Abstract
Differential regulation of telomerase reverse transcriptase (TERT) contributes to the distinct aging and tumorigenic processes in humans and mice. Here, we report that the hTERT gene was strongly repressed during differentiation of human cells, whereas modest mTERT expression was detected in terminally differentiated and post-mitotic cells. The stringent hTERT repression depended on the native chromatin environment because transiently transfected hTERT promoters were not repressed in differentiated cells. Conversely, the transiently transfected mTERT core promoter was repressed during cell differentiation, suggesting that the repression of mTERT promoter did not require its endogenous chromatin structures. To understand the mechanisms of this differential regulation, we examined chromatin structures of the endogenous TERT loci during cell differentiation. In both human and mouse cells, repression was accompanied by the loss of multiple DNase I hypersensitive sites at the TERT promoters and their upstream regions, revealing positions of potential regulatory elements. Interestingly, the hTERT locus was located within a nuclease-resistant chromatin domain in human cells, whereas a corresponding chromatin domain was not detected for the mTERT locus. Taken together, our study indicated that, unlike the repression of mTERT gene, the condensed native chromatin environment of hTERT locus was central to its silencing during cell differentiation.
INTRODUCTION
Telomeres serve as essential protective caps of linear chromosomal ends in all eukaryotic cells (1). In the absence of telomere maintenance, progressive telomere shortening accompanied by cell division leads to chromosomal instability and proliferative senescence (2). In stem cells and immortal cell lines, telomeric TTAGGG repeats are replenished by telomerase, a ribonucleoprotein reverse transcriptase complex containing a template RNA subunit (TER) and a catalytic protein subunit (TERT). In most cells, the transcription of TERT gene is limiting and correlates with telomerase activity (3,4).
Telomerase is highly regulated during development and cell differentiation (5–7). Telomerase expression is a defining feature of pluripotent stem cells and most somatic stem cells, as it is required for the self-renewal capability of stem cells. Telomerase activity is also detected during embryonic development. In most human adult somatic tissues, however, both telomerase activity and the hTERT protein are either undetectable or expressed at an extremely low level (8–10). Incidentally, differentiated human somatic cells, such as fibroblasts, never undergo spontaneous immortalization under in vitro culture conditions. Given that ectopic expression of hTERT leads to immortalization of many human cells (3,11), it is not surprising that the transcription of hTERT gene is tightly repressed in these cells. In fact, we have previously demonstrated that the hTERT gene was embedded in a condensed chromatin domain in several human cell lines and normal human fibroblasts, providing a mechanism for its tight regulation in human somatic cells (12).
In mice, most somatic tissues express a detectable amount of mTERT mRNA, albeit at lower levels than in embryonic tissues and cancer cells (6,9). Consequently, mouse cells have much longer telomeres and do not undergo telomere-dependent proliferative senescence. This interspecies difference likely contributes to some of the distinct characteristics of aging and cancers in humans and mice, and therefore has important implications in using mice as models of human diseases (13–15). The molecular basis of differential TERT expression in humans and mice is only beginning to be addressed (16).
Terminal differentiation of animal cells has been studied extensively in cell lines of myeloid lineages, such as human HL60, U937 and murine M1 cells (17–19). Repression of hTERT and telomerase expression during differentiation was demonstrated in HL60 and U937 cell lines (19–21). Recently, we have demonstrated that a transgenic hTERT reporter locus was silenced during in vitro differentiation of mouse embryonic stem cells (ESCs), in a similar way as the native hTERT gene in human cells (22). In contrast, less is known about the mTERT regulation, besides the reports showing that mTERT is downregulated during differentiation of mouse ESCs and C2C12 myoblasts (7,23).
Here, the repression of hTERT and mTERT genes in differentiating cells was studied and directly compared in the context of their respective native chromatin environments. The experiments indicated that, unlike repression of the mTERT gene, the native chromatin environment played a key role in the silencing of hTERT gene.
MATERIALS AND METHODS
Cell culture and differentiation
Human HL60 and U937 cells were obtained from American Type Culture Collection and cultured in RPMI1640 containing 10% fetal bovine serum. Differentiation of HL60 and U937 cells was induced by treatment with 2.5% dimethyl sulfoxide (DMSO) for 4 days and 10 ng/ml 12-0-tetradecanoyl-1-phorbol-13-acetate (TPA) for 24 h, respectively. Mouse myeloid cell line M1 and normal human foreskin fibroblasts (NHFs) were grown in DMEM with 10% FBS. Differentiation of M1 cells was induced by incubation with 100 ng/ml recombinant human IL-6 (Chemicon) for 5 days. Growth conditions of mouse ESCs and its differentiated derivatives were described previously (22).
Plasmids and transient transfections
Firefly luciferase reporters pYF3, pYF10 and pBTdel408 contained genomic sequences –208 bp to +1 bp, –1396 bp to +1 bp and –461 bp to –50 bp, respectively, relative to the hTERT translational initiation codon (4,12). pSKmTERT-Fluc contained a 432-bp promoter sequence upstream of the mTERT initiation codon, whereas pTOPO-CRR9-Fluc contained a 265-bp promoter sequence upstream of the mCRR9 initiation codon. pBT-255, containing a 295-bp hTERT promoter, and its mutant derivatives pBT-255-hM12, -hM13 and -hM14, were kindly provided by Dr Horikawa (9). The Firefly luciferase reporters were cotransfected into HL60 cells with pRL-SV40 using Lipofectamine™ LTX and PLUS™ reagent (Invitrogen). Cells were split into two equal portions 6 h after transfection, one half being treated with 2.5% DMSO to induce differentiation and the other half maintained in the proliferative condition. Transfections were performed in triplicates and cells were harvested 2 days after transfection. Dual luciferase assays (Promega) were performed and Firefly luciferase activities were normalized to Renilla luciferase activities.
Quantative RT-PCR analyses
cDNAs were synthesized from 1 µg total RNA with an oligo(dT) primer using the SuperScript First Strand Synthesis system (Invitrogen) (4). PCR reactions were carried out in triplicates for each cDNA sample in the ABI PRISM 7300 System (Applied Biosystems). Primer and probe sequences are listed in Supplementary Table 1.
DNase I sensitivity assays
Nuclei were isolated from proliferating or differentiated cells (4) and treated with 0, 2, 4, 8 and 16 U/ml of DNase I (Promega) at 37°C for 20 min. Genomic DNA was extracted from treated nuclei and 10 µg DNA was digested with restriction enzymes followed by Southern analyses. Full-length and DNase I hypersensitive bands were detected by indirect end labeling using probes labeled with P32-dCTP and quantified by phosphoimaging analysis. General DNase I sensitivities were calculated by the equation S = log(Xd/Xi)/log(Cd/Ci)xT, where X and C were test and control band intensities for the initial (i) or digested (d) samples, respectively, and T represented the size ratio of control fragment to test fragment (24). Probes for human and mouse sequences are listed in Supplementary Tables 2 and 3, respectively.
