Abstract
Nucleosome depletion at transcription start sites (TSS) has been documented genome-wide in multiple eukaryotic organisms. However, the mechanisms that mediate this nucleosome depletion and its functional impact on transcription remain largely unknown. We have studied these issues at human MHC class II (MHCII) genes. Activation-induced nucleosome free regions (NFR) encompassing the TSS were observed at all MHCII genes. Nucleosome depletion was exceptionally strong, attaining over 250-fold, at the promoter of the prototypical HLA-DRA gene. The NFR was induced primarily by the transcription factor complex that assembles on the conserved promoter-proximal enhancer situated upstream of the TSS. Functional analyses performed in the context of native chromatin demonstrated that displacing the NFR without altering the sequence of the core promoter induced a shift in the position of the TSS. The NFR thus appears to play a critical role in transcription initiation because it directs correct TSS positioning in vivo. Our results provide support for a novel mechanism in transcription initiation whereby the position of the TSS is controlled by nucleosome eviction rather than by promoter sequence.
INTRODUCTION
The basic building block of eukaryotic chromatin is the nucleosome, which consists of 147 bp of DNA wrapped around an octamer of histones H3, H4, H2A and H2B. Packaging of DNA into nucleosomes creates a restrictive environment that reduces accessibility of DNA to factors mediating chromatin templated processes such as transcription. Dynamic structural rearrangements that render the chromatin permissive for transcription are therefore intimately associated with the regulation of gene expression. In agreement with the restrictive impact of chromatin on transcription, large scale mapping studies examining nucleosome occupancy in the genomes of yeast, chicken, Drosophila melanogaster and human have revealed that many genes in these organisms tend to exhibit a depletion of nucleosomes at their transcription start site (TSS) and core promoter (1–6). Reduced nucleosome occupancy has also been observed at distal enhancers controlled by specific transcription factors (7,8). The average nucleosome depletion revealed by these studies ranged from <1.5-fold to >4-fold depending on the species, genes examined and methods used.
Several mechanisms have been proposed to be responsible for nucleosome depletion at the TSS/promoter, including sequence-dependent structural features of the DNA (9–11), assembly of the preinitiation complex (PIC) containing general transcription factors and Pol II (2,6), the binding of sequence-specific transcription factors (5,12,13), and the recruitment of chromatin modifying and remodeling factors (14,15). However, for the majority of genes, particularly in higher eukaryotic organisms, the critical parameters responsible for nucleosome eviction from the TSS/promoter have not been characterized in vivo. Furthermore, in most cases the functional impact of nucleosome eviction on gene expression has not been defined. It is for instance not known whether nucleosome eviction from the TSS is generally a prerequisite for, or a consequence of, PIC assembly and transcription. We have therefore examined the mechanisms responsible for nucleosome eviction, and the importance of this event for the activation of transcription, in a genetically and biochemically well characterized human system, namely expression of the family of genes encoding major histocompatibility complex class II (MHCII) molecules (16–19).
MHCII molecules are heterodimeric cell-surface glycoproteins that play a pivotal role in the immune system because they present peptides to the antigen receptor (TCR) of CD4+ T cells. The recognition of MHCII-peptide complexes by the TCR guides the development of CD4+ T cells in the thymus and the initiation, regulation and development of T cell-dependent immune responses in the periphery. Humans have six to seven gene coding for the α and β chains of three ‘classical’ MHCII isotypes, HLA-DR, HLA-DP and HLA-DQ. There are four additional genes coding for the α and β chains of two ‘non-classical’ MHCII molecules, HLA-DO and HLA-DM, which are accessory molecules required for loading peptides onto the classical MHCII molecules. All these MHCII genes are clustered together in the MHCII sub-region of the MHC locus on the short arm of chromosome 6. Classical and non-classical MHCII genes are expressed in a tightly co-regulated manner (16–19). Constitutive expression is restricted to specialized cells of the immune system, including thymic epithelial cells, dendritic cells, macrophages and B cells. Other cell types do not express MHCII genes unless they are stimulated with interferon-γ (IFNγ).
The molecular machinery that regulates MHCII expression has been exceptionally well defined thanks to the elucidation of the genetic defects responsible for the Bare Lymphocyte Syndrome (BLS), a hereditary immunodeficiency disease resulting from mutations in genes encoding transcription factors that are essential for MHCII expression (16). One of these factors, the class II transactivator (CIITA), is a transcriptional coactivator that is exquisitely specific for the activation of MHCII genes (20,21). CIITA serves as the master regulator of MHCII genes and is expressed in a cell-type-specific and IFNγ-inducible manner dictating the constitutive and inducible pattern of MHCII expression (16–19). CIITA is recruited to MHCII promoters by means of protein–protein interactions with a multi-protein ‘enhanceosome’ complex that assembles on a characteristic enhancer known as the S-Y module, which consists of four conserved sequence elements called the S, X, X2 and Y boxes (16–19). This enhanceosome complex consists of the X-box-binding-factor Regulatory Factor X (RFX), the X2-box-binding-factor cAMP Responsive Element-binding protein (CREB), and the Y-box-binding-factor Nuclear Factor Y (NF-Y) (Figure 1A).
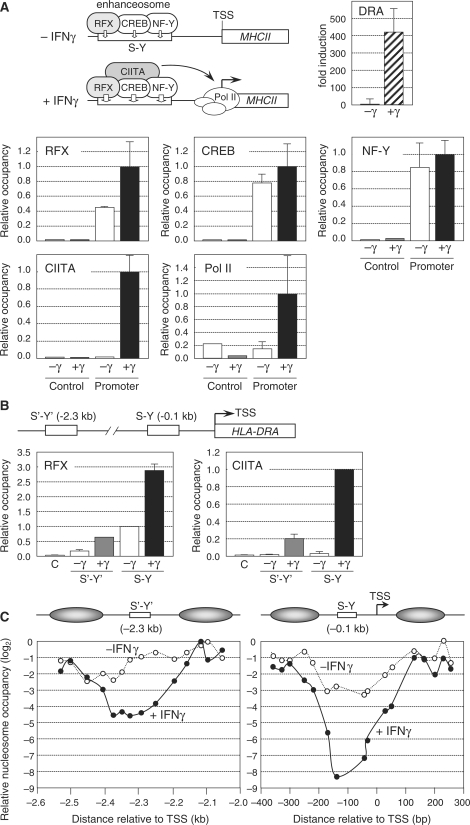
Figure 1.
Nucleosome eviction at the HLA-DRA gene in IFNγ-induced cells. (A) HLA-DRA mRNA expression was measured by qRT–PCR in uninduced Me67.8 cells (−γ) and in Me67.8 cells induced for 24 h with IFNγ (+γ). Occupation of the HLA-DRA promoter by RFX, CREB, NF-Y, CIITA and Pol II was analyzed by ChIP in uninduced Me67.8 cells (−γ) and in Me67.8 cells induced for 24 h with IFNγ (+γ). An upstream region (control) was used to verify specificity. Occupancy is expressed relative to the value observed at the promoter in IFNγ-induced cells. The means and standard deviations derived from three independent experiments are shown. A schematic summary of MHCII-promoter occupation in uninduced and IFNγ-induced cells is represented. (B) The HLA-DRA gene contains both a promoter-proximal S-Y enhancer (situated at −0.1 kb) and a distal S′-Y′ enhancer (situated at −2.3 kb). Occupation of the S′-Y′ and S-Y enhancers by RFX and CIITA was analyzed by ChIP in uninduced Me67.8 cells (−γ) and in Me67.8 cells induced for 24 h with IFNγ (+γ). An upstream region (C) was used to control for specificity. Occupancy by RFX is expressed relative to the value observed at the S-Y enhancer in uninduced cells. Occupancy by CIITA is expressed relative to the value observed at the S-Y enhancer in induced cells. The means and standard deviations derived from three independent experiments are shown. (C) Nucleosome occupancy in the S′-Y′ (left) and S-Y/TSS (right) regions was measured by qMNAse-ChIP in uninduced Me67.8 cells (−IFNγ) and in Me67.8 cells induced for 24 h with IFNγ (+IFNγ). Results were generated using overlapping amplicons of which the centres are represented by dots. Results were normalized with respect to MNAse-treated genomic DNA, are expressed on a log2 scale relative to the position at which maximum occupation was observed, and represent the mean of three independent experiments. Schematic representations of the NFRs and their flanking nucleosomes are provided. Distance in base pair relative to the TSS is indicated. Primer pairs are indicated in Supplementary Table 1.
