Abstract
The transcriptional program for a gene consists of the promoter necessary for recruiting RNA polymerase along with neighboring operator sites that bind different activators and repressors. From a synthetic biology perspective, if the DNA-binding specificity of these proteins can be changed, then they can be used to reprogram gene expression in cells. While many experimental methods exist for generating such specificity-altering mutations, few computational approaches are available, particularly in the case of bacterial transcription factors. In a previously published computational study of nitrogen oxide metabolism in bacteria, a small number of amino-acid residues were found to determine the specificity within the CRP (cAMP receptor protein)/FNR (fumarate and nitrate reductase regulatory protein) family of transcription factors. By analyzing how these amino acids vary in different regulators, a simple relationship between the identity of these residues and their target DNA-binding sequence was constructed. In this article, we experimentally tested whether this relationship could be used to engineer novel DNA–protein interactions. Using Escherichia coli CRP as a template, we tested eight designs based on this relationship and found that four worked as predicted. Collectively, these results in this work demonstrate that comparative genomics can inform the design of bacterial transcription factors.
INTRODUCTION
DNA encodes not just the gene but also the program for expression. At the level of transcription, a given gene's program consists of the promoter sequences necessary for recruiting RNA polymerase along with cis-regulatory sequences specific for different transcriptional activators and repressors (1). In bacteria, these regulatory proteins bind specific DNA sequences, also known as operator sites, typically proximal to the promoter. The sequence of the operator site determines which activators and repressors regulate the activity of the promoter. In order for this regulation to work, the proteins regulating a given promoter must bind specifically to the associated operator sites, otherwise aberrant regulation will occur. Understanding the molecular basis for this recognition and specificity has been the focus of innumerable studies [cf. (2–4)]. This information can potentially be used to change the DNA-binding specificity of transcription factors, enabling the reprogramming of gene expression in cells with applications, for example, in synthetic biology and metabolic engineering (5–11). As a result, an active area of protein engineering has been to identify mutations that alter the DNA-binding specificity of these transcription factors (12). While a number of experimental methods exist for generating such specificity-altering mutations (13–17), few computational approaches exist, particularly in the case of bacterial transcription factors. One approach is the use of comparative genomics as a tool for altering the DNA-binding specificity of transcription factors in bacteria.
There are currently over 700 sequenced bacterial genomes, and thousands more are in the sequencing pipeline (18). Through the use of comparative genomics, we can inform transcription factor design using the large pool of genetic diversity contained within these data sets. In particular, by studying the co-variation of transcription factors with their target DNA-binding sites, we may be able to correlate how certain amino-acid sequences determine which DNA sequences the protein binds within a family of regulators, despite the fact that there is no general ‘recognition code’ for protein–DNA interactions across families (19). Along these lines, in a previous comparative study of nitrogen oxide metabolism in bacteria (20), we found that a small number of amino-acid residues determine the specificity of regulators within the CRP (cAMP receptor protein)/FNR (fumarate and nitrate reductase regulatory protein) family of transcription factors (21). Specifically, we predicted that the three amino-acid residues in Escherichia coli CRP (Arg180/Glu181/Arg185) making direct contact with DNA bases in the major groove are sufficient for determining the specificity of regulators within this family of proteins (22). By analyzing how these amino acids vary in different regulators, a simple correlation between the identity of these residues and their target DNA-binding sequence was constructed. These correlations were interesting because they suggested similarity in the binding mode among the different regulators within the abundant CRP/FNR family. The idea of a family-specific binding mode is consistent with theoretical and empirical work (2,23), including a recently described method that successfully predicted transcription factor specificities across families of regulators using structural data from a small number of homologs (24) and a method that identified CRP-binding sites in E. coli using structural information (25). From a protein-engineering viewpoint, the correlations suggested that designs focused on only these three residues may be sufficient for altering DNA-binding specificity.
In this study, we experimentally tested these correlations from our previous, purely computational genomics study (20) in order to see whether such an approach would be effective for bacterial transcription factor engineering. As only a few amino acids were predicted to determine specificity and the CRP/FNR family is quite large, it was not immediately obvious that such an approach would work. Using E. coli CRP as a template, we generated eight different variants based on these correlations and then determined whether these variants could bind their cognate operator sequence and regulate transcription. In all cases, the variants involved substitutions within the Arg180/Glu181/Arg185 amino-acid triad of CRP along with the corresponding changes to the CRP operator sequence in the lacZ promoter. Of the eight, four CRP variants were able to bind their new operator sequences and activate transcription. Furthermore, to the best of our knowledge, none of these four mutations had previously been isolated despite extensive work over the years on the binding mechanism of CRP. In addition, these results appear to be the first where results from computational analysis have been used to design bacterial transcription factors. Collectively, these results suggest that comparative genomics can inform the design of proteins, transcription factors in particular.
MATERIALS AND METHODS
Bacterial strains, media and growth conditions
All cloning steps were performed either in the E. coli strains DH5α or XL1-Blue supercompetent cells (Stratagene). Gene expression experiments were performed either in E. coli strain MG1655 or an isogenic derivative where the crp gene was deleted. The Δcrp strain (Δcrp::FRT) was made using the gene inactivation method of Datsenko and Wanner with pKD4 as the template and the primers CRP_F (5′-ATT CAT AAG TAC CCA TCC AAG AGC ACG CTT ATT CAC CAG GGT GTA GGC TGG AGC TGC TTC-3′) and CRP_R (5′-CAG CAT CTT CAG AAT GCG TCC CAC GGT TTC ACG AGA ACA GCA TAT GAA TAT CCT CCT TAG-3′) (26). Prior to removal of the antibiotic resistance marker, the constructs were moved into a clean, wild-type background (MG1655) by P1vir transduction. Removal of the kanamycin resistance gene was achieved by passing pCP20 through the strain. E. coli was grown in Luria–Bertani (LB) broth at 37°C. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 17 μg/ml; and kanamycin, 40 μg/ml. All enzymes were obtained from New England Biolabs or Stratagene. The inducer atc was used at a concentration of 20 ng/ml.
Construction of crp mutants
The crp gene was amplified by PCR using genomic E. coli MG1655 DNA as the template with primers 5′-CCA CAT CCT GAC GCC CTT TT-3′ and 5′-CCG TAC CAG AGA GTG CCC AA-3′ (genomic region 3483651–3485261). The resulting PCR fragment was then inserted into the pCR®2-TOPO® plasmid using the TOPO cloning procedure as described by the manufacturer's protocols (Invitrogen), yielding the plasmid pTOPO-crp. Site-directed mutagenesis of the crp gene using pTOPO-crp as a template was accomplished using either enzymatic inverse PCR (EIPCR) (27) or QuikChange® Site-Directed Mutagenesis Method Kit (Stratagene). For construction of the operator mutations, the plasmid, pd2EGFP (BD Biosciences Clontech), was used as the template. This plasmid contains the d2EGFP variant of the green fluorescent protein gene under the control of the lacZ promoter. EIPCR was used to introduce the mutations into the CRP operator site. The primers are listed in Table S1. To minimize the possibility of unwanted mutations, strains harboring the crp mutations were propagated and maintained in M9 minimal media supplemented with 1% glucose. In addition, they were repeatedly sequenced in order to test for any possible mutations.
