Abstract
Recent advances in DNA-sequencing technology have made it possible to obtain large datasets of small RNA sequences. Here we demonstrate that not all non-perfectly matched small RNA sequences are simple technological sequencing errors, but many hold valuable biological information. Analysis of three small RNA datasets originating from Oryza sativa and Arabidopsis thaliana small RNA-sequencing projects demonstrates that many single nucleotide substitution errors overlap when aligning homologous non-identical small RNA sequences. Investigating the sites and identities of substitution errors reveal that many potentially originate as a result of post-transcriptional modifications or RNA editing. Modifications include N1-methyl modified purine nucleotides in tRNA, potential deamination or base substitutions in micro RNAs, 3′ micro RNA uridine extensions and 5′ micro RNA deletions. Additionally, further analysis of large sequencing datasets reveal that the combined effects of 5′ deletions and 3′ uridine extensions can alter the specificity by which micro RNAs associate with different Argonaute proteins. Hence, we demonstrate that not all sequencing errors in small RNA datasets are technical artifacts, but that these actually often reveal valuable biological insights to the sites of post-transcriptional RNA modifications.
INTRODUCTION
Deep sequencing methodologies such as pyrosequencing (1) have enabled extensive exploration of small RNA transcriptomes (2–4). Small RNAs, a term previously reserved to describe what is now known as tRNAs (5,6), evolved to describe RNA 18–30 nt in length (7–9), such as micro RNAs, which are important for gene regulation (10,11). Deep sequencing projects identifying small RNAs can generate datasets containing hundreds of thousands of sequences. RNA sequences not perfectly matching the genome from these large datasets are often discarded as these mismatched sequences are attributed to experimental sequencing errors.
Random sequencing errors can arise from either the pyrosequencing procedures or during the reverse transcription of small RNAs. A variant of Moloney Murine Leukemia Virus Reverse Transcriptase termed SuperScript II is often used to generate cDNAs from small RNAs (3). The error rate of this reverse transcriptase is reported to be ∼1/15 000 (12–14). Of the 1/15 000 sequencing errors, two-third consist of insertions or deletions and one-third substitutions as determined in a lacZ forward mutation frequency assay (15). Additional errors can arise during the base calling of raw sequencing data whether the intrinsic program supplied with a pyrosequencing machine or a probabilistic model presented by Vacic and colleagues (16) is used. Both algorithms are functionally similar to the Sanger-sequencing base calling program Phred (17,18). Overall there is a 3.3% error rate for insertions and deletions and a 0.5% error rate for substitutions using pyrosequencing as determined during re-sequencing the Mycoplasma genitalium genome (1).
We hypothesize that some sequence discrepancies between small RNA-sequencing datasets and the genomic sequence may have a biological origin. Cloning and sequencing of post-transcriptionally modified RNAs may result in a variety of sequencing discrepancies observed with high-throughput DNA-sequencing technologies when compared to genomic DNA sequences. To identify sites of post-transcriptional modification, we predict that sequence mismatches originating from post-transcriptional modifications will be repeatedly observed at single sites with high frequencies in contrast to the more random occurrences of conventional or technical sequencing errors.
The presence of base modifications to micro RNAs has broad implications regarding their function. Modifications to micro RNAs may potentially alter which mRNAs are targeted for post-transcriptional regulation or the modifications could alter micro RNA biogenesis. Examples of micro RNA modifications have already been reported where an adenosine deaminases acting on RNA (ADARs) has been identified to act on pri-miR-142 (19) and there are reports on 3′ uridylation of small RNAs (20–22).
To detect sites of post-transcriptional modifications within large datasets of small RNA sequences, discarded data containing sequences that did not exactly match the genome of origin from two different small RNA cloning and sequencing projects were analyzed. The discarded dataset were 3852 small RNA sequences from Oryza sativa (3) and 193 024 small RNA sequences from Arabidopsis thaliana (23). A third dataset comprised of various A. thaliana small RNAs co-immunoprecipitated with anti-Argonaute1 (AGO1), AGO2, AGO4 and AGO5 antibodies was used to determine Argonaute specificity shifts of modified micro RNAs (24).
As a positive control for post-translational modifications we computationally analyzed highly modified tRNAs (25) from O. sativa and A. thaliana. Investigations of small RNAs typically use size fractionated samples from total RNA isolations, which typically contain some tRNA fragments. The source of these tRNA fragments 15–30 nt in length is unclear, whether they are simple breakdown products or a result of a biological event as seen in some starvation related pathways (26). We demonstrate that some apparent sequencing errors actually correspond to post-transcriptional modifications of tRNAs. Furthermore, we find evidence for tissue-specific RNA editing of micro RNAs and other modifications affecting Argonaute complex preference of micro RNAs.
MATERIALS AND METHODS
Small RNA datasets
The rice dataset was a gift from Peter Unrau (Simon Fraser University) and is described in detail in a publication by Morin et al. (3). The Arabidopsis dataset was a generous gift from Ramya Rajagopalan (University of Wisconsin-Madison) and David Bartel (Whitehead Institute/MIT/HHMI) and is described in detail in a publication by Rajagopalan et al. (23). Mi and colleagues generously deposited all their raw data of various ago-co-immunoprecipitated small RNAs from A. thaliana into the GEO public database with the accession number GSE10036 (24).
Ebbie-(mis)match
Detailed information on the algorithm is provided in the Supplementary Data. The source code and a compiled version of Ebbie-(mis)match (Ebbie-MM) and Ebbie-MM-ago are available under the General Public License II at http://www.bioinformatics.org/ebbie/.