RFX is a central component of the enhanceosome complex. It is composed of three subunits, RFX5, RFXAP and RFXANK (16). As for CIITA, mutations in each of the three RFX subunits give rise to the BLS disease (16,22–25). RFX is essential for MHCII expression because it nucleates assembly of the enhanceosome complex by binding cooperatively with CREB and NF-Y (16–19,26,27). Thus, in the absence of a functional RFX complex, enhanceosome assembly, CIITA recruitment and MHCII gene activation are abolished.
Mechanisms implicated in MHCII gene activation by the enhanceosome and CIITA include the recruitment of chromatin-remodeling factors, histone-modifying complexes, PIC components and transcription-elongation factors (28–32). However, the role of nucleosome eviction at MHCII promoters has not been addressed. We demonstrate here that all MHCII promoters exhibit an exceptionally strong depletion of nucleosomes at their TSS and adjacent S-Y modules in vivo. Nucleosome eviction is induced mainly by assembly of the MHCII enhanceosome, although it is further consolidated by the recruitment of CIITA. Finally, we show that formation of the nucleosome free region (NFR) at MHCII promoters appears to be a critical event because it determines correct positioning of the TSS in vivo. The latter finding suggests that eviction of nucleosomes from the promoter can replace the requirement for core-promoter sequences in directing initiation at the correct TSS.
MATERIALS AND METHODS
Cells
The Me67.8 melanoma cell line, the wild-type B cell line Raji, the CIITA-deficient B cell mutant RJ2.2.5, the RFXANK-deficient B cell mutant BLS1 and the RFX5-deficient B cell mutant SJO have been described previously (20,22,24), and were grown in RPMI+ Glutamax medium (Invitrogen) supplemented with 10% fetal calf serum and antibiotics. Me67.8 cells were induced with 200 U/ml of human IFNγ (Invitrogen).
qRT–PCR
Quantifications of HLA-DRA mRNA expression and nascent transcript abundance were performed by qRT–PCR as described (33,34).
Mononucleosome preparations
Cells were treated with 1% formaldehyde for 8 min at room temperature. Crosslinking was stopped by addition of 0.2 M glycine. Cells were lysed for 10 min at 4°C in cold lysis buffer (50 mM HEPES–KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, 0.3 M sucrose, 2 μg/ml aprotinin and leupeptin). Nuclei were pelleted through lysis buffer containing 0.9% sucrose and washed in TE (10 mM Tris–HCl pH 8.0, 1 mM EDTA) containing 200 mM NaCl, 0.5 mM EGTA, 2 μg/ml aprotinin and leupeptin. Purified nuclei from 3 × 106 cells were resuspended in Micrococcal Nuclease (MNAse) Buffer (MNB: 10 mM Tris–HCl pH 7.4, 15 mM NaCl, 60 mM KCl, 3 mM CaCl2) and digested with 6–10 U MNAse (Fermentas) for 40 min at 37°C. Digestions were stopped by adding 10 mM EDTA and 10 mM EGTA. MNAse treated nuclei were then used for chromatin immunoprecipitation (ChIP) experiments (see below). The extent of MNAse digestion was verified by extraction of the DNA and analysis by agarose gel electrophoresis (Supplementary Figure 2).
ChIP
MNase-treated nuclei from 3 × 106 cells were incubated in 300 μl of MNB containing 20 μl CL-4B beads (Amersham) diluted two times in Dilution Incubation Buffer (DIB: 0.2 M HEPES pH 7.9, 2 M NaCl, 0.02 M EDTA, 200 μg/ml herring sperm DNA, 1 mg/ml BSA, 10 mM EDTA, 10 mM EGTA) containing 1% Triton X-100, 0.1% Na-DOC and 1× Complete Protease Inhibitor Cocktail (Roche) for 30 min at room temperature with rotation. Samples were cleared by centrifugation for 10 min at maximum speed in a microfuge and supernatants were incubated O/N at 4°C with 700 μl of MNB containing 10 mM EDTA, 10 mM EGTA and antibodies. Histone antibodies were obtained from Abcam (core H3, Ab1791; H3K4Me3, Ab8580) or Upstate (H3Ac, 06-599; H4Ac, 06-866). BRG1 antibodies were obtained from Upstate (07-478) or Santacruz (sc-10768). H2A.Z antibodies were from Abcam (Ab4174). Samples were incubated with 15 μl of protein A FF sepharose beads (GE Healthcare) diluted two times in DIB for 90 min at room temperature and washed twice with IP Buffer 1 (0.2 M HEPES pH 7.9, 0.2 M NaCl, 0.02 M EDTA, 0.1% Na-DOC, 0.1% SDS, 10 mM EDTA, 10 mM EGTA), twice with IP Buffer 2 (0.5 M NaCl, 0.2 M HEPES pH 7.9, 0.02 M EDTA, 0.1% Na-DOC, 0.1% SDS, 10 mM EDTA, 10 mM EGTA) twice with IP Buffer 3 (20 mM Tris–HCl pH 8.0, 0.25 M LiCl, 0.5% Na-DOC, 0.5% NP-40, 10 mM EDTA, 10 mM EGTA), and once with IP Buffer 4 (10 mM Tris–HCl pH 8.0, 0.1% NP-40, 10 mM EDTA, 10 mM EGTA). Immunoprecipitated chromatin fragments were eluted for 10 min at 65°C with 100 mM Tris–HCl pH 8.0, 1% SDS, 10 mM EDTA, 10 mM EGTA, and crosslinks were reversed O/N at 65°C. DNA was extracted, precipitated and resupended in TE.
Classical ChIP experiments using sonicated chromatin and antibodies against CIITA, RFX5, NF-Y (Diagenode, pAb-TFNYB), CREB (Santacruz, sc-186, sc-58), RNA Pol II (Abcam, Ab817), core H3, H4Ac, H3Ac, H3K4Me3, BRG1 and H2A.Z were performed as described above except that samples were washed with IP Buffers 1 and 2 containing 1% Triton instead of SDS, with IP Buffer 3 containing 0.25% Na-DOC instead of NP-40, and without adding 10 mM EDTA and 10 mM EGTA to the buffers.
Quantitative PCR (qPCR)
qPCR was performed using the iCycler iQ Real-time PCR Detection system (Biorad) and a SYBRGreen-based kit for quantitative PCR (iQ Supermix Biorad). Primers are listed in Supplementary Table 1. Amplification specificity was controlled by gel electophoresis and melting curve analysis. Results were quantified using a standard curve generated with serial dilutions of input DNA. All PCR amplifications were performed in triplicate.
ChIP–chip experiments
ChIP–chip experiments were performed with a custom NimbleGen array of our own design. The latter carries all unique sequences from the entire extended human MHC (7.7 Mb on chromosome 6; genomic coordinates 26.1 Mb to 33.8 Mb on hg17) as well as a number of other selected control regions (total of 0.9 Mb). These genomic regions were covered at high density with overlapping Tm-matched oligonucleotides (∼50 bp long) spaced such that their 5′ ends are situated ∼10 bp apart. Genomic input DNA and H3-ChIP samples prepared from MNAse-treated chromatin were used directly for probe preparation without amplification. DNA labeling (4 μg for each sample), hybridization and array scanning were performed by NimbleGen Systems.
Nucleosome depletion profiles were generated by collecting the log2-ratios for each Nimblegen oligonucleotide probe into a grid of 30 bp windows relative to the positions of the TSS. The log2-ratios within the same window were averaged and the ‘fold change’ was computed as 1 divided by the exponentiation of the averaged log-ratios.
To generate a list of non-MHCII genes that are present on the Nimblegen array and highly expressed in Raji B cells, mappings of Affymetrix U133-PLUS-2 probe sets were intersected with the genomic regions covered by the Nimblegen array using Galaxy (35). From the resulting 444 genes represented by probes on both the Affymetrix and Nimblegen arrays, the 50 genes expressed most strongly in Raji cells (Supplementary Table 2) were defined on the basis of their average expression level in microarray datasets for Raji cells (GEO GDS596) (36).