Construction of the TetR-regulated expression plasmid
In order to construct an inducible expression vector, the crp gene was introduced into the restriction sites EcoRI and BamHI of a pPROTet.E plasmid derivative (Clontech Laboratories, Inc.) under the control of the strong promoter pLtetO-1, resulting in the plasmids pCRP-x, where x denotes the allele. In the absence of TetR, the pLtetO-1 promoter is constitutively active. In order to regulate promoter activity, the tetR gene from transposon Tn10 was amplified using the primers TetR_F (ACC AGC GGC CGC AAG GAG ATA GAA ATG ATG TCT AGA TTA GAT AA) and TetR_R (ATC ATT AAT TTA AGA CCC ACT TTC AGA TT) and then inserted into the NotI and AseI restriction sites of pPROTet.E. As TetR represses expression from the pLtetO-1 by binding to an operator sequence within the promoter and sterically inhibiting RNA polymerase, this promoter is inactive in the absence of atc inducer. In the presence of inducer, TetR no longer binds the promoter and inhibits expression. The high copy number ColE1 origin of replication of pPROTet.E was also replaced with the medium copy number p15a origin of replication from the plasmid pZA31 by swapping the fragments generated by the restriction sites of AvrII and SacI in order to make the pCRP plasmids compatible with pd2EGFP.
Library construction and screening
Randomization of the middle six positions within the CRP operator site (positions 9, 10 and 11) was achieved using EIPCR with the degenerate primers. In the case of the Om5 reporter, the target of the screens, the primers 5′-GGA AAG GTC TCA TGT AAN NNN NNT TAC AGA TTA GGC AC-3′ and 5′-GAC TAG GTC TCA TAC AGT AAT TGC GTT GCG C-3′ were used. A similar procedure was used to randomize positions 5, 7 and 8 in the Owt reporter. To screen for operator sequences that we activated by one of the CRP variants, cells transformed with the appropriate plasmids were plated, allowed to grow overnight, and screened for GFP expression using UV light. Fluorescent colonies were then picked, sequenced and analyzed using the methods described below.
Fluorescence and cellular growth measurements
To measure the gene expression of the green fluorescent protein (GFP) and the cell growth in E. coli, cultures were grown overnight in 3 ml of LB in test tubes with the appropriate antibiotics and inducers with constant shaking at 37°C. End-point fluorescent and OD600 nm measurements were taken in a Tecan Safire2 microplate reader, where 100 μl of culture was first transferred to a 96-well microplate. The excitation wavelength was 488 nm, and the emission wavelength was 520 nm. Bandwidth was specified at 10 nm, and the gain was set to 45. Four readings were taken in each well, and all of the measurements were the result of three independent growth experiments.
RESULTS
Results from comparative analysis of CRP/FNR family of transcription factors
Following our previous comparative genomics analysis (20), we identified eight regulator–operator cognate sequence pairs for mutagenesis and subsequent genetic analysis (Tables 1 and 2). Five of the regulators (CRP1-4′) have cognate operator sites that have been experimentally characterized, and binding sites or profiles were obtained from the RegTransBase database of literature-culled protein–DNA interactions (28). Three of the regulators (CRP5-7) had binding-site predictions based on computational analysis (20,29) that were used to design their corresponding operator sites. Om5 is based on the consensus sequence of 22 predicted binding sites for HcpR across lostriduim and Treponema species, Om6 matches a single predicted binding site for HcpR in Porphiromonas gingivalis, and Om7 is based on the consensus sequence for 17 predicted binding sites for CooA in two Desulfovibrio species.
Table 1.
Operator site mutations
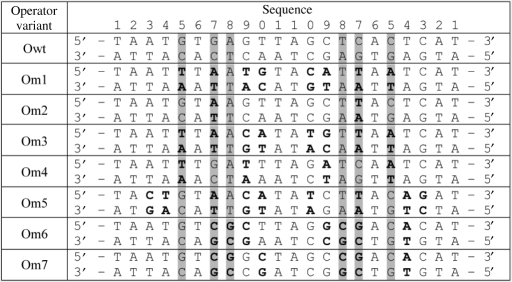 |
Bolded bases denote mutations. The shaded columns denote the bases that made direct contact to amino-acid side chains in wild-type CRP. Based on the structure of DNA-bound CRP, Arg180 contacts the guanine at position 5, Glu181 contacts cytosine at position 5, and Arg185 contacts the guanine at position 7 and thymine at position 8 (22).
Table 2.
CRP mutations
 |
Bolded residues denote mutations. The designations CRP4 and CRP4′ are used to emphasize that both mutations are predicted to bind the same Om4 operator site.
Motivated by the success of the comparative study (20) in predicting binding specificity from a small number of residues, we made the simplifying assumption that the mode of binding is identical within the CRP family of regulators studied. Although our original study (20) identified two amino-acid positions, corresponding to Arg180/Glu181 in the wild-type E. coli CRP (CRPwt), as sufficient for predicting specificity, a third position, corresponding to Arg185, makes significant contact within the major groove and thus was also targeted for mutagenesis. Our strategy was to modify only these three residues, keeping the remainder of the CRP protein intact in an attempt to maintain its stability and effector-binding properties. A multiple sequence alignment (Figure 1) provided a mapping from regulators with different specificities within the CRP family to specificity-determining residues in CRPwt. We note that the mapping from PrfA (CRP3) is ambiguous since there is a single amino-acid insertion between the two clusters of conserved residues. Nonetheless, we attempted to transfer PrfA-like specificity into CRP by anchoring our alignment on the position corresponding to Arg185 in CRPwt, as this site is both well-conserved across the family and is involved in significant major groove interactions in CRPwt.
Figure 1.
Multiple sequence alignment of the DNA-recognition helix within candidate proteins from the CRP/FNR family of transcription factors. Residues in CRP known to make specific contacts within DNA bases (22) are highlighted in black. The specificity-determining residues from Rodionov and coworkers (20) correspond to columns 4, 5 and 10. Note that the specificity determining residues for PrfA are somewhat ambiguous due to an insertion in the alignment relative to CRPwt. The numbers in parentheses denote the region of the protein shown in the alignment and the mutant column designates the corresponding CRP mutation used in this study.
Reporter system
As an indirect measure of CRP-DNA binding, we employed a transcriptional fusion between the lacZ promoter and the green fluorescent protein (GFP). The lacZ promoter requires CRP for expression (30). Therefore, by measuring fluorescence, we could determine whether different CRP variants were able to bind their cognate operator site and activate the lacZ-gfp transcriptional fusion. To test the suitability of this reporter, we measured fluorescence in wild-type (crp+) and Δcrp (crp−) cells (Figure S1). In wild-type cells, the lacZ-gfp reporter is active whereas in the crp− background it is inactive. Furthermore, we were able to complement the Δcrp mutant by expressing CRP from an atc-inducible promoter (see ‘Materials and Methods’ section for details). When atc was added to the growth media, near (70%) wild-type levels of expression were observed. Weak expression, on the other hand, was observed in the absence of atc due to the inducible promoter being slightly leaky. We also expressed CRP from the atc-inducible promoter in wild-type cells where the native crp locus was intact. In the absence of atc, a moderate inhibition of expression relative to wild type occurred for reasons unknown. Conversely, in the presence of atc, a moderate enhancement of expression was observed. Thus, we were able to conditionally activate expression of the lacZ-gfp transcriptional fusion in a crp– background by expressing CRP from a regulated promoter. This atc-inducible promoter was used in all subsequent studies involving the different CRP variants.
