Summary
Mammalian SR proteins are a family of reversibly phosphorylated RNA binding proteins primarily studied for their roles in alternative splicing. While budding yeast lack alternative splicing, they do have three SR-like proteins: Npl3, Gbp2, and Hrb1. However, these have been best characterized for their roles in mRNA export, leaving their potential roles in splicing largely unexplored. Here we combined high-density genetic interaction profiling and genome-wide splicing-sensitive microarray analysis to demonstrate that a single SR-like protein, Npl3, is required for efficient splicing of a large set of pre-mRNAs in Saccharomyces cerevisiae. We tested the hypothesis that Npl3 promotes splicing by facilitating co-transcriptional recruitment of splicing factors. Using chromatin immunoprecipitation, we showed that mutation of NPL3 reduces the occupancy of U1 and U2 snRNPs at at genes whose splicing is stimulated by Npl3. This result provides strong evidence that an SR protein can promote recruitment of splicing factors to chromatin.
Introduction
The basic components of the splicing machinery have been conserved between budding yeast and mammals, including five snRNAs and >80 proteins (Jurica and Moore, 2003). The major differences between these organisms reflect the prominence of alternative splicing in higher eukaryotes, which relies on a family of splicing regulators termed SR proteins (Blencowe et al., 1999; Bourgeois et al., 2004; Hertel and Graveley, 2005). The 10 known family members contain one or more N-terminal RNA Recognition Motifs (RRM) and a C-terminal domain rich in alternating serine-arginine dipeptides (RS/SR) that are subject to reversible serinephosphorylation. SR proteins are generally thought to function by binding to exonic sequences adjacent to sub-optimal splice sites and promoting the recruitment of U1 and U2 snRNPs.
In yeast, only a handful of spliced genes contain more than one intron and the majority of splice sites adhere to a strict consensus (http://www.yeastgenome.org). The lack of opportunity for alternative splicing has promoted the widespread belief that yeast also lack SR proteins (Blencowe et al., 1999; Shen and Green, 2006). Yet Npl3, Hrb1 and Gbp2 share the basic domain structure of canonical SR proteins (Gilbert et al., 2001; Hacker and Krebber, 2004) and Npl3 and Gbp2 can be serine-phosphorylated by a kinase, Sky1 (Gilbert et al., 2001; Lukasiewicz et al., 2007), which has high structural similarity to mammalian SR Protein Kinase 1 (SRPK1; Siebel et al., 1999).
Npl3 and Gbp2 were originally identified as mRNA export factors (Kadowaki et al., 1994; Lee et al., 1996; Singleton et al., 1995; Windgassen and Krebber, 2003), a function that has now been documented for several mammalian SR proteins (Huang et al., 2003; Huang and Steitz, 2001). More recently Npl3 has been implicated in transcription elongation, termination/3′ end processing (Bucheli and Buratowski, 2005; Bucheli et al., 2007; Dermody et al., 2008; Wong et al., 2007) and translation (Windgassen et al., 2004). Potential roles for these SR-like proteins in pre-mRNA splicing have not yet been systematically tested.
Here we use a combination of genetic, molecular and biochemical approaches to demonstrate that one of the three SR-like proteins, Npl3, is required for the efficient splicing of a large subset of pre-mRNAs. Moreover we provide in vivo evidence that SR proteins facilitate co-transcriptional binding of early splicing factors to chromatin.
Results
Npl3 is required for the splicing of a subset of pre-mRNAs
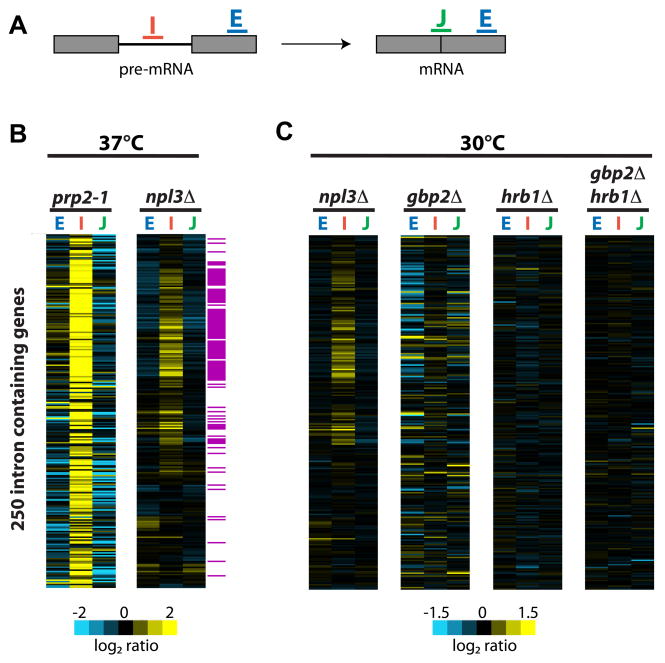
To test whether SR-like proteins are involved in pre-mRNA splicing in Saccharomyces cerevisiae, we employed splicing-specific microarrays (Pleiss et al., 2007b). These microarrays contain oligos that hybridize to exon 2 (E), the intron (I), and the exon:exon junction (J) to detect and differentiate total mRNA, pre-mRNA, and mature mRNA, respectively (Figure 1A). We performed competitive hybridization in which cDNA from one deletion mutant (npl3Δ, gbp2Δ or hrb1Δ) and from an isogenic WT strain were differentially labeled and mixed, then hybridized to the microarray. Shown for comparison is the splicing profile of a strain expressing a mutant form of the canonical splicing factor PRP2, prp2-1 (Figure 1B; courtesy of Pleiss et al., 2007b). Mutation of PRP2 causes a strong splicing defect as seen by a global accumulation of pre-mRNA (I) and a decrease in mature mRNA (J). The npl3Δ strain also shows accumulation, albeit to a lesser extent than prp2-1, of a large subset of pre-mRNAs consisting primarily of ribosomal protein gene (RPG) transcripts (Figure 1B; highlighted in purple). We used quantitative PCR analysis to confirm these results for seven of the 69 transcripts affected (Supplemental Figure 1). By contrast, deletion of either GBP2 or HRB1 alone or in combination (gbp2Δhrb1Δ) does not affect splicing (Figure 1C). Notably, ~80% of the most strongly affected pre-mRNAs identified in our splicing microarray analysis overlap with those found for npl3Δ in a recent analysis of pre-mRNA and mRNA levels in 80 different gene expression mutants using a similar splicing microarray platform (gbp2Δ and hrb1Δ were not tested in that study; Burckin et al., 2005). Our data strongly suggest that of the three SR-like proteins, only Npl3 is involved in splicing.