Luciferase constructs and reporter gene assays
The HLA-DRA promoter—Firefly Luciferase cassette from the pDRAprox vector (34) was cloned into the pRRLSIN lentiviral vector (http://tronolab.epfl.ch) by replacing the EcoRV-SalI fragment containing PGK-GFP. The BglII site in the HLA-DRA promoter was used to insert random sequences between the HLA-DRA S-Y module and the TSS. Inserted sequences corresponded to NCBI Build 36.1 coordinates: chr6:26500854-26500892 (DRA+40), chr6:26500854-26500934 (DRA+80), chr6:26500854-26501002 (DRA+150a) and chr6:26492247-26492395 (DRA+150b). Virus production and transductions were performed as described (http://tronolab.epfl.ch). Transient transfections were performed using lipofectamin 2000 (Invitrogen). Luciferase activity was measured with a Dual luciferase reporter gene system (Promega). Results were normalized with respect to both total protein (quantified by Advance protein assay reagent from Cytosqueleton) and the number of vector integrations in the transduced cells (quantified by qPCR).
RNAse protection assays (RPA)
A fragment spanning the TSS of the DRA luciferase constructs was subcloned into a Bluescript vector (Stratagene). Probe preparation, hybridization, RNAse digestion and analysis by gel electrophoresis were done as previously described (20). Signals were detected using a Cyclone (Packard) phosphorimager and quantified using the OptiQuant software (Packard).
RESULTS
Nucleosome eviction at the HLA-DRA gene in IFNγ-induced cells
To analyze chromatin structure at MHCII promoters we first studied IFNγ-induced MHCII gene activation in the Me67.8 melanoma cell line. These cells were chosen because they exhibit robust and reproducible IFNγ-induced MHCII gene activation (Figure 1A), and have been used extensively in previous studies on MHCII expression. Quantitative chromatin immunoprecipitation (qChIP) experiments demonstrated that the enhanceosome complex containing RFX, CREB and NF-Y assembles at the promoter-proximal S-Y enhancer of the prototypical MHCII gene HLA-DRA in non-induced Me67.8 cells (Figure 1A). As documented previously (33,37,38), treatment of these cells with IFNγ induced the expression of CIITA, which was recruited to the HLA-DRA S-Y enhancer by binding to the enhanceosome complex (Figure 1A). The recruitment of CIITA stabilized binding of the enhanceosome and induced the recruitment of Pol II (Figure 1A and B). Stabilization of the enhanceosome by CIITA was most evident when binding of RFX was assessed (Figure 1A and B). However, this stabilization concerns the entire enhanceosome complex, since in vivo footprint experiments have demonstrated that occupation of the entire S-Y module is increased by treatment of cells with IFNγ (39,40).
The HLA-DRA gene contains a second S-Y enhancer (denoted S′-Y′) situated ∼2.3-kb upstream of the TSS (Figure 1B) (34). The pattern of binding of the enhanceosome (as assessed by binding of RFX) and CIITA to this distal S′-Y′ enhancer in uninduced and induced Me67.8 cells was very similar to that observed for the proximal S-Y enhancer (Figure 1B and data not shown) (34,41). However, the level of enhanceosome and CIITA occupation was lower at the upstream S′-Y′ enhancer (Figure 1B).
IFNγ-induced activation of the HLA-DRA gene has been shown to lead to an increase in histone H4 acetylation (H4Ac) over a large >5-kb upstream domain encompassing both the S′-Y′ and S-Y modules (33). We observed that this increase in H4Ac was consistently lower at the S-Y and S′-Y′ modules (Supplementary Figure 1A). To determine whether this lower level of H4Ac might reflect a reduction in nucleosome density we performed qChIP experiments with antibodies directed against unmodified histone H3. Nucleosome density in IFNγ-induced Me67.8 cells was indeed found to be significantly lower at the S-Y and S′-Y′ modules (Supplementary Figure 1A). We also analyzed micrococcal nuclease (MNAse) sensitivity of the chromatin at different positions within the HLA-DRA upstream region. Sensitivity to digestion with MNAse was greatest at the S-Y and S′-Y′ modules in IFNγ-treated Me67.8 cells (Supplementary Figure 1A). Taken together, these findings suggested that nucleosomes are depleted at the HLA-DRA S-Y and S′-Y′ modules in IFNγ induced Me67.8 cells. This was consistent with earlier studies showing that IFNγ-induced HLA-DRA gene expression is associated with the appearance of two DNAse I hypersensitive sites at positions flanking the promoter-proximal S-Y module (42).
To map nucleosome-depleted regions more precisely, we performed qChIP experiments with H3 antibodies and MNAse-treated chromatin (qMNAse-ChIP). Digestion conditions were chosen such that conversion to mono-nucleosomes was almost complete (Supplementary Figure 2). Results were quantified by real time PCR using a series of overlapping amplicons. Two NFRs of ∼200–300 bp in size were revealed in IFNγ-induced Me67.8 cells (Figure 1C). One was centred on the distal S′-Y′ module. The second spanned the proximal S-Y module and TSS. Relative to flanking regions, nucleosome density was reduced up to 20-fold (log2 < −4) at the S′-Y′ enhancer and more than 250-fold (log2 < −8) at the S-Y/TSS region. The difference in the extent of nucleosome depletion (∼10-fold) between the S′-Y′ and S-Y/TSS regions (Figure 1C) correlated well with the relative level of occupation of these two regions by RFX and CIITA (Figure 1B).
Several lines of evidence indicated that the strong reduction in H3-qMNAse-ChIP signals observed within the S′-Y′ and S-Y/TSS regions reflects nucleosome depletion rather than an artefact resulting from preferential sensitivity of the DNA sequences within these regions to digestion with MNAse. First, a strong reduction in nucleosome density at these regions was also revealed by classical H3-ChIP experiments using sonicated chromatin instead of MNAse treated chromatin (Supplementary Figure 1A). Second, the chromatin structure in these regions was highly sensitive even to mild digestion with MNAse (Supplementary Figure 1B). Third, these regions were not degraded preferentially upon digestion of naked DNA with MNAse (data not shown). Finally, these regions were not digested preferentially when the H3-qMNAse-ChIP experiments were performed with chromatin from RFX-deficient cells, in which nucleosome eviction is abrogated because enhanceosome assembly and CIITA recruitment are abolished (see below and Figure 2C).
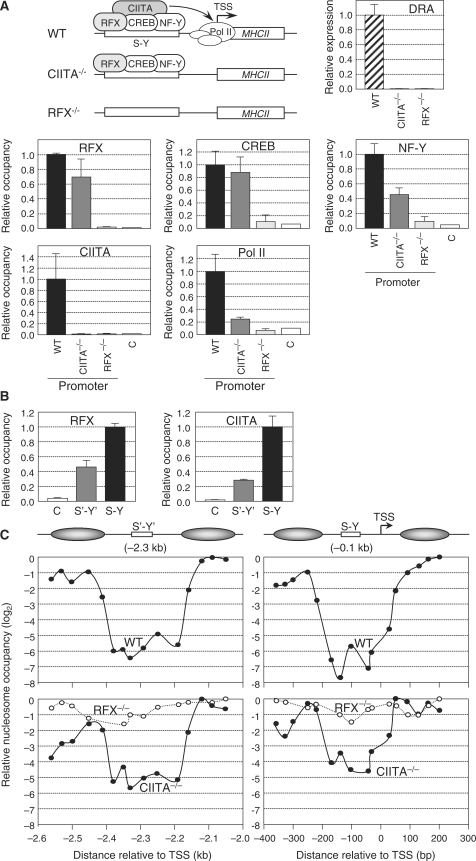
Figure 2.