This assay is more stringent than a DNA-binding assay because a positive read out requires both DNA binding and activation of transcription; our assay does not distinguish between mutations that can bind their cognate operator sequence but fail to activate transcription from those that simply do not bind their cognate operator sequence. However, as mutations were made only to the three amino-acid residues in CRP that directly contact base pairs in the major groove, these changes are unlikely to affect activation, although the possibility cannot be discounted. For an example of the latter, these mutations could somehow affect the ability of CRP to contact RNA polymerase by disrupting the mode of DNA binding. In addition, these mutations may affect protein stability or have some other, unknown biochemical effect. Despite these potential limitations, this assay is ideally suited for our ultimate goal of re-engineering the specificity of CRP, namely to use these mutated transcription factors for reprogramming gene expression.
Analysis of CRP and operator site mutations in isolation
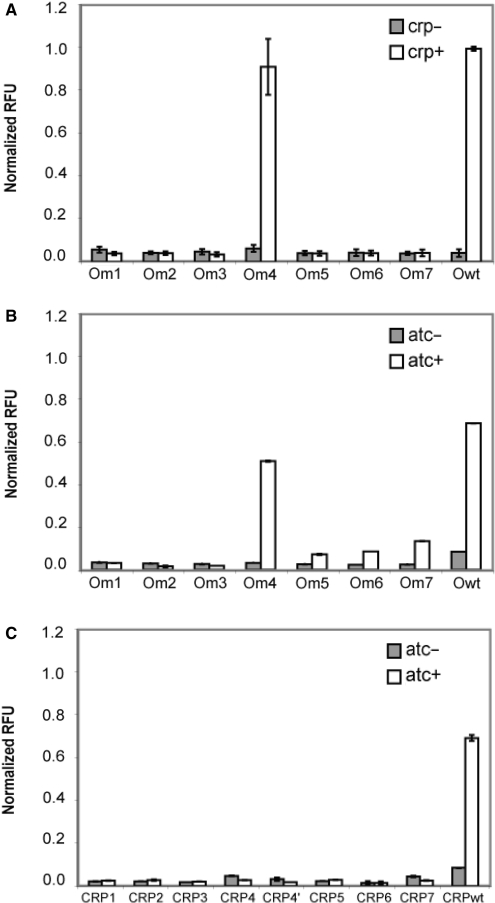
Based on the results from the computational analysis (Table 1), mutations were made to the crp gene and CRP operator site within the lacZ-gfp reporter as described in the ‘Materials and Methods’ section. To facilitate comparisons, the values in all subsequent figures are normalized to the relative fluorescence values measured for the lacZ-gfp reporter with a wild-type operator site in a wild-type (crp+) background. We first tested whether the lacZ-gfp reporter with mutations to the CRP operator site would be active in wild-type (crp+) and the Δcrp (crp−) strains (Figure 2A). In the crp+ background, all of the reporters with mutated operator sites were inactive with the exception of the Om4 reporter. Of the bases that directly interact with amino-acid side chains, only position 5 has been changed in Om4, where the guanine was mutated to thymine (Table 1). This guanine is known to form hydrogen bonds with the side chains of Arg180. Previously, Zhang and Ebright observed a 4-fold reduction in gene expression when position 5 was changed to a thymine, similar to Om4, and wild-type CRP was expressed from a plasmid in an otherwise crp– background (31). We, on the other hand, observed no decrease in gene expression with a thymine at position 5 when wild-type CRP was expressed from its native locus. Note that the wild-type guanine at position 9 has also been changed to a thymine in Om4. In the crp− background, all of the promoters, as expected, were inactive. These results also indicate that no other protein binds to these mutant operator sites and activates transcription.
Figure 2.
(A) Expression of lacZ-gfp transcriptional fusion with mutated operator sites in wild-type and Δcrp cells. (B) Expression of lacZ-gfp transcriptional fusion with mutated operator sites in Δcrp cells when wild-type CRP is ectopically expressed from an atc-inducible promoter. (C) Expression of lacZ-gfp transcriptional fusion with wild-type operator site in Δcrp when the different CRP variants are ectopically expressed from an atc-inducible promoter. All expression values were normalized relative to the lacZ-gfp transcriptional fusion in a wild-type (crp+) background.
We also tested the operator mutations when wild-type CRP (designated CRPwt in the figures) was expressed from the atc-inducible promoter in an otherwise crp− background (Figure 2B). Consistent with our previous results, all of the reporters were inactive in the absence of atc. In the presence of atc, only the reporters with the wild type and Om4 operator site were active. In the case of Om4, we observed slightly weaker expression relative to Owt, which was more in agreement with the results of Zhang and Ebright (31). We do note that weak expression was observed for the reporters with operator sites Om5, Om6 and Om7 in the presence of atc, roughly the same level observed for the wild-type operator (Owt) in the absence of atc. The mutations in Om5 involve, amongst others, changing the guanine at position 7 to an adenine whereas the mutations in Om6 and Om7 both involve, amongst others, changing the guanine and adenine at positions 7 and 8 to a cytosine and guanine, respectively. As the two bases at positions 7 and 8 form hydrogen bonds with the side chains of Glu181 and Arg185 in the case of wild-type CRP, the similar behavior of Om6 and Om7 is expected (22). Overall, these results are consistent with the results in the crp+ background, with the exception of weak activation in the cases of Om5, Om6 and Om7. We cannot explain why weak activation is observed when CRP is expressed ectopically whereas no activation is observed when CRP is expressed from its native locus.
Finally, we tested the ability of the different CRP mutants to activate the wild-type (Owt) reporter in a crp− background (Figure 2C). Of the eight CRP variants, none were capable of activating the wild-type reporter. Ebright and colleagues previously analyzed a Val181 substitution, the same as our CRP2 (32). Consistent with our results, where CRP2 does not activate the Owt reporter, they demonstrated that this mutation eliminates the interaction between the side chain and the guanine at position 7, resulting in a 10-fold reduction in binding affinity.