RESULTS
Repression of TERT genes during cell differentiation
In order to understand the molecular mechanisms of differential TERT expression in human and mouse cells, we analyzed and compared hTERT and mTERT mRNA levels in a set of matched human and mouse cells. The levels of TERT mRNAs were measured by quantitative RT-PCR and normalized to 18S rRNA. The hTERT mRNA level in human newborn foreskin fibroblasts (NHFs) was barely detectable and it was at least three orders of magnitude lower than in human embryonic stem cell line (hESC) H9 and human leukemic HL60 cells (Table 1). The hTERT expression was higher in human embryonic lung fibroblast IMR90 cells than in NHFs, but its level was still about 100-fold lower than that in H9 hESCs. In contrast, significant mTERT expression was detected in mouse embryonic fibroblasts (MEFs) and its level was one-third of that in mouse embryonic stem cell line (mESC) HM1 and 1/6 of that in mouse leukemic M1 cells (Table 1). Because human and mouse sequences might be amplified with different efficiencies, it is difficult to directly compare TERT mRNA levels in human and mouse cells. Embryonic stem cells are pluripotent cells capable of differentiating into all cell types in adult organisms. They are immortal normal cells that express high levels of telomerase and TERT mRNAs. Therefore, assuming that TERT expression was similar in undifferentiated human and mouse ESCs, the hTERT mRNA levels in NHF and IM90 cells were about 1000- and 30-fold lower than the mTERT mRNA level in MEFs, respectively. The higher hTERT expression in IMR90 cells, as compared to NHF cells, was likely due to its embryonic origin (8) rather than in vitro passaging, because NHF cells used in this study were of earlier passages (population doublings or PDLs 10–15) than IMR90 cells (PDLs 25–30). Thus, hTERT expression was significantly lower in human fibroblasts than its counterpart in mouse fibroblasts.
Table 1.
Relative levels of TERT expression in human and mouse cells
| Comparison | Relative ratio of hTERT expressiona | Comparison | Relative ratio of mTERT expressiona |
|---|---|---|---|
| NHF/H9-hESC | 0.0004 ± 0.0001 | MEF/HM-1mESC | 0.34 ± 0.06 |
| NHF/HL60 | 0.0001 ± 0.0001 | MEF/M1 | 0.15 ± 0.03 |
| IMR90/H9-hESC | 0.012 ± 0.001 | ||
| F2-1d/F2-1mESCb | 0.0016 ± 0.0020 | F2-1d/F2-1mESC | 0.17 ± 0.01 |
| F9-1d/F2-9mESCb | 0.0016 ± 0.0021 | F9-1d/F2-9mESC | 0.11 ± 0.03 |
NHF, normal human newborn foreskin fibroblasts. IMR90, normal human embryonic lung fibroblasts. HL60, a human promyelocytic leukemic line. U937, a human monoblast line. M1, a mouse myeloblast cell line. hESC, human embryonic stem cell line H9. mESC, mouse embryonic stem cell line HM-1. F2-1 and F2-9, two independent mESC lines containing a chromosomally integrated BAC reporter. F2-1d and F2-9d, osteogenic cells differentiated from F2-1 and F2-9, respectively.
aThe levels of TERT mRNA and 18S rRNA were determined by real-time RT-PCR. Each TERT mRNA level was normalized to the 18S rRNA level in the same cells.
bA BAC reporter was stably integrated into F2-1 and F2-9 mESCs (22). Within the reporter, a Firefly luciferase and a Renilla luciferase open reading frame were inserted into the hTERT and hCRR9 translational initiation codon, respectively. Because transcription from the hCRR9 promoter did not change significantly during osteogenic differentiation of mESCs, the activity of hTERT promoter was determined as the ratio of Firely to Renilla luciferases.
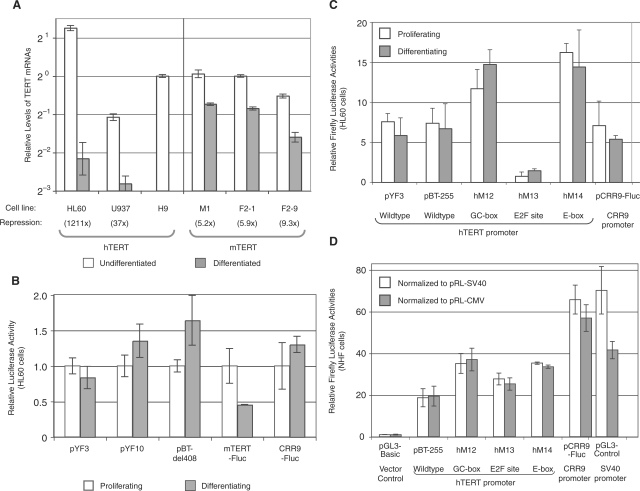
We next examined TERT repression during in vitro differentiation of several human and mouse cell lines. Human acute promyelocytic leukemic HL60 cells were differentiated into granulocyte-like cells by DMSO treatment and monoblast U937 cells were induced to differentiate into monocytic cells by treatment with TPA (Supplementary Figure S1). As shown in Figure 1A, the expression of hTERT gene, after normalized to that of 18S rRNA, was downregulated by more than 1000-fold following induction of HL60 cell differentiation. In U937 cells, the hTERT mRNA level was reduced 37-fold upon induction of monocytic differentiation. A direct comparison of hTERT mRNA in these two cell lines revealed that proliferating U937 cells expressed about 100-fold less hTERT mRNA than proliferating HL60 cells and the hTERT mRNA expression was low in both differentiated HL60 and U937 cells. Therefore, the hTERT transcription was strongly repressed during differentiation of both HL60 and U937 cells.
Figure 1.
Transcriptional regulation of the TERT genes during cell differentiation. (A) TERT mRNA levels in undifferentiated and differentiated human and mouse cells. Total RNAs were harvested from undifferentiated and differentiated cells and cDNAs were synthesized by reverse transcription. The level of each TERT mRNA was determined by real-time PCR analyses and normalized to 18S rRNA. The relative levels of TERT mRNAs were presented as averages of triplicate reactions in a log2 scale, relative to the hTERT and mTERT levels in undifferentiated hESC H9 and mESC F2-1 cells, respectively. The experiments were performed at least twice and one representative experiment is shown here. The numbers in parentheses are fold decreases of TERT mRNA down-regulation during differentiation. (B, C) Activities of transiently transfected TERT promoters in HL60 cells. HL60 cells were transfected with Firefly luciferase reporters containing the hTERT promoter, the mTERT promoter and the mCRR9 promoter, followed by treatment with and without 2.5% DMSO (differentiating and proliferating, respectively) for 2 days. Firefly luciferase activities were normalized to Renilla luciferase activities from a cotranfected pRL-SV40 control plasmid. (D) Activities of transiently transfected hTERT promoter in NHF cells. NHF cells were transfected with plasmids and luciferase activities were determined 2 days after transfection. The Firefly luciferase activities were normalized to Renilla luciferase activities from a cotransfected pRL-SV40 (white bars) or pRL-CMV (gray bars) control plasmid. All RT-PCR and reporter assays were set up in triplicates and average values were presented. Descriptions of cell lines are shown in Table 1 legend.
Like HL60 cells, mouse leukemic myeloblast M1 cells undergo terminal differentiation into monocytes and granulocyte-like cells upon exposure to IL-6. The differentiation of M1 cells is accompanied by induction of several differentiation-specific genes and processes, such as MyD expression, growth arrest, cell attachment and macrophage maturation (18). Proliferating M1 cells expressed a high level of mTERT mRNA, similar to that in undifferentiated mESC clones (F2-1 and F2-9, Figure 1A). However, in contrast to hTERT expression, mTERT expression in M1 cells decreased only 5-fold following a 5-day differentiation process induced by IL-6 (Figure 1A). Because only terminally differentiated M1 cells attached to tissue culture dishes (17), IL-6-treated cells were washed prior to harvest to eliminate any possible partially differentiated non-adherent cells. All differentiated M1 cells exhibited mature monocyte-like morphology as determined by May–Grünwald-Giemsa staining (Supplementary Figure S1), indicating that the moderate mTERT repression was not due to incomplete differentiation.
