Abstract
Inteins are genetic elements, inserted in-frame into protein-coding genes, whose products catalyze their removal from the protein precursor via a protein-splicing reaction. Intein domains can be split into two fragments and still ligate their flanks by a trans-protein-splicing reaction. A bioinformatic analysis of environmental metagenomic data revealed 26 different loci with a novel genomic arrangement. In each locus, a conserved enzyme coding region is broken in two by a split intein, with a free-standing endonuclease gene inserted in between. Eight types of DNA synthesis and repair enzymes have this ‘fractured’ organization. The new types of naturally split-inteins were analyzed in comparison to known split-inteins. Some loci include apparent gene control elements brought in with the endonuclease gene. A newly predicted homing endonuclease family, related to very-short patch repair (Vsr) endonucleases, was found in half of the loci. These putative homing endonucleases also appear in group-I introns, and as stand-alone inserts in the absence of surrounding intervening sequences. The new fractured genes organization appears to be present mainly in phage, shows how endonucleases can integrate into inteins, and may represent a missing link in the evolution of gene breaking in general, and in the creation of split-inteins in particular.
INTRODUCTION
Protein chains are often non-contiguously encoded on the DNA. This is best known in eukaryotic genes which are interrupted by spliceosomal introns, and also in microbial and organellar genomes that include group-I and -II self-splicing introns (1). These genetic elements are removed from the transcribed RNA, yielding a contiguous RNA message and subsequent protein molecule. An additional type of intervening sequences, termed ‘intein’, is processed at the protein level (2). Inteins are selfish genetic elements which are inserted in-frame in protein coding regions of diverse prokaryotes, unicellular eukaryotes and organelles. Inteins and intein-like domains are translated with their host proteins and then post-translationally catalyze their removal from the protein precursor by a protein-splicing reaction (3,4).
Protein-coding genes of one species are sometimes broken into two (or more) separate genes in other species. The broken genes code for separate protein chains that typically interact with each other to form the mature protein molecule (5–7). For example, Nanoarchaeum equitans contains a dozen broken enzymes, whose break sites lie between functional domains of the encoded proteins (8). Similarly, the B-type DNA polymerase of Methanobacterium thermoautotrophicum, is encoded on two different genes (9). In other cases, transcripts of broken genes can undergo trans-RNA splicing into one mRNA molecule, which is then translated into a contiguous protein molecule (10). This splicing process might represent an intermediate step in the breaking of genes (11).
Intein-containing genes can be broken into two fragments within the intein domain, and still create the mature protein product by a trans-protein splicing reaction (i.e. protein ligation) (12). Each of the broken genes codes for a separate polypeptide: one consisting of the N-terminal part of the host followed by the N-terminal part of the intein, and the other consisting of the C-terminal part of the intein followed by the C-terminal part of the host. The two intein parts, termed ‘split-intein’ (13), associate tightly and specifically with each other leading to a protein-splicing reaction, which ligates the two host protein parts in trans. Protein-splicing is typically a robust and rapid reaction, needing no cofactors or external energy sources. Split-inteins can be engineered synthetically, but also occur naturally. Split-inteins were readily created in the lab from different natural contiguous inteins, with several different breaking points (14–16). Naturally split-inteins have been identified in only two contexts. In the first, the alpha catalytic subunit of replicative DNA polymerase (dnaE) in a large number of diverse cyanobacteria species is coded on different genes that each includes one part of a split-intein (12,17,18). Similarly, the B-type DNA polymerase of N. equitans is coded on two genes, each includes a split-intein part (19). All the cyanobacterial broken dnaE genes include very similar split-inteins at the same integration point. This, and the phylogenetic analysis of the DnaE and intein sequences, suggest that these broken genes originated from a single, irreversible breaking event. This breaking event should be a complex process—to avoid dysfunctional intermediates it needs to create stable protein products, a new transcription termination site and a functional promoter. It is unclear how cells can survive with dysfunctional intermediates, since the replicative DNA polymerase is an essential protein needed in each cell cycle (12).
Recently, two types of proteins were found to be broken into a two genes arrangement with an intervening probable homing endonuclease. Karam, Petrov and co-workers found the gp43 B-type DNA polymerases of several T4-like phages to be broken into two genes separated by 2–3000 bases. In one case the intervening region includes a GIY-YIG putative homing endonuclease (20,21). Edgell and co-workers described a novel broken gene organization, regulation and function (22,23), where the alpha catalytic subunit of ribonucleotide reductase (NrdA) in the Aeh1 T-even bacteriophage is coded on two separate genes. The genes are located within one operon, with the gene coding for the nrdA 5′ region present upstream of the gene for the 3′ region. Between these two genes is a third gene coding for a HNH-type putative homing endonuclease. The genes are co-transcribed but each has different regulatory regions. The homing endonuclease gene is regulated on both transcription and translation levels, limiting its translation to late phage-infection time points. The two nrdA genes are under separate control and generate two protein chains that form an active ribonucleotide reductase together with the NrdB subunit. Comparison with other T-even phages showed that this arrangement is likely to have been formed by an invasion of a free standing homing endonuclease into a nrdA gene.
Homing endonucleases occur in different prokaryotes, phages, and organelles. They are encoded within both group I and group II introns, as auxiliary protein domains incorporated within intein scaffolds, and as free standing genes (24). In all cases they have been shown to promote the duplication of their genes and their surrounding mobile elements into specific integration points (homing). Most often the integration point of a homing endonuclease is also its recognition and cleavage sequence. Homing endonucleases mediate the invasion of their coding regions into unoccupied alleles, spreading the elements they are present in by horizontal gene transfers. Most inteins have a homing endonuclease domain of one of a few types, but some are missing them altogether. Thus, it seems that inteins acquired homing endonuclease domains several times in evolution, possibly by invasions of the intein coding regions by homing endonuclease genes (25).
The creation of broken genes is not well understood, but probably involves different mechanisms of gene fission and duplication. Gene breaking ultimately creates separately encoded protein chains, which must associate into one functional protein. Complementation of the protein's function by these individually translated peptides should satisfy a number of key requirements. Foremost is their assembly to reconstitute the original protein function. However, the proper transcription, translation, solubility, cellular targeting and correct folding of the intermediate protein chains are also crucial. Since dysfunctional intermediates of proteins might be lethal to the organism, broken genes should also be efficiently co-regulated.
In this work, we describe the discovery and analysis of a novel genomic arrangement of broken genes from microbial environmental metagenomic data. Each pair of gene parts is located in one locus, with a split-intein and a free-standing homing-endonuclease gene occurring in between the broken gene coding regions. We term this organization ‘fractured genes’, since a gene is broken into two genes that are still retained in the original orientation and locus of the parent gene. Twenty-six examples of this arrangement, with different types of fractured host genes, split-inteins and homing-endonucleases were identified. In two other examples either the split-intein or the homing endonuclease was absent. We describe the identification of these loci, and analyze both the global features of this arrangement, and the nature of fractured proteins, split-inteins and the endonucleases [including a predicted new structural homing endonuclease family, similar to very short patch repair (Vsr) endonucleases]. Finally, we discuss why this arrangement seems most common in phages and present an evolutionary model for this type of gene fracturing.
MATERIALS AND METHODS
Source of sequences
GOS metagenomic dataset (26), including phase I (Halifax to Galapagos islands) and South Pacific samples upto Rangirora Atoll, was downloaded from the Camera site (camera.calit2.net).
