Abstract
Structural integrity as well as mechanical stability of the parts of a molecular motor are crucial for its function. In this study, we used high-resolution force spectroscopy by atomic force microscopy to investigate the force-dependent opening kinetics of the neck coiled coil of Kinesin-1 from Drosophila melanogaster. We find that even though the overall thermodynamic stability of the neck is low, the average opening force of the coiled coil is >11 pN when stretched with pulling velocities >150 nm/s. These high unzipping forces ensure structural integrity during motor motion. The high mechanical stability is achieved through a very narrow N-terminal unfolding barrier if compared with a conventional leucine zipper. The experimentally mapped mechanical unzipping profile allows direct assignment of distinct mechanical stabilities to the different coiled-coil subunits. The coiled-coil sequence seems to be tuned in an optimal way to ensure both mechanical stability as well as motor regulation through charged residues.
Keywords: atomic force microscopy, neck coiled coil, unzipping, molecular motor
Biological motor proteins are nanoscopic machines that convert chemical energy into directed movement. Many members of the kinesin and myosin motor families are double-headed with the 2 heads joined by a double-helical coiled coil. In the case of the molecular motor kinesin, various studies have shown that for processive movement along microtubular tracks, a communication between the 2 heads through mechanical strain is essential (1–4). A recent study by Yildiz et al. (5) has shown that transmission of the strain generated by neck-linker docking can be critically affected by inserting artificial peptides between neck-linker and neck, thus introducing slack into the connection between the 2 heads. In turn, those findings suggest that a functional processive kinesin motor requires both heads to be joined rigidly together and that, in a working molecular motor, the structural integrity of the coiled-coil neck must be preserved. Substantial coiled-coil unwinding would cause a loss of the head-to-head communication and thus interfere with proper motility. In addition, it has been suspected that kinesin's neck may be able to temporarily shift out-of-register, a phenomenon possibly related to the limping behavior of kinesin (6). Moreover, interaction studies with the kinesin neck coiled coil have indicated that this structural element also plays an essential role for motor regulation (7, 8).
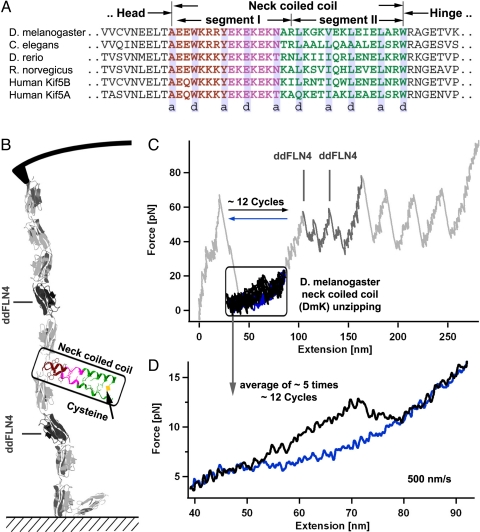
Kinesin-1's neck coiled coil contains highly unconventional sequence elements (9, 10) that seem to be conserved in Kinesin-1 motor proteins from bilateral animals (Fig. 1A) (11). Both structurally and thermodynamically, the Kinesin-1 neck can be divided into a highly conserved N-terminal segment I (brown and pink in Fig. 1A) containing amino acid residues that are atypical for coiled-coil structures, and a less conserved segment II that forms a canonical coiled coil with a classic knob-into-hole structure (green in Fig. 1A) (12, 13). Segment I is characterized by a so-called N-terminal hydrophobic collar, containing solvent-exposed bulky hydrophobic amino acid residues (brown in Fig. 1A) followed by a highly charged EKEK motif (pink in Fig. 1A) (12). How can those putatively destabilizing sequence elements be reconciled with the requirement of high mechanical stability of the kinesin neck? In this study, we used single molecule force spectroscopy by atomic force microscopy (AFM) to measure the mechanical stability of the neck coiled coil of kinesin from Drosophila melanogaster. We find that the unconventional N-terminal parts of the neck coiled coil, despite their relatively low thermodynamic stability, exhibit a surprising mechanical stability. The associated energy barriers are localized close to the N terminus and make this coiled coil mechanically as stable as conventional coiled coils if load is applied on the timescale of the kinetic cycle.