Nucleosome eviction at the HLA-DRA gene in B cells. (A) HLA-DRA mRNA expression was measured by qRT–PCR in WT, CIITA-deficient and RFX-deficient B cell lines. Occupation of the HLA-DRA promoter by RFX, CREB, NF-Y, CIITA and Pol II was analyzed by ChIP in wild-type Raji B cells (WT), CIITA-deficient cells (CIITA−/−) and RFX-deficient cells (RFX−/−). An upstream region (C) was used to determine background binding. Occupancy is expressed relative to the value observed at the promoter in WT cells. The means and standard deviations derived from three independent experiments are shown. A schematic summary of MHCII-promoter occupation in WT, CIITA-deficient and RFX-deficient cells is shown. (B) Occupation of the HLA-DRA S′-Y′ and S-Y enhancers by RFX and CIITA was analyzed by ChIP in Raji B cells. An upstream region (C) was used to determine background binding. Occupancy is expressed relative to the value observed at the S-Y enhancer. The means and standard deviations derived from three independent experiments are shown. (C) Nucleosome occupancy in the S′-Y′ (left) and S-Y/TSS (right) regions was measured by qMNAse-ChIP in wild-type Raji B cells (WT, top panels), CIITA-deficient cells (CIITA−/−, bottom panels), and RFX-deficient cells (RFX−/−, bottom panels). Results were generated using overlapping amplicons of which the centres are represented by dots. Results were normalized with respect to MNAse treated genomic DNA, are expressed on a log2 scale relative to the position at which maximum occupation was observed, and represent the mean of three independent experiments. Schematic representations of the NFRs and their flanking nucleosomes are provided. Distance in base pair relative to the TSS is indicated. Primer pairs are indicated in Supplementary Table 1.
Nucleosome depletion was also evident, albeit less strongly, at the S′-Y′ enhancer (2–4-fold reduction) and S-Y/TSS region (8-fold reduction) in uninduced Me67.8 cells (Figure 1C), despite the absence of CIITA expression and recruitment (Figure 1B). The pre-existence of these NFRs in uninduced cells suggested that enhanceosome assembly was sufficient to induce nucleosome eviction, although maximal nucleosome depletion required IFNγ induced CIITA recruitment and/or enhanceosome stabilization by CIITA.
Nucleosome eviction at the HLA-DRA gene in B cells
To extend our analysis of nucleosome eviction at the HLA-DRA gene to another cell type we turned to B cells, which express MHCII genes in a constitutive manner. qChIP experiments performed with the B cell line Raji confirmed that the HLA-DRA-promoter region is occupied constitutively by Pol II, CIITA, and the three enhanceosome components RFX, CREB and NF-Y (Figure 2A). The distal S′-Y′ enhancer also exhibited constitutive occupation by the enhanceosome (assessed by binding of RFX) and CIITA (Figure 2B) (34). As observed in IFNγ-induced cells, binding of the enhanceosome and CIITA was stronger to the S-Y enhancer than to the S′-Y′ enhancer (Figure 2B) (34).
qChIP studies performed with B cells have shown that the levels of H4Ac at the HLA-DRA gene is lowest at the S′-Y′ and S-Y modules (34). MNAse sensitivity of the chromatin was also enhanced at these regulatory sequences in B cells (Supplementary Figure 3). Finally, B cells exhibited two constitutive DNAse I hypersensitive sites at positions flanking the S-Y module of the HLA-DRA gene (Supplementary Figure 4) (42). Taken together these results indicated that B cells also exhibit nucleosome depletion at the HLA-DRA S′-Y′ and S-Y/TSS regions.
To map the nucleosome-depleted regions in B cells we performed qMNAse-ChIP experiments with H3 antibodies. This revealed the presence of ∼200–300 bp NFRs at the S′-Y′ and S-Y/TSS regions of the HLA-DRA gene (Figure 2C). Relative to flanking sequences, nucleosome density was reduced over 50-fold (log2 < −6) at the S′-Y′ enhancer and more than 100-fold (log2 < −7) at the S-Y/TSS region. The minor difference (∼2-fold) in the extent of nucleosome depletion between the S′-Y′ and S-Y/TSS regions (Figure 2C) correlated well with the relative level of occupation of these two regions by RFX and CIITA (Figure 2B).
Dominant role of the enhanceosome in mediating nucleosome eviction
To distinguish between the roles of enhanceosome binding, CIITA recruitment, PIC assembly and active transcription in promoting nucleosome eviction at the HLA-DRA gene we analyzed nucleosome occupancy at the S′-Y′ and S-Y/TSS regions in CIITA-deficient and RFX-deficient B cells. In the CIITA-deficient cells, enhanceosome binding was normal but the absence of CIITA completely abrogated PIC assembly and transcription (Figure 2A) (34). On the other hand, in RFX-deficient cells the upstream regulatory region remained unoccupied because enhanceosome assembly and CIITA recruitment are strictly dependent on an intact RFX complex (Figure 2A) (34). qMNAse-ChIP experiments demonstrated that strong nucleosome depletion was retained at the S′-Y′ and S-Y/TSS regions in CIITA-deficient cells (Figure 2C) despite the absence of CIITA, PIC assembly and transcription (Figure 2A and B). In contrast, no significant nucleosome depletion was evident at the S′-Y′ and S-Y/TSS regions in RFX-deficient cells (Figure 2C). These results are consistent with the finding that the DNAse I hypersensitive sites flanking the HLA-DRA S-Y module were detected in CIITA-deficient cells but not in RFX-deficient cells (Supplementary Figure 4). Taken together, these results indicated that nucleosome eviction at the HLA-DRA gene in B cells is mediated mainly by assembly of the DNA-bound enhanceosome complex. CIITA recruitment, PIC assembly and/or ongoing transcription do not seem to play a major role. However, these processes may—as observed in IFNγ-induced cells—enhance nucleosome eviction, since nucleosome depletion is slightly stronger (∼2–4-fold) in wild-type B cells than in CIITA-deficient cells (Figure 2C). These results also demonstrated that formation of the NFR spanning the TSS is on its own not sufficient to promote PIC assembly and transcription, because the latter processes do not occur in CIITA-deficient cells (Figure 2A). This is consistent with previous studies demonstrating that PIC assembly and transcription of the HLA-DRA gene are strictly dependent on CIITA.
Independence of nucleosome eviction on chromatin modification and remodeling
It has been reported in other systems that nucleosome eviction from promoters may be dependent on the introduction of histone modifications, such as histone acetylation (43). The activation of MHCII genes is well known to be associated with the acetylation of histones H3 and H4 (H3Ac and H4Ac and the tri-methylation of lysine 4 of H3 (H3K4Me3) (29,33,34,41,44,45). To determine whether these modifications might be required for nucleosome eviction we examined whether their introduction correlates with establishment of the NFR at the promoter of the HLA-DRA gene. Compared to wild-type B cells, the H3Ac, H4Ac and H3K4Me3 modifications were reduced as strongly in CIITA-deficient B cells as in RFX-deficient B cells (Figure 3), despite the fact that nucleosome eviction at the promoter is essentially complete in the former but abolished in the latter (Figure 2C).
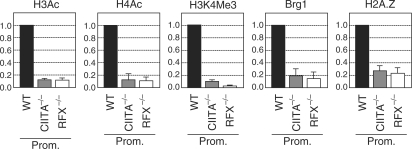
Figure 3.
Nucleosome eviction does not correlate with chromatin modification, BRG1 recruitment or H2A.Z deposition. The levels of H4Ac, H3Ac, H3K4Me3, BRG1 recruitment and H2A.Z deposition at the HLA-DRA promoter were analyzed by qChIP in wild-type Raji B cells (WT), CIITA-deficient B cells (CIITA−/−) and RFX-deficient B cells (RFX−/−). Results were corrected for nucleosome density using antibodies directed against unmodified histone H3, and were normalized relative to a control position at the TBP promoter. Results are expressed relative to the values observed in wild-type cells, and represent the means and standard deviations derived from three independent experiments. Primers used are indicated in Supplementary Table 1.
BRG1—the ATPase subunit of human SWI/SNF chromatin remodeling complexes—required for MHCII gene activation and is recruited to MHCII promoters by interactions with CIITA and RFX (32). We therefore assessed whether BRG1 recruitment might be required for establishment of the NFR at the HLA-DRA promoter. There was no correlation between BRG1 recruitment and establishment of the NFR, since BRG1 association was eliminated to the same extent in CIITA-deficient and RFX-deficient B cells (Figure 3).
NFRs marking yeast promoters are flanked by nucleosomes containing the histone variant H2A.Z (13,46,47). Nucleosomes containing H2A.Z are also enriched at the 5′ end of active genes in Drosophila and humans (2,6,46,48). H2A.Z incorporation has moreover been suggested to destabilize nucleosomes (46). We therefore determined whether H2A.Z deposition might be required for establishment of the NFR at the HLA-DRA promoter. H2A.Z deposition at the promoter was indeed found to be associated with HLA-DRA gene activation in B cells (Figure 3) and IFNγ-induced Me67.8 cells (data not shown). There was however no correlation between H2A.Z deposition and establishment of the NFR, since H2A.Z was lost to the same extent in CIITA-deficient and RFX-deficient B cells (Figure 3).