RESULTS
In principle, there are several possible origins for sequencing errors. Besides technological artifacts, there may also be biological reasons for sequencing errors. Cloning and sequencing of post-transcriptionally modified RNAs may result in a variety of sequencing discrepancies when compared to genomic DNA sequences. To prove our hypothesis that ‘sequencing errors’ are not random technical events but rather have biological significance, we obtained two datasets comprised of small RNA sequences that did not match their genome of origin. The first dataset originated from O. sativa and is comprised of 7790 sequences, of which 3852 did not match the O. sativa genome (3). The second dataset comprised of 193 024 sequences from A. thaliana, all of which could not be aligned perfectly to the A. thaliana genome (23). Both datasets contained only non-redundant sequences together with their cloning frequency. For this report, these two datasets are referred to the rice and Arabidopsis datasets, respectively. The Arabidopsis dataset contained an additional level of biological information in that it is a compilation of sequences originating from different plant tissues (F: flower, R: root, S: seed, Q: silique). One criterion to evaluate whether sequencing errors are technical artifacts or have biological significance is the occurrence of mismatches overlapping in homologous sequences.
Ebbie-(mis)match: premise and algorithm description
To identify single nucleotide mismatches from large sets of DNA sequencing data, as can be readily generated by pyrophosphate DNA sequencing platforms, a computer algorithm was developed. As an extension to Ebbie (27), the algorithm was named Ebbie-(mis)match (Ebbie-MM) (for algorithm availability and description see ‘Materials and Methods’ section and Supplementary Data). In light of large datasets of small RNA sequences, it is natural to use the short oligonucleotide alignment program (SOAP) presented by Li and colleagues to align the small RNA sequences to a database (28). Li and colleagues impressively demonstrated that their algorithm could be three orders of magnitude faster than BlastN when aligning 10 million small RNA sequences to a 5 MB region of the human genome. However, when aligning our Arabidopsis dataset of 193 024 small RNAs to all A. thaliana predicted tRNAs, SOAP detected only 457 1-nt-mismatched alignments versus BlastN aligned 1000. Additionally, SOAP does not align any single nucleotide mismatched small RNA sequences if the reference database is comprised of small RNA sequences 15–30 nt in length, e.g. mature micro RNAs. As our focus is on tRNA fragments and micro RNAs, we implemented Ebbie-MM using BlastN (29). The objective of Ebbie-MM is to identify sequences with single nucleotide discrepancies with respect to the reference database, determine and count the nature of the single nucleotide mismatch, and record the alignment between query and subject. Details of the algorithm as well as benchmarking parameters are listed in the Supplementary Data (Supplementary Figure 1, Supplementary Tables 1 and 2).
Genomic survey of single nucleotide mismatched sequences
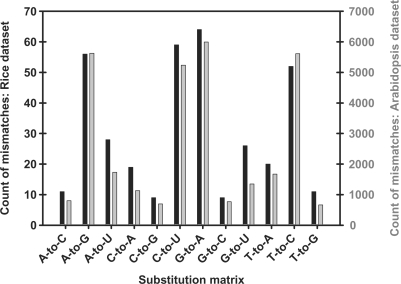
Both the rice and Arabidopsis datasets of unmatched sequences were compared to their respective genomes using Ebbie-MM. Ebbie-MM analysis of the rice dataset comprising of 3852 unmatched sequences resulted in the identification of 364 (9.45% of input) sequences contained single nucleotide mismatches with respect to the genome. The single nucleotide discrepancies can be grouped by the identity of the substitution. An A-to-G substitution is where a genomically encode adenosine is identified as a guanosine during sequencing. The observed occurrence for each possible substitution is graphically depicted as histogram in Figure 1 for the rice dataset (black bars). The prominent substitutions observed are A-to-G, C-to-U, G-to-A and T-to-C. The Arabidopsis 193 024 sequence dataset was also analyzed using Ebbie-MM, Figure 1 (grey bars) graphically portrays the observed occurrences of each type of nucleotide substitution with roughly 16% of all non-matching small RNA sequences are single nucleotide mismatches (number of single nucleotide mismatches: 31‱291 or 16.21%). The most frequent substitutions were A-to-G, C-to-U, G-to-A and T-to-C, all of which were frequent substitutions observed in the rice dataset. In concert with the rice dataset, all rare substitutions were rare substitutions in the Arabidopsis dataset with exact numbers given in Supplementary Table 3. From this data, it appears that the nature of substitution error are not distributed randomly, but rather show a consistent pattern found in both datasets, even though the datasets are 50-fold different in size.
Figure 1.
Histogram displaying the nature of single nucleotide mismatches when comparing of the rice dataset (black bars) and the Arabidopsis dataset (grey bars) to their respective genomes. Substitutions listed on the abscissa are from DNA (genome) to RNA (small RNA sequence). Note that both histograms virtually overlap, despite a discrepancy of 50-fold in the input datasets.
Survey of single nucleotide mismatches in tRNA fragments
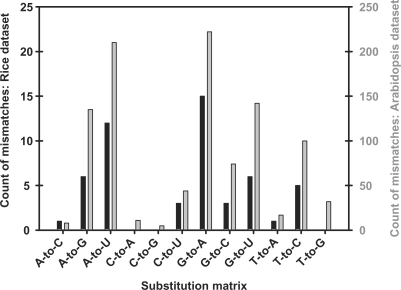
As tRNAs are composed of many non-canonical RNA bases, they are well suited as positive controls to further investigate a link between post-transcriptional modifications, and nucleotide substitution frequencies shown in Figure 1. To compare the small RNA datasets to the respective tRNAs, all the tRNAs encoded in the O. sativa and A. thaliana genomes were identified using tRNA-scan-SE 1.21 (30). Alternatively, predicted tRNA genes are now available from http://gtrnadb.ucsc.edu/ (31). Using the rice dataset as input and all predicted O. sativa tRNAs as the reference database, Ebbie-MM identified 52 single nucleotide mismatches (or 1.35%). The substitution histogram is shown in Figure 2 (black bars). There were prevalent substitutions, such as A-to-U and G-to-A, and rare substitutions, such as C-to-A and C-to-G. As these are sparse statistics, the Arabidopsis dataset was also compared to all predicted A. thaliana tRNAs using Ebbie-MM. Our algorithm detected 1000 (or 0.52%) single nucleotide mismatches with the resulting substitution matrix shown in Figure 2 (grey bars). The substitution matrixes of the rice and Arabidopsis datasets are virtually super-imposable, even though the Arabidopsis dataset is 50-fold larger than the rice dataset. A complete list of all base substitutions from the rice and Arabidopsis datasets is given in Supplementary Table 4.