Figure 1. Deletion of NPL3, but not GBP2 or HRB1, causes an accumulation of pre-mRNA as determined by microarray analysis.
(A). Schematic of oligos on microarrays. (B). Splicing profile of prp2-1 and npl3Δcompared to WT grown at 37°C. Transcripts that encode the ribosomal protein genes, as determined by Gene Ontology Function annotation (structural constituent of the ribosome; http://www.yeastgenome.org/), are highlighted in purple. (C). Splicing profiles of each of the SR protein mutants, npl3Δ, gbp2Δ, and hrb1Δ and the double mutant gbp2Δ hrb1Δ compared to WT, all grown at 30°C. Each horizontal line in the splicing profile represents of one gene. Exon2 oligo (E), intron oligo (I), exon junction oligo (J). Gene order along the Y-axis is identical for all arrays in B–C.
The pre-mRNA accumulation observed in npl3Δ could be due to an indirect affect of Npl3′s role in mRNA export and transcription termination/3′ end processing. This is unlikely, however, as a number of other export mutants (sub2-1, sub2-85, mex67-5, yra1-1) and 3′end processing mutants (rna14-64, brr5-1, paf1Δ) do not display defects in splicing (Burckin et al., 2005; Pleiss et al., 2007b). Moreover, we compared the splicing profile of npl3Δ to those in our previously published set of RNA processing mutants (Pleiss et al., 2007b) and found that it most closely correlates with splicing factor mutants (TLK, G. Whitworth, and CG, unpublished data), consistent with the observations of Burckin et al. (Burckin et al., 2005).
The observed accumulation of pre-mRNA in the npl3Δ strain could also be due to an effect on pre-mRNA decay. Recent studies have shown that strains harboring deletions of factors involved in nuclear or cytoplasmic RNA decay, such as Rrp6, Ski2, or Rai1 do not result in an accumulation of RPG pre-mRNAs (Burckin et al., 2005; Pleiss et al., 2007a). Thus, the pre-mRNA accumulation observed in the npl3Δ strain is unlikely to be due to a block in pre-mRNA decay. To ensure that our observations are not due to indirect effects involving the nonsense-mediated mRNA decay (NMD) pathway, which has been shown to affect the abundance of unspliced pre-mRNAs (Sayani et al., 2008), we repeated our npl3Δ versus WT comparisons in strains entirely deficient for NMD (upf1Δ, He and Jacobson, 1995), and still observed the accumulation of pre-mRNAs in npl3Δ cells (npl3Δ upf1Δ versus upf1Δ; Supplemental Figure 2).
Npl3 exhibits genetic interactions with the splicing machinery
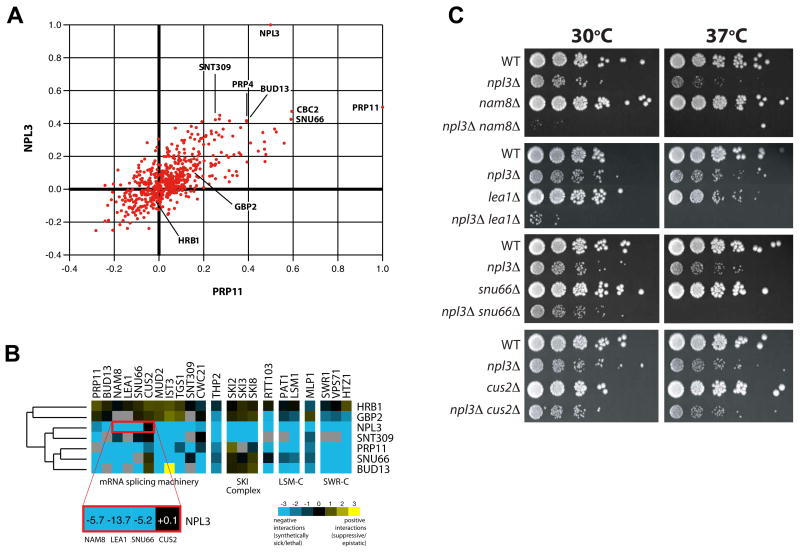
To further characterize the roles of yeast SR-like proteins, we included the npl3Δ, gbp2Δ and hrb1Δ strains in our “RNA processing” E-MAP (Epistatic-Mini Array Profile; Wilmes et al., this issue). An E-MAP describes all pair-wise genetic interactions within a defined subset of genes in a quantitative manner, allowing for identification of both negative/aggravating (e.g. synthetic sick) and positive/alleviating (e.g. suppressive or epistatic) interactions (Collins et al., 2007; Schuldiner et al., 2005). The RNA processing E-MAP is comprised of 552 distinct mutants involved in RNA-related processes, including pre-mRNA splicing. Mutants displaying similar genetic interaction profiles are often functionally related (Collins et al., 2007; Pan et al., 2006; Schuldiner et al., 2005; Tong et al., 2004). We found that the genetic interaction profile generated by deletion of NPL3, but not HRB1 or GBP2, is most similar to mutants of known pre-mRNA splicing factors (Figure 2A), suggesting that Npl3 functions with these factors in vivo. For example, NPL3 and PRP11 display genetic interaction profiles highly similar to each other and to other well-characterized mRNA splicing factors (SNU66, BUD13, PRP4, and SNT309; Figure 2A). Inspection of the individual genetic interactions between these mutants revealed negative interactions with many splicing factors (Figure 2B), particularly factors important for spliceosome assembly, such as the U1 snRNP factor Nam8 (Gottschalk et al., 1998), the commitment complex factor Mud2 (Abovich et al., 1994; Rain and Legrain, 1997), and the U2 snRNP factor Lea1 (Caspary and Seraphin, 1998). NPL3 and PRP11 also display negative interactions with RNA degradation factors (e.g. SKI complex and LSM-C) and with the SWR1 chromatin-remodeling complex (e.g. Swr1, Vps71 and Htz1). Notably, GBP2 and HRB1 do not display negative genetic interactions with splicing factors (Figure 2B and Supplemental Figure 3), further confirming that they are not involved in pre-mRNA splicing.