Analysis tools
Data from each GOS sampling location was searched for inteins using BLAST (27) for sequence-to-sequence searches with intein sequence queries (e.g. from http://www.neb.com/neb/inteins.html), and using BLIMPS (28) for blocks-to-sequence with intein conserved blocks (25). This initial screen identified hundreds of reads coding for potential inteins. Paired and nearly identical reads were assembled using BLAST and CAP3 (29) programs, respectively. The resulting sequences were further analyzed specifically searching for new types of split-inteins by examining the positions of the inteins in the open reading frames (ORFs) and the nature of these and surrounding ORFs. Vsr-like putative homing endonucleases in non-intein contexts were searched for by BLAST, using the sequences found in the split-intein loci, and by BLIMPS, using the conserved motifs of these sequences (Figure 3). Probable Vsr repair endonucleases were identified and eliminated from this set by searching it with Vsr repair endonucleases using BLAST. The remaining sequences were further analyzed searching for separated parts of known protein types on both sides of the Vsr-like ORF.
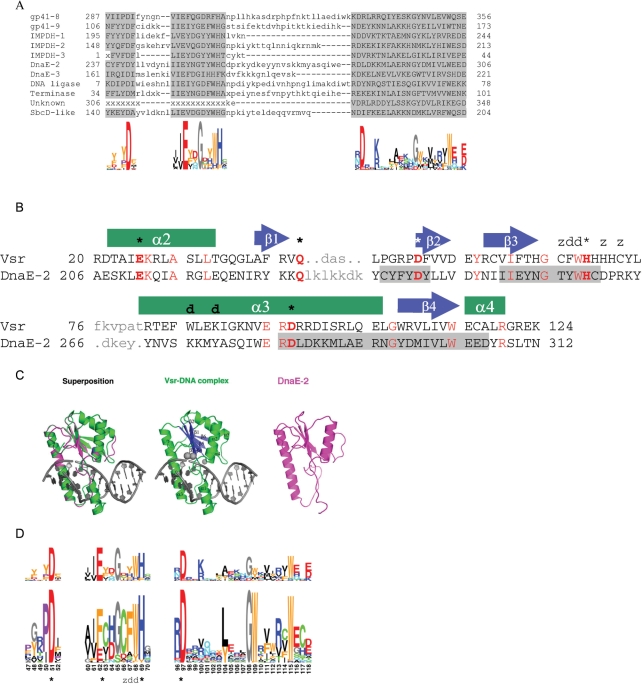
Figure 3.
Sequence features of the Vsr-like putative homing endonuclease family and its similarity to Vsr repair endonucleases. (A) Conserved sequence motifs of Vsr-like putative homing endonucleases, and their sequence logos. (B) Structure based alignment of DnaE-2 locus Vsr-like putative homing endonuclease with E.coli Vsr repair endonuclease (Vsr; PDB code 1CW0). Residues that were modeled in similar positions and backbone conformations after sequence threading and energy minimization are shown in upper case; unaligned sequence regions are shown in grey lowercase. Identical residues are highlighted in red. Vsr active site residues are marked by asterisks, DNA binding residues are marked by ‘d's, and Zinc binding residues are marked by ‘z's. The secondary structure of Vsr is shown above its sequence. Conserved sequence motifs of DnaE-2 are marked as in A. The Phyre server Z-score for this alignment was 5 × 10−14. (C) Predicted structure of DnaE-2 locus Vsr-like protein positions 196–315, and its similarity to structure of Vsr (D) Motif to motif alignment of the Vsr-like putative homing endonucleases (top) with Vsr repair endonuclease (bottom). Alignments of the second and third (rightmost) aligned blocks are significant, expected to occur by chance 2e−2 and <5e−7. The first aligned block has a non-significant score since it is shorter and less conserved. Nevertheless, the alignment is probably genuine since the corresponding Vsr and Vsr-like regions were also found aligned in sequence to sequence, sequence to multiple alignment, and structure threading alignments. Functional residues of the Vsr endonuclease are marked as in B.
Multiple sequence alignments of split-inteins and Vsr-like domains were constructed with MEME (30), MACAW (31) and Dialign (32) programs. The MEME program was also used to search for conserved nucleotide motifs in the hairpin regions upstream of the endonuclease ORFs. Ka/Ks ratio analysis was done using the SELECTON server (selecton.tau.ac.il (33)]. RNA secondary structures were identified using the Vienna package (RNAfold) via http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi using default parameters, and visualized using PseudoViewer [http://pseudoviewer.inha.ac.kr].
Structural models
Similarity of the Vsr-like homing endonuclease to known structures was found using the Phyre server (34), and a structure model was created with the Rosetta structure prediction program suite using the K*Sync alignment method (35), with PDB structure 1CW0 as a template. For split-inteins structural models we used the I-Tasser server (36) for initial alignment and model creation. Sequences were submitted fused, and the resulting models were subsequently separated, by removing N-terminal Methionine of the C-terminal part of the split-intein. These initial models were subsequently refined with the standard Rosetta interface refinement protocol [The same protocol was used in (37), a review of the underlying mechanism can be found in (38)] that minimizes all degrees of freedom at the interface (rigid body orientation, backbone and side-chain dihedral angle) to remove clashes. In order to evaluate additional possibilities for salt bridges across the interface, we further repacked the interface side chains, using the existing side chains and an extended rotamer library for all side chains with at least 10 neighbors (Rosetta option—ex1 -ex2 -ex3 -ex4 -use_input_sc -extra_chi_cutoff 10). This created slightly more salt bridges. Additional modeling with an energy function that includes a Generalized Born term to account explicitly for the electrostatic contribution to total energy did not change significantly the results, suggesting that the main effect of the salt bridges across the interface is long-range electrostatic steering in protein–protein association rather than in the details of the final complex conformation.
Definition of putative salt bridges
Possible salt bridges were defined as opposite charge atoms contacts across the interface; different distance cutoffs were evaluated and resulted in similar results (e.g. 4.5 Å, 5.0 Å, 6.0 Å): The results described here are from an analysis with a 5.0 Å cutoff.
RESULTS
A new gene-in-pieces arrangement
Analyzing Global Ocean Sampling environmental metagenomic sequence data [GOS (26)] we identified a novel genomic arrangement common to several different genes. In each locus, the coding region of a conserved enzyme is fractured in two by a split-intein, with an endonuclease gene in between the two coding regions. All three coding regions are coded on the same DNA strand, where the N-terminal part of the enzyme is followed in-frame by the N-terminal part of a split-intein; next is a non-coding region (27–51 bases) followed by the endonuclease gene; and finally the C-terminal split-intein, in-frame with the C-terminal part of the enzyme (Figure 1 and Supplementary Table S1).
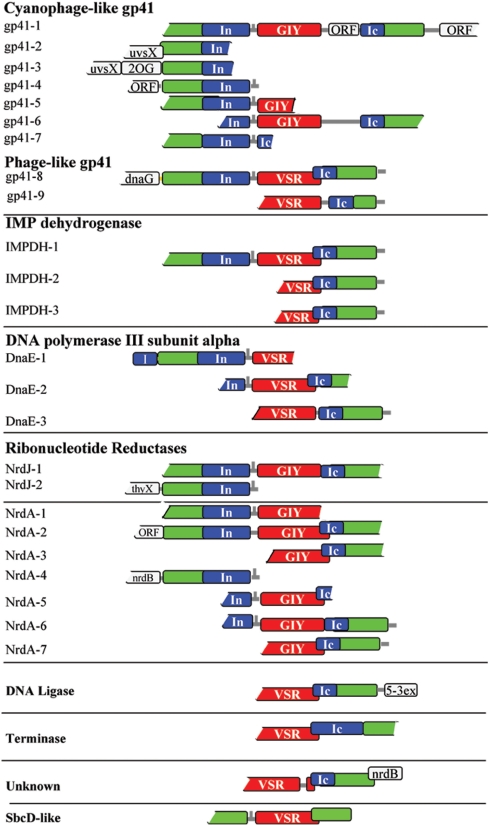
Figure 1.