Figure 2.
Histogram displaying the nature of single nucleotide mismatches when comparing both datasets to their respective tRNA datasets. Histogram of substitutions for the rice dataset (black bars) and the Arabidopsis dataset (grey bars) being matched to their respective tRNAs.
Sequencing errors in tRNA align due to non-canonical bases
To visualize the sequence alignments, sequence logos and the theory of information content in multiple sequence alignment first described by Schneider and colleagues (32,33) is used. The authors convert an entropy level into the height of a letter on a 2-bit scale for RNA alignments; whereby high entropy in a position of a sequence logo results in a smaller height of a letter and vice versa. We used WebLogo (34), an implementation of this sequence theory to generate the sequence logos presented here.
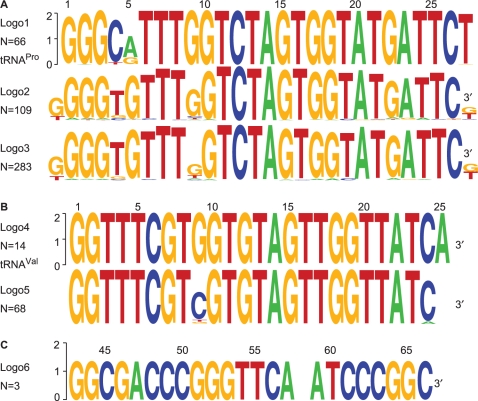
To identify homologous sequences, we further scrutinized the Ebbie-MM analysis of the Arabidopsis dataset against the predicted A. thaliana tRNA database (histogram of the analysis was shown in Figure 2). One of the single nucleotide mismatch sequences, F40955, was annotated as the 5′ end of tRNAPro. To distinguish sequencing errors in the small RNA sequence alignment from natural variations of the 66 predicted tRNAPro from A. thaliana, we aligned all tRNAPro gene sequences and generated the sequence logo in Figure 3A, logo 1. The different genomic loci show a degree of variation most obvious in positions 4, 5, 27 and 28. Then, we searched the Arabidopsis dataset for homologues of the small RNA F40955 sequence and detected an additional 108 unique sequences from all tissue types in the Arabidopsis dataset. The sequence logo of multiple sequence alignments of all 109 sequences is shown in Figure 3A, logo 2. In addition to the genomic loci variations, sequence logo 2 reveals an additional variation at position 9. In the alignment of genomic loci of tRNAPro, position 9 is always a G, whereas in the alignment of homologous sequences not matching their genome of origin, position 9 shows a high degree of entropy. The degree of uncertainty is even more compounded when the cloning frequency of all 109 small RNAs is considered, resulting in a multiple sequence alignment of 283 sequences as shown in Figure 3A, logo 3. In addition to this perturbation in the sequence logo at position 9, further data analysis of other tRNA sequences reveals a similar perturbation to the sequence logo at position 9 of tRNAVal (Figure 3B).
Figure 3.
Sequence logos of tRNA fragments. (A) Logo1: sequence logo resulting from the alignment of the 5′ terminus of all predicted tRNAPro from A. thaliana Logo2: alignment of homologous non-redundant sequences annotating to the 5′ terminus of tRNAPro. Logo3: same as Logo2 but the input data contains the cloning frequencies of all the homologous small RNAs. Note the degree of uncertainty increases in position 9 of Logo2 when compared to Logo3. (B) Logo4: sequence logo resulting from the alignment of the 5′ terminus all predicted tRNAVal from A. thaliana. Logo5: alignment of homologous sequences, including their cloning frequency, annotated to the 5′ terminus of tRNAGly. Note the overwhelming sequence variance in position 9 of Logo5. Position 9 of many tRNAs are modified to generate a N1-methyl-guanosine. (C) From the rice dataset, three small RNA sequences, each cloned once, were annotated as tRNAGlu. The variable position at nucleotide 58 is often modified to N1-methyl-adenosine in many tRNAs.
From the limited rice dataset, an example in which three homologous sequences aligned to the 3′ end of tRNAGlu was identified. The site of the overlapping sequencing error in the RNAs were determined to be position 58 of the tRNA and the resultant sequence logo is shown in Figure 3C, logo 6.
Post-transcriptional modification of small RNAs
In contrast to tRNA modifications, which are ubiquitous and typically quantitative, there are less abundant modifications of other RNAs, such as RNA-editing events due to enzymatic deamination catalyzed by CDARs (35) and ADARs (36). Deamination of cytosine to uracil (C-to-U) by a CDAR or deamination of adenosine to inosine (A-to-I) by an ADAR are also predicted to result in a discrepancy between the genomic DNA sequence and the sequenced RNA. Unlike tRNA modifications, some RNA-editing events due to deamination are only partially complete with both the modified and unmodified RNAs coexisting within a cell (35,36). RNA-editing events have been observed in plant organelles (37,38), but to our knowledge not outside the organelles. In the case of micro RNAs, such deamination events, especially in the 5′-seed region, could potentially change the target mRNA (39).