Downregulation of mTERT expression also occurs during differentiation of pluripotent mouse ESCs (7,22). We examined the mTERT expression in mESC clones F2-1 and F2-9 and their osteogenic cell-derived differentiated F2-1d and F2-9d cells (22). As shown in Figure 1A, F2-1d and F2-9d cells expressed 6- and 9-fold less mTERT mRNA than the parental F2-1 and F2-9 ESCs, respectively. F2-1 and F2-9 ESCs were two independent HM1 clones containing a stably integrated human bacterial artificial chromosome (BAC) reporter, in which a Firefly luciferase ORF and a Renilla luciferase ORF were inserted into the hTERT and hCRR9 translational initiation codons, respectively (22). Because transcription from the hCRR9 promoter did not change during differentiation of mESCs into osteogenic cells (22), expression of Firefly luciferase from the hTERT promoter was normalized to Renilla luciferase expressed from the hCRR9 promoter. Transcription from the hTERT promoter in differentiated F2-1d and F2-9d cells was about 1/600 of that in the parental F2-1 and F2-9 ESCs, as measured by luciferase activities (Table 1) and quantitative RT-PCR (data not shown). Therefore, mTERT expression was downregulated upon in vitro differentiation of mouse cells, but its repression was much less stringent than that of the hTERT promoter.
To determine if proximal promoter sequences accounted for the differential TERT repression during cell differentiation, we performed transient transfection experiments in HL60 cells using Firefly luciferase plasmid reporters containing TERT promoters. Six hours after transfection, HL60 cells were treated with or without DMSO for 2 days to induce differentiation (Figure 1B). Whereas the endogenous hTERT mRNA was completely repressed within 24 h following DMSO treatment (data not shown), plasmids pYF3 and pYF10, containing a 208- bp and a 1.4-kb human genomic sequence upstream of the hTERT initiation codon, respectively, maintained similar levels of Firefly luciferase expression 2 days after induction of differentiation. pBTdel-408, containing a sequence –462-bp to –51-bp relative to the hTERT initiation codon but lacking the downstream E-box, expressed a slightly higher level of Firefly luciferase activity in differentiating HL60 cells than in proliferating cells. On the other hand, pmTERT-Fluc, including a 432-bp mouse genomic sequence upstream of the mTERT initiation codon, was downregulated by about 2-fold. As controls, both the CRR9 promoter and the SV40 promoter were expressed at a similar level in proliferating and differentiating HL60 cells (Figure 1B and data not shown). These results indicated that transiently transfected plasmid reporters containing up to 1.4-kb hTERT promoter sequences were not repressed during HL60 cell differentiation.
Horikawa and colleagues (9) recently identified several consensus elements important for hTERT repression in normal human fibroblasts within the hTERT core promoter, including a nonconserved GC-box, an E2F consensus site and the downstream E-box. We have obtained and tested a subset of hTERT reporter plasmids used in their studies. These reporters included pBT-255, which contained a 295-bp hTERT core promoter, and its mutant derivatives with mutations at the nonconserved GC-box (pBT-255-hM12), the E2F consensus site (pBT-255-hM13) and the conserved downstream E-box (pBT-255-hM14) (9) Similar to pYF3 and pYF10, the hTERT promoter in pBT-255 was similarly active in proliferating and differentiating HL60 cells (Figure 1C). Also similar to the results in NHF cells published by Horikawa and colleagues (9,25), mutations at the nonconserved GC-box (pBT-255-hM12) and the conserved downstream E-box (pBT-255-hM14) led to an ∼2-fold increase in the hTERT promoter activity in HL60 cells. Surprisingly, a mutation at the E2F consensus site (pBT-255-hM13) resulted in an almost complete loss of the promoter activity, suggesting the involvement of E2F family transcription factors in hTERT transcription in HL60 cells. However, all of these hTERT plasmid reporters, pBT-255 and its mutant derivatives, had similar activities in both proliferating and differentiating cells, indicating that none of the aforementioned potential regulatory elements were sufficient to repress the hTERT promoter during HL60 cell differentiation.
To determine whether a transiently transfected hTERT promoter is repressed in fully differentiated cells, in which the endogenous hTERT gene is silenced, the hTERT reporter activities were directly compared with those of constitutively active CRR9 promoter and SV40 promoter in NHF cells. As shown in Figure 1D, Firefly luciferase activity expressed from the wild-type hTERT core promoter was about 20-fold higher than that of the control vector, pGL3-Basic, and 30–50% of those from the CRR9 and SV40 promoters. This result was consistent with our previously published data that the hTERT promoter activity was 30–50% of that of the CMV promoter in telomerase-negative cells (4). In addition, mutation of the non-conserved GC-box resulted in about 2-fold increase of the hTERT promoter activity, whereas mutation of the E2F consensus site did not abolish the promoter activity in NHF cells, supporting the previously published results by Horikawa et al. (9).
In summary, transiently transfected hTERT promoter was unable to undergo repression upon cell differentiation, in contrast to chromosomally integrated BAC DNAs containing the hTERT locus (9,22). These results suggested that, whereas E2F family transcription factors are important for hTERT promoter activity in HL60 cells, the repression of hTERT promoter during cell differentiation was chromatin dependent. Distal elements and/or chromatin environment might be essential for hTERT silencing. In contrast, moderate repression of the mTERT promoter during cell differentiation did not appear to require its native chromatin environment.
Mapping DNase I hypersensitive sites in human cells
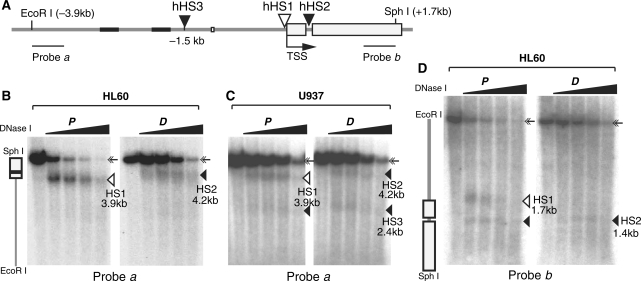
Transcriptional regulation is associated with chromatin structural alterations, such as the appearance of DNase I hypersensitive sites (DHSs). Such sites are discontinuities of nucleosomal arrays that often result from interactions between regulatory proteins and cis-elements. To determine the chromatin structural changes that accompanied hTERT repression, we examined the DNase I sensitivity of the proximal promoter region between EcoRI (–3.9 kb) and SphI (+1.7 kb) sites by Southern blotting and indirect end-labeling. Using a probe specific for the upstream end of this region (probe a), a prominent band (3.9 kb) was detected in proliferating HL60 cells, in addition to the full-length genomic fragment of 5.6 kb (Figure 2B, left panel). This 3.9-kb band was present only in DNase I-treated nuclei and disappeared at higher concentrations of the nuclease, indicative of a DHS band corresponding to the hHS1 that we reported previously in several other telomerase-positive human cell lines (12) (Figure 2A). In differentiated HL60 cells, however, the 3.9-kb band shifted upward to 4.2 kb and had a reduced intensity (Figure 2B, right panel). When the same blot was hybridized with the downstream probe (probe b), two DHS bands (1.4 kb and 1.7 kb) were seen near the promoter region in proliferating HL60 cells (Figure 2D, left panel). The 1.7-kb band, corresponding to the hHS1, disappeared upon cell differentiation and thereby correlated with hTERT transcriptional repression. The 1.4-kb size of the second band indicated the presence of a novel DHS within intron 1 of the hTERT gene (hHS2). This DHS, however, was present in both proliferating and differentiated HL60 cells.