Schematic representation of fractured gene arrangements. Genomic arrangement of 27 loci, assembled from GOS reads, and grouped according to the type of the enzyme host. Protein coding regions are shown as rectangles, with the enzyme hosts in green, split-intein parts in blue, and endonucleases in red. N-terminal intein half (In) and C-terminal intein half (Ic); free-standing homing endonuclease domains: GIY-YIG (GIY), very short repeat like (Vsr). Abbreviations for other gene names are specified in the text. Possible hairpin structures are marked as vertical lines on the 5′ untranslated regions of endonuclease genes. Coding frames overlap are marked by offset of overlapping coding regions.
We found 26 different loci with this arrangement, distributed in seven diverse and distant bio-geographic GOS sampling locations: the Delaware Bay estuary; the Gulf Stream Off Nags Head, North Carolina; Off Key West; Gulf Of Mexico; freshwater Lake Gatun in the Panama Canal; Punta Cormorant hypersaline lagoon in the Galapagos Islands and the Rangirora South Pacific coral reef atoll (data sets GS11, GS13, GS15, GS16, GS20, GS33 and GS51, respectively of the GOS project).
Eight different types of house-keeping enzymes involved in various forms of DNA synthesis and repair were present in these loci [gp41 DNA helicase, Inosine-5′-monophosphate dehydrogenases (IMPDH), DnaE polymerase catalytic subunit, ribonucleotide reductase catalytic subunits NrdA and NrdJ, DNA Ligase and phage terminase]. Different types of split-inteins are present in each of these enzyme hosts. The free standing endonuclease genes are also of different types (representing either the GIY-YIG family, or novel ‘Vsr-like’ endonucleases described below). By nature of the environmental metagenomic data, most of the loci are missing one or both of their termini. Nevertheless, five loci were assembled with both N- and C-terminal regions of the host regions, together with complete split-intein parts and the endonuclease gene (Figure 1, gp41-1, gp41-8, IMPDH-1, NrdJ-1, NrdA-2). One gp41 locus was found without a homing endonuclease between the split-intein parts (Figure 1, gp41-7). All 27 loci include putative transcriptional regulatory elements. The control regions, different split-inteins and endonucleases are described in the next sections.
The fractured enzyme types
The fractured genes all encode conserved proteins with essential cellular functions (Figure 1 and Supplementary Table S1). All these enzymes and enzyme subunits, except for dnaE, were previously not known to include split inteins. The dnaE gene is already observed in a broken form in cyanobacteria, containing split-inteins but in a different integration point than the one reported here, with the broken genes present in separate loci (12). All the fractured enzymes that we identified have microbial or phagic homologs, and contain the conserved and catalytic motifs typical to each type (data not shown).
Seven loci of fractured gp41 DNA helicase proteins were found in two different GOS sampling locations (Lake Gatun and Off Nags Head). One locus could be assembled to include the complete 5′ and 3′ regions of gp41, the split-intein and the intervening endonuclease (gp41-1). The other loci are missing parts of either their 5′ or 3′ ends. The protein sequences from all loci are highly similar to each other (89–100%). Examination of the DNA sequences of all seven loci showed more synonymous substitutions over non-synonymous (Ka/Ks < 1, data not shown), indicating purifying selection of these genes. All the gp41 proteins are fractured in the exact same point (corresponding to position 356 of cyanophage P-SSM2 gp41, gi:61806058). This suggests that genes gp41-1-7 are different alleles of the fractured gp41 gene. Three of the loci contain upstream ORFs of proteins present in cyanophages [UvsX, a T4-like RecA-like recombination protein; and a 2-oxoglutarate oxygenase, 2OG (Figure 1)]. The gp41 protein sequences from all loci are most similar (>85%) to gp41 proteins from Myoviridae T4-like cyanophages like P-SSM2, syn9 and P-SSM4. The gp41-7 locus has the same genomic arrangement but is missing the endonuclease gene.
Two additional loci of phage-like gp41 proteins (gp41-8 and 9) were found in two GOS locations (Lake Gatun and Punta Cormorant). One includes the complete sequences of the fractured host and intein, with an intervening putative homing endonuclease gene. This locus also includes an ORF similar to DnaG primases, upstream of the N-part of the host. The intein integration points of these probable gp41 proteins are different from each other and from the integration point of the cyanophage-like gp41 proteins (from gp41-1 to 7). The putative homing-endonucleases of the two groups of gp41 proteins (cyanophage-like and phage-like) are also of two different types (see below).
We identified nine loci of fractured ribonucleotide reductase (RNR) class I (NrdA) and II (NrdJ) catalytic subunits. We identified seven NrdA subunits (NrdA-1–7), from three GOS locations (Lake Gatun, Punta Cormorant and Rangirora Atoll), and two NrdJ proteins (NrdJ-1 and 2) from Lake Gatun. The fractured NrdA enzymes are most similar to the corresponding enzymes from cyanophages. NrdA and NrdJ are homologous to each other. However, while the integration points are identical within each enzyme class, they are different between them. The NrdA integration point is directly N-terminal to the class I and II RNRs thiyl radical active site. This is the most frequent insertion point of contiguous inteins in these RNRs (www.neb.com/neb/inteins.html, bioinfo.weizmann.ac.il/∼pietro/inteins/). The NrdJ integration point is one residue C-terminal to the NrdA integration point. This intein insertion point was not previously observed.
IMPDHs are another type of nucleotide metabolism enzyme that we found to be fractured by endonucleases. IMPDH catalyzes a rate-limiting reaction in de novo biosynthesis of guanine nucleotides. Three loci were identified from two different GOS locations (Lake Gatun and Delaware Bay), with similar sequences of IMPDH proteins, fractured by inteins at the same integration point, and with an intervening putative homing endonucleases.
DNA polymerase III is a complex, multi-chain enzyme responsible for replicative DNA synthesis in bacteria. We identified three loci where the DnaE catalytic (alpha) subunit of this polymerase includes a split-intein and a putative free-standing homing endonuclease (from Rangirora Atoll, Lake Gatun, and the Gulf Of Mexico). The split-inteins integration points are different from each other and from the known cyanobacterial DnaE split-intein integration point (www.neb.com/neb/inteins.html bioinfo.weizmann.ac.il/∼pietro/inteins/). One of the loci (DnaE-1) includes a C-terminal part of an additional intein, embedded in-frame in the DnaE host.
Three additional loci were identified. A DNA ligase with a split-intein was identified in one locus from Lake Gatun. The sequence was most similar to a putative DNA ligase from enterobacteriophage Felix 01 (NCBI gi:38707859). Downstream of the DNA ligase is a gene with a 5′–3′ exonuclease domain (Figure 1, 5-3ex). An additional locus from Lake Gatun was similar to DNA packaging large subunit terminase (gp17) of Myoviridae viruses. Finally, the same loci arrangement was identified in a protein gene with an unknown function, present immediately upstream of an nrdB gene.
Identification of new types of split-inteins
The 27 split-inteins loci which we describe here contain five pairs of full-length split-inteins, five pairs of truncated split-inteins, and 17 individual split-intein halves (Figure 1 and Supplementary Figure S1). The split-inteins include the six conserved protein-splicing motifs of the HINT (Hog/Intein) family (Figure 2A and B). They contain the five catalytic active-site residues, required for protein-splicing (N-terminal Cys1, Thr79, His82, and C-terminal Asn36, Cys37; coordinates on the DnaE split-intein). These split-inteins are of new types, not closely related to other recognized split and regular inteins. The protein sequences of the new split-inteins are only 40% similar to known cyanobacterial naturally split-inteins (17). The length of the N-terminal split-intein parts vary from 88 to 105 residues, while the C-terminal parts vary from 33 to 54 residues. One split-intein (Terminase, Figure 1) is fractured at an unusual point, resulting in a long (122 residues) C-terminal part. This part seems to not only include the two C-terminal protein-splicing motifs but also all N-terminal motifs, except N1 (Supplementary Figure S1).