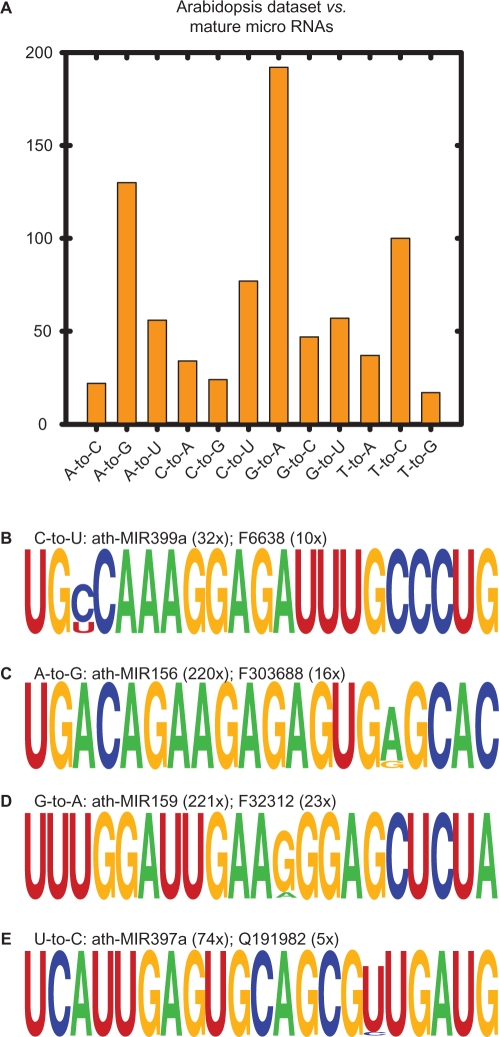
Analysis of both datasets against the genome reveal that C-to-U and A-to-G (A-to-I modifications are predicted to be observed as an A-to-G substitution) substitutions were very common (Figure 1). To explore if we could detect possible enzymatic deaminations, the rice and Arabidopsis datasets were compared with Ebbie-MM to their respective mature micro RNAs as recorded in MirBASEv12 (40). The statistical output of Ebbie-MM for the rice dataset against all O. sativa micro RNAs contained 52 single nucleotide substitutions (Supplementary Table 5). The Arabidopsis dataset was also compared to their respective micro RNAs and the histogram of the substitution frequency is shown in Figure 4A. Of the 793 single nucleotide substitutions, the most frequent substitutions were A-to-G (16.4%), G-to-A (24.2%), C-to-U (9.7%) and T-to-C (12.6%). A-to-G and C-to-U could be explained by deamination of A and C, respectively. The histogram cannot distinguish between apparent spontaneous and enzymatic deamination. To determine whether the observed substitutions are site specific and thus possible enzymatic deamination events, the frequency of the observed micro RNA-editing event was compared to the frequency of the apparent parent micro RNA (23). Furthermore, all small RNAs in question were searched against the A. thaliana genome using Ebbie-MM. The latter search was done as single nucleotide mismatches when aligned to a small database, such as the mature micro RNA database, could be misleading and an alternative alignment is found elsewhere in the genome. Thus, only single nucleotide mismatched sequences with identical mutations in the same location, regardless of comparing the small RNA to the micro RNA database or the genome, were considered further. The cloning frequencies and sequence alignment of all candidate sequences that meet our stringent parameters are listed in Supplementary Table 6 with some potential examples for deamination are shown in Figure 4B and C. Additional examples of G-to-A and T-to-C substitutions as shown in Figure 4D and E.
Figure 4.
Comparing the Arabidopsis dataset to confirmed mature micro RNAs. (A) Substitution histogram of single nucleotide mismatch small RNA sequences. Examples of potential micro RNA editing that are observed as C-to-U (B), A-to-G (C), G-to-A (D) and U-to-C (E) are shown with the number of time the parental micro RNA or the modified sequence was cloned. The first letter of the modified sequence indicates the tissue of origin, F: flower and Q: silique. Additional examples and their cloning frequencies are given in the Supplementary Table 6.
Poly-U extensions of micro RNAs
In the literature, there are reports on 3′ uridylation of small RNAs (20–22). As these additions of uracil are post-transcriptional modifications, it should be possible to detect these in the Arabidopsis dataset. For this analysis, all reported micro RNAs identified from different plant tissues from Rajagopalan and colleagues were obtained as a dataset (23) and compared to the matching Arabidopsis dataset comprised of sequences not matching the genome. First SOAP (28) was used for analysis, but obtained zero single mismatched alignments comparing the two small RNA databases to each other. Thus, BlastN (29) was used for the alignments with an e-value cut off of 0.001. Only positive hits with a ratio of ≥0.2 (count of small RNA/count of micro RNA) were considered as significant hits.
Sequence alignments of the small RNAs extracted from different plant tissues revealed the addition of one or more uridines to the 3′ terminus of several micro RNAs as shown in Table 1. RNA extracted from A. thaliana flower tissue, two micro RNAs, ath-MIR171b and ath-MIR319a, were found in nearly equal proportions of unmodified and 3′ uridinylated forms in flower tissue with ratios of 1.01 and 0.808, respectively. Two small RNAs R133686 and R107534 were also identified that annotate to the 5′ of ath-MIR397a, but with the 3′ terminal 4 nt removed and replaced with a 3 or 4-nt long poly-U-track, respectively. Furthermore, sequence F42411 was annotated as a variant of ath-MIR408 with two additional U attached to the 3′ terminus and one residue removed from the 5′ terminus when compared to the apparent parent ath-MIR408. Surprisingly, F42411 was cloned from flower tissue 91 times, while the apparent parent ath-MIR408 was cloned only 25 times in the same tissue.
Table 1.