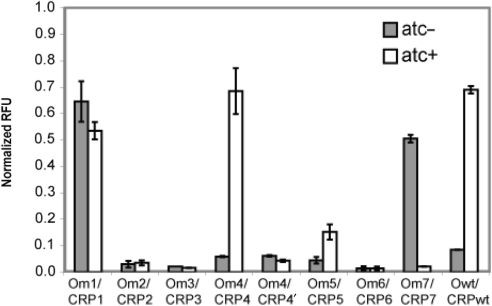
Results from pairing cognate CRP and operator mutations
Next we tested the computational predictions by pairing the different CRP mutations with their cognate promoters in an otherwise crp− background (Figure 3). Of the eight CRP mutations predicted from the analysis, four were able to activate the reporters containing their cognate operator site, three strongly and one weakly. In the case of CRP1, strong activation was observed both in the presence and absence of atc inducer. Note that this mutant involves the most severe changes, replacing the wild-type Arg180–Glu181–Arg185 triad with a His180–Val181–Lys185 triad. The fact that this mutation (CRP1) was able to activate its cognate promoter (Om1) in the absence of the atc inducer indicates that binding was particularly strong. As we have noted, weak expression of the crp gene still occurs in the absence of atc due to ‘leakiness’ in the inducible system. Therefore, the apparently strong affinity between CRP1 and Om1 appears to compensate for reduced expression in the absence of atc. In the case of CRP4, we observed that it was able to bind to Om4 and activate the lacZ-gfp transcriptional reporter in a dose-dependent manner, similar to wild-type but unlike CRP1–Om1 pair. This mutant involves a Pro180 substitution. Also, recall that wild-type CRP was able to bind to and activate the Om4 reporter whereas CRP4 was unable to do the same with the wild-type reporter. These results suggest that the Pro180 substitution increases the specificity of binding to Om4.
Figure 3.
Results obtained when pairing the lacZ-gfp transcriptional fusion with the cognate CRP mutation ectopically expressed from an atc-inducible promoter. All expression values were normalized relative to the lacZ-gfp transcriptional fusion in a wild-type (crp+) background.
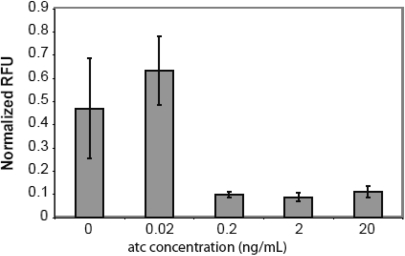
In the case of CRP7 and Om7, we observed activation only in the absence of atc inducer. This mutant consists of a Gln181/Thr185 double substitution. Note that as the Om7 reporter is inactive in a crp– background, this result means that weak expression of CRP7 is capable of strongly activating the Om7 reporter. Further investigation demonstrated that the CRP7–Om7 combination is only active at low levels of atc inducer (Figure 4). At higher levels, no activation is observed. Furthermore, there is a moderate decrease (25%) in cell density at higher atc concentrations (results not shown), suggesting that the CRP7-Om7 pairing may be toxic to cells under strong induction conditions. Note that the other pairings, assuming they were capable of activation, yielded dose-dependent activation of gene expression with respect to increasing atc concentrations (results not shown). Strangely, wild-type CRP, which weakly activates the Om7 reporter, does not have this inverse response or any effect on cell density. Why the Om7 reporter coupled with CRP7 exhibits this behavior is not known. We note that no decrease in viability or gene expression due to atc was observed with the other reporters.
Figure 4.
Expression with the CRP7–Om7 pair at varying levels of atc induction. As a reference, all other results involve atc induction at 20 ng/ml. All expression values were normalized relative to the lacZ-gfp transcriptional fusion in a wild-type (crp+) background.
Finally, we also observed weak activation in the case of the CRP5/Om5 pair at levels roughly 25% wild type. CRP5 involves a Pro181 substitution. All other CRP mutations were unable to bind to their cognate operator sequence and activate transcription of the lacZ-gfp reporter. Overall, these results indicate that between 37% (3/8) and 50% (4/8) mutations work as predicted, depending on one's measure of success.
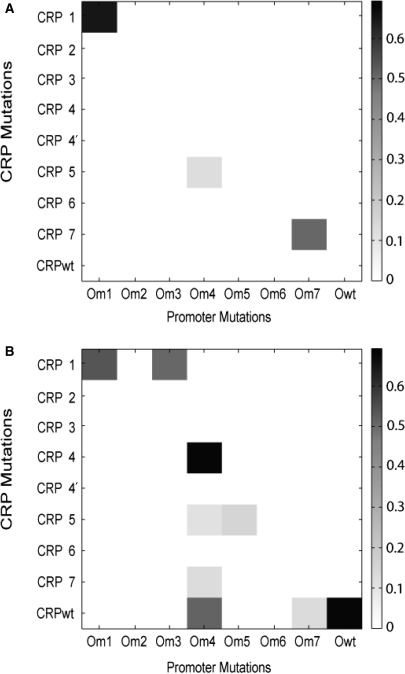
Results from pairing CRP and operator mutations
We also tested specificity by pairing all CRP mutations against all operator site mutations (Figure 5). In the absence of atc inducer, we observed strong activation by the CRP6–Om7 pair and weak activation by the CRP5–Om4 pair. In the case of Om7, this operator site mutation was also activated by CRP7 in the absence of inducer as shown previously. As a comparison, CRP6 involves an Ala181/Arg185 substitution whereas CRP7 involves a Gln181/Thr185 substitution. The Om7 mutation involves changing the wild-type guanine and adenine at positions 7 and 8 to cytosine and guanine, respectively. Recall, these two bases are the ones predicted to interact with the residues at positions 181 and 185. Thus, the Ala181 and Arg185 substitutions have the same effect as the Glu181 and Thr185 ones despite different chemistries. Furthermore, Om6 also involves the same changes at positions 7 and 8, yet neither CRP6 nor CRP7 is able to activate expression when paired with it, either in the presence or absence of atc inducer. The only differences between Om6 and Om7 are the bases at positions 9 and 10 (Table 1), which do not make direct contact with the amino-acid side chains in CRPwt. Therefore, we conclude that these non-specific mutations prevent both CRP6 and CRP7 from binding to the mutated operator site Om6.
Figure 5.
Results obtained when pairing all CRP mutations against all operator site mutations. (A) Results in the absence of atc; (B) Results in presence of atc. All values were normalized relative to the lacZ-gfp transcription fusion in a wild-type (crp+) background. To improve contrast, all values less than 10% the expression of the lacZ promoter in a wild-type background were displayed as having zero expression. The standard deviation for all pairs was less than ten percent (results not shown).
In the presence of atc, we observed both strong and weak activation with a number of non-canonical pairs. In the case of CRP1, it was able to activate promoters with the non-canonical Om3 reporter in addition to the canonical Om1 reporter. Both Om1 and Om3 are identical at positions 5, 7 and 8, the bases that make direct contact with the amino-acid side chains at residues 180, 181 and 185 in CRPwt. The only difference between these two operator mutations are at positions 9 and 10, the bases thought to have non-specific interactions with CRP. Note that CRP1 does not activate the Om3 reporter in the absence of atc, unlike the CRP1–Om1 pair. Therefore, these differences at positions 9 and 10 have a strong effect on affinity (thymine and guanine for Om1 versus cytosine and adenine for Om3); for Om1 only a small amount of CRP1 is necessary for activation whereas for Om3 a lot is needed.