Figure 2. NPL3 is genetically implicated in mRNA splicing.
(A). Correlation plot of correlation coefficients generated from comparison of the genetic profiles from npl3Δ or prp11-DAmP to all other profiles in the RNA processing E-MAP (Wilmes et al., this issue). (B). Several individual genetic interactions are common to NPL3 and genes encoding other mRNA splicing factors (SNT309, PRP11, SNU66, BUD13). Negative interactions are blue while positive ones are yellow. (C). Serial dilutions of WT, single and double mutant strains grown at 30°C and 37 °C.
We reproduced a number of these genetic interactions using traditional growth assays (Figure 2C), confirming that npl3Δ is synthetic sick or lethal with deletions of non-essential splicing factors. Interestingly, these interactions are specific for a subset of spliceosome assembly factors; for example, cus2Δ (involved in U2 snRNA folding; Yan et al., 1998) does not interact with npl3Δ (Figure 2B and 2C). We also noted synthetic sick interactions between npl3Δ and components important for the catalytic steps of splicing (Figure 2B and 2C), such as SNU66, a component of the triple snRNP (Stevens et al., 2001). Overall, our genetic analyses strongly support a role for Npl3, but not Gbp2 or Hrb1, in mRNA splicing.
Npl3 physically associates with spliceosome assembly factors
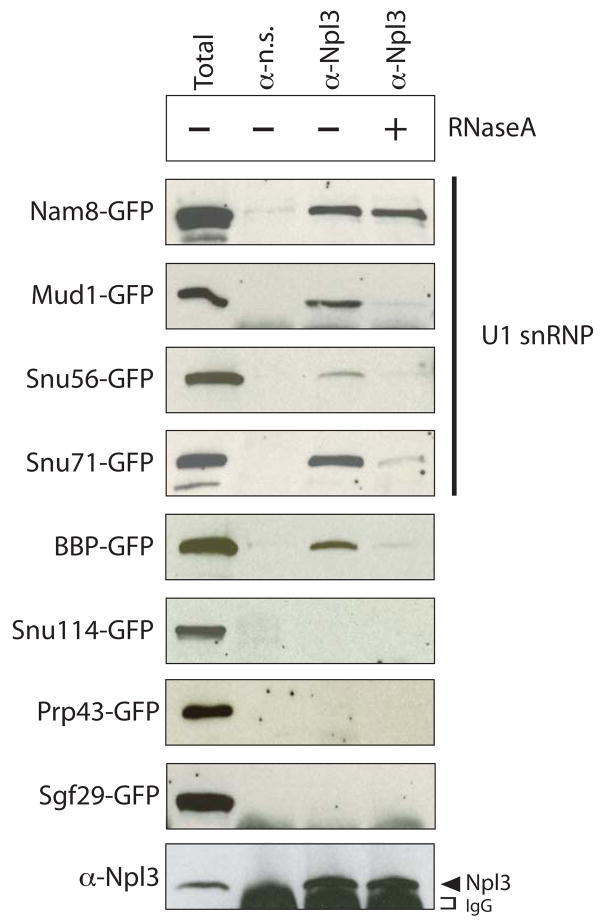
In mammals, SR proteins facilitate spliceosome assembly by interacting with SR-related splicing factors, such as the U1 snRNP protein U1-70K (Blencowe et al., 1999; Bourgeois et al., 2004). We tested whether Npl3 can physically associate with U1 snRNP proteins and the Branchpoint Binding Protein (BBP; Berglund et al., 1997) using co-immunoprecipitation assays (Figure 3). We co-immunoprecipitated proteins from strains carrying GFP-tagged U1 snRNP factors (Nam8, Mud1, Snu56, and Snu71; Gottschalk et al., 1998) and GFP-tagged BBP using α-Npl3 antibodies. The α-Npl3 antibody immunoprecipitated all of these proteins (Figure 3), whereas a non-specific antibody did not. To test whether the interactions are mediated by RNA we treated the extracts with RNase A prior to immunoprecipitation. Notably, only the Nam8:Npl3 interaction was RNase A insensitive, suggesting that this interaction is not dependent on RNA, while other U1 components interact indirectly via RNA.
Figure 3. Npl3 physically interacts with U1 snRNP and the branchpoint binding protein, BBP.
