino acids at positions 11 and 25 for the two participants with nonsubtype B virus did not change the predicted phenotype of any sequences from I2 (subtype F) but decreased the percentage of predicted X4 phenotypes proportionally in both sputum- and PBMC-derived sequences from F1 (subtype D).
Discussion
During suppressive ART, features of HIV-1 sequences indicative of increased viral replication were more common in viruses derived from sputum compared to PBMC. Specifically, sputum-derived HIV-1 env had a greater mean divergence from the ancestor of infection, an increase in the frequency of drug resistance mutations in HIV-1 RT, and a trend to greater X4 genotype. These changes are characteristic of ongoing viral replication in untreated persons [33,34]. Detection of evolution in two genes (pol and env) under different selective forces (antiretrovirals and host immunity) strengthens our evidence for increased viral replication in the sputa-derived viral sequences.
A different rate of viral replication in the respiratory tract cells relative to the blood is a straightforward explanation for the observed differences. However, unequal selective pressure and rates of HIV-1 infected cell turnover may also have contributed to the disparity. In this cross-sectional study, we cannot determine when or where these differences originated. Longitudinal studies should provide insight into whether viral replication and/or selection are ongoing during suppressive ART.
Inflammatory cytokines are associated with increased HIV-1 replication [35], and may have contributed to the differences between viruses we amplified from the sputum and blood, as well as those with pulmonary infections in previous studies [23,24,26]. We reason that increased immune activation in cells within the respiratory tract mucosa and associated lymphoid tissue, even in the absence of respiratory symptoms, could enhance viral replication and/or the proliferation of infected cells compared to the blood.
Given that macrophages were more prevalent than lymphocytes in all sputa, and that the HIV-1-DNA loads we measured in sputa were similar to published levels in purified alveolar macrophages [24,36], it is tempting to speculate that the observed differences are due to HIV-1 from alveolar macrophages. However, recently published data suggests that the bulk of HIV-1 DNA in bronchoalveolar lavage specimens comes from lymphocytes [37,38]. This suggests that the differences in HIV-1 sequences we derived from sputum versus blood may be due to sampling a different population of cells associated with the respiratory tract mucosa.
In summary, increased viral divergence and drug resistance in sequences derived from sputa compared to the blood indicates relatively greater HIV-1 replication in the respiratory tract. Longitudinal characterization of viral evolution by sputa cell-type should determine whether specific respiratory tract cells provide a sanctuary from ART and allow low-level viral replication during ART that appears suppressive by blood tests.
Acknowledgments
Acknowledgment of Financial Support: This work was supported by the Foster Foundation (LMF) and the NIH, including R21 AI058723 (LMF), K23 AI058683-01 (NHT), K23 K23AI077357-01 and T32 HDO07233-24 (TAW), University of Washington CFAR P30 AI27757, and the General Clinical Research Center at the University of Washington M01RR-00037.
Footnotes
Conflict of Interest: None of the authors have a commercial or other association that might pose a conflict of interest.
Previous presentations: This work was presented in part at The XV International HIV Drug Resistance Workshop, 13 – 17, June 2006, Sitges, Spain (Abstract #56) and The 14th Conference on Retroviruses and Opportunistic Infections, 25 – 28, February 2007, Los Angeles, CA (Abstract #632).
Authors’ contributions: N.H.T and L.M.F designed the research; T.A.W., N.H.T., A.J.M., K.M.M., and L.M.F conducted clinical study; T.A.W., N.H.T., J.K.M., M.X., and L.M.F. performed laboratory research; T.A.W., N.H.T., J.K.M., G.H.L., J.I.M., and L.M.F. conducted phylogenetic analysis; T.A.W. and L.M.F analyzed the data; and T.A.W. and L.M.F. prepared the manuscript.