In the case of Om4, we observed strong activation by CRP4 and CRPwt and weak activation by CRP5 and CRP7. Om4 is unique among the operator site mutations in that positions 7 and 8 are unchanged. All other operator sites involve changes at positions 7 or 8. Both CRP4 and CRPwt, which activate Om4 the strongest, are unchanged at Glu181 and Arg185, the residues that interact directly with the bases at positions 7 and 8. Note that CRP4′ is also unchanged at these two positions, though it is unable to activate Om4. The difference is the Val180 substitution in the case of CRP4′ versus a Pro180 substitution in the case of CRP4. Despite both CRP4 and CRPwt being able to activate the Om4 reporter, CRP4 is unable to activate the Owt reporter whereas, obviously, CRPwt can. These results suggest that Pro180 side chain is unable to form hydrogen bonds with the wild-type guanine at position 5 or that it disrupts the α-helical structure, altering the protein's binding mode. However, both the wild-type Arg180 and mutant Pro180 side chains can form hydrogen bonds with the mutated thymine at position 5 in the case of Om4. Finally, the weak activators of the Om4 reporter, CRP5 and CRP7, have unchanged, wild-type Arg180 and Arg185 residues. Therefore, the Glu181 side chain appears to be necessary for strong activation. Note that CRP5 is able to weakly activate Om4 in both the presence and absence of atc inducer. The results suggest that CRP5 is able to bind the Om4 site strongly. However, CRP5 is unable to strongly activate transcription, suggesting its mode of binding may be altered.
Optimizing designs using simple genetic screen
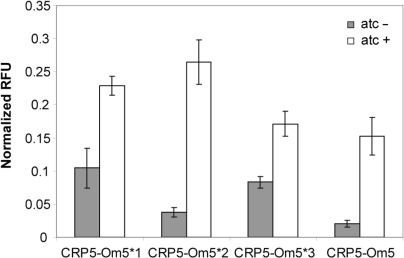
Three of the four CRP mutants were capable of activating reporters containing a cognate operator site at roughly wild-type levels. The fourth (CRP5/Om5), however, could activate its reporter at a level only 15% of wild type. We hypothesized that weak activation may be due to limitations in the ability of the computational analysis to resolve the consensus operator site. In particular, there are a number of non-specific interactions that may not be resolved solely through sequence analysis. To test whether we could improve activation by CRP5, we randomized the middle six positions in Om5 (positions 9, 10 and 11) and then screened for increased activity. In our simple screen, we were able to isolate three variants with increased activity with respect to the canonical Om5 reporter (Figure 6). While these results still do not approach wild-type levels, we are encouraged because our screen is far from comprehensive, and they demonstrate that the computational designs can further be improved using directed evolution-based approaches. In particular, the Om5*2 reporter resulted in roughly a 60% increase in activation by CRP5. Likely, further increases could be obtained in a more comprehensive screen, where multiple positions would be randomized. We note that we also tried screening for operator sequences that would be activated by the CRP mutants that failed to activate their cognate sequence (CRP2, CPR3, CRP4′ and CRP6) by randomizing positions 5, 7 and 8 in Owt. However, we did not find sequences that were activated by these CRP variants in similar screens.
Figure 6.
Results obtained from screen of randomized operator sites. In these experiments, we screened for increased CRP5 activation of Om5 reporter by randomizing positions 9, 10 and 11 in the operator sequences. The resulting sequences isolated in the screen were as follows: Om5*1 (TCCGGT), Om5*2 (CAGTGA), Om5*2 (GCTGGA) and Om5 (CATATC).
DISCUSSION
In this study, we explored the use of comparative genomics as a tool for transcription factor engineering. Based on correlations derived from a previous study of nitrogen oxide metabolism in bacteria (20), we experimentally tested eight different mutations for their ability to change DNA-binding specificity using CRP as the template. In all cases, the mutations were made to a triad of amino acids (Arg180/Glu181/Arg185) that are known to directly contact DNA bases within the major groove. These three amino acids alone were predicted to be sufficient for DNA-binding specificity within the CRP/FNR family. For each set of mutations made to CRP, we also made a corresponding set of mutations to the CRP operator site within the lacZ promoter. Of the eight CRP variants, four were able to bind their cognate operator sequence and activate transcription of the lacZ promoter. Though the results, in general, are less dramatic than the wild-type CRP/Owt pair, they provide excellent targets for subsequent refinement by directed evolution and other more traditional methods. Along these lines, we were able to demonstrate that we could improve activation by the CRP5/Om5 pair by screening for operator sequences with select positions randomized. While this screen was limited, it nonetheless demonstrates that further refinement is possible.
Utilizing genomic data to inform protein engineering is not a new idea. For example, the active site in an enzyme can often be determined within a multiple sequence alignment by identifying conserved residues (33) or specificity-determining conserved within functional subfamilies (34,35) within a multiple sequence alignment. In the case of transcription factors, however, a multiple sequence alignment often will not suffice, as the DNA-binding sequence must also be considered in the analysis. In particular, identifying the specificity determining residues is often not sufficient for design purposes. Rather, we seek to identify the specific amino acids that bind different DNA sequences. Therefore, the new idea in this work is to use mutual information between transcription factors and their target DNA-binding sequences to inform protein engineering. In many regards, the computational approach used to generate the predictions tested here is similar to those used to study interacting proteins (36–39). These approaches work under the assumption that any mutation to a specificity-determining residue on one binding partner must be matched by a compensating mutation to a specificity-determining residue on the other binding partner. By studying the co-variation of residues among binding partners in a given family of proteins, one can identify the specificity-determining residues and then apply the information to inform protein engineering. Of notable significance is the recent work by Skerker and colleagues (38), where they were able to utilize these data to change the specificity of the EnvZ histidine kinase for its target response regulator. In conjunction with the analogous work presented here, these results demonstrate how purely genomic-based approaches can inform the re-engineering of protein interactions.
One limitation of the use of genomic-based approaches for transcription factor engineering is our ability to identify the target DNA-binding sequences and also discriminate among the potentially large number of DNA-sequences that these proteins can bind to. In the case of the work by Skerker and colleagues, the advantage of their system is that histidine kinase-response regulator pairs can often be inferred directly through their proximity in the genome, as they both typically reside in the same operon (40). Furthermore, most histidine kinases interact exclusively with a single response regulator. In the case of transcription factors, identifying the target DNA-binding sequence is often impossible unless other data are available. The results used in this work were obtained from a comparative study of nitrogen oxide metabolism that integrated multiple data from both experimental and computational analysis (20). For an arbitrary family of transcription factors, such results may not always be forthcoming. Furthermore, there is always a degree of uncertainty, often unquantifiable, associated with the identification of target-binding sites. Finally, with regards to specificity, many transcription factors are known to regulate multiple target genes. For example, CRP is estimated to regulate approximately 200 promoters in E. coli and other relative organisms (41). This promiscuity adds an extra degree of complexity, as the protein/operator site pairs often cannot directly be assigned and instead consensus sequences must be estimated. Despite these challenges, our results demonstrate the utility of these approaches for bacterial transcription factor engineering.