Co-immunoprecipitation assays using α-Npl3 antibodies to pull down proteins from the indicated GFP-tagged strains. Blots were probed with α-GFP antibodies. The sensitivity of the interaction to RNase A (shown in right-most lane, + RNaseA) was determined by treating lysates with RNase A prior to immunoprecipitation. A total sample for each lysate (1/60th of input) is shown in the first lane. Bottom panel is a representative blot probed with α-Npl3. α n.s = non-specific antibody.
Npl3 did not co-immunoprecipitate Snu114, a U5 snRNP factor (Bartels et al., 2002), Prp43, which is important for spliceosome disassembly (Arenas and Abelson, 1997; Sawa and Shimura, 1991), or an unrelated factor involved in histone modification, Sgf29 (Figure 3; Sanders et al., 2002). To confirm that Snu114 and Prp43 do not physically interact with Npl3 we also performed the reciprocal immunoprecipitation experiments, pulling down GFP-Nam8, GFP-Snu114 or GFP-Prp43 using α-GFP antibodies and then detecting Npl3 using α-Npl3 antibodies. Npl3 co-purified with Nam8 but not with either Snu114 or Prp43 (data not shown). These results suggest that Npl3 specifically interacts with factors involved in early steps of spliceosome assembly, consistent with a role for Npl3 in recruiting splicing factors to pre-mRNA.
Npl3 is required for the association of early splicing factors with chromatin
Mammalian SR proteins are likely to recruit splicing factors co-transcriptionally as they associate with sites of ongoing transcription and interact with splicing factors (Blencowe et al., 1999; Bourgeois et al., 2004). However, direct evidence for this is lacking. Npl3 associates with chromatin during transcription (Lei et al., 2001; Lei and Silver, 2002) and is therefore at the right time and place to facilitate co-transcriptional recruitment of splicing factors to chromatin. To test this we utilized a chromatin immunoprecipitation (ChIP) assay to compare the association of GFP-tagged Mud1 (U1 snRNP) with chromatin in the WT versus the npl3Δ strain. We found that U1 snRNP association was significantly decreased in the npl3Δ strain; however, we noted that the RNA polymerase (pol) II signal was also decreased (often by >25%; TLK and CG; unpublished observation). Accurate comparison of ChIP results relies on the ability to rule out indirect changes attributable to defects in transcription. Therefore we chose to utilize a temperature sensitive npl3-1 mutant (Henry et al., 1996), which consistently showed similar RNA pol II ChIP profiles when compared to WT (Supplemental Figure 4A) and, like npl3Δ, displays a splicing defect (although to a lesser extent than npl3Δ; Supplemental Figure 1 and 4B). We compared the association of a panel of HA-tagged splicing factors with a set of ribosomal protein genes in WT and the npl3-1 strains. Importantly, we confirmed that the HA-tagged protein levels were equivalent in these strains (Supplemental Figure 4C).
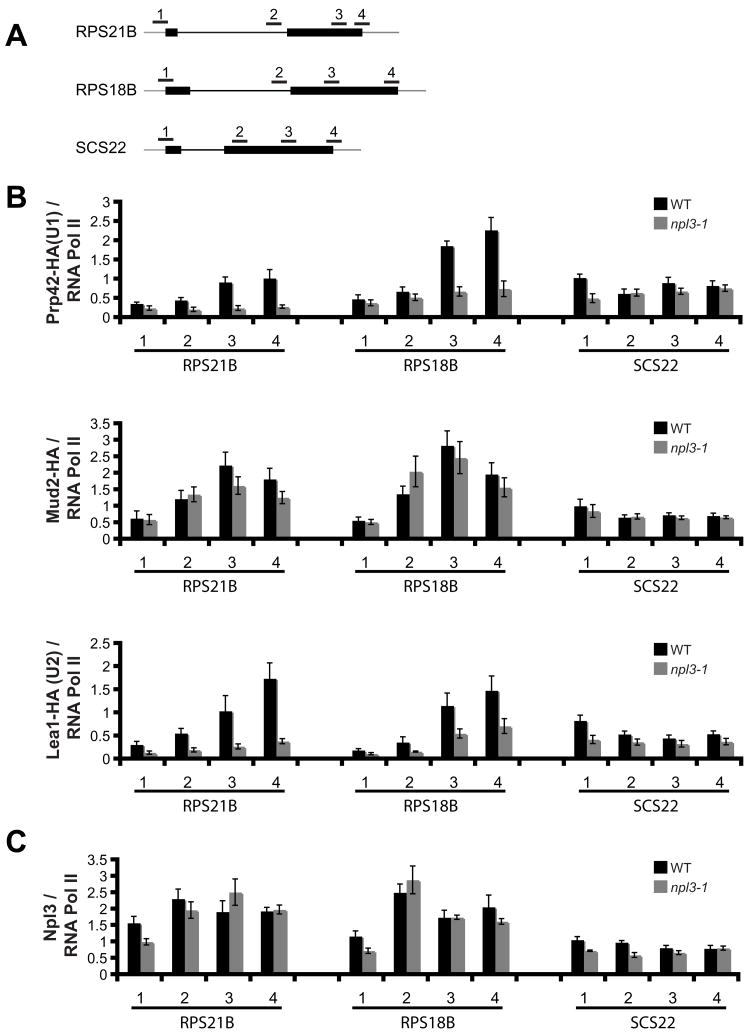
In yeast, Prp42 (U1 snRNP), Lea1 (U2 snRNP), and Mud2 (U2AF) are co-transcriptionally associated with chromatin (Gornemann et al., 2005; Kotovic et al., 2003; Lacadie and Rosbash, 2005) and recent whole genome ChIP-chip analysis has revealed the Prp42-HA and Lea1-HA ChIP profiles for every gene (Tardiff et al., 2006). We used ChIP to analyze the association of Prp42, Mud2, and Lea1 with two RPGs whose splicing is inhibited in the npl3-1 strain (RPS21B and RPS18B), and with SCS22, which is unaffected (Supplemental Figures 1 and 4B). For each transcript tested we used four primer sets: one that amplifies the 5′UTR/beginning of the first exon (Figure 4A, primer set 1); one that amplifies either within the intron or just inside the second exon (primer set 2); one that amplifies further downstream in exon 2 (primer set 3); and one that amplifies the end of exon 2 and the 3′UTR (primer set 4). We measured changes in the accumulation of the HA-tagged proteins relative to RNA pol II by comparing the HA ChIP signal to the RNA pol II ChIP signal (Figure 4B, e.g. Prp42-HA/RNA Pol II).