Fig. 1.
Experimental method for the mechanical coiled-coil unzipping. (A) Alignment of different Kinesin-1 neck sequences. (B) Schematic view of the experimental setup. The coiled coil (in this case the D. melanogaster neck coiled coil) is inserted between globular ddFLN1–5 domains, which act as handles for the proper attachment to the AFM. (C) Representative force trace of the whole protein construct. The saw-tooth pattern at forces of 50 pN corresponds to the unfolding of single ddFLN1–5 domains, whereas coiled-coil unzipping occurs in the low force regime (framed). (D) Averaged forward (black) and backward (blue) force trace obtained by averaging traces from 5 independent experiments where every experiment contains ≈12 forward and backward cycles.
Results and Discussion
Mechanical Stability of the Wild-Type Neck Coiled Coil.
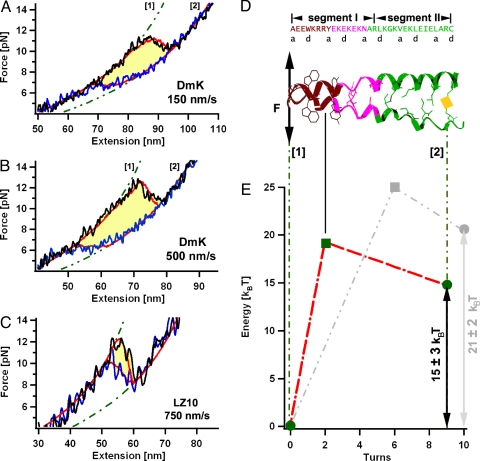
A scheme of our force-spectroscopy experiment is described in Bornschlögl et al. (14) and is shown in Fig. 1B. In brief, 5 Ig domains from Dictyostelium discoideum filamin (ddFLN1–5) were fused to the N terminus of the coiled-coil structures and serve as handles for attachment of the coiled coil to both AFM tip and glass substrate. In a typical force extension measurement (Fig. 1C), unfolding of the ddFLN1–5 handles results in a characteristic saw-tooth pattern indicating that a single molecule has attached in the desired unzipping geometry (for more details see Materials and Methods). Preceding the ddFLN1–5 unfolding pattern, unzipping of the coiled coil can be observed at very low forces and short extension (framed in Fig. 1C). To prevent complete separation of the coiled-coil strands, we introduced a disulfide cross-link at its C-terminal end via a cysteine (Fig. 1B). For the improvement of the signal-to-noise ratio, every coiled-coil structure was consecutively unzipped (black) and rezipped (blue) ≈12 times (Fig. 1C). The obtained force vs. distance traces for unfolding and refolding were averaged. Further averaging of such traces obtained from ≈5 different molecules increased the force resolution to <1 pN (Fig. 1D). In a first set of experiments, we obtained unzipping/rezipping force profiles of the D. melanogaster kinesin neck coiled coil (DmK) at 2 different pulling velocities (Fig. 2 A and B). We find that DmK starts to unfold mechanically at forces of 11–13 pN, whereas rezipping of the neck coiled coil only occurs if force is reduced to <6 pN. The hysteresis between unfolding and refolding cycles (yellow area in Fig. 2 A and B) acts as a measure for the energy dissipated to the heat bath per unzipping/rezipping cycle (≈15 kBT), indicating that unzipping/rezipping occurs far from equilibrium at the measured pulling velocities. This behavior is clearly distinct from the canonical coiled-coil behavior of a leucine zipper (14). For comparison, a force profile for unzipping/rezipping of the GCN4 variant LZ10 is shown in Fig. 2C. Though LZ10 unzipping/rezipping was performed at higher velocity (750 nm/s), a hysteresis between unzipping and rezipping is only barely observable (≈4 kBT). This small hysteresis indicates that LZ10 unzipping occurs very close to equilibrium. Toward lower pulling velocities the hysteresis will even vanish. From the speed-dependent force profiles, the underlying energy landscape of coiled-coil unzipping (see Fig. 2E) can be reconstructed using a nonequilibrium Monte Carlo simulation (15). For the simulation we consider a 2-state system that can switch with force-dependent rates kon/off[i](F) between the opened state [1] and the closed state [2]. By adjusting the rates so that the simulation results fit