References
- 1.Gallant JE, DeJesus E, Arribas JR, et al. Tenofovir DF, emtricitabine, and efavirenz vs. zidovudine, lamivudine, and efavirenz for HIV. N Engl J Med. 2006;354:251–260. doi: 10.1056/NEJMoa051871. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett JA, Fath MJ, Demasi R, et al. An updated systematic overview of triple combination therapy in antiretroviral-naive HIV-infected adults. Aids. 2006;20:2051–2064. doi: 10.1097/01.aids.0000247578.08449.ff. [DOI] [PubMed] [Google Scholar]
- 3.Sharkey ME, Teo I, Greenough T, et al. Persistence of episomal HIV-1 infection intermediates in patients on highly active antiretroviral therapy. Nat Med. 2000;6:76–81. doi: 10.1038/71569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu T, Muthui D, Holte S, et al. Evidence for human immunodeficiency virus type 1 replication in vivo in CD14(+) monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy. J Virol. 2002;76:707–716. doi: 10.1128/JVI.76.2.707-716.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun TW, Nickle DC, Justement JS, et al. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. J Clin Invest. 2005;115:3250–3255. doi: 10.1172/JCI26197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tobin NH, Learn GH, Holte SE, et al. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: expression of archival virus and replication of virus. J Virol. 2005;79:9625–9634. doi: 10.1128/JVI.79.15.9625-9634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey JR, Sedaghat AR, Kieffer T, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80:6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Persaud D, Ray SC, Kajdas J, et al. Slow human immunodeficiency virus type 1 evolution in viral reservoirs in infants treated with effective antiretroviral therapy. AIDS Res Hum Retroviruses. 2007;23:381–390. doi: 10.1089/aid.2006.0175. [DOI] [PubMed] [Google Scholar]
- 9.Ramratnam B, Mittler JE, Zhang L, et al. The decay of the latent reservoir of replication-competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged antiretroviral therapy. Nat Med. 2000;6:82–85. doi: 10.1038/71577. [DOI] [PubMed] [Google Scholar]
- 10.Chun T-W, Nickle DC, Justement JS, et al. HIV infected individuals receiving effective antiretroviral therapy for extended periods of time continually replenish their viral reservoir. J Clin Invest. 2005;115:3250–3255. doi: 10.1172/JCI26197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun TW, Davey RT, Jr, Ostrowski M, et al. Relationship between preexisting viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti retroviral therapy. Nat Med. 2000;6:757–761. doi: 10.1038/77481. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, Chung C, Hu BS, et al. Genetic characterization of rebounding HIV-1 after cessation of highly active antiretroviral therapy. J Clin Invest. 2000;106:839–845. doi: 10.1172/JCI10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dybul M, Daucher M, Jensen MA, et al. Genetic characterization of rebounding human immunodeficiency virus type 1 in plasma during multiple interruptions of highly active antiretroviral therapy. J Virol. 2003;77:3229–3237. doi: 10.1128/JVI.77.5.3229-3237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veazey RS, DeMaria M, Chalifoux LV, et al. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 15.Li Q, Duan L, Estes JD, et al. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005;434:1148–1152. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- 16.Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature. 2005;434:1093–1097. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- 17.Guadalupe M, Sankaran S, George MD, et al. Viral suppression and immune restoration in the gastrointestinal mucosa of human immunodeficiency virus type 1-infected patients initiating therapy during primary or chronic infection. J Virol. 2006;80:8236–8247. doi: 10.1128/JVI.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belmonte L, Olmos M, Fanin A, et al. The intestinal mucosa as a reservoir of HIV-1 infection after successful HAART. AIDS. 2007;21:2106–2108. doi: 10.1097/QAD.0b013e3282efb74b. [DOI] [PubMed] [Google Scholar]
- 19.Anderson PL, Zheng JH, King T, et al. Concentrations of zidovudine- and lamivudine-triphosphate according to cell type in HIV-seronegative adults. AIDS. 2007;21:1849–1854. doi: 10.1097/QAD.0b013e3282741feb. [DOI] [PubMed] [Google Scholar]