Figure 4. Mutation of Npl3 results in decreased association of U1 and U2 snRNP with chromatin.
(A). Schematic diagram of the ribosomal protein genes and the primer sets used in the chromatin immunoprecipitation (ChIP) assays. (B). ChIPs were carried out using α-HA or α-RNA pol II antibodies in a Prp42-HA strain, Mud2-HA strain, or Lea1-HA strain, grown at 33 °C. Shown are the average amounts of HA-tagged protein bound relative to RNA pol II bound. (C). ChIPs were carried out using α-Npl3 or α-RNA pol II antibodies in a Prp42-HA strain grown at 33°C. Shown are the average amounts of Npl3 bound relative to RNA pol II bound. Error bars represent +/− SEM for each strain and primer set, n=3–4 biological replicates.
If Npl3 is required for U1 snRNP, Mud2, or U2 snRNP association with chromatin, mutation of NPL3 could confer two possible outcomes. First, it might alter the accumulation of these factors, resulting in a change in ChIP signal amplitude. Alternatively, mutation of NPL3 might delay the association of these factors, which would manifest as a change in distribution or position along the gene. We therefore monitored whether there is a change in the accumulation or distribution of Prp42-HA, Mud2-HA and Lea1-HA association with these genes in the npl3-1 strain. Strikingly, accumulation of Prp42-HA and Lea1-HA on chromatin was significantly decreased for RPS21B and RPS18B at primer sets 2–4 (Figure 4B), suggesting that functional Npl3 is required for maximal association of these proteins with chromatin at regions beyond the promoter. Conversely, the binding of Prp42-HA and Lea1-HA to the SCS22 gene was not significantly impaired, with the exception of a slight decrease at the promoter (Figure 4B). Although the co-transcriptional splicing factor association was low at SCS22, there was still significant enrichment over intergenic regions. In contrast to Prp42-HA and Lea1-HA, the Mud2-HA distribution was not strongly altered in the npl3-1 strain at any of the three genes (Figure 4B). Taken together, these data suggest that Npl3 functions to facilitate the co-transcriptional binding of U1 and U2 snRNP, but not Mud2, with target genes.
We also monitored the association of Npl3 itself with chromatin relative to RNA pol II by comparing the Npl3 ChIP signal to the RNA pol II ChIP signal (Figure 4C). Npl3 has been shown to bind promoter regions (Lei et al., 2001; Lei and Silver, 2002) and indeed we found that to be true for RPS21B, RPS18B, and SCS22. For RPS21B and RPS18B, Npl3 association increases within the intron (primer set 2) and remains associated in the downstream regions. In contrast, association of Npl3 with SCS22 does not change along the length of the gene. Notably, the increase in Npl3 association (primer set 2) with RPS21B and RPS18B appears to precede that of U1 and U2 snRNPs (primer set 3), but not that of Mud2, consistent with a role for Npl3 in facilitating U1 and U2 snRNP binding. The similar ChIP profiles of Npl3 in WT and npl3-1 is congruent with a previous report showing that the npl3-1 mutation does not impair RNA binding (Rollenhagen et al., 2007)
Finally, we tested whether Npl3 can influence U1 and U2 snRNP association with other RPGs whose splicing is affected by mutation of NPL3 (Supplemental Figures 1 and 4B). For all of the RPGs tested, Prp42-HA and Lea1-HA association was significantly impaired (1.8–5.6 fold) in the npl3-1 mutant compared to WT (Supplemental Figure 5), while Mud2-HA association was not. These data demonstrate that functional Npl3 is required for full U1 and U2 snRNP association with chromatin, and strongly suggest that SR proteins mediate efficient co-transcriptional binding of splicing factors to pre-mRNA.
Discussion
We have presented evidence that of the three SR-like proteins in budding yeast only Npl3 is involved in splicing. Deletion or mutation of NPL3 inhibits splicing of a large subset of transcripts comprised mainly of RPGs. Moreover, quantitative genetic profiling shows that NPL3, unlike GBP2 or HRB1, is most highly correlated with splicing factor genes (e.g. PRP11). In keeping with these genetic interactions, we demonstrate that Npl3 can co-immunoprecipitate components acting early in the splicing pathway, including BBP and U1 snRNP proteins. This is consistent with previous observations linking Npl3 to U1, BBP and Mud2 (Gavin et al., 2002; Gottschalk et al., 1998; Tardiff et al., 2007).
What is the molecular basis of Npl3′s role in splicing? Mammalian SR proteins influence splicing by promoting the usage of non-consensus splice sites, typically by binding to adjacent exonic sequences (Blencowe et al., 1999; Bourgeois et al., 2004; Hertel and Graveley, 2005). They can do so by either interacting directly with another SR-like splicing factor, such as U1-70K or U2AF, or by promoting a sub-optimal snRNA:pre-mRNA base-pairing interaction. Importantly, recent work in S. cerevisiae demonstrates that tethering of a canonical mammalian SR domain to an artificial pre-mRNA improves splicing when, and only when, splice site pairing is suboptimal, suggesting that this mechanism is conserved in yeast (Shen and Green, 2006). However, most of the splice sites in the npl3Δ-sensitive RPGs are canonical, suggesting alternative mechanisms for Npl3-stimulated splicing of the RPGs.