Taken together, these findings imply that BRG1 recruitment, H2A.Z deposition and H3Ac, H4Ac or H2K4Me3 are not essential for nucleosome eviction at the HLA-DRA promoter in B cells. The same conclusion was arrived at by studying the relationship between these processes in IFNγ-induced Me67.8 cells (data not shown). This suggests that enhanceosome assembly may be sufficient by itself to induce nucleosome eviction.
Nucleosome eviction occurs at all MHCII promoters
All MHCII genes have an S-Y module situated at a very similar distance (50–100 bp) upstream of their TSS. NFRs similar in position and size to the one observed at the HLA-DRA gene would therefore be expected to unmask the TSS at all MHCII genes (Figure 4A). The generation of a NFR encompassing the TSS might thus be a conserved and functionally important role of the promoter-proximal S-Y module. To confirm this we quantified nucleosome occupancy by qMNAse-ChIP at the promoters of several additional MHCII genes (HLA-DPA, HLA-DPB, HLA-DQA, HLA-DQB and HLA-DRB1). Suitable primers could not be designed for the other MHCII genes. At all genes tested, a strong (>20- to 50-fold) degree of nucleosome depletion was evident at their S-Y modules in Raji B cells (Figure 4B).
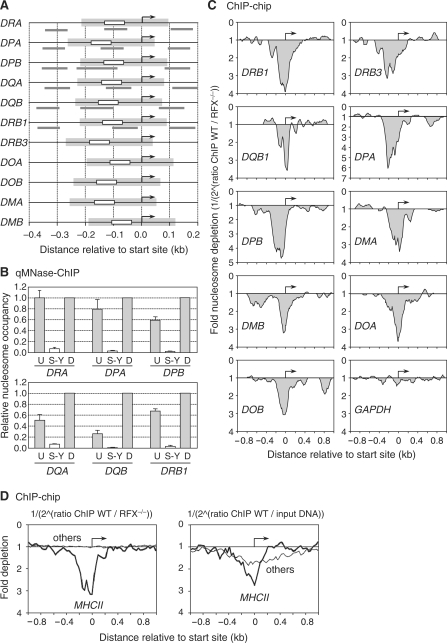
Figure 4.
Nucleosome eviction from the S-Y/TSS region occurs at all MHCII genes. (A) MHCII promoters were aligned relative to their TSSs (arrows). Positions of the S-Y modules (white boxes), predicted NFRs (gray boxes) and amplicons (gray lines) used in panel B are indicated. The sizes and positions of the predicted NFRs were extrapolated from the NFR that was mapped at the DRA promoter. (B) Nucleosome occupancy in the vicinity of the S-Y/TSS regions of the HLA-DRA, HLA-DPA, HLA-DPB, HLA-DQA, HLA-DQB and HLA-DRB1 genes was measured by qMNAse-ChIP in Raji B cells. Primer pairs were situated upstream of (U), within (S-Y) and downstream of (D) the S-Y/TSS NFRs. Results were normalized with respect to MNAse treated genomic DNA, expressed relative to the value obtained at the downstream position, and show the means and standard deviations derived from three independent experiments. Amplicons used are depicted in panel A and indicated in Supplementary Table 1. (C) Nucleosome occupancy at the indicated MHCII genes was analyzed by ChIP–chip using antibodies directed against unmodified histone H3 and MNAse treated chromatin from wild-type Raji B cells (WT) and RFX-deficient B cells (RFX−/−). The results are represented as the fold nucleosome depletion in WT relative to RFX−/− cells. The NFRs and TSSs are indicated by gray shading and arrows, respectively. (D) The average nucleosome density observed at the TSS of MHCII genes (thick lines) was compared with that found at 50 other genes present on the array and expressed most strongly in B cells (thin profiles). Results represent the average fold depletion observed in ChIP samples from wild-type Raji cells (WT) relative to ChIP samples from RFX-deficient (RFX−/−) cells (left) or input DNA (right). The TSSs are indicated by arrows.
We next performed ChIP-chip (ChIP-on-microarray) experiments using a high density microarray carrying the entire human MHC as well as several control loci (see Materials and methods section). ChIP samples were prepared using H3 antibodies and MNAse treated chromatin from Raji B cells. Probes prepared from these H3-ChIP samples were hybridized to the microarrays in conjunction with control probes prepared from either MNAse treated genomic DNA or H3-ChIP samples derived from RFX-deficient B cells. The latter control was used to assess the dependence of nucleosome depletion on assembly of the S-Y-specific regulatory machinery. Strong nucleosome depletion at the S-Y/TSS region was observed at all MHCII genes (Figure 4C and D), as well as at the MHCII-associated invariant chain gene (CD74), which also contains a promoter proximal S-Y module (Supplementary Figure 5). Nucleosome depletion was evident when signals obtained with the Raji H3-ChIP probes were compared with control signals obtained both with the H3-ChIP probes derived from RFX-deficient cells (Figure 4C and D) and with input DNA (Figure 4D). In the former comparison, a NFR spanning the TSS was evident at all MHCII genes but not at non-MHCII genes present on the array, indicating that it is induced by assembly of the MHCII-specific regulatory machinery on the S-Y module. In the latter comparison, nucleosome depletion at the TSS was observed at both MHCII genes and non-MHCII genes exhibiting strong expression in B cells (Supplementary Table 2). However, the depletion was on the average significantly stronger and more focused on the TSS at MHCII genes than at non-MHCII genes (Figure 4D). Analysis of individual genes indicated that the wider and more mitigated pattern of nucleosome depletion observed at non-MHCII genes resulted from considerable inter-gene variability in the extent of nucleosome loss and in the position and size of the NRFs (Supplementary Figure 5).
Nucleosome eviction controls TSS selection
A luciferase reporter gene assay was developed to study the function of nucleosome eviction from the TSS of the HLA-DRA gene. We generated HLA-DRA-promoter driven luciferase constructs in which the distance between the S-Y module and the TSS was progressively increased by the insertion of unrelated sequences (Figure 5A). To study the function of these constructs in the context of chromatin, they were integrated into the genome of Me67.8 cells using lentiviral vectors. qCHIP experiments demonstrated that displacing the S-Y module away from the TSS did not affect the binding of RFX or CIITA (Figure 5B). This suggested that nucleosome eviction at the S-Y module was likely to be reproduced in the reporter gene constructs.
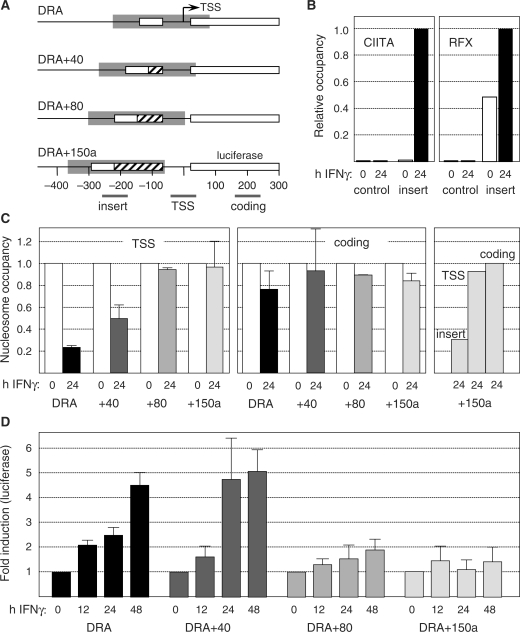
Figure 5.