the experimental data, we can deduce the underlying energy landscape (for more details see Materials and Methods). By using the energy landscape, as shown in red in Fig. 2E, we were able to fully reproduce the experimental DmK coiled-coil force profiles (see red lines in Fig. 2 A and B). Surprisingly, the barrier width for unzipping (the distance in turns between the minima, shown as a green dot in Fig. 2E, at state [1] and the maxima, shown as a green square in Fig. 2E) lies at 2 opened turns, very close to the N terminus within the segment I containing noncanonical coiled-coil amino acid residues. Consequently, the opening of only 2 N-terminal coiled-coil turns will destabilize the complete neck and lead to unfolding. Mechanically, even though unfolding forces are similar, the DmK coiled coil is hence a more rigid structure as compared with the canonical coiled coil of LZ10, where the transition state is only reached after the opening of 6 turns (15) (see gray energy landscape in Fig. 2E). Both the energy barrier and the equilibrium free energy of the DmK are lower than those in LZ10. In contrast to LZ10, DmK unzipping occurs much further away from equilibrium and hence forces will drop at lower pulling velocities. However, even at pulling speeds as low as 150 nm/s, DmK is still mechanically as resistant as the LZ10 leucine zipper pulled at even 750 nm/s. Apparently, the DmK coiled coil can compensate its lower energy barrier by short-range interactions close to the N terminus that lead to the same mechanical stability as observed for the softer LZ10.
Fig. 2.
Mechanical unzipping profiles and mapped energy landscapes of the kinesin neck (DmK) and a leucine zipper. (A and B) Averaged forward (black) and backward (blue) force traces pulled at 150 nm/s (A) and 500 nm/s (B) measured on the DmK coiled coil. Red lines are the results of the kinetic Monte Carlo simulation. Green dotted lines represent the WLC elasticity (19) for polypeptides with fixed contour length. (C) For comparison, the averaged force traces from a canonical coiled coil (LZ10) at velocities of 750 nm/s are shown. (D) Sequence of the DmK-neck coiled coil aligned to the structure model of the comparable neck coiled coil from Rattus norwegicus (13). The C-terminal half (segment II) shows the canonical knob-into-holes scheme, whereas within segment I noncanonical amino acids are found. (E) Energy landscape of the DmK coiled coil (red) and the LZ10 coiled coil (gray) used for the Monte Carlo simulation.
The N-Terminal Segment I Is a Domain-Like Substructure.
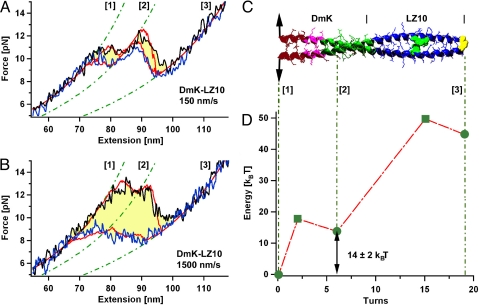
The apparent 2-state behavior we observe for DmK, where the entire coiled coil is acting as one unit being either in the totally folded or the totally unfolded state, does not reflect the sequential subdivision of DmK into 2 distinct segments (see Fig. 1A), from which one could expect an intermediate state at the boundary position of segment I and segment II. A possible explanation for the observed 2-state behavior could lie in the energetic costs of the nucleation seed associated with formation of coiled-coil structures (16–18). Even though segment II is a perfect coiled-coil forming sequence, it may just be too short to form a dimer stable enough to be observed in our mechanical assay after segment I has unfolded, thus leading to a cooperative 2-state rupture of the whole neck. To test this hypothesis and gain further insight into the mechanical substructure of the kinesin neck, we extended the canonical coiled coil of segment II by fusing an LZ10 zipper to its C terminus (see Fig. 3C), resulting in the elongated coiled coil DmK-LZ10.