We hypothesize that Npl3 may increase the splicing efficiency of the RPGs by recruiting splicing factors to chromatin during transcription, thereby coupling splicing with transcription. Co-transcriptional coupling of spliceosome assembly likely results in more efficient splicing, as suggested by recent experiments in yeast in which splicing was artificially uncoupled from transcription (Tardiff et al., 2006). Mammalian SR proteins might serve to couple transcription to splicing as they are present at sites of actively transcribed genes in vivo, and interact with a variety of splicing factors (Blencowe et al., 1999; Bourgeois et al., 2004; Hertel and Graveley, 2005). This model is supported by recent work using a coupled in vitro system, which showed that enhancement of splicing by SR proteins only occurs when they are present at the commencement of transcription, but not if they are added after transcription is complete (Das et al., 2007). Moreover, it was shown that SR proteins and U1 snRNPs are specifically co-immunoprecipitated by RNA pol II, supporting a model where SR proteins facilitate splicing by enabling co-transcriptional recruitment of U1.
Npl3 is recruited early during transcription (Lei et al., 2001; Lei and Silver, 2002) and can be co-purified with RNA pol II (Dermody et al., 2008; Lei et al., 2001; Tardiff et al., 2007); therefore it is perfectly poised to facilitate co-transcriptional recruitment of the spliceosome. Here we have shown that Npl3 co-immunoprecipitates U1 snRNP proteins and BBP. Most importantly, we have used ChIP to demonstrate that mutational inactivation of NPL3 results in significant decreases in the association of U1 and U2 snRNPs with intron-containing genes whose efficient splicing is dependent on Npl3. While we cannot rule out that the affect of Npl3 on U1 and U2 chromatin association is indirect (e.g. via destabilizing U1 or U2 snRNP), our genetic and biochemical data strongly support a model in which Npl3 couples transcription to splicing by interacting with U1 and facilitating its binding to chromatin. To our knowledge, this is the first demonstration that SR protein function is required for efficient binding of splicing factors to chromatin. Interestingly, we have recently identified numerous genetic interactions between npl3Δ and mutants in histone modification factors (TLK and CG, unpublished data), further suggesting a link between Npl3 and chromatin.
Our observation that Npl3 is required for efficient splicing of RPGs fits well with several other findings. First, Npl3 was previously shown to preferentially co-immunopurify RPG transcripts (Kim Guisbert et al., 2005). Second, recent ChIP-chip experiments also show enrichment of Npl3 among RPGs (Yu et al., 2004), including RPGs whose splicing is impaired in npl3Δ. It is presently unclear what directs the specificity of Npl3 for RPGs. While an RNA binding site for Npl3 has not yet been defined, recent work suggests that it binds preferentially to GU- or GC-rich regions (K. Kim Guisbert and CG, unpublished data; Deka et al., 2008). Alternatively, Npl3 could be recruited to RPGs via an interaction with another factor that interacts specifically either with RPGs directly, or with the nascent RPG transcripts; extension of our recent observation that NPL3 interacts genetically with chromatin-modifying factors may provide important insight into this possibility (TLK and CG, unpublished data).
While the preceding discussion emphasizes the role of the yeast SR protein in coupling transcription to U1 and U2 snRNP recruitment, our data also suggest that a subset of Npl3 target pre-mRNAs may employ a mechanism akin to that generally observed for mammalian SR protein target pre-mRNAs. Npl3 is physically associated with Nam8, which is essential for the splicing of certain meiosis-specific pre-mRNAs (Spingola and Ares, 2000) that are distinguished by their non-consensus 5′ splice sites (Davis et al., 2000; Engebrecht et al., 1991; Leu and Roeder, 1999). Indeed, we have found that mutation of NPL3 inhibits splicing of meiosis-specific pre-mRNAs (TLK and CG, unpublished data). Thus future studies of Npl3 should provide important insight into SR protein function both in splice site selection and in the coupling of pre-mRNA splicing to transcription.
Experimental Procedures
Yeast strains
Strains were constructed as described in Supplemental Experimental Procedures and are listed in Supplemental Table 1. GFP-tagged strains are from the yeast GFP-tagged strain collection (Huh et al., 2003).
Growth analysis
Yeast cultures were grown to mid-log (OD600 0.5–0.8) and diluted to OD600 0.1, then further diluted by serial 5-fold dilutions, spotted on YEPD plates and incubated at 30°C or 37°C for 3 days. Analysis and generation of the E-MAP data was performed as previously described (Wilmes et al., this issue, Collins et al., 2007; Collins et al., 2006; Schuldiner et al., 2005).
Microarray and QPCR Analysis
All microarray analyses were carried out as previously described (Pleiss et al., 2007a) with the following modifications. Yeast cultures were grown to mid-log in 50 ml YEPD at 30°C. Where indicated the cultures were shifted to the non-permissive temperature (either 33°C or 37°C) for 30 min. Each microarray profile shown is an average of n=2–6 biological replicates. Gene axis was ordered by hierarchical clustering of the npl3Δ data, using the C Clustering Library version 1.32 (de Hoon et al., 2004). Data were clustered using average linkage and Pearson correlation as the distance measure. Figure 1 and Figure S2 are displayed using the resulting gene axis. All microarray data are available at the Gene Expression Omnibus (GEO) repository at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo); accession number GSE11491.