Nucleosome displacement from the TSS is required for IFNγ-induced activation of the HLA-DRA gene. (A) Schematic representation of the wild type and mutated DRA promoter—luciferase constructs. Each construct was transduced into Me67.8 cells using lentiviral vectors. Maps show relative positions of the S-Y modules (open boxes), TSSs (arrows), inserted sequences (crosshatched boxes, +40, +80 and +150a), predicted NFRs, and amplicons used in (B and C). The amplicons were specific for the insert situated adjacent to the S-Y module, the TSS and the luciferase coding region of the constructs. The sizes and positions of the predicted NFRs were extrapolated from the NFR found at the endogenous DRA promoter. (B) Binding of RFX and CIITA to the S-Y module of the integrated DRA+150a construct (insert) and to a negative control region (control) were analyzed by qChIP in uninduced and IFNγ-induced Me67.8 cells. Results were expressed relative to the level of binding observed at the S-Y module in IFNγ-induced cells. (C) Nucleosome occupancy at the TSS, within the downstream luciferase coding region and near the S-Y module (insert) of the integrated constructs were analyzed by qMNAse-ChIP prior to and after induction for 24 h with IFNγ. Results are expressed relative to uninduced cells and show the mean and standard deviations derived from three independent experiments. The amplicons used are indicated in panel A and in Supplementary Table 1. (D) Luciferase activity was measured in Me67.8 cells that had been transduced with each construct and induced with IFNγ for the indicated times. Results are represented as fold induction relative to uninduced cells, and show the means and standard deviations derived from three independent experiments.
The sizes of the inserted sequences were chosen such that the TSS would be expected to remain within the predicted NFR (DRA+40 construct) or be masked by the first downstream nucleosome (DRA+80 and DRA+150a constructs) (Figure 5A). To verify this prediction, nucleosome occupancy at the TSS of the integrated constructs was measured by qMNAse-ChIP prior to and after induction with IFNγ. The results confirmed that IFNγ-induced eviction of nucleosomes from the TSS was reproduced in the wild-type DRA construct, retained in the DRA+40 mutant, but lost in the DRA+80 and DRA+150a mutants (Figure 5C, left panel). The nucleosome depletion observed in the DRA and DRA+40 constructs was specific to their TSS regions, since no significant reduction in nucleosome occupancy was evident in the downstream luciferase coding region (Figure 5C, middle panel). We next confirmed that nucleosome eviction is actually retained in the vicinity of the S-Y module of the reporter gene constructs. Compared to the TSS and coding regions, a clear reduction in nucleosome occupation was observed near the S-Y module of the DRA+150a construct in IFNγ-induced cells (Figure 5C, right panel). Taken together, these results confirmed that displacement of the S-Y module away from the TSS led to displacement of the TSS outside of the NFR.
We next measured IFNγ-induced luciferase activity in the transduced cells (Figure 5D). Luciferase activity was induced with similar efficiencies in cells transduced with the wild type and DRA+40 constructs. In contrast, the induction of luciferase activity was significantly reduced in cells transduced with the DRA+80 and DRA+150a constructs. This reduction was independent of the inserted sequence as it was observed with two unrelated inserts (Supplementary Figure 6A and B). Similar results were obtained following transduction of the constructs into B cells (Supplementary Figure 6C). These results indicated that displacement of the TSS outside of the NFR correlated with impaired luciferase reporter gene activity.
To determine whether impaired luciferase reporter gene activity was a consequence of reduced transcription initiation we performed RNAse protection assays (RPA) with a probe that overlaps the 5′ end of the luciferase gene and the TSS derived from the HLA-DRA promoter (Figure 6). In cells transduced with the wild-type construct, the 5′ end of most IFNγ-induced transcripts mapped to the expected TSS (Figure 6A–C, transcript c). Only a minor fraction of the transcripts initiated at other positions (Figure 6A–C, transcripts a, b and d). IFNγ-induced initiation at the major TSS was completely abolished in cells carrying the DRA+150a construct (Figure 6A–C). Instead, a reduced but significant level of incorrectly initiated transcription was induced. These transcripts were initiated within the NFR now situated upstream of the normal TSS (Figure 6A–C, transcript a).
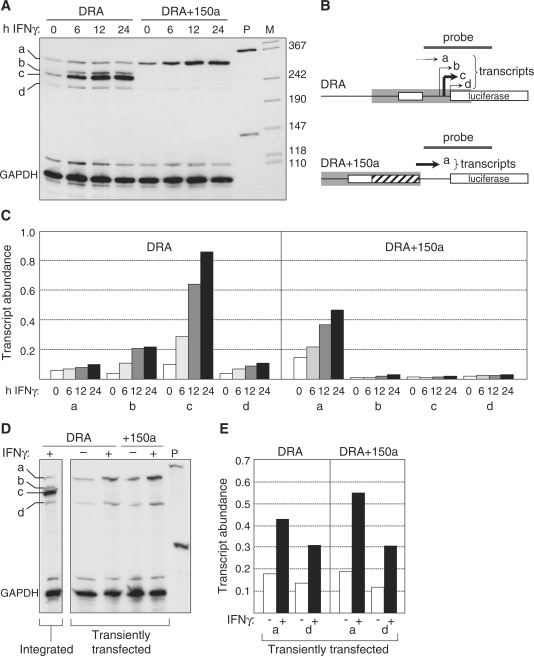
Figure 6.
Nucleosome eviction is essential for faithful TSS selection. (A) Expression of the DRA and DRA+150a constructs was assessed by RPA in stably-transduced Me67.8 cells induced with IFNγ for the indicated times. Transcripts corresponding to major (c) and minor (a, b, d) TSSs are indicated (left). GAPDH mRNA was used as control. P, non-digested probes. M, size marker (in bp). (B) Schematic representation of the results shown in panel A. Positions of the probe and IFNγ-induced transcripts a–d are indicated above the maps of the two constructs. The maps show the S-Y modules (open boxes), the inserted sequence (crosshatched box) in DRA+150a, and the predicted positions of the NFR (gray boxes). (C) Quantification of the a, b, c and d transcripts levels derived from the DRA and DRA+150a constructs in stably-transduced cells induced with IFNγ for 0, 6, 12 or 24 h. Transcript abundance is expressed relative to GAPDH mRNA. (D) Expressions of the DRA and DRA+150a constructs were assessed by RPA in transiently-transfected Me67.8 cells. Left panel shows the band pattern obtained with the integrated DRA construct in stably-transduced Me67.8 cells. Cells were uninduced (−) or induced with IFNγ for 24 h (+). P, non-digested probes. Bands are labeled a–d as in panel A. GAPDH mRNA was used as control. (E) Quantification of transcripts a and d levels derived from the DRA and DRA+150a constructs in transiently-transfected Me67.8 cells. Cells were uninduced (−) or induced with IFNγ for 24 h (+). Transcript abundance is expressed relative to GAPDH mRNA.
Upstream displacement of the major TSS in the DRA+150a construct suggested that TSS selection was determined either by the distance from the S-Y module or the position of the NFR. To distinguish between these two possibilities, we mapped by RPA the 5′ ends of the transcripts that are derived from transiently-transfected DRA and DRA+150a constructs, at which a normal chromatin structure is not expected to be established. The major TSS observed for the stably-integrated wild-type DRA construct was not used at all in transiently transfected cells (Figure 6D). Instead two of the minor TSSs were used predominantly (Figure 6D and E, transcripts a and d). Furthermore, the abnormal pattern of TSS selection observed for the wild-type DRA construct was not altered by insertion of the 150-bp spacer in the DRA+150a construct (Figure 6D and E). Taken together, these results demonstrate that faithful TSS selection requires the establishment of a normal chromatin environment and that selection of the major TSS is determined by the position of the NFR rather than simply by distance from the S-Y module.
DISCUSSION
Our results demonstrate that an exceptionally strong depletion of nucleosomes from the promoter and TSS is a unique feature associated with transcriptional activation of all classical and non-classical MHCII genes. This nucleosome eviction event is induced primarily by assembly of the multiprotein enhanceosome complex on the conserved S-Y regulatory modules of MHCII genes, and it leads to the formation of a 200–300 bp NFR that unmasks the TSS. The generation of this NFR appears to be critical for transcription initiation at MHCII genes in vivo because it controls correct positioning of the TSS.