Figure 2.
DHSs at the proximal region of the hTERT promoter in proliferating (P) and differentiated (D) human cells. HL60 and U937 cells were treated with DMSO for 4 days and TPA for 24 h, respectively. Nuclei were isolated from proliferating and differentiated cells and treated with 0, 2, 4, 8 and 16 U DNase I per ml. Genomic DNAs were extracted, digested with EcoRI (nt –3926) and SphI (nt +1682), and subjected to Southern analysis. (A) A diagram of the human genomic region containing the hTERT promoter. Gray lines represent intergenic genomic DNA and introns, with black portions depicting short repetitive DNA sequences. Rectangles represent exons 1 and 2 of the hTERT gene. The position of hHS1, which appears only in proliferating cells, is shown by an open triangle. Closed triangles indicate the positions of DHSs (hHS2 and hHS3) that are present in both proliferating and differentiated cells. The positions of probes a and b are indicated by two horizontal bars. Nucleotide positions are relative to the transcription start site (TSS, +1). (B) Detection of DHSs in HL60 cells with probe a. A vertical diagram of the hTERT proximal region is shown on the left. (C) Analysis of DHSs in U937 cells with probe a. (D) Determination of DHSs in HL60 cells with probe b. The 5.6-kb full-length genomic band is indicated by double arrows. DHS bands are marked by triangles and their sizes are as indicated.
In proliferating U937 cells, the 3.9-kb DHS band was also detected by probe a, and this band similarly shifted upward upon differentiation, indicating the presence of both hHS1 and hHS2 (Figure 2C). In addition, a 2.4-kb weak DHS band was detected in both proliferating and differentiated U937 cells with probe a, suggesting the presence of a DHS (hHS3) positioned at about 1.5 kb upstream of the transcription start site. hHS3 was also found in HeLa cells and several SV40-transformed human fibroblast lines (12), but it was barely detected in HL60 cells. Therefore, multiple DHSs were identified within a 2-kb region covering the hTERT promoter in HL60 and U937 cells. hHS1, which overlapped with the hTERT core promoter and was present specifically in hTERT-expressing proliferating cells, likely corresponded to an assembly site for RNA polymerase II and general transcription factors. hHS2 and hHS3, on the other hand, were found in both proliferating and differentiated cells and were candidates of regulatory elements involved in hTERT regulation in these cells.
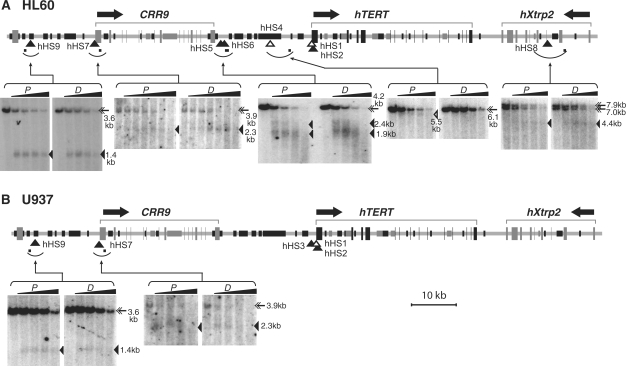
To identify distal regulatory elements, a genomic region (∼125 kb) that included the entire hTERT gene, its upstream and downstream neighboring loci, hCRR9 and hXtrp2, were examined for the presence of DHSs. Besides DHSs proximal to the hTERT promoter, six other DHSs were identified within this genomic region in HL60 cells (Figure 3A). hHS4, a weak DHS, was mapped to 9.5 kb upstream of the hTERT promoter and appeared only in proliferating cells, whereas hHS5 and hHS6, which were located 1.5 kb and 2.0 kb downstream of the 3′ end of hCRR9 gene, respectively, were present in both proliferating and differentiated HL60 cells. hHS7, coincided with the hCRR9 core promoter, was present in both proliferating and differentiated cells, consistent with its constitutive transcription in these cells. Two other constitutive DHSs, hHS8 and hHS9, were located in the middle of hXtrp2 gene and about 12 kb upstream of the hCRR9 promoter, respectively. In U937 cells, there were only two additional constitutive DHSs (hHS7 and hHS9), located at and upstream of the hCRR9 promoter, respectively (Figure 3B). U937 cells generally had fewer and weaker DHSs than HL60 cells, coincident with the lower hTERT expression level in these cells.
Figure 3.
DNase I hypersensitive sites at the hTERT locus and neighboring loci in HL60 cells (A) and U937 cells (B). HL60 and U937 cells were treated with DMSO for 4 days and TPA for 24 h, respectively. Nuclei were isolated and digested with 0, 2, 4, 8 and 16 U/ml DNase I in all panels except for the hHS7 panel of HL60 cells, in which 0, 0.5, 1, 2, 4, 8 and 16 U/ml DNase I were used. Genomic DNAs were extracted and digested with restriction enzymes, followed by Southern blotting and indirect labeling of genomic DNA bands. The top portion of each panel set is a schematic diagram of the genomic region containing the hTERT, hCRR9 and hXtrp2 loci. Horizontal arrows above the diagrams indicate the direction of transcription for each gene. Exons are depicted as tall rectangles and vertical lines. Black portions of intergenic and intronic sequences correspond to short repetitive sequences and dark gray portions represent mini-satellite sequences. Arcs below genomic sequences indicate chromosomal fragments that were examined by Southern analyses and small horizontal bars within the arcs denote the positions of probes for indirect labeling of the hypersensitive bands. In the Southern autoradiographs, proliferating cells (P) are shown on the left panels and differentiated cells (D) are on the right panels for each examined genomic region. Full-length genomic fragments are marked by double arrows. Open triangles point to DHS bands that appeared only in proliferating cells. The positions of these DHSs are also designated by open triangles in the genomic diagrams. Closed triangles indicate the hypersensitive bands and their genomic positions for constitutive DHSs that were present in both proliferating and differentiated cells. The double full-length genomic DNA bands in the hHS8 panel were caused by polymorphic mini-satellite sequences within the restriction fragment in HL60 cells. The sizes of full-length fragments and DHS bands were labeled on the right side of each panel set. Restriction digestions of genomic DNAs are as follows: hHS4, EcoRI; hHS5 and hHS6, EcoRI/SphI; hHS7, SacI; hHS8, SphI; and hHS9, EcoRI/SphI.