Examples of 3′ uridylated stable micro RNAs with cloning frequencies (Count)
| ID | Count | Sequence | Ratio |
|---|---|---|---|
| ath-MIR171b | 82 | UUGAGCCGUGCCAAUAUCACG | |
| F71514 | 83 | UUGAGCCGUGCCAAUAUCACGU | 1.012 |
| ath-MIR319a | 26 | UUGGACUGAAGGGAGCUCCCU | |
| F23837 | 21 | UUGGACUGAAGGGAGCUCCCUU | 0.808 |
| ath-MIR397a | 27 | UCAUUGAGUGCAGCGUUGAUG | |
| R133686 | 11 | UCAUUGAGUGCAGCGUUUUU | 0.407 |
| ath-MIR397a | 27 | UCAUUGAGUGCAGCGUUGAUG | |
| R107534 | 6 | UCAUUGAGUGCAGCGUUUUUU | 0.222 |
| ath-MIR408 | 25 | AUGCACUGCCUCUUCCCUGGC | |
| F42411 | 91 | -UGCACUGCCUCUUCCCUGGCUU | 3.640 |
| ath-MIR408 | 25 | AUGCACUGCCUCUUCCCUGGC | |
| F102722 | 7 | -UGCACUGCCUCUUCCCUGGCUUU | 0.280 |
Micro RNA modifications can alter AGO-complex specificity
Deletions of the 5′ nucleotide have intriguing biological consequences. The sequenced RNA, F42411, has a single nucleotide removed on the 5′ and two U attached to its 3′ terminal when compared to the apparent parent micro RNA ath-MIR408, see Table 1. We term this modification −1+UU for the remainder of the manuscript. According to Mi and colleagues, who recently published a study in which they co-immunoprecipitated small RNAs using anti AGO1, AGO2, AGO4 or AGO5 antibodies, it was determined that the identity of the 5′ termini of a small RNA governs their association with a specific AGO complex (24). Small RNAs with a terminal adenosine most often reside in AGO2 and AGO4 complexes, while small RNAs with a terminal uracil most often reside in AGO1 complex. The modifications identified by our analysis generates the hypothesis that the unmodified ath-MIR408 would be found selectively in an AGO2 complex, while the post-transcriptional modified RNA would be found in an AGO1 complex.
To test this hypothesis of altered AGO targeting, we searched the AGO-dataset (24) for ath-MIR408 and F42411 sequences and if present, identify which AGOs they are associated with. Ath-MIR408 and F42411 from the Arabidopsis dataset were identified in the AGO-dataset, which is supportive of F42411 being a bona fide RNA. Further computational analysis to identify which AGO the two RNAs are associated with remarkably demonstrate a clear shift in AGO binding. The ath-MIR408 micro RNA was almost exclusively (96.8% or 549/567) found in the AGO2 complex, while F42411 identified mostly in the AGO1 complex (91.3% or 21/23) (Figure 5A). These results illustrate the biological importance of the −1+UU modification as it causes a shift in AGO-complex preference from AGO2 to AGO1 for ath-MIR408 and F42411, respectively. It was noted that the cloning frequency and therefore ratio of micro RNA to modified micro RNA is not identical between the Arabidopsis dataset and the AGO-dataset. Discrepancies may be attributed the F42411 sequence arising solely from flower tissue in the Arabidopsis dataset, while Mi and colleagues used whole plants for their studies (23,24).
Figure 5.
Examples of micro RNAs with 5′ deletions and 3′ uridine additions. Micro RNAs with these modifications vary in distribution in different AGO complexes. (A) Ath-MIR408 and F42411 were initially detected in the Arabidopsis dataset, while the cloning frequencies for the different AGO complexes are from the AGO-dataset. (B) Using Ebbie-MM-ago, ath-MIR822 was determined to reside almost exclusively in the AGO2 complex, while its modified variant is found equally in AGO2 and AGO4 complexes. More examples are given in Supplementary Table 7.
F42411 is only one example in which a micro RNA is cleaved on its 5′ terminus by one nucleotide and the 3′ terminus is appended with two uridines. To identify additional examples, we compared all A. thaliana micro RNAs reported in MirBASEv12 (40) with the AGO-dataset (24) using a Perl script termed Ebbie-MM-ago. This script removes the 5′ nucleotide of the known micro RNA and then adds two uridines to the 3′ termini of the sequence. After the modification, the Mi-dataset is searched for matches to the modified sequence. The search identified several other candidate sequences. For example, the unmodified ath-MIR822 micro RNA was mostly cloned (1065/1097) from the AGO1 complex with a few occurrences (8/1097) from the AGO4 complex, while the modified ath-MIR822 (−1MIR822+UU) sequence was cloned from both the AGO1 complex (120/245) and the AGO4 complex (120/245) as seen in Figure 5B. We also find examples, in which the modified micro RNA is detected more frequently than the apparent parent micro RNA, e.g. the modified ath-miR156g was identified 10 times more often in the AGO1 complex than the unmodified parental ath-miR156g sequence. Additional sequences and exact cloning frequencies of the mentioned examples are listed in Supplementary Table 7.
DISCUSSION
Investigating discarded sequencing datasets from small RNA-sequencing projects, it was determined that sequences not aligning to the genome are not all a result of random sequencing errors or a result of technical artifacts, but many are of biologically relevant origin. Post-transcriptional modifications and RNA editing can generate RNAs that, when cloned and sequenced, can result in discrepancies when compared to the genomic sequence or origin. By identifying and characterizing the single nucleotide mismatch discrepancies between small RNA datasets and the genomic sequence of origin, it is possible to identify some sites of modification and obtain some insight into the basis of the modifications. The foundation for identifying RNA modifications and editing is that the large datasets provided by deep sequencing projects enable multiple sequence alignments and mismatch frequencies, which then facilitate the identification of site-specific modifications from random errors.