Genome-wide studies have indicated that nucleosome depletion at the core promoter and TSS is a widespread feature of many genes in all eukaryotic organisms analyzed. In yeast, nucleosome depletion at the core promoter and TSS is typically greater than 3–4-fold (4,5). However, in the chicken, Drosophila and human, nucleosome density at the promoter and TSS is on the average reduced by 50% or less when examining large sets of genes (1,6,49). This low magnitude is likely to result from several parameters, including inter-gene variability in the precise position and size of the NRFs, the fraction of the cells in which a given gene exhibits the NFR and the stability of nucleosome loss over time. In this respect, MHCII genes stand out in that the NFRs found at their promoters are remarkably homogeneous with respect to their size and position, and exhibit a particularly marked nucleosome loss, attaining greater than 250-fold when quantified by qMNAse-ChIP. The latter implies that the NFRs observed at MHCII genes are maintained stably over time and in the majority of cells. At least three mechanisms have been proposed to be implicated in the depletion of nucleosomes from gene regulatory regions. First, DNA sequence itself can be an important parameter favoring nucleosome eviction because nucleosome stability is strongly influenced by specific sequence-dependent structural features of DNA. Intrinsic nucleosome positioning signals embedded in the DNA have been proposed to play a major role in determining nucleosome occupancy in vivo (9). However, other studies have concluded that, although a subset of nucleosomes may be positioned by the underlying DNA sequence, the majority are not (10). We found that an intrinsic nucleosome positioning code does not seem to play a dominant role in inducing nucleosome depletion at MHCII promoters, since no significant nucleosome depletion was evident in RFX-deficient cells, where the MHCII-specific regulatory machinery can not assemble. We can however not exclude the possibility that intrinsic destabilizing sequences situated within MHCII promoters may facilitate nucleosome displacement induced by binding of the MHCII-specific regulatory machinery. A second parameter that has been implicated in nucleosome eviction from promoters is PIC assembly. The potential importance of this mechanism is supported by genome wide studies demonstrating the existence of a correlation between the extent of nucleosome depletion and either Pol II occupancy or transcriptional activity (2,6). This mechanism does again not appear to be critical at MHCII promoters, where nucleosome eviction occurs normally in CIITA-deficient cells despite the absence of Pol II recruitment and transcription. Finally, specific transcription factor-binding sites, have been found to correlate with nucleosome eviction in vivo (5,12,13), suggesting that certain transcription factor complexes may gain access to DNA by excluding nucleosomes. The latter is consistent with in vitro studies illustrating that nucleosomes can be destabilized or excluded by cooperative binding of transcription factors (50–52). This third mechanism is critical at MHCII promoters, where nucleosome eviction requires assembly of the MHCII enhanceosome complex.
The nucleosome eviction mechanism documented here differs strikingly from the situation reported for the interferon β (IFNβ) gene (53). Assembly of the IFNβ enhanceosome occurs within a pre-existing NFR that is flanked by two nucleosomes positioned by their underlying DNA sequences. The nucleosome situated downstream of the NFR masks the promoter and is induced to slide further downstream by the bend introduced in the DNA by binding of TFIID. Nucleosome mobilization at the IFNβ promoter is thus triggered by partial PIC assembly rather than directly by binding of the enhanceosome complex. At MHCII genes on the other hand, nucleosome eviction is independent of PIC assembly and is instead induced by binding of the enhanceosome complex.
Assembly of the MHCII enhanceosome complex requires cooperative binding between RFX, CREB and NF-Y. It remains to be established how these three factors contribute, respectively, to nucleosome eviction. Several lines of evidence suggest that NF-Y may play a critical role in this process. NF-Y consists of three subunits, NF-YA, NF-YB and NF-YC (27). NF-YB and NF-YC contain histone-fold domains exhibiting striking homology to H2B and H2A, respectively (27). NF-YB-NF-YC dimers interact with DNA in a manner analogous to H2A-H2B dimers (54), suggesting that NF-Y might compete with nucleosomes for access to DNA. Furthermore, binding of NF-Y induces a ∼60–80° bend in the DNA (55,56), a structural deformation that could contribute to nucleosome destabilization. Finally, NF-Y can bind in vitro to the mouse MHCII Eα promoter in the context of nucleosomal DNA irrespectively of the position of its target site relative to the nucleosome (57). Taken together, these features of NF-Y suggest that it might play a key role in displacing nucleosomes from MHCII promoters. Interestingly, an NF-Y-binding site (CCAAT box) is found in numerous genes at an upstream position (–60 to –100 bp) very similar to that observed in MHCII genes (58). It is consequently tempting to speculate that nucleosome eviction from the TSS may be a widespread function of NF-Y.
The functional importance of nucleosome eviction from promoters has been addressed for only very few genes in vivo. The most well documented example is activation of the yeast PHO5 promoter. At the PHO5 gene, activator-induced disassembly of nucleosomes at the promoter is required for transcriptional activation (59–61). However, the precise activation step regulated by nucleosome depletion has not been defined in this system. We show here that nucleosome eviction from MHCII promoters is critical for determining the position of the TSS. Our results demonstrate that the major TSS of the HLA-DRA gene is only used in the context of chromatin. Furthermore, this major TSS is no longer used when it is displaced downstream of the NFR, and is instead replaced by a cryptic upstream TSS situated within the NFR. The position of the NFR is thus more critical for TSS selection than the underlying DNA sequence itself. This suggests that the NFR induced at MHCII promoters actually replaces the requirement for core-promoter sequences by restricting access to DNA to a precisely delimited region. These findings provide an explanation for the previously puzzling observation that MHCII genes do not have typical core promoters defined by a TATA box, an initiator element and/or other motifs implicated in PIC assembly, despite the fact that they have a precisely positioned TSS (62–64). This had led to the speculation that MHCII genes might rely on an alternative mechanism for proper positioning of the TSS (64). Our results indeed provide direct evidence for an alternative mechanism in which selection of the major TSS is guided by the generation of a strong NFR rather than simply by the sequence of the core promoter.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation [3100A0-105895]; the Geneva Cancer League; the Swiss Multiple Sclerosis Society and the National Center of Competence in Research on Neural Plasticity and Repair (NCCR-NEURO). Funding for open access charge: Swiss National Science Foundation [3100A0-105895].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Michel Strubin, and to all members of the laboratory, for constructive discussions. We thank P. Salmon (Geneva, Switzerland) for providing lentiviral vectors.
REFERENCES
- 1.Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- 2.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmid CD, Bucher P. ChIP-Seq data reveal nucleosome architecture of human promoters. Cell. 2007;131:831–832. doi: 10.1016/j.cell.2007.11.017. author reply 832–833. [DOI] [PubMed] [Google Scholar]
- 4.Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL. Global nucleosome occupancy in yeast. Genome Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao H, Kim A, Song SH, Dean A. Enhancer blocking by chicken beta-globin 5'-HS4: role of enhancer strength and insulator nucleosome depletion. J. Biol. Chem. 2006;281:30573–30580. doi: 10.1074/jbc.M606803200. [DOI] [PubMed] [Google Scholar]
- 8.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 9.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peckham HE, Thurman RE, Fu Y, Stamatoyannopoulos JA, Noble WS, Struhl K, Weng Z. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 12.Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 13.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 15.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 16.Reith W, Mach B. The bare lymphocyte syndrome and the regulation of mhc expression. Annu. Rev. Immunol. 2001;19:331–373. doi: 10.1146/annurev.immunol.19.1.331. [DOI] [PubMed] [Google Scholar]
- 17.Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109(Suppl.):S21–S33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 18.Reith W, LeibundGut-Landmann S, Waldburger JM. Regulation of MHC class II gene expression by the class II transactivator. Nat. Rev. Immunol. 2005;5:793–806. doi: 10.1038/nri1708. [DOI] [PubMed] [Google Scholar]