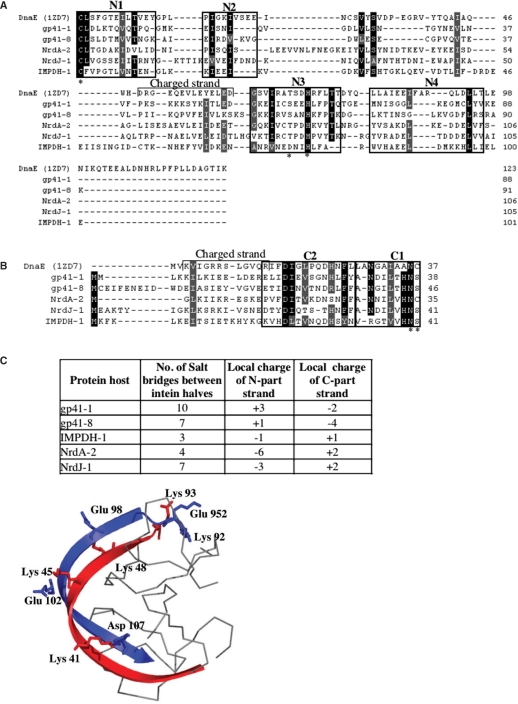
Figure 2.
Sequence features of the new split-inteins. Multiple sequence alignment of N-terminal (A) and C-terminal (B) halves of full-length split-inteins. Conserved motifs of the HINT protein-splicing family are boxed and labeled, and active-site residues are marked with an asterisk. The sequence alignment was refined based on structural modeling with the cyanobacterial DnaE split-intein as a template (PDB code: 1ZD7). Sequences are named after their protein host. (C) Electrostatic characteristics of full-length split-inteins. The number of salt-bridges was calculated from the modeled tertiary structures, and is indicated together with the local charges of the two interacting beta-strands of the intein-halves. (D) Illustration of salt-bridges at the interaction interface between the N-terminal (blue) and C-terminal (red) halves, in the modeled structure of gp41-1. The two longest anti-parallel beta-strands of the intein molecule are shown in cartoon representation.
Several inteins are integrated in new protein hosts, not known to contain inteins (i.e. Terminase, DNA ligase, IMPDH). In other cases, inteins were found in new integration points of known hosts (i.e. DnaE, NrdJ). Allelic split-inteins, those integrated within the same protein host type and integration point, are very similar to each other (Supplementary Figure S1). This may suggest that the split-inteins were present in an ancestral locus, which further diverged.
To further examine the sequence-to-function features of the new split-inteins, five full-length split-intein pairs were selected from different loci (gp41-1, gp41-8, IMPDH-1, NrdA-2 and NrdJ-1) for a structural analysis. The N- and C-terminal sequence parts of the each split-intein were joined to form a contiguous intein, and the overall structure of each intein was modeled based on the available crystal structure of a joined DnaE split-intein (PDB code: 1ZD7). All five split-inteins fit the intein structure without major clashes, despite their sequence variations. We previously suggested that the charge distribution along the two long anti-parallel beta-strands of the associated split-inteins molecule has an important role in the electrostatic interaction between the two split-intein parts (17). Examination of the five intein models showed that there are 3–6 salt-bridges in the predicted interaction interface between the N- and the C-intein parts (Figure 2C and D, Supplementary Table S2). Additionally, strands at the interface between the two fractured parts have opposite local net charges, where in two loci (gp41-1 and gp41-8) the N-intein was positive and the C-intein was negative, while in three examples (IMPDH-1, NrdA-2 and NrdJ-1) the N-intein was negative and the C-intein was positive.
A new predicted family of homing-endonucleases
Two different families of free standing homing endonuclease ORFs were found in between the two halves of the fractured genes. The first corresponds to the well-characterized GIY-YIG homing endonucleases. These are frequent in some phages and organelles of unicellular organisms, as a free-standing enzyme or within group-I introns (24). GIY-YIG homing endonucleases are bi-partite proteins, with the DNA cleaving active site domain separated by linker from the DNA-binding substrate recognition domain. DNA-binding domains of GIY-YIG homing endonucleases include small NUclease-associated MOdular Domains (NUMOD) that were predicted and shown to be specific DNA-binding regions (39–41). NUMOD motifs appear together in different combinations in GIY-YIG and HNH homing endonucleases (39–41).
A second type of a putative homing endonuclease family appeared in 12 fractured genes loci. Its sequences were similar to each other and included three short conserved sequence motifs (Figure 3A). Six of the sequences were significantly similar to Vsr DNA G:T-mismatch endonucleases (42) by sequence to sequence (blastp e-values <1e−3 when searching the NCBI nr protein database) or sequence to multiple sequence alignment comparisons (data not shown). Structure-based sequence threading of several of these sequences also identified them as containing the core protein fold corresponding to the Vsr catalytic region (Figure 3B and C). The three conserved motifs of the sequences we found were also similar to motifs of the Vsr family (Figure 3D). These Vsr motifs include three invariant and well-conserved catalytic-site residues (E. coli Vsr protein Asp51, His69 and Asp 97), two of the DNA major-groove intercalating residues (E. coli Vsr protein Phe67 and Trp68), and one of its three Zinc-coordinating residues (Figure 3B and C). An additional apparent catalytic residue, E. coli Vsr protein Gln42, is in a less-conserved sequence region but is generally maintained as a Gln or Lys residue (data not shown). Our sequence and structure comparisons thus show that the Vsr-like proteins we found have conserved residues corresponding to the active site and DNA-intercalating residues of Vsr endonucleases.
Vsr endonucleases have a type II restriction enzyme topology, but their active sites and hypothesized catalytic mechanism has significantly diverged from the canonical ‘PD-(D/E)xK’ motif associated with that enzyme scaffold, including the use of an activated histidine as a general base (42). The Vsr-like ORFs appear to have maintained most of the features of this unique active site arrangement, although at least one additional strongly conserved acidic residue in the active site region (an invariant glutamate at the position corresponding to Phe62 in Vsr) may indicate further subtle divergence in catalytic mechanism, possibly relating to the difference in activity between repair and homing endonucleases.
No significant similarity was found to the Vsr repair endonuclease residues, at amino-acid positions that recognize and bind the T:G mismatched base pair in its DNA substrate (43) (Figure 3A). However, two Vsr-like proteins each contain a tandem repeat of ‘NUMOD-3’ motifs, which were previously shown in the structure of the I-TevI homing endonuclease to be a sequence-specific DNA-binding helix (40,43). This motif is located N-terminal to the Vsr-like catalytic domain (IMPDH-1 residues 41–54 and 69–82; IMPDH-2 residues 52–65 and 71–84).
We thus conclude that the ORFs that we found in fractured genes loci likely represent the first recognized examples of a new type of Vsr-like putative homing endonucleases. This family is likely to have a type-II restriction enzymes topology and to utilize the Vsr repair endonucleases catalytic mechanism (Mg–water clusters). In contrast to Vsr endonucleases, these proteins seem unlikely to incorporate a structural Zn ion. The family probably has separate DNA binding and catalytic domains, like the GIY-YIG and HNH homing endonucleases. This is the second example of a type-II PD-(D/E)xK containing enzyme that has been observed to function as a homing endonuclease [the first being the I-Ssp6803I endonuclease that drives group I intron insertion into tRNA genes in cyanobacteria (44)]. However, the extreme divergence between that endonuclease and the Vsr-like homing endonucleases described here indicate that their domain organization and DNA-recognition mechanisms are likely to be quite different from one another.
To examine the generality of these putative homing endonucleases we searched sequence databases for loci where a Vsr-like gene is inserted within a protein-coding region, but with no flanking intein sequences. In one case, a locus from Lake Gatun GOS data includes two ORFs that are similar to N- and C-terminal halves of SbcD-like DNA-repair exonucleases, and contain a Vsr-like ORF between them (Figures 1 ‘SbcD-like’ and 4B). The genomic arrangement is identical to the other loci with Vsr-like ORFs, except for the absence of a split-intein. Comparing the concatenated protein sequence of the two SbcD-like ORFs also showed that they each have an inserted region next to their fracturing point. The 25 C-terminal residues of the upstream ORF and the nine N-terminal residues of the downstream ORF are flanked by consecutive conserved motifs of the SbcD-like proteins, but themselves are absent from these proteins (Supplementary Figure S2).