Application of the Ebbie-MM algorithm to the discarded sequence datasets from small RNA-sequencing projects determined that substitution errors are not distributed randomly (as seen in Figures 1, 2 and 4A). Technical ‘sequencing errors’ may be random or reflect the fidelity of the enzymes used in the cloning and sequencing of the small RNAs. The reverse transcriptase, SuperscriptII (Invitrogen, USA), often used for the cloning of small RNAs (3,41) most commonly makes T-to-G and C-to-A substitution errors which are infrequently observed in our data (Figures 1 and 2).
tRNA modifications
Analysis of tRNA fragments found in the small RNA sequence datasets clearly demonstrate the application sequence mismatch alignments provided by Ebbie-MM to identify post-transcriptional modifications (Figure 3). The rational for the data is that the reverse transcriptase is unable to accurately incorporate thymine or cytosine when the RNA base is modified to either a N1-methyl-adenosine or N1-methyl-guanosine, respectively.
Identification of the modifications at position 9 of A. thaliana tRNAPro and tRNAVal (Figure 3A and B) are in agreement with reports that position 9 is commonly modified to N1-methyl-guanosine in eukaryotic tRNAs (43). Recently, the methyltransferase responsible for the modification at position 9 of tRNAGly in Saccharomyces cerevisiae was identified as Trm10p (43). Searching GenBank for Trm10p homologues in A. thaliana, a putative (Guanine-1)-methyltransferase with the accession number AT5G47680 was identified. The Saccharomyces and Arabidopsis enzymes exhibit 33% (83/247) sequence identity and 54% (134/247) sequence similarity. Furthermore, Barciszewska and colleagues demonstrated that the N1-methyl-guanosine modification occurs in plants by detecting the modification in wheat-germ tRNAArg (44).
It has been demonstrated that tRNAGlu in some species has a N1-methyl-adenosine at position 58 (45,46), which is in agreement with the sequence logo for tRNAGlu from rice (Figure 3C). A homologue of the methyltransferase Gcd14p, is found in O. sativa annotated as an unknown protein (GENE ID: OSJNBb0026L04.5) with 37% (99/264) sequence identity and 52% (138/264) sequence similarity.
The data clearly demonstrates the power of large sequencing datasets to identify post-transcriptional modifications. We are limited to detecting modifications that alter the base pairing properties of the RNA. Sequencing data cannot determine the chemical identity of post-transcriptional modifications, but may be inferred by homology. Nonetheless, deep sequencing techniques provide unique opportunities to map post-transcriptional modifications. It would be intriguing to analyze purified intact tRNAs by pyrophosphate sequencing to obtain an essentially complete tRNA dataset of tRNA modifications that are detectable by reverse transcription.
Micro RNA base modifications
Micro RNA post-transcriptional modifications have broad implications as the modifications may alter targeting, inactivate or alter stability of the RNA. Recently, Kuchenbauer and colleagues detected 3300 isomiRs, which are variants of micro RNAs that do not match the genome of origin, when analyzing sequencing data obtained by Illumina massive parallel-sequencing platform (4). Using the same sequencing platform, Reid and colleagues find strong evidence for let-7-editing events (47). These examples contribute further to the notion that ‘sequencing errors’ are often the result of post-transcriptional modifications of RNA, not simply technical artifacts.
Analysis by Ebbie-MM identified examples of micro RNAs with site-specific occurrences of all four of the most common observed base substitutions, C-to-U, A-to-G, G-to-A and T-to-C (Figure 4A). Using RNA secondary structure prediction via energy density minimization (42), we predicted the secondary structure of all micro RNAs shown in Figure 4B and could not detect any secondary structure in the majority of sequences. Thus, we rule out that these substitutions are due to hindering or inhibition of the reverse transcriptase during conversion of RNA to DNA. We postulate that the C-to-U and A-to-G-sequencing substitutions may be attributed to (at least in part) C-to-U and A-to-I deaminations. These modifications can be catalyzed by CDARs (35) or ADARs (36), respectively. There has already been a report describing adenosine deamination of an adenosine in a micro RNA from mice pri-miR-142 (19). Our results indicate that this may be a much more frequent micro RNA processing event.
C-to-U RNA editing has been demonstrated in plant mitochondria (37) and other plant organelles (38). However, we find evidence for C-to-U RNA editing in micro RNAs. It seems unlikely that micro RNAs are imported and edited in plant organelles, as there is no evidence for argonaute proteins or other RISC components in organelles. Another cause for C-to-U editing could be a spontaneous deamination of cytidine (48), which is predicted to occur randomly. However, as seen in Figure 4B, the C-to-U conversion is directed to position 3 of ath-MIR399a with one-third of all sequences being edited. Further examples are listed in Supplementary Table 6. Therefore, we conclude there must be a C-to-U-editing enzyme CDAR (35) acting on micro RNAs in either the nucleus or cytoplasm. Using BlastP, we searched for yeast cytidine deaminase CDD1 (GeneBankID: NP_013346), a reported CDAR, in the A. thaliana protein databank and found putative cytidine deaminase CDA6 (NP_194690.1) with identities 37/130 (28.5%) and similarity 58/130 (44.6%). Although no concrete evidence of A. thaliana CDAR activity has been verified in vitro, the evidence of frequent site-specific micro RNA C-to-U editing in vivo provides strong support for future investigations of CDAR activity in A. thaliana. Similar to CDAR activity, ADAR activity has been reported in plant organelles (37,38), but it remains elusive how small RNAs not associated with organelles are edited by an apparent ADAR activity in the nucleus or cytoplasm of the plant cell.
Micro RNA nucleotide modification
Evidence for 5′ and 3′ post-transcriptional processing of micro RNAs was found in the small RNA datasets, with six examples of the Arabidopsis dataset listed in Table 1. Our data with the 3′ addition of uridines extends previous findings by other research groups (20–22). The cloning frequencies we report for post-transcriptionally uridylated micro RNAs are much higher than those reported by others (20,24). We reason that our reported cloning frequency is higher due to small RNAs isolated from individual plant tissues, e.g. root or flower, whereas other large scale sequencing projects used total plant RNA from A. thaliana. The tissue-specific micro RNAs will be diluted in bulk micro RNAs extracted from the whole plant, explaining the discrepancy between the findings of different research groups.