- 19.Boss JM, Jensen PE. Transcriptional regulation of the MHC class II antigen presentation pathway. Curr. Opin. Immunol. 2003;15:105–111. doi: 10.1016/s0952-7915(02)00015-8. [DOI] [PubMed] [Google Scholar]
- 20.Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency. Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 21.Krawczyk M, Seguin-Estevez Q, Leimgruber E, Sperisen P, Schmid C, Bucher P, Reith W. Identification of CIITA regulated genetic module dedicated for antigen presentation. PLoS Genet. 2008;4:e1000058. doi: 10.1371/journal.pgen.1000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steimle V, Durand B, Barras E, Zufferey M, Hadam MR, Mach B, Reith W. A novel DNA binding regulatory factor is mutated in primary MHC class II deficiency (Bare Lymphocyte Syndrome) Genes Dev. 1995;9:1021–1032. doi: 10.1101/gad.9.9.1021. [DOI] [PubMed] [Google Scholar]
- 23.Durand B, Sperisen P, Emery P, Barras E, Zufferey M, Mach B, Reith W. RFXAP, a novel subunit of the RFX DNA binding complex is mutated in MHC class II deficiency. EMBO J. 1997;16:1045–1055. doi: 10.1093/emboj/16.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masternak K, Barras E, Zufferey M, Conrad B, Corthals G, Aebersold R, Sanchez JC, Hochstrasser DF, Mach B, Reith W. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat. Genet. 1998;20:273–277. doi: 10.1038/3081. [DOI] [PubMed] [Google Scholar]
- 25.Nagarajan UM, Louis-Plence P, DeSandro A, Nilsen R, Bushey A, Boss JM. RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, an MHC class II immunodeficiency. Immunity. 1999;10:153–162. doi: 10.1016/s1074-7613(00)80016-3. [DOI] [PubMed] [Google Scholar]
- 26.Moreno CS, Beresford GW, Louis-Plence P, Morris AC, Boss JM. CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity. 1999;10:143–151. doi: 10.1016/s1074-7613(00)80015-1. [DOI] [PubMed] [Google Scholar]
- 27.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 28.Mahanta SK, Scholl T, Yang FC, Strominger JL. Transactivation by CIITA, the type II bare lymphocyte syndrome- associated factor, requires participation of multiple regions of the TATA box binding protein. Proc. Natl Acad. Sci. USA. 1997;94:6324–6329. doi: 10.1073/pnas.94.12.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spilianakis C, Kretsovali A, Agalioti T, Makatounakis T, Thanos D, Papamatheakis J. CIITA regulates transcription onset viaSer5-phosphorylation of RNA Pol II. EMBO J. 2003;22:5125–5136. doi: 10.1093/emboj/cdg496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanazawa S, Okamoto T, Peterlin BM. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity. 2000;12:61–70. doi: 10.1016/s1074-7613(00)80159-4. [DOI] [PubMed] [Google Scholar]
- 31.Mudhasani R, Fontes JD. The class II transactivator requires brahma-related gene 1 to activate transcription of major histocompatibility complex class II genes. Mol. Cell Biol. 2002;22:5019–5026. doi: 10.1128/MCB.22.14.5019-5026.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mudhasani R, Fontes JD. Multiple interactions between BRG1 and MHC class II promoter binding proteins. Mol. Immunol. 2005;42:673–682. doi: 10.1016/j.molimm.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 33.Rybtsova N, Leimgruber E, Seguin-Estevez Q, Dunand-Sauthier I, Krawczyk M, Reith W. Transcription-coupled deposition of histone modifications during MHC class II gene activation. Nucleic Acids Res. 2007;35:3431–3441. doi: 10.1093/nar/gkm214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masternak K, Peyraud N, Krawczyk M, Barras E, Reith W. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat. Immunol. 2003;4:132–137. doi: 10.1038/ni883. [DOI] [PubMed] [Google Scholar]
- 35.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muhlethaler-Mottet A, Di Berardino W, Otten LA, Mach B. Activation of the MHC class II transactivator CIITA by interferon-γ requires cooperative interaction between Stat1 and USF-1. Immunity. 1998;8:157–166. doi: 10.1016/s1074-7613(00)80468-9. [DOI] [PubMed] [Google Scholar]
- 38.Krawczyk M, Leimgruber E, Seguin-Estevez Q, Dunand-Sauthier I, Barras E, Reith W. Expression of RAB4B, a protein governing endocytic recycling, is co-regulated with MHC class II genes. Nucleic Acids Res. 2007;35:595–605. doi: 10.1093/nar/gkl980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villard J, Muhlethaler-Mottet A, Bontron S, Mach B, Reith W. CIITA-induced occupation of MHC class II promoters is independent of the cooperative stabilization of the promoter-bound multi-protein complexes. Int. Immunol. 1999;11:461–469. doi: 10.1093/intimm/11.3.461. [DOI] [PubMed] [Google Scholar]
- 40.Westerheide SD, Louis-Plence P, Ping D, He XF, Boss JM. HLA-DMA and HLA-DMB gene expression functions through the conserved S-X- Y region. J. Immunol. 1997;158:4812–4821. [PubMed] [Google Scholar]
- 41.Krawczyk M, Peyraud N, Rybtsova N, Masternak K, Bucher P, Barras E, Reith W. Long distance control of MHC class II expression by multiple distal enhancers regulated by regulatory factor X complex and CIITA. J. Immunol. 2004;173:6200–6210. doi: 10.4049/jimmunol.173.10.6200. [DOI] [PubMed] [Google Scholar]
- 42.Gönczy P, Reith W, Barras E, Lisowska-Grospierre B, Griscelli C, Hadam MR, Mach B. Inherited Immunodeficiency with a defect in a Major Histocompatibility Complex class II promoter-binding protein differs in the chromatin structure of the HLA-DRA gene. Mol. Cell Biol. 1989;9:296–302. doi: 10.1128/mcb.9.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma N, Nyborg JK. The coactivators CBP/p300 and the histone chaperone NAP1 promote transcription-independent nucleosome eviction at the HTLV-1 promoter. Proc. Natl Acad. Sci. USA. 2008;105:7959–7963. doi: 10.1073/pnas.0800534105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beresford GW, Boss JM. CIITA coordinates multiple histone acetylation modifications at the HLA- DRA promoter. Nat. Immunol. 2001;2:652–657. doi: 10.1038/89810. [DOI] [PubMed] [Google Scholar]
- 45.Zika E, Ting JP. Epigenetic control of MHC-II: interplay between CIITA and histone-modifying enzymes. Curr. Opin. Immunol. 2005;17:58–64. doi: 10.1016/j.coi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 46.Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16:166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 49.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat. Rev. Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morse RH. Nucleosome disruption by transcription factor binding in yeast. Science. 1993;262:1563–1566. doi: 10.1126/science.8248805. [DOI] [PubMed] [Google Scholar]
- 51.Workman JL, Kingston RE. Nucleosome core displacement in vitro via a metastable transcription factor-nucleosome complex. Science. 1992;258:1780–1784. doi: 10.1126/science.1465613. [DOI] [PubMed] [Google Scholar]
- 52.Owen-Hughes T, Workman JL. Remodeling the chromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J. 1996;15:4702–4712. [PMC free article] [PubMed] [Google Scholar]
- 53.Lomvardas S, Thanos D. Nucleosome sliding via TBP DNA binding in vivo. Cell. 2001;106:685–696. doi: 10.1016/s0092-8674(01)00490-1. [DOI] [PubMed] [Google Scholar]
- 54.Romier C, Cocchiarella F, Mantovani R, Moras D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003;278:1336–1345. doi: 10.1074/jbc.M209635200. [DOI] [PubMed] [Google Scholar]
- 55.Ronchi A, Bellorini M, Mongelli N, Mantovani R. CCAAT-box binding protein NF-Y (CBF, CP1) recognizes the minor groove and distorts DNA. Nucleic Acids Res. 1995;23:4565–4572. doi: 10.1093/nar/23.22.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liberati C, di Silvio A, Ottolenghi S, Mantovani R. NF-Y binding to twin CCAAT boxes: role of Q-rich domains and histone fold helices. J. Mol. Biol. 1999;285:1441–1455. doi: 10.1006/jmbi.1998.2384. [DOI] [PubMed] [Google Scholar]
- 57.Motta MC, Caretti G, Badaracco GF, Mantovani R. Interactions of the CCAAT-binding trimer NF-Y with nucleosomes. J. Biol. Chem. 1999;274:1326–1333. doi: 10.1074/jbc.274.3.1326. [DOI] [PubMed] [Google Scholar]
- 58.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 59.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell. 2004;14:667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 60.Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 61.Straka C, Horz W. A functional role for nucleosomes in the repression of a yeast promoter. EMBO J. 1991;10:361–368. doi: 10.1002/j.1460-2075.1991.tb07957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mantovani R, Tora L, Moncollin V, Egly JM, Benoist C, Mathis D. The major histocompatibility complex (MHC) Ea promoter: sequences and factors at the initiation site. Nucleic Acids Res. 1993;21:4873–4878. doi: 10.1093/nar/21.21.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Viville S, Jongeneel V, Koch W, Mantovani R, Benoist C, Mathis D. The E alpha promoter: a linker-scanning analysis. J. Immunol. 1991;146:3211–3217. [PubMed] [Google Scholar]
- 64.Benoist C, Mathis D. Regulation of major histocompatibility complex class-II genes: X, Y and other letters of the alphabet. Annu. Rev. Immunol. 1990;8:681–715. doi: 10.1146/annurev.iy.08.040190.003341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.