In another case, Vsr-like ORFs are present in two group I introns of recA genes. One of these introns is found in a gene fragment from a whole genome shotgun sequencing project of Bacillus cereus (NCBI accession NZ_ABDA01000203.1). The fragment includes part of a recA 5′ region and a group I intron with a complete Vsr-like ORF. This region is 94% identical to a B. anthracis recA gene with an active group I intron (45) and identical recA loci from several other sequenced Bacilli (data not shown). However, the B. cereus loci we found also has an 884 bases insert with the Vsr-like ORF (Supplementary Figure S3A). Similarly, a second group I intron with a Vsr-like ORF is found at the same integration point in recA gene of B. thuringiensis phage 0305φ8-36 (46) (Supplementary Figure S3A). Both Vsr-like ORFs are inserted in the loop of the introns P1 secondary structure region (Supplementary Figure S3B).
Thus, the Vsr-like homing endonuclease family appears to enjoy evolutionary success, in terms of widespread inclusion within multiple fractured gene arrangements and host targets, similar to that observed for previously characterized mobile endonuclease families.
Nucleotide features of free-standing homing-endonucleases
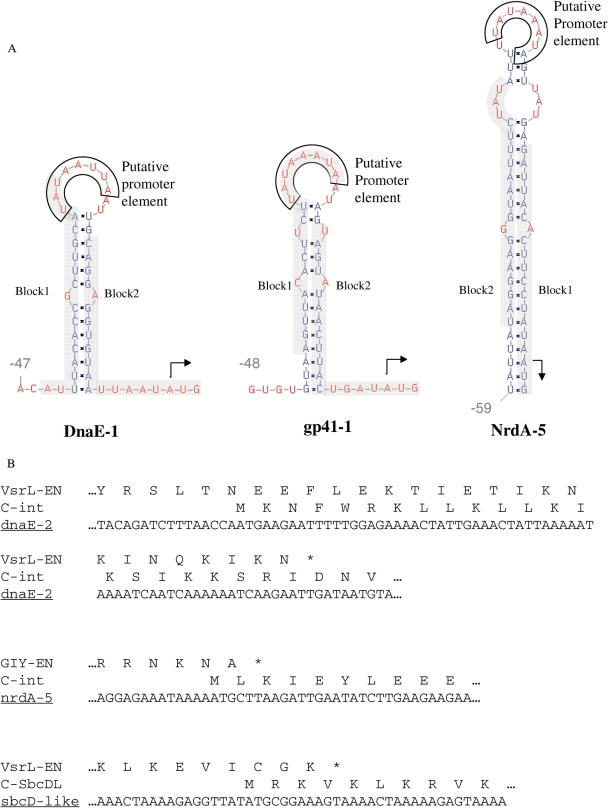
Gene control elements were identified for several members of each type of intervening homing endonuclease genes described in this study. In 15 out of 16, 5′ untranslated regions of the endonucleases we identified RNA hairpin structures upstream of initiation codons (Figures 1 and 4A). This may be a translational regulatory hairpin structure known from various phage genes, including homing endonucleases (22,47–49). The hairpins we found are in fractured gene loci with different protein host types, including gp41, IMPDH, DnaE, NrdJ and NrdA. Ten of these hairpins also include a conserved sequence of Aeh1 phage late promoter element (TATAAATA), which was recently shown to inhibit the translation of a gene-fracturing endonuclease gene, by sequestering its ribosome binding site (22,23). Analysis of the endonuclease 5′ untranslated regions revealed two conserved motifs (Figures 4A and Supplementary Figure S4), which usually complement each other on the stem of the conserved hairpin structures.
Figure 4.
Nucleotide features of endonuclease genes. (A) RNA hairpin structures at the 5′ untranslated region of endonuclease ORFs in the gp41-1 (representing the very similar sequences of gp41-1–7), nrdA-5 and DnaE-1 gene loci. Initiator codons are marked by arrows, conserved putative T4 late promoter elements are boxed, and conserved sequence motifs (Supplementary Figure S4) are highlighted in grey. The expected values for motifs 1 and 2 are 1.7−10 and 9.9−3, respectively. RNA structures were calculated using the Vienna package (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi), and sequence motifs were identified using the MEME program. (B) Overlapping#protein coding regions of endonuclease 3′ termini and the 5′ termini of their downstream genes.
Additionally, 15 loci included an overlapping region of 6–69 bp between the coding regions of the probable endonuclease C-termini and the downstream N-termini of the split-intein part (Figures 1 and 4B). This overlap is present in both types of endonucleases, and is independent on the presence of a split-intein part in the fractured gene, also occurring in the putative sbcD DNA-repair gene that we found, as well as in the Aeh1 fractured nrdA found by Gibb and Edgell (22).
DISCUSSION
New genes can be formed by fusion and fission of existing genes (50). These processes enable the creation of genes with new functions or control features. The new genes can supplement or replace the genes they were created from. Such gene forming events are probably ongoing throughout genome evolution. However, their intermediate stages are rarely observed. Gene fission should create substantial new gene control and protein coding features, and these are unlikely to be optimal at first. The scarcity of observed gene fission intermediate stages is probably due to their removal or quick replacement by better-suited gene versions.
In this article we present a large number of probable intermediate stages in a particular type of gene fission event. The gene organizations we found most likely resulted from genes that were broken up in two in their intein protein-splicing elements by homing endonucleases. The new resulting genes are present in the same locus, and could still use some of the gene control elements of the parent gene, together with control elements brought in by the invading homing endonuclease. The final protein product of the fractured genes would be identical to that of the parent gene since the split-intein parts can ligate their host flanks. This is a remarkable scenario, where one disrupted gene locus produces two transcripts translated into two protein precursors, that are finally joined to create the exact product of the parent gene (and of its intein-less gene predecessor). The key for enabling this is the flexibility of inteins to carry out protein splicing reactions either in cis as one molecule, or in trans when present as two separate intein halves. A possible alternative explanation is translational bypassing where internal RNA regions between two translated stretches are untranslated by the ribosome (51). Nevertheless, known untranslated regions are tens of bases long while the intervening regions we observed are hundreds of bases long.
Our findings show that gene fracturing by intein-targeted homing endonuclease invasions is a highly modular process. The fractured genes final product, split-inteins, and invading homing endonucleases, can each be of different kinds. This shows the generality of the process and hints at its ubiquity, at least in some species or ecosystems. Perhaps this process has not been observed in previously determined sequence data because of insufficient sequencing coverage and/or sequencing that was limited to organisms from a very small fraction of environments (ecosystems). Supporting the first possibility is our identification of the process in the massive amount of Global Ocean Sampling data. The second possibility is supported by the origin of this data from certain aquatic environments, and its relative high enrichment in viral sequences (52).
The tremendous diversity of the Global Ocean Sampling data is coupled to its extreme fragmentary coverage. Most of our observed fractured gene loci are thus missing parts of their genes and other fractured gene loci that we did not observe are likely to exist, perhaps with different settings such as other host genes and homing endonuclease types. It is also possible that some of the other hundreds of partial intein-loci we observed in the data (not shown) are parts of fractured genes. These possibilities could be resolved with re-sequencing and re-sampling of Global Ocean Sampling and other environmental sites.