Fig. 3.
Mechanical unzipping profiles and mapped energy landscapes of an elongated coiled coil containing the kinesin neck. (A) Averaged forward (black) and backward (blue) force traces of the DmK-LZ10 coiled coil obtained at a pulling velocity of 150 nm/s. Red lines represent the results of the Monte Carlo simulation. Between totally closed state [1] and totally opened state [3], an intermediate state with 6 ± 1 opened turns [2] is resolvable. (B) Averaged force traces at 1,500 nm/s. (C) Schematic structure of the DmK-LZ10 coiled-coil construct. The colors characterize the hydrophobic collar region (brown), the EKEK-motif (pink), the canonical segment II (green), and the canonical LZ10 sequence (blue). (D) Underlying energy landscape used for Monte Carlo simulations. We find, in agreement with the data for the DmK construct, a narrow N-terminal energy barrier at 2 ± 1 opened turns. The position of the intermediate state lays with 6 opened turns close to the boundary between segments I and II of the DmK coiled coil.
Averaged force-extension traces of DmK-LZ10 are shown in Fig. 3A for a pulling velocity of 150 nm/s. A new intermediate state (state [2] in Fig. 3A) can now be observed both in the unzipping trace (Fig. 3A, black) and rezipping trace (Fig. 3A, blue). The length gain upon unzipping determined by worm-like chain (WLC) elasticity (19) (Fig. 3A, green dotted lines) allows us to conclude that 6 N-terminal turns have opened in the intermediate state [2]. The boundary between opened and closed turns lies very close to the boundary of segments I and II. Apparently, segment II has merged seamlessly with the attached LZ10 zipper whereas segment I constitutes a separate domain. This result now directly corroborates the division of DmK into the 2 distinct segments proposed earlier (13). The intermediate state [2] is also populated during the rezipping process. This becomes obvious in Fig. 3A (blue trace) where 2 distinct hystereses can be observed (shaded in yellow). The first hysteresis marks formation of the nucleation seed for the canonical coiled coil ([3] → [2]) whereas formation of segment I leads to another hysteresis ([2] → [1]).
Modeling the data obtained at the different pulling velocities of 150 nm/s and 1,500 nm/s (Fig. 3 A and B) by using a nonequilibrium Monte Carlo simulation of a 3-state system again allows us to extract the underlying energy landscape. Consistent with the results obtained from the DmK construct, we find a very early barrier located only 2 turns from the N terminus (see Fig. 3D). The response to different pulling velocities of the rigid segment I with its very narrow barrier of 2 turns can now be directly compared with the softer canonical coiled coil comprising the segment II and LZ10. Although at 150 nm/s the transition [1] → [2] i.e., opening of segment I, starts at lower forces of ≈11 pN, this force increases to 13 pN at 1,500 nm/s (compare Fig. 3 A and B). Such a strong dependence on pulling velocity is characteristic for narrow barriers.
In contrast, opening of segment II together with LZ10 (transition [2] → [3]) occurs at nearly identical forces at both velocities, a consequence of the wide barrier (9 turns, see Fig. 3D). A simple estimate using the well known approximations for speed-dependence of unbinding forces based on Bell's model (20) leads to similar pulling-speed-dependent force increases.