The biological significance of 3′ uridylation of micro RNA is uncertain, but may be involved in micro RNA turnover. In the case of U6 snRNA, 3′ uridylation is part of the regeneration process after exonucleolytic processing (49) effectively stabilizing the 3′ uridylated RNA. On the other hand, 3′ uridylation of mRNAs has a destabilizing effect (50). The observation that 3′ uridylation of micro RNAs is blocked by a 2′-O-methyl moiety (20), the inhibition of small RNA degrading nuclease (SDN) by 2′-O-methyl moiety on the 3′ terminus (51), the ubiquitous nature of a 3′ terminal 2′-O-methyl moiety (41), and the relative low abundance of 3′ uridylated micro RNAs in the Arabidopsis dataset as well as observed by others (20–22) all point to 3′ uridylation of micro RNAs as degradation signal. However, the abundance of 3′ uridylated micro RNAs presented in Table 1 can not be ignored. We acknowledge that these six examples of stable 3′ uridylated micro RNAs are the minority of micro RNAs expressed in their respective tissues. If the exosome was inactive or repressed in these tissues, we would have expected to find more significant examples of 3′ uridylated micro RNAs. Therefore, there may be a specialized role for these small RNAs presented in Table 1.
Exploring the notion of an alternative specialized role for stable 3′ uridylated micro RNAs, we identified that the combination of 5′ deletion and 3′ uridine addition of micro RNAs alters the preference of AGO association as outlined in Figure 5. Our proposed model is strongly supported by the data reported by Mi and colleagues whom first identified the micro RNA sequence preference by the different AGO proteins in plants (24). Their sequencing project of RNAs co-immunoprecipitated with different AGOs revealed the strong 5′ bias towards the sorting of small RNAs into AGO complexes and confirmed the bias by site-directed mutagenesis of the 5′ nucleotide of a micro RNA. By searching the AGO-specific-sequencing data, we identified several examples of 5′- and 3′-edited micro RNAs. Remarkably, the micro RNA / −1+UU micro RNA pair were identified in different AGO complexes (Figure 5). Most intriguingly is the shift of MIR822 predominantly residing in the cytoplasmic AGO1 complex, to −1MIR822+UU shifting to nucleolar AGO4 complex which is implemented in silencing of chromosomal DNA (52–54). The micro RNA is not only cleaving the mRNAs in the cytoplasm, but the same micro RNA transcript, −1+UU modified, potentially silences the genomic loci of the corresponding mRNA. Additional sequences and details are listed in Supplementary Table 7. Thus, like in the case of U6 snRNA (49), some cases of 3′ terminal uridylation in conjunction with a 5′ nucleotide removal may leads to stabilization of a micro RNA.
In closing, we conclude that small RNA sequences from deep sequencing projects that do not match their genome of origin, often disregarded as sequencing errors, hold valuable biological information. This was demonstrated by identifying overlapping sequencing errors due to non-canonical RNA bases in tRNA fragments and micro RNA modifications possibly due to enzymatic deamination or other enzymatic activity. Additionally, 3′-uridinylated sequences have been identified as reported by others, but we have additionally identified a novel subset with 5′ deletions which results in sorting into different Argonaute protein complexes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Research Fellowship from the Alberta Cancer Research Institute [to H.A.E.]. Operating grants from the Natural Sciences and Engineering Research Council of Canada and the Alberta Cancer Research Institute [to R.P.F.]. Funding for open access charge: Alberta Cancer Research Institute.
Conflict of interest statement. None declared
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Ramya Rajagopalan (University of Wisconsin-Madison), David Bartel (Whitehead Institute/MIT/HHMI) and Peter Unrau (Simon Fraser University) for their generous gift of the non-redundant dataset which contained small RNA sequences not matching the A. thaliana and O. sativa genomes. We also thank Todd Lowe (University of California, Santa Cruz) for providing a very valuable perl script parsing the output of tRNA-scan-SE into a FASTA formatted list of tRNAs. We thank Luc Berthiaume (University of Alberta) for critically reading the manuscript. We also thank the current members of the Research Support Group of the Academic Information and Communication Technologies at the University of Alberta for technical support using the Linux cluster and numerical servers. Special thanks to Kay C. Wiese (Simon Fraser University), members of the Bioinformatics Research Lab (Simon Fraser University) and the Proteome Analyst group (University of Alberta) for their support in the development of the Ebbie-MM algorithm.