Homing endonuclease invasions also fracture genes without inteins. Edgell and co-workers reported an invasion of an HNH-type putative homing endonuclease into the nrdA gene of Aeh1 bacteriophage, and showed that the two formed NrdA parts can create a functional ribonucleotide reductase (22,23). Karam, Petrov and coworkers reported a case where the a GIY-YIG type putative homing endonuclease is in between two parts of a fractured gp43 B-type DNA polymerase of Aeromonas bacteriophage 25 (20,21). We also found an example where a putative homing endonuclease fractured an intein-less gene. The homing endonucleases in these two examples are of different kinds, HNH, GIY-YIG and Vsr-like types. The latter is a new type, first described here. It is related to the well studied Vsr short-patch DNA repair enzymes (42). We show that Vsr-like putative homing endonucleases are also found in group-I introns. All this further illustrates the duality of several nuclease domains that are present in DNA repair and maintenance enzymes, and in homing or restriction endonucleases. This protein domain modularity was previously observed for GIY-YIG domains in UvrC subunits of (A) BC exonucleases (53) and HNH in certain MutS subunits of DNA mismatch repair enzymes (54).
Protein–protein interaction between the two broken parts of split-inteins was suggested to be tuned by electrostatics of their two longest beta-strands (17). The previously known cyanobacterial naturally split-inteins have opposite charges on these strands (the N-intein is negative and the C-intein is positive). The new split-intein pairs in this work further support our previous suggestion of split-intein parts protein–protein interaction by complementary local charge distribution. This includes their breaking in the same region, between motifs N4 and C2, and the charge distribution of each split-intein part. Such electrostatic patches may be of use for designing proteins, and to increase the binding affinity between protein fragments.
All split-inteins found in this work, except the terminase, and all previously described natural split-inteins are broken at the same region, where all known homing endonuclease domains are also present (55). Thus, although inteins can be synthetically broken in other regions (56), the homing endonuclease insertion region seems to have some advantage for successful intein breaking and homing endonuclease domain recruitment. The one exception we found shows another break point.
The new Vsr-like type of putative homing endonuclease is another example of the remarkable modularity of nuclease domains. Many such domains are present in nucleases, restriction endonucleases and homing endonucleases (24). Most inteins have a homing endonuclease domain of different types (i.e. LAGLI-DADG and HNH). The two types of likely homing endonuclease that we observed in the fractured gene loci (GIY-YIG and Vsr-like) are not known to be present in inteins. Nevertheless, our findings suggest that LAGLI-DADG and HNH could fracture genes, and a way that inteins could acquire such, and other, endonuclease domains (see below).
Gene-breaking model and evolution of split-inteins
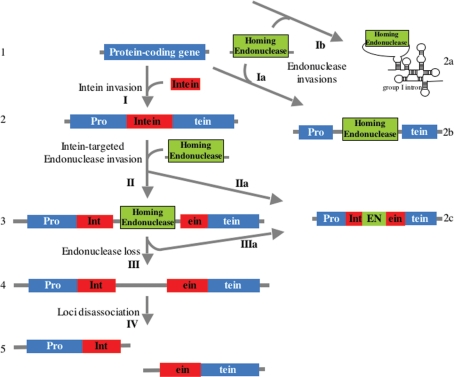
The genomic arrangement described in this work provides missing intermediates in the evolution of split-inteins from typical contiguous inteins. Gene breaking may have evolved stepwise as shown in Figure 5. Ancestral inteins are assumed to have transposed into, or otherwise be present in, protein coding regions, without requiring an intein endonuclease domain (step I) (57,58). Invasion of a free standing endonuclease, another kind of mobile element, into an intein containing gene (step II) would result in fractured loci such as those observed in this work (Figure 1). Note that the endonuclease integration point is within the intein-coding region, which is distinct from the intein insertion point. It is also possible that the endonuclease would have been integrated in frame inside the intein to create the endonuclease domains observed within most inteins [steps IIa or IIIa; e.g. (57)], either directly in the invasion step or subsequently by loss of the free-standing endonuclease flanks. The entire free standing endonuclease gene could also be lost leading to a gene arrangement which we also observed (step III; Figure 1 locus gp41-7; note that the hairpin structure found upstream of the putative homing endonucleases in all loci is present in that locus upstream of the C-terminal gene part). Loss of the free-standing endonuclease could eventually lead to genomic uncoupling of the N- and C-host gene parts (step IV). This situation, with each gene part in a different locus was previously described in the broken cyanobacterial DnaE and N. equitans DNA polymerases. Free standing endonucleases could also invade intein-less genes (step Ia), as observed by Edgell and co-workers and by us, and invade group I introns (step Ib), as previously observed for various known homing endonucleases (24) and as observed here for the new Vsr-like putative homing endonucleases.
Figure 5.
A model for gene breaking by intein-targeted homing endonuclease invasions. Observed genomic organizations are numbered in Arabic numbers, and putative evolutionary processes are marked with arrows and Roman numbers. Protein coding genes can be invaded by an in-frame intein domain (I). Invasion of the intein domain gene by an endonuclease gene can fracture the gene in two (II), or insert the endonuclease gene in frame within the intein (IIa). Invasion of an endonuclease gene into an intein-less gene can fracture that gene (Ia). Such an invasion can also insert the endonuclease gene into a group I intron (Ib). Loss of the endonuclease gene from the split-intein locus (III) may be followed by dislocation of the two fractured gene parts into two separate loci (IV). Loss of the flanks of an endonuclease gene in a fractured gene locus could insert the endonuclease coding region in-frame into the intein, recreating a contiguous gene (IIIa).
In our model the invading endonuclease gene brings along not only its own gene control regions but also ones required by the fractured genes. The translation stop codon and transcription stop signals of the upstream gene are created by the 5′ end of the invading element. The initiator Met codon of the downstream gene is created by the 3′ end of the invading element. Supporting this last suggestion is the overlap between the coding sequences of endonucleases and their downstream gene (Figure 4B). This coupling would reduce the amount of acceptable mutations in the overlap region. Preserving a protein coding start site at its 3′ end is likely to enhance the invasion success of the endonuclease element. The transcription signals for the downstream gene could be present in the 3′ end of the invading element or be in the endonuclease gene promoter. In this last scheme a multiple-genes transcript would include the coding region of the endonuclease, followed by that of the downstream gene. The hairpins immediately upstream of endonuclease coding regions (Figure 4A) are similar to the one found to attenuate the translation of the Aeh1 nrdA endonuclease (22), and thus they could control the translation of their downstream genes.
In contrast to endonuclease fracturing of protein-coding genes, homing endonuclease invasions into group I introns often utilize the conserved sequences of the introns as contiguous regions of their distal coding regions [Supplementary Figure S3A; (59–62)]. This difference is likely due to the distinct outcomes of the invasion events. In gene breaking events several control elements and coding regions must be inserted for the invasion to succeed. When a homing endonuclease invasion results in an additional domain in a self-processing intron or intein, there is no additional transcript of the host gene. The endonuclease ORFs will just require an upstream element to control their translation. Moreover, in non-split inteins and in some introns the endonuclease is transcribed and translated with the host (60). Thus, such successful invasions do not require sequences beyond the coding regions of the invading endonuclease.
The split-inteins loci that we identified are likely to be of phage and virus (viral) origins. This is supported by sequences closely related to viral ones (e.g. the gp41 helicases), by gene organizations that appear in phages (e.g. the DNA ligase and exonuclease locus), and by occurrence of phage gene types (e.g. DNA packaging large subunit terminase). This could partially explain the occurrences of split-inteins in DNA synthesis and repair enzymes. Phage proteins are extremely diverse and usually not well conserved (63). This impedes the propagation and survival of intein elements that invade and survive best in highly conserved integration points (57,64,65). Phage DNA synthesis and repair enzymes are better conserved and offer ‘safe havens’ and more constant targets for inteins. Another explanation is the one proposed by Liu (58). Activity of mature DNA synthesis and repair enzymes can counteract the detrimental effects of gene homing by the inteins that were expressed with these enzymes.