Surprisingly, the equilibrium energy required for opening the 6 N-terminal turns of segment I within the elongated DmK-LZ10 construct is 14 kBT (Fig. 3D). This indicates that, despite its nonideal sequence pattern, segment I still provides a considerable thermodynamic stability. This finding is consistent with a study by Tripet et al. (12), who reported that much of the overall thermodynamic stability of the kinesin neck can be attributed to residues within segment I. As described above, based on its sequence peculiarities, segment I can be subdivided in a hydrophobic collar region and the repetitive EKEK sequence. However, the charged residue found at the d position in the EKEK sequence (see Fig. 1A) acts in an extremely destabilizing manner within a canonical coiled coil (9, 21). Consequently, the observed overall stability of segment I may only be achieved if the low stability of the EKEK region is overcompensated by the hydrophobic collar. Thus, only the specific combination of the hydrophobic collar with the unstable EKEK region may lead to a stable segment I.
Segment I Is a Nondividable Subdomain.
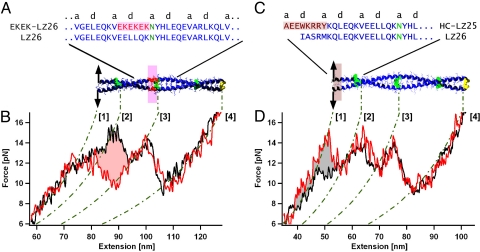
To test this hypothesis, we put both the isolated hydrophobic collar region as well as the isolated EKEK region into a canonical coiled-coil background and investigated their local influence on the total free energy. In the first construct EKEK-LZ26 (see Fig. 4A), we replaced one heptad of a triple leucine zipper LZ26 (described in ref. 14) by the EKEK motif. A direct comparison of unzipping traces obtained from EKEK-LZ26 and an unmodified LZ26 is shown in Fig. 4B, red and black traces, respectively. Consistent with its noncanonical nature, introduction of the EKEK sequence into the canonical LZ26 coiled coil leads to a drastic local destabilization of 10.6 ± 1.5 kBT (red shaded area in Fig. 4B).
Fig. 4.
Energetic contributions of single subfragments from the kinesin neck within a canonical coiled coil. (A) Amino acid sequence and schematic structure of the LZ26 coiled coil with inserted EKEK-sequence (red). (B) Averaged forward force trace of the resulting EKEK-LZ26 coiled coil (red), compared to averaged forward force trace of the unmodified LZ26 coiled coil (black). The shaded area indicates a local decrease in equilibrium energy of 10.6 ±1.5 kBT. (C) Amino acid sequence and schematic structure of the LZ26 coiled coil, where the N terminus is replaced by the hydrophobic collar sequence from DmK (brown). (D) Average forward force trace of the resulting HC-LZ25 coiled coil (red) compared with the unmodified LZ26 forward trace (black). The positive free energy of the hydrophobic collar sequence was calculated from the gray shaded area as 10 ± 1.5 kBT.
Similarly, we investigated the influence of the 8 amino acid hydrophobic collar sequences on the stability of the LZ26 coiled coil. By fusing the hydrophobic collar sequence to an LZ25 coiled coil we obtained the HC-LZ25 construct (see. Fig. 4C). A direct comparison between the profiles of LZ26 and HC-LZ25 is shown in Fig. 4D, black and red curves, respectively. The gray shaded area indicates a stabilization energy of 10 ± 1.5 kBT provided by the hydrophobic collar sequence.