Mapping DHSs in mouse cells
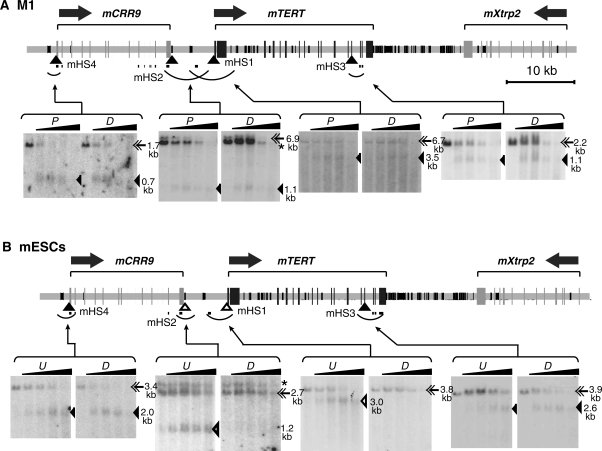
The mouse genomic region containing the mTERT gene is arranged in the same order as the hTERT gene in human cells. However, the intergenic region 5′ to the mTERT gene is only 6.3 kb in length, compared to 22.5 kb of the corresponding human region. In contrast, the 3′ intergenic region is about 15 kb, larger than its human counterpart (7.0 kb). We mapped DHSs within a 90-kb genomic region that included mCRR9, mTERT and mXtrp2 loci in M1 cells. Four DHSs were identified; of which mHS1 and mHS4 coincided with the transcription start sites of mTERT and mCRR9 genes, respectively (Figure 4A). mHS2 was positioned about 6 kb upstream of the mTERT promoter and near the 3′ end of the mCRR9 gene and mHS3 was mapped to exon 13 of the mTERT gene. All these DHS sites were present in both proliferating and differentiated M1 cells, consistent with the results that the mTERT gene was only moderately downregulated and the mCRR9 gene was constitutively expressed during M1 cell differentiation.
Figure 4.
DNase I hypersensitive sites in the genomic region containing the mTERT gene in myeloid leukemia M1 cells (A) and mouse embryonic stem cells F2-9 (B). For M1 cells, differentiation was induced by treating cells with IL-6 for 5 days. F2-9 was a mouse embryonic stem cell line derived from HM-1 cells. F2-9d cells were derived from osteogenic cells differentiated from F2-9ESCs. The performance of experiments and diagrams are similar to those described in Figure 3. A fragment of mCRR9 cDNA was used for the indirect labeling of mHS2. This probe also hybridized to extra bands indicated by asterisks (*) on the autoradiographs. Restriction digestions of genomic DNAs from M1 cells are: mHS1, DraI; mHS2 and mHS3, XbaI; mHS4, HincII. Restriction digestions for F2-9 and F2-9d cells are as follows: mHS1, EcoRI/EcoRV; mHS2, EcoRI/HincII; mHS3, SphI/EcoRI; mHS4, DraI. For M1 cells: P, proliferating cells; D, differentiated cells. For F2-9 cells: U, Undifferentiated F2-9ESCs; D, differentiated F2-9d cells.
In mouse F2-9ESCs, all four DHSs identified in M1 cells were also present, mapping to the same positions on the mouse genome (Figure 4B). The different sizes of these DHS bands on the Southern blots, as compared to Figure 4A, resulted from the use of different restriction enzymes. However, in F2-9d cells derived from differentiated F2-9ESCs, the DHS site mHS1 at the promoter disappeared, consistent with lower mTERT expression level in these cells. Interestingly, mHS2, located at mTERT 5′ intergenic region, was also absent in F2-9d cells (Figure 4B). In contrast, both mHS3 and mHS4 were detected in F2-9d cells. Therefore, formation of mHS1 and mHS2 correlated with high level of mTERT transcription in F2-9 cells.
General DNase I sensitivity of the hTERT and mTERT genes
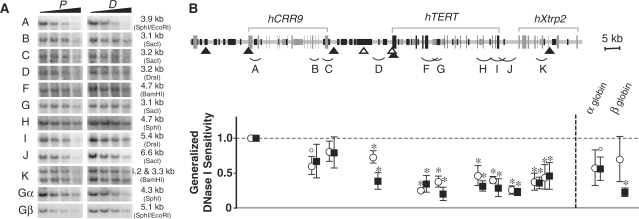
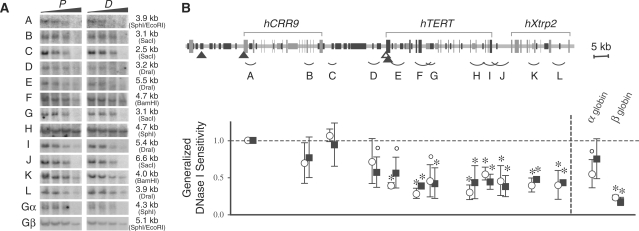
Previously, we demonstrated that the hTERT locus was embedded in a large condensed chromatin domain in HeLa, HEK293, and human fibroblast cell lines (12). Such a chromatin domain would provide a repressive chromatin environment for the tight regulation of hTERT gene in human somatic cells. Here, we examined general DNase I sensitivity of the 110-kb genomic region containing the hCRR9, hTERT and hXtrp2 loci in HL60 and U937 cells. For accurate assessment, genomic fragments that were similar in sizes and devoid of DHSs were examined for their sensitivities to the nuclease, measured by digestion rates of genomic fragments as functions of DNase I concentration (Figures 5 and 6). The hTERT and hXtrp2 genes were more resistant to nuclease digestion than the hCRR9 locus in HL60 and U937 cells. The general DNase I sensitivities of most chromosomal fragments, with the exception of fragment D in HL60 cells, were similar in proliferating and differentiated cells. Fragment D, located about 2.5 kb upstream of the hTERT promoter, became significantly more resistant to the nuclease digestion as HL60 cells differentiated, correlating with the strong hTERT repression during HL60 cell differentiation. α- and β-globin loci were used as internal controls. In both differentiated HL60 and U937 cells, the β-globin locus was highly resistant to DNase I digestion as was previously reported (26). Therefore, as in fibroblasts and epithelial cell lines, the hTERT gene was embedded in a repressive chromatin domain in HL60 and U937 cells and this condensed domain persisted throughout terminal differentiation.
Figure 5.
General DNase I sensitivity of the genomic region containing hCRR9, hTERT and hXtrp2 loci in proliferating and differentiated HL60 cells. For differentiation, HL60 cells were treated with DMSO for 4 days. (A) Southern blots of genomic fragments following DNase I digestion. Nuclei isolated from proliferating and differentiated HL60 cells were treated with increasing concentration of DNase I (2, 4, 8 and 16 U/ml). Genomic DNAs were digested with restriction enzymes and analyzed by Southern analyses using probes specified in Supplementary Table 2. Restriction enzymes used for genomic DNA digestion and sizes of genomic fragments are indicated on the right side of each panel set. The two bands of fragment K were caused by a polymorphic mini-satellite sequence. Gα, α-globin; Gβ, β-globin. (B) The upper diagram is a schematic display of the genomic sequence. The chart below shows the calculated general DNase I sensitivities of the chromosomal fragments corresponding to their positions (A–K), as described in ‘Materials and Methods’ section. Closed triangles depict DHSs present in both proliferating and differentiated cells, whereas open triangles indicate DHSs present only in proliferating cells. Genomic fragments that have been examined by DNase I digestion were indicated by arcs below the diagram. The values in the chart are averages of the lanes treated with 4, 8 and 16 U/ml DNase I, relative to fragment A (designated as 1.0). Open circles and closed squares represent proliferating cells (P) and differentiated cells (D), respectively. ° and * indicate P < 0.05 and P < 0.01, relative to fragment A, by Student's t-test. There were no statistical differences in general DNase I sensitivities between corresponding chromatin fragments in proliferating and differentiated HL60 cells, except for fragment D, for which P = 0.03.