For QPCR, cDNA was generated as described previously (Pleiss et al., 2007a) with changes noted in the Supplemental Experimental Procedures. Primers are listed in Supplemental Table 2. Relative copy numbers were derived from duplicate measurements from a single biological sample. The percent unspliced RNA = relative copies intron/relative copies Exon2 × 100 (Supplemental Figure 1).
Co-immunoprecipitation assays
GFP-tagged strains (Huh et al., 2003) were grown to mid-log at 25°C. Cells were harvested at 3000 rpm, washed in TBS, and resuspended in IP buffer (50 mM Tris pH 7.4, 125 mM KCl, 0.1% NP40) plus protease inhibitors (PIs; 1μg/mL antipain, 1μg/ml leupeptin, 1μg/mL aprotonin, 1μg/mL pepstatin, and 100 μM phenylmethanesulphonylfluoride (PMSF), Sigma) and 12 units/mL RNasin Ribonuclease Inhibitor (Promega N2511). Cells lysates were generated by bead-beating followed by centrifugation at 10,000 RCF. For RNase A treated lysates, 200 μL of lysate was incubated with 5 mg RNase A (Sigma 6513) for 10 min. at room temperature prior to addition to the immunoprecipitation reaction. Protein G Beads (10 μL; Amersham) were pre-incubated with 7 μL of either polyclonal α-Npl3 (Siebel and Guthrie, 1996) or a non-specific antibody directed against the Brr2 protein (A. Kutach and CG; unpublished information) for 1 hr. Cell lysate (200 μL/IP) was added and incubated for 1–2 hrs at 4°C while rocking. IPs were washed in 4 × 1ml IP buffer plus PIs and RNasin, resuspended in 40 μL 1x Laemmli sample buffer, boiled 5 mins, and 20 μL of sample was separated by 10% SDS-PAGE and probed by Western Blot with either monoclonal α-GFP (Roche 1814460) or polyclonal α-Npl3 (Siebel and Guthrie, 1996). Total samples equivalent to 1/60th of the input were analyzed in parallel.
Chromatin Immunoprecipitation Assay
ChIPs were performed as previously described (Strahl-Bolsinger et al., 1997), with modifications noted in the Supplemental Experimental Procedures. Primers are listed in Supplemental Table 2. Each IP sample was run in duplicate, the calculated relative amounts were averaged and normalized to an averaged relative amount for an intergenic region. Each IP was then normalized to the average total input sample. The values reported in Figure 4B and Supplemental Figure 5 are the averaged α-HA IP/α-RNA pol IP and those reported in Figure 4C are the averaged α –Npl3 IP/α-RNA pol IP.
Supplementary Material
Acknowledgments
We thank K. Neugebauer and P. Silver for yeast strains; G. Whitworth for assistance with microarray analysis, D. Cameron and the Guthrie lab for critical reading of the manuscript, the authors of Wilmes et al. (this issue) for sharing data, M. Shales for Figure 2A and 2B generation. T.L.K was supported by a postdoctoral fellowship from the American Cancer Society and C.G is an American Cancer Society Research Professor of Molecular Genetics. This work was funded by N.I.H grant GM21119.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abovich N, Liao XC, Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- Arenas JE, Abelson JN. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci U S A. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C, Klatt C, Luhrmann R, Fabrizio P. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO Rep. 2002;3:875–880. doi: 10.1093/embo-reports/kvf172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ, Bowman JA, McCracken S, Rosonina E. SR-related proteins and the processing of messenger RNA precursors. Biochem Cell Biol. 1999;77:277–291. [PubMed] [Google Scholar]
- Bourgeois CF, Lejeune F, Stevenin J. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog Nucleic Acid Res Mol Biol. 2004;78:37–88. doi: 10.1016/S0079-6603(04)78002-2. [DOI] [PubMed] [Google Scholar]
- Bucheli ME, Buratowski S. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. Embo J. 2005;24:2150–2160. doi: 10.1038/sj.emboj.7600687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheli ME, He X, Kaplan CD, Moore CL, Buratowski S. Polyadenylation site choice in yeast is affected by competition between Npl3 and polyadenylation factor CFI. Rna. 2007;13:1756–1764. doi: 10.1261/rna.607207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong JL, Chang TH, Squazzo S, Hartzog G, Ares M., Jr Exploring functional relationships between components of the gene expression machinery. Nat Struct Mol Biol. 2005;12:175–182. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- Caspary F, Seraphin B. The yeast U2A′/U2B complex is required for pre-spliceosome formation. Embo J. 1998;17:6348–6358. doi: 10.1093/emboj/17.21.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007 doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi M, Krainer A, Gygi S, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Davis CA, Grate L, Spingola M, Ares M., Jr Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res. 2000;28:1700–1706. doi: 10.1093/nar/28.8.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Deka P, Bucheli ME, Moore C, Buratowski S, Varani G. Structure of the yeast SR protein Npl3 and Interaction with mRNA 3′-end processing signals. J Mol Biol. 2008;375:136–150. doi: 10.1016/j.jmb.2007.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS ONE. 2008;3:e3273. doi: 10.1371/journal.pone.0003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht JA, Voelkel-Meiman K, Roeder GS. Meiosis-specific RNA splicing in yeast. Cell. 1991;66:1257–1268. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gilbert W, Siebel CW, Guthrie C. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. Rna. 2001;7:302–313. doi: 10.1017/s1355838201002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot HV, Mann M, Seraphin B, Rosbash M, Luhrmann R, Fabrizio P. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. Rna. 1998;4:374–393. [PMC free article] [PubMed] [Google Scholar]
- Hacker S, Krebber H. Differential export requirements for shuttling serine/arginine-type mRNA-binding proteins. J Biol Chem. 2004;279:5049–5052. doi: 10.1074/jbc.C300522200. [DOI] [PubMed] [Google Scholar]