Our results also uncovered some surprising results, highlighting our limited understanding of even simple protein–DNA interactions. When CRP7 was paired with the Om7 reporter, expression was induced at low levels of CRP expression and repressed at high. This reporter was also toxic at high levels of CRP7 expression. Moreover, the reporter was not active in a crp− background and showed weak, dose-dependent behavior with wild-type CRP, suggesting a complex, concentration-dependent interaction between this regulator–operator pair. In addition, significant cross-talk was observed in the case of wild-type CRP, which activated reporters with Om4 and Om7 in addition to the wild-type reporter, whereas the mutant regulators displayed far less promiscuity. From an evolutionary perspective, this cross-talk is not entirely unexpected; most of the regulator–operator pairs were taken from different species, so there may be no explicit evolutionary pressure to avoid crosstalk. Because large regulons such as the one dictated by CRP include an enormous diversity of promoters, the duplication and specialization of regulators could be a general mechanism in the evolution of regulatory pathways (42), especially given the observation that birth and evolutionary turnover of regulatory sites may occur at a very fast rate even under relatively weak selection (43,44).
An additional puzzle concerns the role of bases within the operator sequence that do not make direct contact with amino-acid side chains. Previous work has established that only positions 5, 7 and 8 make direct contact with amino-acid side chains (22). In addition to these bases, our results demonstrate that the so-called, non-specific bases also affect binding and specificity. For example, both Om1 and Om3 are identical at positions 5, 7 and 8, yet their response to CRP1 is different. In the case of Om1, CRP1 binds this site so strongly that it is able to activate transcription both in the presence and absence of atc inducer. However, in the case of Om3, CRP3 is able to activate transcription only in the presence of atc (i.e. at high levels of expression). Similarly, Om2 and Om5 are identical at positions 5, 7 and 8, yet wild-type CRP is only able to activate promoters with Om5, albeit weakly. Finally, in the case of Om5, we were able to improve the ability of CRP5 to activate these promoters by modifying positions 9, 10 and 11. Collectively, these results show that these ‘non-specific’ bases likely do make specific interactions, though the mode may be quite complex. Moreover, as we focused only on the residues that make specific contact and saw weaker activation in general than wild type, future endeavors will likely need to consider optimizing non-specific binding as these interactions may be needed to facilitate and/or compensate for changes to the core contacts.
We note that CRP2 was previously identified by Ebright and colleagues in a genetic selection for CRP mutants that were able to bind the lacZ promoter with an adenine or thymine at position 7 within the CRP operator site, a condition that Om2 satisfies (16). In addition to the valine substitution, they also found a lysine and leucine. Subsequent analysis demonstrated that the Val181 (and Leu181) substitution was unable to distinguish between different bases at position 7, resulting in roughly a 10-fold decrease in binding affinity relative to wild type (32). As discussed (Figure 3), we found that CRP2 was unable to activate transcription of the lacZ promoter involving the Om2 operator site, results that correspond with their in vitro binding analysis.
In the context of transcription factor engineering, we have shown that comparative genomics can be used to computationally isolate mutations that alter DNA-binding specificity. Previously, in the case of bacteria, these designs have resulted from randomized screens. With regards to applications, engineered transcription factors with novel DNA-binding specificity can greatly facilitate the design of synthetic gene circuits, as they expand the number of components available to build these circuits. One challenge in constructing these gene circuits is that the designs are limited by the number of components available that do not interfere with host physiology. Engineering such orthogonal components has been central focus in the nascent field of synthetic biology (45–47). In addition, these engineered transcription factors provide additional tools for fine tuning gene expression with cells, a key task in uncovering new regulation and also for potentially designing new therapeutics approaches.
We note that one limitation of the approach explored in this work is that it does not provide information regarding the strength of the protein–DNA interaction. The analysis is based simply on correlations derived from sequence analysis and provides no information regarding binding energies. With regards to gene regulation, the final product is ultimately linked to the template protein. In our case, where the template is CRP, the natural product is a transcriptional activator. However, CRP is also a transcription repressor for a number of promoters [cf. (48,49)], so it can potentially be used to engineer repressors with novel specificity. In addition, the approach tested in this work can be applied to other families of transcription factors, including repressors.
To what extent is there a ‘code’ for transcription factor specificity? Our previous study suggested a three amino-acid code may be sufficient for inferring specificity in the CRP/FNR family of regulators. The results reported here highlight the importance of a combined computational and experimental approach: the amino acids sufficient for inferring the specificity of known regulators, but insufficient to design novel regulators. Thus, the residues at these positions may constrain binding to a small number of possible operator sites, even if they contribute only a fraction of the total energy of binding. For example, in our previous study, we found that the identity of two residues (positions 180 and 181) were sufficient to predict binding specificity (i.e. each unique combination of residues at these positions mapped to a unique binding site). However, a careful inspection of the CRP–DNA co-crystal structure indicates contacts between position 185 and the major groove, motivating us to include position 185 in our redesign experiments.
In a broader evolutionary context, our results show that it is possible to create orthogonal regulatory pathways after a surprisingly small number of mutational steps. Novel regulatory pathways are thought to evolve through gene duplication events (50), horizontal gene transfer (51), changes in the specificity of regulators (52) and site turnover (53,54) yielding rewiring of regulatory pathways (55). In order for these new pathways to form, transcription factors must mutate so that they no longer regulate their old target genes but instead target new ones. Sometimes we may observe early stages of this process; some examples of recently duplicated E. coli regulators partially sharing their binding sites are UxuR/ExuR (56,57), GalS/GalR (58,59)and NarL/NarP (60–63). In the case of regulators from the CRP/FNR family (and likely other helix-turn-helix transcription factors), specificity is predominantly determined by a core set of residues. The limited number of residues implies that these regulatory networks are quite plastic, as only a few mutations either to the protein or operator sites are necessary to change specificity and introduce new regulation or rewire existing networks. One open question is why there is only a small set of possible motifs observed within this family. Is this a structural constraint or are entirely new specificities possible within this family?
In conclusion, we have shown experimentally that comparative genomics can be used to inform transcription factor engineering. To date, bacterial transcription factor engineering has exclusively utilized direct evolution/random mutagenesis or domain swapping (12). In particular, few computational approaches exist particularly in the case of bacterial transcription factors. Based on the computational analysis of co-variation between the specificity-determining residues within the DNA-recognition helix and the cognate, consensus binding sequence, we were able to engineer novel DNA-binding specificity in CRP. In fact, four out of eight designs worked as predicted with no subsequent refinement. Likely, the application of these computational approaches for engineering novel specificity in other proteins will also utilize directed evolution for subsequent refinement. One question might then be why employ computational approaches at all. As our results showed, many of the designs that actually worked involved multiple mutations to both the CRP protein and operator site. Searching such a large space of possible sequences is extremely labor intensive. The computational approach described in this work can focus mutagenesis to a core set of targets, greatly reducing the number of mutants needed to screen and also expanding the range of likely targets.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation CAREER Award CBET-0644744 [to C.V.R.] and grants from the U.S. Department of Energy Genomics:GTL program [to E.J.A.]. Partially support from the Howard Hughes Medical Institute (55005610), the Russian Foundation of Basic Research (08-04-01000), and the program ‘Molecular and Cellular Biology’ of the Russian Academy of Sciences [D.R. and M.G.]. Funding for open access charge: National Science Foundation CAREER CBET-0644744.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Alexandra Rakhmaninova, Desislava Miteva and Vita Stepanova for sharing unpublished results.