Figure 6.
General DNase I sensitivity of the genomic region containing hCRR9, hTERT and hXtrp2 loci in proliferating and differentiated U937 cells. For differentiation, U937 cells were treated with TPA for 24 h. (A) Southern blots of genomic fragments. (B) A schematic display of the human genomic region and calculated general DNase I sensitivities of genomic fragments shown in (A). The symbols are the same as in Figure 5. There were no statistical differences in general DNase I sensitivities between corresponding chromatin fragments in proliferating and differentiated U937 cells.
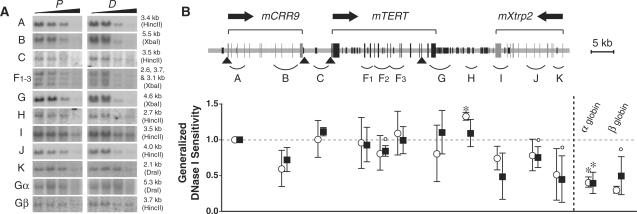
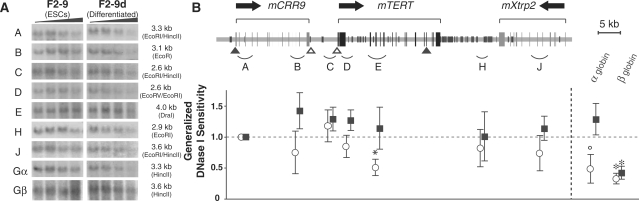
In mouse cells, a 75-kb genomic region that contained mCRR9, mTERT and mXtrp2 loci, was measured for general nuclease sensitivity. The mTERT gene, its 5′ and 3′ intergenic regions (fragments C to H) showed similar DNase I sensitivities compared to the mCRR9 locus (fragments A and B) in both proliferating and differentiated M1 cells (Figure 7). In F2-9 ESCs and F2-9d cells, although fewer genomic fragments were quantified, the mTERT and mXtrp2 loci in general were no more resistant to nuclease digestion than the mCRR9 locus (Figure 8). Thus, a condensed chromatin domain at the mTERT locus did not exist in mouse cells. As we reported previously, the transgenic hTERT locus in the chromosomally integrated BAC reporter was more resistant to DNase I digestion than the hCRR9 locus in both F2-9 and F2-9d cells, recapitulating the native hTERT locus in human cells (22). Hence, we concluded that the endogenous mTERT gene was not located within a repressive chromatin domain, unlike the hTERT gene in human cells.
Figure 7.
General DNase I sensitivity of the genomic region containing mCRR9, mTERT and mXtrp2 loci in proliferating and differentiated mouse M1 cells. For differentiation, M1 cells were treated with IL-6 for 5 days. (A) Southern blots of genomic fragments. The three bands in F1-3 were recognized by the same mTERT cDNA probe: F1, 2.6 kb; F2, 3.7 kb; F3, 3.1 kb. (B) The upper portion is a diagram of the mouse genomic region including mCRR9, mTERT and mXtrp2 loci. The lower panel shows the calculated general DNase I sensitivities of genomic fragments matching to their positions A to K in the upper diagram, relative to fragment A (1.0). The symbols in this figure are the same as in Figure 5.
Figure 8.
General DNase I sensitivity of mCRR9, mTERT and mXtrp2 loci in mouse embryonic stem cells F2-9 and differentiated derivative F2-9d cells. (A) Southern blots of genomic fragments. (B) A schematic display of the mouse genomic region and calculated general DNase I sensitivities of genomic fragments shown in A. The symbols are the same as in Figures 5–7.
DISCUSSION
It was shown that hTERT expression was lower in the majority of somatic tissues than mTERT (6,9). In the current study, we provided evidence that compared to mTERT, hTERT was strongly repressed in differentiated human somatic cells and this stringent repression depended on its native chromatin conformation. Differentiated human cells, including fibroblasts and differentiated granulocytes and monocytic cells, expressed very low levels of hTERT mRNA compared to undifferentiated human cells, such as embryonic stem cells. In newborn foreskin fibroblast cells, the hTERT mRNA was barely detectable and its level was estimated to be at least 1000-fold lower than in hESC H9 cells and leukemic HL60 cells (Table 1). Embryonic fibroblast IMR90 cells expressed slightly more hTERT mRNA, but still 100-fold less than in H9 and HL60 cells. This low hTERT expression might still be required for maintaining telomere integrity in normal human fibroblasts (10).
The hTERT expression appeared to be inversely correlated to the status of cell differentiation. For example, HL60 cells differentiate to either granulocyte-like cells or monocyte/macrophage-like cells (17), whereas U937 cells are already committed to the monocytic lineage (27). Accordingly, proliferating HL60 cells expressed about 100-fold more hTERT mRNA than U937 cells (Figure 1A). Following terminal differentiation, hTERT transcription was repressed to very low levels in both HL60 and U937 cells. Thus, silencing of the hTERT gene in most human somatic cells likely resulted from strong transcriptional repression of the hTERT gene during cell differentiation.
Like HL60 cells, mouse M1 cells are myeloblasts that undergo differentiation into granulocyte- and monocyte-like cells upon IL-6 treatment (28). However, mTERT expression in post-mitotic differentiated M1 cells was still one-fifth of those in proliferating M1 cells. Similarly, MEFs contained a significant level of mTERT mRNA compared with undifferentiated mESCs and M1 cells. Recently, we have published the study of a chromosomally integrated hTERT reporter locus, in which the hTERT promoter was repressed by several orders of magnitudes when mouse ESCs were differentiated into osteogenic cells (22). In these same cells, mTERT expression was downregulated by only 6–9-fold (Table 1). Taken together, mTERT silencing was not tightly linked to terminal differentiation as it was for hTERT in human cells.
Several aspects of the endogenous loci need to be addressed in order to explain the differential repression of hTERT and mTERT genes. Interactions between transcription factors and the promoter elements might be different for hTERT and mTERT genes. c-Myc, Sp-1, and E2F factors were thought to be the main transcription activators to regulate the hTERT core promoter. The downstream E-box and the E2F consensus site have been shown to participate in both positive and negative regulation of telomerase (25,29,30). Switch of occupancy by Myc/Max to Mad1/Max at the hTERT core promoter was proposed to be responsible for the downregulation of hTERT transcription during HL60 cell differentiation (20). In this report, we demonstrated that, unlike the endogenous hTERT promoter, transiently transfected hTERT promoter reporters were not silenced in NHF cells and during HL60 cell differentiation. Mutation of the downstream E-box resulted in a moderate increase of the hTERT promoter activity, whereas mutation of the E2F consensus site significantly decreased its activity. These data suggested that, although both E-box and E2F sites are important regulators of the hTERT promoter activity, presence of these sites are insufficient for the repression of hTERT promoter during differentiation of HL60 cells. Furthermore, these two sites are conserved at hTERT and mTERT promoters, raising the possibility that additional mechanisms contribute to the differential expression of hTERT and mTERT in human and mouse cells.
It was previously reported that several binding elements near or within the hTERT core promoter were involved in hTERT repression in human cells. Particularly, a nonconserved GC-box and upstream AP-1 consensus sites were shown to mediate the suppression of the hTERT core promoter in transiently transfected reporter assays in NHF cells (9,16). Our data confirmed these results and showed that mutation of the nonconserved GC-box resulted in a 2-fold increase in hTERT promoter activity in transient transfection assays in both NHF and HL60 cells. These data suggested that, whereas the GC-box and the downstream E-box negatively regulated the hTERT core promoter, these elements were unable to silence the hTERT promoter in differentiated cells without an appropriate chromatin context.