- He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- Henry M, Borland CZ, Bossie M, Silver PA. Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics. 1996;142:103–115. doi: 10.1093/genetics/142.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel KJ, Graveley BR. RS domains contact the pre-mRNA throughout spliceosome assembly. Trends Biochem Sci. 2005;30:115–118. doi: 10.1016/j.tibs.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff AM. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Guisbert K, Duncan K, Li H, Guthrie C. Functional specificity of shuttling hnRNPs revealed by genome-wide analysis of their RNA binding profiles. Rna. 2005;11:383–393. doi: 10.1261/rna.7234205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotovic KM, Lockshon D, Boric L, Neugebauer KM. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol Cell Biol. 2003;23:5768–5779. doi: 10.1128/MCB.23.16.5768-5779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA: 5′ss base pairing in yeast. Mol Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- Lei EP, Krebber H, Silver PA. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 2001;15:1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Silver PA. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes Dev. 2002;16:2761–2766. doi: 10.1101/gad.1032902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu JY, Roeder GS. Splicing of the meiosis-specific HOP2 transcript utilizes a unique 5′ splice site. Mol Cell Biol. 1999;19:7933–7943. doi: 10.1128/mcb.19.12.7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasiewicz R, Velazquez-Dones A, Huynh N, Hagopian J, Fu XD, Adams J, Ghosh G. Structurally unique yeast and mammalian serine-arginine protein kinases catalyze evolutionarily conserved phosphorylation reactions. J Biol Chem. 2007;282:23036–23043. doi: 10.1074/jbc.M611305200. [DOI] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Rapid, trascript-specific changes in splicing in response to environmental stress. Mol Cell. 2007a;27:928–937. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript Specificity in Yeast pre-mRNA Splicing Revealed by Mutations in Core Spliceosomal Components. PLoS. 2007b;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain JC, Legrain P. In vivo commitment to splicing in yeast involves the nucleotide upstream from the branch site conserved sequence and the Mud2 protein. Embo J. 1997;16:1759–1771. doi: 10.1093/emboj/16.7.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollenhagen C, Hodge CA, Cole CN. Following temperature stress, export of heat shock mRNA occurs efficiently in cells with mutations in genes normally important for mRNA export. Eukaryot Cell. 2007;6:505–513. doi: 10.1128/EC.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol Cell Biol. 2002;22:4723–4738. doi: 10.1128/MCB.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa H, Shimura Y. Requirement of protein factors and ATP for the disassembly of the spliceosome after mRNA splicing reaction. Nucleic Acids Res. 1991;19:6819–6821. doi: 10.1093/nar/19.24.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayani S, Janis M, Lee CY, Toesca I, Chanfreau GF. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol Cell. 2008;31:360–370. doi: 10.1016/j.molcel.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Shen H, Green MR. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes Dev. 2006;20:1755–1765. doi: 10.1101/gad.1422106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel CW, Feng L, Guthrie C, Fu XD. Conservation in budding yeast of a kinase specific for SR splicing factors. Proc Natl Acad Sci U S A. 1999;96:5440–5445. doi: 10.1073/pnas.96.10.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel CW, Guthrie C. The essential yeast RNA binding protein Np13p is methylated. Proc Natl Acad Sci U S A. 1996;93:13641–13646. doi: 10.1073/pnas.93.24.13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton DR, Chen S, Hitomi M, Kumagai C, Tartakoff AM. A yeast protein that bidirectionally affects nucleocytoplasmic transport. J Cell Sci. 1995;108(Pt 1):265–272. doi: 10.1242/jcs.108.1.265. [DOI] [PubMed] [Google Scholar]
- Spingola M, Ares M., Jr A yeast intronic splicing enhancer and Nam8p are required for Mer1p-activated splicing. Mol Cell. 2000;6:329–338. doi: 10.1016/s1097-2765(00)00033-2. [DOI] [PubMed] [Google Scholar]
- Stevens SW, Barta I, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. Biochemical and genetic analyses of the U5, U6, and U4/U6 × U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. Rna. 2001;7:1543–1553. [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Tardiff DF, Abruzzi KC, Rosbash M. Protein characterization of Saccharomyces cerevisiae RNA polymerase II after in vivo cross-linking. Proc Natl Acad Sci U S A. 2007;104:19948–19953. doi: 10.1073/pnas.0710179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Lacadie SA, Rosbash M. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell. 2006;24:917–929. doi: 10.1016/j.molcel.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Wilmes et al., this issue.
- Windgassen M, Krebber H. Identification of Gbp2 as a novel poly(A)+ RNA-binding protein involved in the cytoplasmic delivery of messenger RNAs in yeast. EMBO Rep. 2003;4:278–283. doi: 10.1038/sj.embor.embor763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windgassen M, Sturm D, Cajigas IJ, Gonzalez CI, Seedorf M, Bastians H, Krebber H. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol Cell Biol. 2004;24:10479–10491. doi: 10.1128/MCB.24.23.10479-10491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CM, Qiu H, Hu C, Dong J, Hinnebusch AG. Yeast cap binding complex impedes recruitment of cleavage factor IA to weak termination sites. Mol Cell Biol. 2007;27:6520–6531. doi: 10.1128/MCB.00733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Perriman R, Igel H, Howe KJ, Neville M, Ares M., Jr CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol Cell Biol. 1998;18:5000–5009. doi: 10.1128/mcb.18.9.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu MC, Bachand F, McBride AE, Komili S, Casolari JM, Silver PA. Arginine methyltransferase affects interactions and recruitment of mRNA processing and export factors. Genes Dev. 2004;18:2024–2035. doi: 10.1101/gad.1223204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.