- 20.Richman DD, Kornbluth RS, Carson DA. Failure of dideoxynucleosides to inhibit human immunodeficiency virus replication in cultured human macrophages. J Exp Med. 1987;166:1144–1149. doi: 10.1084/jem.166.4.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perno CF, Svicher V, Schols D, Pollicita M, Balzarini J, Aquaro S. Therapeutic strategies towards HIV-1 infection in macrophages. Antiviral Res. 2006;71:293–300. doi: 10.1016/j.antiviral.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 22.Crowe S, Zhu T, Muller WA. The contribution of monocyte infection and trafficking to viral persistence, and maintenance of the viral reservoir in HIV infection. J Leukoc Biol. 2003;74:635–641. doi: 10.1189/jlb.0503204. [DOI] [PubMed] [Google Scholar]
- 23.Itescu S, Simonelli PF, Winchester RJ, Ginsberg HS. Human immunodeficiency virus type 1 strains in the lungs of infected individuals evolve independently from those in peripheral blood and are highly conserved in the C-terminal region of the envelope V3 loop. Proc Natl Acad Sci U S A. 1994;91:11378–11382. doi: 10.1073/pnas.91.24.11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakata K, Weiden M, Harkin T, Ho D, Rom WN. Low copy number and limited variability of proviral DNA in alveolar macrophages from HIV-1-infected patients: evidence for genetic differences in HIV-1 between lung and blood macrophage populations. Mol Med. 1995;1:744–757. [PMC free article] [PubMed] [Google Scholar]
- 25.Atkins M, Strappe P, Kaye S, et al. Quantitative differences in the distribution of zidovudine resistance mutations in multiple postmortem tissues from AIDS patients. J Med Virol. 1998;55:138–146. doi: 10.1002/(sici)1096-9071(199806)55:2<138::aid-jmv10>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 26.White NC, Israel-Biet D, Coker RJ, Mitchell DM, Weber JN, Clarke JR. Different resistance mutations can be detected simultaneously in the blood and the lung of HIV-1 infected individuals on antiretroviral therapy. J Med Virol. 2004;72:352–357. doi: 10.1002/jmv.20010. [DOI] [PubMed] [Google Scholar]
- 27.Stebbing J, Gazzard B, Douek DC. Where does HIV live? N Engl J Med. 2004;350:1872–1880. doi: 10.1056/NEJMra032395. [DOI] [PubMed] [Google Scholar]
- 28.Pavord ID, Pizzichini MM, Pizzichini E, Hargreave FE. The use of induced sputum to investigate airway inflammation. Thorax. 1997;52:498–501. doi: 10.1136/thx.52.6.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodrigo AG, Goracke PC, Rowhanian K, Mullins JI. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res and Hum Retrovir. 1997;13:737–742. doi: 10.1089/aid.1997.13.737. [DOI] [PubMed] [Google Scholar]
- 30.Jensen MA, van’t Wout AB. Predicting HIV-1 coreceptor usage with sequence analysis. AIDS Rev. 2003;5:104–112. [PubMed] [Google Scholar]
- 31.Frenkel LM, Wang Y, Learn GH, et al. Multiple viral genetic analyses detecting low-level HIV-1 replication during effective HAART. J Virol. 2003;77:5721–5730. doi: 10.1128/JVI.77.10.5721-5730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruff CT, Ray SC, Kwon P, et al. Persistence of wild-type virus and lack of temporal structure in the latent reservoir for human immunodeficiency virus type 1 in pediatric patients with extensive antiretroviral exposure. J Virol. 2002;76:9481–9492. doi: 10.1128/JVI.76.18.9481-9492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shankarappa R, Margolick JB, Gange SJ, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cozzi-Lepri A, Phillips AN, Ruiz L, et al. Evolution of drug resistance in HIV-infected patients remaining on a virologically failing combination antiretroviral therapy regimen. AIDS. 2007;21:721–732. doi: 10.1097/QAD.0b013e3280141fdf. [DOI] [PubMed] [Google Scholar]
- 35.Abel K, Rocke DM, Chohan B, Fritts L, Miller CJ. Temporal and anatomic relationship between virus replication and cytokine gene expression after vaginal simian immunodeficiency virus infection. J Virol. 2005;79:12164–12172. doi: 10.1128/JVI.79.19.12164-12172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewin SR, Kirihara J, Sonza S, Irving L, Mills J, Crowe SM. HIV-1 DNA and mRNA concentrations are similar in peripheral blood monocytes and alveolar macrophages in HIV-1-infected individuals. AIDS. 1998;12:719–727. doi: 10.1097/00002030-199807000-00008. [DOI] [PubMed] [Google Scholar]
- 37.Brenchley J, Knox K, Asher A, et al. High frequencies of polyfunctional HIV-specific T cells are associated with preservation of mucosal CD4 T cells in bronchoalveolar lavage. Mucosal Immunology. 2008;1:49–58. doi: 10.1038/mi.2007.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Twigg HL, Weiden M, Valentine F, et al. Effect of Highly Active Antiretroviral Therapy on Viral Burden in the Lungs of HIV-Infected Subjects. Journal of Infectious Disaeses. 2008;197:109–116. doi: 10.1086/523766. [DOI] [PubMed] [Google Scholar]