Compared to earlier reports suggesting that the hTERT promoter reporters were inactive in telomerase-negative cells, we found that the transiently transfected hTERT promoters were active in NHFs and differentiated HL60 cells, in which the endogenous hTERT promoter was repressed. However, we do not believe that our experimental results were significantly different from those of Horikawa et al. (9). We demonstrated that transiently transfected hTERT promoters were less active than the mTERT promoter, as shown by Horikawa et al. (9), but they were not silenced in telomerase-negative fibroblasts (Figure 1D). Our results further indicated that chromatin-dependent repression of the hTERT locus was an important mechanism of differential expression of the TERT genes in human and mouse cells.
The native chromatin environments of the hTERT and mTERT genes likely contribute to their differential regulation (4,31). In this study, we showed that, in HL60 and U937 cells, the endogenous hTERT locus was embedded in a nuclease-resistant chromatin domain and this condensed domain persisted throughout differentiation. However, in mouse M1 cells, F2-9 ESCs, and their differentiated derivatives, such a condensed chromatin domain was not detected for the mTERT locus. In addition, the transgenic hTERT locus in F2-9 mESCs and F2-9d cells adopted a conformation similar to its native locus in human cells (22) and was silenced upon differentiation (Table 1). Furthermore, inhibition of histone deacetylases in telomerase-negative fibroblasts led to an opening of the chromatin and induction of hTERT transcription (12). Together, these results collectively provided evidence for a causal role of the chromatin conformation in tight hTERT repression. In proliferating cells, activation by strong transcription activators such as c-Myc, Sp1, and E2Fs might permit transcription from the hTERT promoter despite repression by this chromatin domain. In fact, the endogenous hTERT promoter is under repression even in telomerase-positive immortal cell lines because inhibition of histone deacetylases resulted in a further increase of hTERT transcription in these cells (data not shown). Conversely, this condensed chromatin domain would cooperate with repressors that were recruited to the promoter during cell differentiation and effectively reduce basal transcription. Therefore, the chromatin environment is likely a crucial factor for the tight hTERT repression in human somatic cells. In contrast, although mTERT expression was moderately repressed during differentiation, this regulation did not appear to require the native chromatin structure.
The present study also identified several novel DHSs within and near both hTERT and mTERT genomic loci. DHSs are often the binding sites for proteins that regulate chromatin structure and/or transcription in vivo. In earlier studies, we showed that hHS1 coincided with the hTERT core promoter and was present in all telomerase-expressing cells but absent in telomerase-negative cells (4,12). Here, we found that the novel hHS2 was mapped to the first intron of hTERT gene, which contained two ETS consensus sites (+288 nt and +390 nt). It was previously reported that these ETS sites mediated the activation of hTERT transcription by ER81, an ETS family transcription factor involved in the oncogenic HER2/Neu-Ras signaling pathway (32). hHS3, located about 1.5 kb upstream of the hTERT transcription start site in U937 cells, overlapped with one of the AP-1 sites identified by Takakura et al. (16). Because hHS2 and hHS3 were present in both proliferating and differentiated cells, it remains to be determined whether these sites play a role in hTERT repression during differentiation. In contrast, hHS4, mapped to about 9.5 kb upstream of the hTERT promoter, appeared only in proliferating HL60 cells. This DHS coincided with an Alu repeat. Interestingly, mHS2, located about 6 kb upstream of the mTERT promoter in F2-9 mESCs and M1 cells, was also mapped to a mouse type II Alu-like SINE repetitive element B2. Alu elements belong to the SINE family (short interspersed nuclear elements) of repetitive elements and may regulate, often negatively, transcription of several genes (33,34). For example, Alu repeats were proposed to be transcriptional silencers of the human erythropoietin receptor gene, the Wilms’ tumor gene WT1, and BRCA2 gene (35–37). However, both hHS4 and mHS2 disappeared upon differentiation, suggesting that formation of these DHSs associated with activation of the TERT promoters. Also, mHS2 appeared to be linked to mTERT transcription but not differentiation per se, as it was detected in both proliferating and terminally differentiated M1 cells (Figure 4C).
In most human cells, including HL60 and U937 cells, transition from the nuclease-sensitive hCRR9 locus to the condensed chromatin domain of hTERT locus occurs at the 3′ end of hCRR9 gene [(12) and Figures 5 and 6], suggesting the presence of a chromatin boundary element within this region. It was previously shown that an E-box binding protein USF1 bound to the 5′HS4 insulator of β-globin domain and acted as a barrier by recruiting histone modifying activities (38). Interestingly, at least two DHSs (hHS5 and hHS6) were identified near the 3′ end of hCRR9 gene in HL60 cells. This region contained a number of potential USF consensus sites and correlated with histone hyperacetylation (data not shown). Thus, it will be interesting to determine if hHS5 and hHS6 function as a barrier to prevent encroachment of heterochromatin from the condensed chromatin domain into the hCRR9 gene.
In summary, our study demonstrated that the hTERT gene was silenced upon cellular differentiation whereas the mTERT expression was only moderately downregulated. The hTERT locus, but not the mTERT gene, was embedded in a condensed chromatin domain. The strong hTERT repression in somatic cells was dependent on its repressive chromatin environment and was not recapitulated by transient transfection of the hTERT promoter. In contrast, the regulation of the mTERT promoter was relatively independent of its native chromatin structure because transiently transfected mTERT promoter was repressed during cell differentiation. Therefore, chromatin environments of the hTERT and mTERT genes played an important role in differential telomerase regulation in human and mouse cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health grant GM071725; and J.Z. is a Research Scholar of American Cancer Society. Funding for open access charge: American Cancer Society.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr. Dan Liebermann for providing M1 cells and the protocol on culturing and differentiating these cells and Dr. Izumi Horikawa for pBT-255 and its mutant derivatives. We thank Drs. Renjith Mathew and Sergei Grigoryev for critically reading the manuscript and Bing Dong for technical assistance. We also thank Anne Stanley, David Stanford, and Joe Bednarczyk of the Macromolecular and Molecular Genetics Core Facilities at Penn State College of Medicine for their excellent services.