Broken genes are more likely to arise in phages and viruses relative to cellular organisms due to several reasons. First, viral broken genes can function sub-optimally or even marginally at first, since the phages and viruses replicate within other cells that can complement suboptimal or missing protein functions. Second, the very large progeny of viruses and phages allow accelerated evolution that eases the formation of novel gene combinations and features. Third, viruses and phages readily exchange gene and gene parts with their hosts and with each other (66). More specific to our case is the ubiquity of homing endonucleases in some phages [e.g. (67,68)]. All this, and enrichment of GOS dataset in viral sequences, lead us to propose that most if not all of homing-endonculease split-intein loci we found originated in phages and viruses. Some are probably still found in these type of organisms and other might have been transferred to cellular species.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: The Leo and Julia Forchheimer Center for Molecular Genetics.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Gil Amitai, Edouard Jurkevitch, Oded Beja, Itai Sharon and the manuscript anonymous reviewers for useful comments and suggestions. SP holds a Hermann and Lilly Schilling Foundation chair. This research was partially funded by the Israel Science Foundation grant number 626/03
REFERENCES
- 1.Haugen P, Simon DM, Bhattacharya D. The natural history of group I introns. Trends Genet. 2005;21:111–119. doi: 10.1016/j.tig.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Perler FB, Davis EO, Dean GE, Gimble FS, Jack WE, Neff N, Noren CJ, Thorner J, Belfort M. Protein splicing elements: inteins and exteins – a definition of terms and recommended nomenclature. Nucleic Acids Res. 1994;22:1125–1127. doi: 10.1093/nar/22.7.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dassa B, Haviv H, Amitai G, Pietrokovski S. Protein splicing and auto-cleavage of bacterial intein-like domains lacking a C′-flanking nucleophilic residue. J. Biol. Chem. 2004;279:32001–32007. doi: 10.1074/jbc.M404562200. [DOI] [PubMed] [Google Scholar]
- 4.Dassa B, Yanai I, Pietrokovski S. New type of polyubiquitin-like genes with intein-like autoprocessing domains. Trends Genet. 2004;20:538–542. doi: 10.1016/j.tig.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 6.Sanjanwala B, Ganesan AT. DNA polymerase III gene of Bacillus subtilis. Proc. Natl Acad. Sci. USA. 1989;86:4421–4424. doi: 10.1073/pnas.86.12.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonner DM, DeMoss JA, Mills SE. The evolution of enzymes. In: Bryson V, Vogel HJ, editors. Evolving Genes and Proteins. Orlando, FL: Academic Press; 1965. pp. 305–318. [Google Scholar]
- 8.Waters E, Hohn MJ, Ahel I, Graham DE, Adams MD, Barnstead M, Beeson KY, Bibbs L, Bolanos R, Keller M, et al. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl Acad. Sci. USA. 2003;100:12984–12988. doi: 10.1073/pnas.1735403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelman Z, Pietrokovski S, Hurwitz J. Isolation and characterization of a split B-type DNA polymerase from the archaeon Methanobacterium thermoautotrophicum deltaH. J. Biol. Chem. 1999;274:28751–28761. doi: 10.1074/jbc.274.40.28751. [DOI] [PubMed] [Google Scholar]
- 10.Bonen L. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion. 2008;8:26–34. doi: 10.1016/j.mito.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Knoop V. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr. Genet. 2004;46:123–139. doi: 10.1007/s00294-004-0522-8. [DOI] [PubMed] [Google Scholar]
- 12.Caspi J, Amitai G, Belenkiy O, Pietrokovski S. Distribution of split DnaE inteins in cyanobacteria. Mol. Microbiol. 2003;50:1569–1577. doi: 10.1046/j.1365-2958.2003.03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H, Hu Z, Liu XQ. Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc. Natl Acad. Sci. USA. 1998;95:9226–9231. doi: 10.1073/pnas.95.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun W, Yang J, Liu XQ. Synthetic two-piece and three-piece split inteins for protein trans-splicing. J. Biol. Chem. 2004;279:35281–35286. doi: 10.1074/jbc.M405491200. [DOI] [PubMed] [Google Scholar]
- 15.Southworth MW, Adam E, Panne D, Byer R, Kautz R, Perler FB. Control of protein splicing by intein fragment reassembly. EMBO J. 1998;17:918–926. doi: 10.1093/emboj/17.4.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills KV, Lew BM, Jiang S, Paulus H. Protein splicing in trans by purified N- and C-terminal fragments of the Mycobacterium tuberculosis RecA intein. Proc. Natl Acad. Sci. USA. 1998;95:3543–3548. doi: 10.1073/pnas.95.7.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dassa B, Amitai G, Caspi J, Schueler-Furman O, Pietrokovski S. Trans protein splicing of cyanobacterial split inteins in endogenous and exogenous combinations. Biochemistry. 2007;46:322–330. doi: 10.1021/bi0611762. [DOI] [PubMed] [Google Scholar]
- 18.Gorbalenya AE. Non-canonical inteins. Nucleic Acids Res. 1998;26:1741–1748. doi: 10.1093/nar/26.7.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi JJ, Nam KH, Min B, Kim SJ, Soll D, Kwon ST. Protein trans-splicing and characterization of a split family B-type DNA polymerase from the hyperthermophilic archaeal parasite Nanoarchaeum equitans. J. Mol. Biol. 2006;356:1093–1106. doi: 10.1016/j.jmb.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 20.Petrov VM, Karam JD. Diversity of structure and function of DNA polymerase (gp43) of T4-related bacteriophages. Biochemistry. 2004;69:1213–1218. doi: 10.1007/s10541-005-0066-7. [DOI] [PubMed] [Google Scholar]
- 21.Petrov VM, Nolan JM, Bertrand C, Levy D, Desplats C, Krisch HM, Karam JD. Plasticity of the gene functions for DNA replication in the T4-like phages. J. Mol. Biol. 2006;361:46–68. doi: 10.1016/j.jmb.2006.05.071. [DOI] [PubMed] [Google Scholar]
- 22.Gibb EA, Edgell DR. Multiple controls regulate the expression of mobE, an HNH homing endonuclease gene embedded within a ribonucleotide reductase gene of phage Aeh1. J. Bacteriol. 2007;189:4648–4661. doi: 10.1128/JB.00321-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedrich NC, Torrents E, Gibb EA, Sahlin M, Sjoberg BM, Edgell DR. Insertion of a homing endonuclease creates a genes-in-pieces ribonucleotide reductase that retains function. Proc. Natl Acad. Sci. USA. 2007;104:6176–6181. doi: 10.1073/pnas.0609915104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 25.Pietrokovski S. Modular organization of inteins and C-terminal autocatalytic domains. Protein Sci. 1998;7:64–71. doi: 10.1002/pro.5560070106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henikoff S, Henikoff JG, Alford WJ, Pietrokovski S. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene. 1995;163:17–26. doi: 10.1016/0378-1119(95)00486-p. [DOI] [PubMed] [Google Scholar]
- 29.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karlin S, Altschul SF. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl Acad. Sci. USA. 1990;87:2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgenstern B. DIALIGN: multiple DNA and protein sequence alignment at BiBiServ. Nucleic Acids Res. 2004;32:W33–W36. doi: 10.1093/nar/gkh373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doron-Faigenboim A, Stern A, Mayrose I, Bacharach E, Pupko T. Selecton: a server for detecting evolutionary forces at a single amino-acid site. Bioinformatics. 2005;21:2101–2103. doi: 10.1093/bioinformatics/bti259. [DOI] [PubMed] [Google Scholar]