REFERENCES
- 1.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- 3.Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ. Comparative analysis of the small RNA transcriptomes of pinus contorta and oryza sativa. Genome Res. 2008;18:571–584. doi: 10.1101/gr.6897308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuchenbauer F, Morin RD, Argiropoulos B, Petriv OI, Griffith M, Heuser M, Yung E, Piper J, Delaney A, Prabhu AL, et al. In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res. 2008;18:1787–1797. doi: 10.1101/gr.077578.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hulsmann WC, Lipmann F. Amino acid transfer from sRNA to microsome. 1. activation by sulfhydryl compounds. Biochim. Biophys. Acta. 1960;43:123–125. doi: 10.1016/0006-3002(60)90414-5. [DOI] [PubMed] [Google Scholar]
- 6.Nathans D, Lipmann F. Amino acid transfer from sRNA to microsome. 2. isolation of a heat-labile factor from liver supernatant. Biochim. Biophys. Acta. 1960;43:126–128. doi: 10.1016/0006-3002(60)90415-7. [DOI] [PubMed] [Google Scholar]
- 7.Lee RC, Ambros V. An extensive class of small RNAs in caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 8.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 9.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 12.Bakhanashvili M, Hizi A. Fidelity of the reverse transcriptase of human immunodeficiency virus type 2. FEBS Lett. 1992;306:151–156. doi: 10.1016/0014-5793(92)80988-s. [DOI] [PubMed] [Google Scholar]
- 13.Bakhanashvili M, Hizi A. Fidelity of the RNA-dependent DNA synthesis exhibited by the reverse transcriptases of human immunodeficiency virus types 1 and 2 and of murine leukemia virus: Mispair extension frequencies. Biochemistry. 1992;31:9393–9398. doi: 10.1021/bi00154a010. [DOI] [PubMed] [Google Scholar]
- 14.Bakhanashvili M, Hizi A. The fidelity of the reverse transcriptases of human immunodeficiency viruses and murine leukemia virus, exhibited by the mispair extension frequencies, is sequence dependent and enzyme related. FEBS Lett. 1993;319:201–205. doi: 10.1016/0014-5793(93)80067-5. [DOI] [PubMed] [Google Scholar]
- 15.Potter J, Zheng W, Lee J. Thermal stability and cDNA synthesis capability of SuperScript™ III reverse transcriptase. Focus J. Invitrogen Publ. 2003;25:19–24. [Google Scholar]
- 16.Vacic V, Jin H, Zhu JK, Lonardi S. A probabilistic method for small RNA flowgram matching. Pac. Symp. Biocomput. 2008:75–86. [PMC free article] [PubMed] [Google Scholar]
- 17.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 18.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 19.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in arabidopsis. Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rissland OS, Norbury CJ. The Cid1 poly(U) polymerase. Biochim. Biophys. Acta. 2008;1779:286–294. doi: 10.1016/j.bbagrm.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 2008;18:1456–1465. doi: 10.1101/gr.075572.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soll D. Enzymatic modification of transfer RNA. Science. 1971;173:293–299. doi: 10.1126/science.173.3994.293. [DOI] [PubMed] [Google Scholar]
- 26.Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in tetrahymena thermophila. J. Biol. Chem. 2005;280:42744–42749. doi: 10.1074/jbc.M510356200. [DOI] [PubMed] [Google Scholar]
- 27.Ebhardt HA, Wiese KC, Unrau PJ. Ebbie: Automated analysis and storage of small RNA cloning data using a dynamic web server. BMC Bioinform. 2006;7:185. doi: 10.1186/1471-2105-7-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li R, Li Y, Kristiansen K, Wang J. SOAP: Short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 29.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan PP, Lowe TM. GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider TD, Stormo GD, Gold L, Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J. Mol. Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- 33.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dance GS, Beemiller P, Yang Y, Mater DV, Mian IS, Smith HC. Identification of the yeast cytidine deaminase CDD1 as an orphan C→U RNA editase. Nucleic Acids Res. 2001;29:1772–1780. doi: 10.1093/nar/29.8.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takenaka M, Verbitskiy D, van der Merwe JA, Zehrmann A, Brennicke A. The process of RNA editing in plant mitochondria. Mitochondrion. 2008;8:35–46. doi: 10.1016/j.mito.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Shikanai T. RNA editing in plant organelles: Machinery, physiological function and evolution. Cell Mol. Life Sci. 2006;63:698–708. doi: 10.1007/s00018-005-5449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Habig JW, Dale T, Bass BL. miRNA editing – we should have inosine this coming. Mol. Cell. 2007;25:792–793. doi: 10.1016/j.molcel.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ebhardt HA, Thi EP, Wang MB, Unrau PJ. Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc. Natl Acad. Sci. USA. 2005;102:13398–13403. doi: 10.1073/pnas.0506597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alkan C, Karakoc E, Sahinalp SC, Unrau P, Ebhardt HA, Zhang KZ, Buhler J. RNA secondary structure prediction via energy density minimization. Res. Comput. Mol. Biol. Proc. 2006;3909:130–142. [Google Scholar]
- 43.Jackman JE, Montange RK, Malik HS, Phizicky EM. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA. 2003;9:574–585. doi: 10.1261/rna.5070303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barciszewska MZ, Keith G, Kubli E, Barciszewski J. The primary structure of wheat-germ transfer rnaarg – the substrate for arginyl-transfer RNAarg-protein transferase. Biochimie. 1986;68:319–323. doi: 10.1016/s0300-9084(86)80029-3. [DOI] [PubMed] [Google Scholar]
- 45.Ozanick SG, Bujnicki JM, Sem DS, Anderson JT. Conserved amino acids in each subunit of the heteroligomeric tRNA m1A58 mtase from saccharomyces cerevisiae contribute to tRNA binding. Nucleic Acids Res. 2007;35:6808–6819. doi: 10.1093/nar/gkm574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reid JG, Nagaraja AK, Lynn FC, Drabek RB, Muzny DM, Shaw CA, Weiss MK, Naghavi AO, Khan M, Zhu H, et al. Mouse let-7 miRNA populations exhibit RNA editing that is constrained in the 5′-seed/cleavage/anchor regions and stabilize predicted mmu-let-7a:MRNA duplexes. Genome Res. 2008;18:1571–1581. doi: 10.1101/gr.078246.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Becker H, LeBlanc JC, Johns HE. The U.V. photochemistry of cytidylic acid. Photochem. Photobiol. 1967;6:733–743. doi: 10.1111/j.1751-1097.1967.tb08738.x. [DOI] [PubMed] [Google Scholar]
- 49.Chen Y, Sinha K, Perumal K, Reddy R. Effect of 3′ terminal adenylic acid residue on the uridylation of human small RNAs in vitro and in frog oocytes. RNA. 2000;6:1277–1288. doi: 10.1017/s1355838200000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science, 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- 51.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 53.Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE. Role of arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol. 2004;14:1214–1220. doi: 10.1016/j.cub.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 54.Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.