REFERENCES
- 1.Greider CW, Blackburn EH. Telomeres, telomerase and cancer. Sci. Am. 1996;274:92–97. doi: 10.1038/scientificamerican0296-92. [DOI] [PubMed] [Google Scholar]
- 2.Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA [see comments] Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 3.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells [see comments] Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 4.Wang S, Zhu J. Evidence for a relief of repression Mmchanism for activation of the human telomerase reverse transcriptase promoter. J. Biol. Chem. 2003;278:18842–18850. doi: 10.1074/jbc.M209544200. [DOI] [PubMed] [Google Scholar]
- 5.Prowse KR, Greider CW. Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc. Natl Acad. Sci. USA. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenberg RA, Allsopp RC, Chin L, Morin GB, DePinho RA. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- 7.Armstrong L, Lako M, Lincoln J, Cairns PM, Hole N. mTert expression correlates with telomerase activity during the differentiation of murine embryonic stem cells. Mech. Dev. 2000;97:109–116. doi: 10.1016/s0925-4773(00)00423-8. [DOI] [PubMed] [Google Scholar]
- 8.Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 9.Horikawa I, Chiang YJ, Patterson T, Feigenbaum L, Leem SH, Michishita E, Larionov V, Hodes RJ, Barrett JC. Differential cis-regulation of human versus mouse TERT gene expression in vivo: identification of a human-specific repressive element. Proc. Natl Acad. Sci. USA. 2005;102:18437–18442. doi: 10.1073/pnas.0508964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masutomi K, Yu EY, Khurts S, Ben-Porath I, Currier JL, Metz GB, Brooks MW, Kaneko S, Murakami S, DeCaprio JA, et al. Telomerase maintains telomere structure in normal human cells. Cell. 2003;114:241–253. doi: 10.1016/s0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- 11.Zhu J, Wang H, Bishop JM, Blackburn EH. Telomerase extends the lifespan of virus-transformed human cells without net telomere lengthening [see comments] Proc. Natl Acad. Sci. USA. 1999;96:3723–3728. doi: 10.1073/pnas.96.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, Zhu J. The hTERT gene is embedded in a nuclease-resistant chromatin domain. J. Biol. Chem. 2004;279:55401–55410. doi: 10.1074/jbc.M411352200. [DOI] [PubMed] [Google Scholar]
- 13.Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. [see comments] Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 14.Hathcock KS, Chiang JY, Hodes RJ. In vivo regulation of telomerase activity and telomere length. Immun. Rev. 2005;205:104–113. doi: 10.1111/j.0105-2896.2005.00267.x. [DOI] [PubMed] [Google Scholar]
- 15.Wright WE, Shay JW. Telomere dynamics in cancer progression and prevention: fundamental differences in human and mouse telomere biology. Nat. Med. 2000;6:849–851. doi: 10.1038/78592. [DOI] [PubMed] [Google Scholar]
- 16.Takakura M, Kyo S, Inoue M, Wright WE, Shay JW. Function of AP-1 in transcription of the telomerase reverse transcriptase gene (TERT) in human and mouse cells. Mol. Cell Biol. 2005;25:8037–8043. doi: 10.1128/MCB.25.18.8037-8043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birnie GD. The HL60 cell line: a model system for studying human myeloid cell differentiation. Brit. J. Cancer –Suppl. 1988;9:41–45. [PMC free article] [PubMed] [Google Scholar]
- 18.Lord KA, Abdollahi A, Hoffman-Liebermann B, Liebermann DA. Dissection of the immediate early response of myeloid leukemia cells to terminal differentiation and growth inhibitory stimuli. Cell Growth Diff. 1990;1:637–645. [PubMed] [Google Scholar]
- 19.Günes C, Lichtsteiner S, Vasserot AP, Englert C. Expression of the hTERT gene is regulated at the level of transcriptional initiation and repressed by Mad1. Cancer Res. 2000;60:2116–2121. [PubMed] [Google Scholar]
- 20.Xu D, Popov N, Hou M, Wang Q, Bjorkholm M, Gruber A, Menkel AR, Henriksson M. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc. Natl Acad. Sci. USA. 2001;98:3826–3831. doi: 10.1073/pnas.071043198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L, Saldanha SN, Pate MS, Andrews LG, Tollefsbol TO. Epigenetic regulation of human telomerase reverse transcriptase promoter activity during cellular differentiation. Genes Chromosomes Cancer. 2004;41:26–37. doi: 10.1002/gcc.20058. [DOI] [PubMed] [Google Scholar]
- 22.Wang S, Hu C, Zhu J. Transcriptional silencing of a novel hTERT reporter locus during in vitro differentiation of mouse embryonic stem cells. Mol. Biol. Cell. 2007;18:669–677. doi: 10.1091/mbc.E06-09-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nozawa K, Maehara K, Isobe K. Mechanism for the reduction of telomerase expression during muscle cell differentiation. J. Biol. Chem. 2001;276:22016–22023. doi: 10.1074/jbc.M011181200. [DOI] [PubMed] [Google Scholar]
- 24.Saitoh N, Bell AC, Recillas-Targa F, West AG, Simpson M, Pikaart M, Felsenfeld G. Structural and functional conservation at the boundaries of the chicken beta-globin domain. EMBO J. 2000;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horikawa I, Cable PL, Mazur SJ, Appella E, Afshari CA, Barrett JC. Downstream E-box-mediated regulation of the human telomerase reverse transcriptase (hTERT) gene transcription: evidence for an endogenous mechanism of transcriptional repression. Mol. Biol. Cell. 2002;13:2585–2597. doi: 10.1091/mbc.E01-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown KE, Amoils S, Horn JM, Buckle VJ, Higgs DR, Merkenschlager M, Fisher AG. Expression of alpha- and beta-globin genes occurs within different nuclear domains in haemopoietic cells. Nat. Cell Biol. 2001;3:602–606. doi: 10.1038/35078577. [DOI] [PubMed] [Google Scholar]
- 27.Harris PE, Ralph P. Human leukemic models of myelomonocytic development: a review of the HL-60 and U937 cell lines. J. Leukocyte Biol. 1985;37:407–422. doi: 10.1002/jlb.37.4.407. [DOI] [PubMed] [Google Scholar]
- 28.Sachs L. The molecular control of blood cell development. Science. 1987;238:1374–1379. doi: 10.1126/science.3317831. [DOI] [PubMed] [Google Scholar]
- 29.Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R. Direct activation of TERT transcription by c-MYC. Nat. Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 30.Crowe DL, Nguyen DC, Tsang KJ, Kyo S. E2F-1 represses transcription of the human telomerase reverse transcriptase gene. Nucleic Acids Res. 2001;29:2789–2794. doi: 10.1093/nar/29.13.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ducrest AL, Amacker M, Mathieu YD, Cuthbert AP, Trott DA, Newbold RF, Nabholz M, Lingner J. Regulation of human telomerase activity: repression by normal chromosome 3 abolishes nuclear telomerase reverse transcriptase transcripts but does not affect c-Myc activity. Cancer Res. 2001;61:7594–7602. [PubMed] [Google Scholar]
- 32.Goueli BS, Janknecht R. Upregulation of the catalytic telomerase subunit by the transcription factor ER81 and oncogenic HER2/Neu, Ras, or Raf. Mol. Cell Biol. 2004;24:25–35. doi: 10.1128/MCB.24.1.25-35.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polak P, Domany E. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics. 2006;7:133. doi: 10.1186/1471-2164-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hasler J, Strub K. Alu elements as regulators of gene expression. Nucleic Acids Res. 2006;34:5491–5497. doi: 10.1093/nar/gkl706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maouche L, Cartron JP, Chretien S. Different domains regulate the human erythropoietin receptor gene transcription. Nucleic Acids Res. 1994;22:338–346. doi: 10.1093/nar/22.3.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hewitt SM, Fraizer GC, Saunders GF. Transcriptional silencer of the Wilms' tumor gene WT1 contains an Alu repeat. J. Biol. Chem. 1995;270:17908–17912. doi: 10.1074/jbc.270.30.17908. [DOI] [PubMed] [Google Scholar]
- 37.Sharan C, Hamilton NM, Parl AK, Singh PK, Chaudhuri G. Identification and characterization of a transcriptional silencer upstream of the human BRCA2 gene. Biochem. Biophys. Res. Commun. 1999;265:285–290. doi: 10.1006/bbrc.1999.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. [see comment] Mol. Cell. 2004;16:453–463. doi: 10.1016/j.molcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.