REFERENCES
- 1.Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 2.Pabo CO, Sauer RT. Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 3.Rhodes D, Schwabe JW, Chapman L, Fairall L. Towards an understanding of protein-DNA recognition. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1996;351:501–509. doi: 10.1098/rstb.1996.0048. [DOI] [PubMed] [Google Scholar]
- 4.Sarai A, Kono H. Protein-DNA recognition patterns and predictions. Annu. Rev. Biophys. Biomol. Struct. 2005;34:379–398. doi: 10.1146/annurev.biophys.34.040204.144537. [DOI] [PubMed] [Google Scholar]
- 5.Kaern M, Blake WJ, Collins JJ. The engineering of gene regulatory networks. Annu. Rev. Biomed. Eng. 2003;5:179–206. doi: 10.1146/annurev.bioeng.5.040202.121553. [DOI] [PubMed] [Google Scholar]
- 6.Haseltine EL, Arnold FH. Synthetic gene circuits: design with directed evolution. Annu. Rev. Biophys. Biomol. Struct. 2007;36:1–19. doi: 10.1146/annurev.biophys.36.040306.132600. [DOI] [PubMed] [Google Scholar]
- 7.Voigt CA. Genetic parts to program bacteria. Curr. Opin. Biotechnol. 2006;17:548–557. doi: 10.1016/j.copbio.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Khosla C, Keasling JD. Metabolic engineering for drug discovery and development. Nat. Rev. Drug Discov. 2003;2:1019–1025. doi: 10.1038/nrd1256. [DOI] [PubMed] [Google Scholar]
- 9.Isalan M, Lemerle C, Michalodimitrakis K, Horn C, Beltrao P, Raineri E, Garriga-Canut M, Serrano L. Evolvability and hierarchy in rewired bacterial gene networks. Nature. 2008;452:840–845. doi: 10.1038/nature06847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ. Programmable cells: interfacing natural and engineered gene networks. Proc. Natl Acad. Sci. USA. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins CH, Yokobayashi Y, Umeno D, Arnold FH. Engineering proteins that bind, move, make and break DNA. Curr. Opin. Biotechnol. 2003;14:665. doi: 10.1016/j.copbio.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Wharton RP, Brown EL, Ptashne M. Substituting an alpha-helix switches the sequence-specific DNA interactions of a repressor. Cell. 1984;38:361–369. doi: 10.1016/0092-8674(84)90491-4. [DOI] [PubMed] [Google Scholar]
- 14.Wharton RP, Ptashne M. Changing the binding specificity of a repressor by redesigning an alpha-helix. Nature. 1985;316:601–605. doi: 10.1038/316601a0. [DOI] [PubMed] [Google Scholar]
- 15.Youderian P, Vershon A, Bouvier S, Sauer RT, Susskind MM. Changing the DNA-binding specificity of a repressor. Cell. 1983;35:777–783. doi: 10.1016/0092-8674(83)90110-1. [DOI] [PubMed] [Google Scholar]
- 16.Ebright RH, Cossart P, Gicquel-Sanzey B, Beckwith J. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein of E. coli. Nature. 1984;311:232–235. doi: 10.1038/311232a0. [DOI] [PubMed] [Google Scholar]
- 17.Beerli RR, Barbas CF., 3rd Engineering polydactyl zinc-finger transcription factors. Nat. Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 18.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2008;36:D475–D479. doi: 10.1093/nar/gkm884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pabo CO, Nekludova L. Geometric analysis and comparison of protein-DNA interfaces: why is there no simple code for recognition? J. Mol. Biol. 2000;301:597–624. doi: 10.1006/jmbi.2000.3918. [DOI] [PubMed] [Google Scholar]
- 20.Rodionov DA, Dubchak IL, Arkin AP, Alm EJ, Gelfand MS. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput. Biol. 2005;1:e55. doi: 10.1371/journal.pcbi.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korner H, Sofia HJ, Zumft WG. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 2003;27:559–592. doi: 10.1016/S0168-6445(03)00066-4. [DOI] [PubMed] [Google Scholar]
- 22.Lawson CL, Swigon D, Murakami KS, Darst SA, Berman HM, Ebright RH. Catabolite activator protein: DNA binding and transcription activation. Curr. Opin. Struct. Biol. 2004;14:10–20. doi: 10.1016/j.sbi.2004.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siggers TW, Silkov A, Honig B. Structural alignment of protein–DNA interfaces: insights into the determinants of binding specificity. J. Mol. Biol. 2005;345:1027–1045. doi: 10.1016/j.jmb.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Morozov AV, Siggia ED. Connecting protein structure with predictions of regulatory sites. Proc. Natl Acad. Sci. USA. 2007;104:7068–7073. doi: 10.1073/pnas.0701356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandel-Gutfreund Y, Baron A, Margalit H. A structure-based approach for prediction of protein binding sites in gene upstream regions. Pac. Symp. Biocomput. 2001:139–150. doi: 10.1142/9789814447362_0015. [DOI] [PubMed] [Google Scholar]
- 26.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stemmer WP, Morris SK. Enzymatic inverse PCR: a restriction site independent, single-fragment method for high-efficiency, site-directed mutagenesis. Biotechniques. 1992;13:214–220. [PubMed] [Google Scholar]
- 28.Kazakov AE, Cipriano MJ, Novichkov PS, Minovitsky S, Vinogradov DV, Arkin A, Mironov AA, Gelfand MS, Dubchak I. RegTransBase – a database of regulatory sequences and interactions in a wide range of prokaryotic genomes. Nucleic Acids Res. 2007;35:D407–D412. doi: 10.1093/nar/gkl865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodionov DA, Dubchak I, Arkin A, Alm E, Gelfand MS. Reconstruction of regulatory and metabolic pathways in metal-reducing delta-proteobacteria. Genome Biol. 2004;5:R90. doi: 10.1186/gb-2004-5-11-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J. Mol. Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 31.Zhang XP, Ebright RH. Identification of a contact between arginine-180 of the catabolite gene activator protein (CAP) and base pair 5 of the DNA site in the CAP-DNA complex. Proc. Natl Acad. Sci. USA. 1990;87:4717–4721. doi: 10.1073/pnas.87.12.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebright RH, Kolb A, Buc H, Kunkel TA, Krakow JS, Beckwith J. Role of glutamic acid-181 in DNA-sequence recognition by the catabolite gene activator protein (CAP) of Escherichia coli: altered DNA-sequence-recognition properties of [Val181]CAP and [Leu181]CAP. Proc. Natl Acad. Sci. USA. 1987;84:6083–6087. doi: 10.1073/pnas.84.17.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livingstone CD, Barton GJ. Identification of functional residues and secondary structure from protein multiple sequence alignment. Methods Enzymol. 1996;266:497–512. doi: 10.1016/s0076-6879(96)66031-5. [DOI] [PubMed] [Google Scholar]