- 34.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 35.Chivian D, Baker D. Homology modeling using parametric alignment ensemble generation with consensus and energy-based model selection. Nucleic Acids Res. 2006;34:e112. doi: 10.1093/nar/gkl480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sammond DW, Eletr ZM, Purbeck C, Kimple RJ, Siderovski DP, Kuhlman B. Structure-based protocol for identifying mutations that enhance protein-protein binding affinities. J. Mol. Biol. 2007;371:1392–1404. doi: 10.1016/j.jmb.2007.05.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das R, Baker D. Macromolecular modeling with rosetta. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 39.Sitbon E, Pietrokovski S. Occurrence of protein structure elements in conserved sequence regions. BMC Struct. Biol. 2007;7:3. doi: 10.1186/1472-6807-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Roey P, Waddling CA, Fox KM, Belfort M, Derbyshire V. Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate. EMBO J. 2001;20:3631–3637. doi: 10.1093/emboj/20.14.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen BW, Landthaler M, Shub DA, Stoddard BL. DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J. Mol. Biol. 2004;342:43–56. doi: 10.1016/j.jmb.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 42.Tsutakawa SE, Morikawa K. The structural basis of damaged DNA recognition and endonucleolytic cleavage for very short patch repair endonuclease. Nucleic Acids Res. 2001;29:3775–3783. doi: 10.1093/nar/29.18.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsutakawa SE, Jingami H, Morikawa K. Recognition of a TG mismatch: the crystal structure of very short patch repair endonuclease in complex with a DNA duplex. Cell. 1999;99:615–623. doi: 10.1016/s0092-8674(00)81550-0. [DOI] [PubMed] [Google Scholar]
- 44.Zhao L, Bonocora RP, Shub DA, Stoddard BL. The restriction fold turns to the dark side: a bacterial homing endonuclease with a PD-(D/E)-XK motif. EMBO J. 2007;26:2432–2442. doi: 10.1038/sj.emboj.7601672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ko M, Choi H, Park C. Group I self-splicing intron in the recA gene of Bacillus anthracis. J. Bacteriol. 2002;184:3917–3922. doi: 10.1128/JB.184.14.3917-3922.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hardies SC, Thomas JA, Serwer P. Comparative genomics of Bacillus thuringiensis phage 0305phi8-36: defining patterns of descent in a novel ancient phage lineage. Virol. J. 2007;4:97. doi: 10.1186/1743-422X-4-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macdonald PM, Kutter E, Mosig G. Regulation of a bacteriophage T4 late gene, soc, which maps in an early region. Genetics. 1984;106:17–27. doi: 10.1093/genetics/106.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McPheeters DS, Christensen A, Young ET, Stormo G, Gold L. Translational regulation of expression of the bacteriophage T4 lysozyme gene. Nucleic Acids Res. 1986;14:5813–5826. doi: 10.1093/nar/14.14.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gott JM, Zeeh A, Bell-Pedersen D, Ehrenman K, Belfort M, Shub DA. Genes within genes: independent expression of phage T4 intron open reading frames and the genes in which they reside. Genes Dev. 1988;2:1791–1799. doi: 10.1101/gad.2.12b.1791. [DOI] [PubMed] [Google Scholar]
- 50.Henikoff S, Greene EA, Pietrokovski S, Bork P, Attwood TK, Hood L. Gene families: the taxonomy of protein paralogs and chimeras. Science. 1997;278:609–614. doi: 10.1126/science.278.5338.609. [DOI] [PubMed] [Google Scholar]
- 51.Herr AJ, Atkins JF, Gesteland RF. Coupling of open reading frames by translational bypassing. Annu. Rev. Biochem. 2000;69:343–372. doi: 10.1146/annurev.biochem.69.1.343. [DOI] [PubMed] [Google Scholar]
- 52.Williamson SJ, Rusch DB, Yooseph S, Halpern AL, Heidelberg KB, Glass JI, Andrews-Pfannkoch C, Fadrosh D, Miller CS, Sutton G, et al. The Sorcerer II Global Ocean Sampling Expedition: metagenomic characterization of viruses within aquatic microbial samples. PLoS ONE. 2008;3:e1456. doi: 10.1371/journal.pone.0001456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kowalski JC, Belfort M, Stapleton MA, Holpert M, Dansereau JT, Pietrokovski S, Baxter SM, Derbyshire V. Configuration of the catalytic GIY-YIG domain of intron endonuclease I-TevI: coincidence of computational and molecular findings. Nucleic Acids Res. 1999;27:2115–2125. doi: 10.1093/nar/27.10.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malik HS, Henikoff S. Dual recognition-incision enzymes might be involved in mismatch repair and meiosis. Trends Biochem. Sci. 2000;25:414–418. doi: 10.1016/s0968-0004(00)01623-6. [DOI] [PubMed] [Google Scholar]
- 55.Paulus H. Protein splicing and related forms of protein autoprocessing. Annu. Rev. Biochem. 2000;69:447–496. doi: 10.1146/annurev.biochem.69.1.447. [DOI] [PubMed] [Google Scholar]
- 56.Ando T, Tsukiji S, Tanaka T, Nagamune T. Construction of a small-molecule-integrated semisynthetic split intein for in vivo protein ligation. Chem. Commun. 2007;47:4995–4997. doi: 10.1039/b712843f. [DOI] [PubMed] [Google Scholar]
- 57.Pietrokovski S. Intein spread and extinction in evolution. Trends Genet. 2001;17:465–472. doi: 10.1016/s0168-9525(01)02365-4. [DOI] [PubMed] [Google Scholar]
- 58.Liu XQ. Protein-splicing intein: Genetic mobility, origin, and evolution. Annu. Rev. Genet. 2000;34:61–76. doi: 10.1146/annurev.genet.34.1.61. [DOI] [PubMed] [Google Scholar]
- 59.Haugen P, Bhattacharya D, Palmer JD, Turner S, Lewis LA, Pryer KM. Cyanobacterial ribosomal RNA genes with multiple, endonuclease-encoding group I introns. BMC Evol. Biol. 2007;7:159. doi: 10.1186/1471-2148-7-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saldanha R, Mohr G, Belfort M, Lambowitz AM. Group I and group II introns. Faseb. J. 1993;7:15–24. doi: 10.1096/fasebj.7.1.8422962. [DOI] [PubMed] [Google Scholar]
- 61.Bonocora RP, Shub DA. A novel group I intron-encoded endonuclease specific for the anticodon region of tRNA(fMet) genes. Mol. Microbiol. 2001;39:1299–1306. doi: 10.1111/j.1365-2958.2001.02318.x. [DOI] [PubMed] [Google Scholar]
- 62.Loizos N, Tillier ER, Belfort M. Evolution of mobile group I introns: recognition of intron sequences by an intron-encoded endonuclease. Proc. Natl Acad. Sci. USA. 1994;91:11983–11987. doi: 10.1073/pnas.91.25.11983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rohwer F, Edwards R. The Phage Proteomic Tree: a genome-based taxonomy for phage. J. Bacteriol. 2002;184:4529–4535. doi: 10.1128/JB.184.16.4529-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ichiyanagi K, Ishino Y, Ariyoshi M, Komori K, Morikawa K. Crystal structure of an archaeal intein-encoded homing endonuclease PI-PfuI. J. Mol. Biol. 2000;300:889–901. doi: 10.1006/jmbi.2000.3873. [DOI] [PubMed] [Google Scholar]
- 65.Duan X, Gimble FS, Quiocho FA. Crystal structure of PI-SceI, a homing endonuclease with protein splicing activity. Cell. 1997;89:555–564. doi: 10.1016/s0092-8674(00)80237-8. [DOI] [PubMed] [Google Scholar]
- 66.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl Acad. Sci. USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bonocora RP, Shub DA. A self-splicing group I intron in DNA polymerase genes of T7-like bacteriophages. J. Bacteriol. 2004;186:8153–8155. doi: 10.1128/JB.186.23.8153-8155.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nolan JM, Petrov V, Bertrand C, Krisch HM, Karam JD. Genetic diversity among five T4-like bacteriophages. Virol. J. 2006;3:30. doi: 10.1186/1743-422X-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.