- 34.Kalinina OV, Mironov AA, Gelfand MS, Rakhmaninova AB. Automated selection of positions determining functional specificity of proteins by comparative analysis of orthologous groups in protein families. Protein Sci. 2004;13:443–456. doi: 10.1110/ps.03191704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirny LA, Gelfand MS. Using orthologous and paralogous proteins to identify specificity-determining residues in bacterial transcription factors. J. Mol. Biol. 2002;321:7–20. doi: 10.1016/s0022-2836(02)00587-9. [DOI] [PubMed] [Google Scholar]
- 36.Szurmant H, Bobay BG, White RA, Sullivan DM, Thompson RJ, Hwa T, Hoch JA, Cavanagh J. Co-evolving motions at protein-protein interfaces of two-component signaling systems identified by covariance analysis. Biochemistry. 2008;47:7782–7784. doi: 10.1021/bi8009604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White RA, Szurmant H, Hoch JA, Hwa T. Features of protein-protein interactions in two-component signaling deduced from genomic libraries. Methods Enzymol. 2007;422:75–101. doi: 10.1016/S0076-6879(06)22004-4. [DOI] [PubMed] [Google Scholar]
- 38.Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L, Shakhnovich EI, Mirny LA. Amino acids determining enzyme-substrate specificity in prokaryotic and eukaryotic protein kinases. Proc. Natl Acad. Sci. USA. 2003;100:4463–4468. doi: 10.1073/pnas.0737647100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng D, Constantinidou C, Hobman JL, Minchin SD. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004;32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lozada-Chavez I, Angarica VE, Collado-Vides J, Contreras-Moreira B. The role of DNA-binding specificity in the evolution of bacterial regulatory networks. J. Mol. Biol. 2008;379:627–643. doi: 10.1016/j.jmb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lassig M. From biophysics to evolutionary genetics: statistical aspects of gene regulation. BMC Bioinformatics. 2007;8(Suppl. 6):S7. doi: 10.1186/1471-2105-8-S6-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mustonen V, Lassig M. Evolutionary population genetics of promoters: predicting binding sites and functional phylogenies. Proc. Natl Acad. Sci. USA. 2005;102:15936–15941. doi: 10.1073/pnas.0505537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arkin AP, Fletcher DA. Fast, cheap and somewhat in control. Genome Biol. 2006;7:114. doi: 10.1186/gb-2006-7-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Channon K, Bromley EH, Woolfson DN. Synthetic biology through biomolecular design and engineering. Curr. Opin. Struct. Biol. 2008;18:491–498. doi: 10.1016/j.sbi.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 47.Chin JW. Modular approaches to expanding the functions of living matter. Nat. Chem. Biol. 2006;2:304–311. doi: 10.1038/nchembio789. [DOI] [PubMed] [Google Scholar]
- 48.Aiba H. Autoregulation of the Escherichia coli crp gene: CRP is a transcriptional repressor for its own gene. Cell. 1983;32:141–149. doi: 10.1016/0092-8674(83)90504-4. [DOI] [PubMed] [Google Scholar]
- 49.Polayes DA, Rice PW, Garner MM, Dahlberg JE. Cyclic AMP-cyclic AMP receptor protein as a repressor of transcription of the spf gene of Escherichia coli. J. Bacteriol. 1988;170:3110–3114. doi: 10.1128/jb.170.7.3110-3114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teichmann SA, Babu MM. Gene regulatory network growth by duplication. Nat. Genet. 2004;36:492–496. doi: 10.1038/ng1340. [DOI] [PubMed] [Google Scholar]
- 51.Price MN, Dehal PS, Arkin AP. Horizontal gene transfer and the evolution of transcriptional regulation in Escherichia coli. Genome Biol. 2008;9:R4. doi: 10.1186/gb-2008-9-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodionov DA, Gelfand MS, Todd JD, Curson AR, Johnston AW. Computational reconstruction of iron- and manganese-responsive transcriptional networks in alpha-proteobacteria. PLoS Comput. Biol. 2006;2:e163. doi: 10.1371/journal.pcbi.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berg J, Willmann S, Lassig M. Adaptive evolution of transcription factor binding sites. BMC Evol. Biol. 2004;4:42. doi: 10.1186/1471-2148-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Price MN, Dehal PS, Arkin AP. Orthologous transcription factors in bacteria have different functions and regulate different genes. PLoS Comput. Biol. 2007;3:1739–1750. doi: 10.1371/journal.pcbi.0030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ravcheev DA, Gerasimova AV, Mironov AA, Gelfand MS. Comparative genomic analysis of regulation of anaerobic respiration in ten genomes from three families of gamma-proteobacteria (Enterobacteriaceae, Pasteurellaceae, Vibrionaceae) BMC Genomics. 2007;8:54. doi: 10.1186/1471-2164-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ritzenthaler P, Blanco C, Mata-Gilsinger M. Interchangeability of repressors for the control of the uxu and uid operons in E. coli K12. Mol. Gen. Genet. 1983;191:263–270. doi: 10.1007/BF00334824. [DOI] [PubMed] [Google Scholar]
- 57.Rodionov DA, Mironov AA, Rakhmaninova AB, Gelfand MS. Transcriptional regulation of transport and utilization systems for hexuronides, hexuronates and hexonates in gamma purple bacteria. Mol. Microbiol. 2000;38:673–683. doi: 10.1046/j.1365-2958.2000.02115.x. [DOI] [PubMed] [Google Scholar]
- 58.Geanacopoulos M, Adhya S. Functional characterization of roles of GalR and GalS as regulators of the gal regulon. J. Bacteriol. 1997;179:228–234. doi: 10.1128/jb.179.1.228-234.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Semsey S, Krishna S, Sneppen K, Adhya S. Signal integration in the galactose network of Escherichia coli. Mol. Microbiol. 2007;65:465–476. doi: 10.1111/j.1365-2958.2007.05798.x. [DOI] [PubMed] [Google Scholar]
- 60.Darwin AJ, Tyson KL, Busby SJ, Stewart V. Differential regulation by the homologous response regulators NarL and NarP of Escherichia coli K-12 depends on DNA binding site arrangement. Mol. Microbiol. 1997;25:583–595. doi: 10.1046/j.1365-2958.1997.4971855.x. [DOI] [PubMed] [Google Scholar]
- 61.Stewart V, Bledsoe PJ. Synthetic lac operator substitutions for studying the nitrate- and nitrite-responsive NarX-NarL and NarQ-NarP two-component regulatory systems of Escherichia coli K-12. J. Bacteriol. 2003;185:2104–2111. doi: 10.1128/JB.185.7.2104-2111.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang H, Gunsalus RP. The nrfA and nirB nitrite reductase operons in Escherichia coli are expressed differently in response to nitrate than to nitrite. J. Bacteriol. 2000;182:5813–5822. doi: 10.1128/jb.182.20.5813-5822.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang H, Gunsalus RP. Coordinate regulation of the Escherichia coli formate dehydrogenase fdnGHI and fdhF genes in response to nitrate, nitrite, and formate: roles for NarL and NarP. J. Bacteriol. 2003;185:5076–5085. doi: 10.1128/JB.185.17.5076-5085.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.