Interestingly, the EKEK motif destabilizes a canonical coiled coil by approximately the same amount as the hydrophobic collar stabilizes it. Consequently, a direct combination of a hydrophobic collar and an EKEK motif, as we find in segment I, should not lead to a thermodynamically stable sequence. However, as can be seen from the energy landscape in Fig. 3D, segment I has a considerable thermodynamic stability of 14 kBT. This thermodynamic stability can only be explained if the hydrophobic collar and EKEK motif make favorable cooperative interactions with each other that are absent if they are introduced into a canonical coiled-coil sequence. Hence, to form a noncanonical stable coiled coil from a highly charged EKEK sequence, the N-terminal addition of another noncanonical motif, the hydrophobic collar sequence, seems to be necessary. In contrast, the addition of 8 amino acids of a canonical knob-into-hole sequence onto the EKEK sequence does not lead to a stable structure (see Fig. S1). Thus, segment I can be seen as a nondividable, cooperative subdomain within the neck coiled coil.
Speed-dependent force profiles of the EKEK-LZ26 construct (see Fig. S2 a and c) provide information about the role the EKEK motif plays for folding of the neck. Apparently, the EKEK motif introduces an additional folding barrier into the canonical coiled coil of LZ26. The same folding barrier was already observable in the DmK-LZ10 construct at the position of the EKEK sequence. Hence, the EKEK motif is, to a large degree, responsible for the relatively slow folding of the complete kinesin neck as compared with canonical sequences (for additional information see SI Text).
Adaptations of the Neck Coiled Coil for Proper Motor Function.
Why has nature chosen the complicated noncanonical amino acid residues of segment I instead of using a canonical knob-into-hole structure? It is important to note that beyond ensuring motility through mechanically stable linkage, the neck region (specifically the charged regions in segment I) is likely involved into regulation of the motor function. Two possible mechanisms for a regulation of motor function through the kinesin neck are discussed in the literature: Either the neck coiled-coil interacts directly with the tail (7, 22) or through the light chains (23). Our measurements put an important constraint on the mechanism of such interactions: Unfolding even of only segment I of the neck coiled coil through interactions with either the tail or the light chains is only possible if this interaction energy exceeds 14 kBT.
For a long time, researchers have speculated that the highly charged unconventional residues found in segment I lead to a mechanically labile structure that can open during motor action, thus providing the necessary flexibility to span the 16-nm separation of adjacent microtubule binding sites (9, 24). However, the discovery of the neck-linker region has rendered such a mechanism obsolete (25, 26). In fact, motor constructs with a covalent cross-link close to the N terminus of the coiled coil showed almost unimpaired motion (27). A recent study has pointed out that, in contrast to earlier belief, structural integrity of the coiled-coil structure providing a rigid link between the 2 heads is in fact essential for proper coordination of the 2 heads (5). Apparently, mechanical stability of the kinesin neck is important for proper function.
The mechanical strain between the heads in the 2-heads-bound stepping intermediate has been estimated to be 12 pN (28). In this geometry, it equals the unzipping force exerted on the neck coiled coil. The DmK neck is able to keep its intact folded structure even under those loads (compare Fig. 2). It is important to note that those forces only act for <10 ms during a kinesin step at physiological ATP concentration (29, 30), whereas at low ATP concentrations kinesin is thought to wait with only one head bound (31). The short transition state position of only 2 turns for unfolding of DmK helps to maintain its structural integrity at forces of ≈12 pN if they are applied only for a short time. We find that the probability for DmK to survive a load of 12 pN for 1 ms in our experimental setup is >97%, and for 10 ms it is still >87%. The average lifetimes for folding and unfolding of the DmK neck as a function of force are given in Fig. S3. Up to forces of 15 pN average lifetimes of the folded neck exceed the time span of 10 ms given by the kinetic cycle of the motor. Hence, neck-opening will happen only rarely in a moving Kinesin-1 motor and proper motility is ensured.
In summary, our results show that the DmK neck sequence has mechanical properties distinct from canonical coiled coils, like the leucine zipper. The unconventional coiled-coil motifs combine a high mechanical stability against unzipping with the possibility to regulate motor function through charged residues.
Materials and Methods
Mechanical Coiled-Coil Unzipping.
To apply mechanical unzipping force to different coiled-coil structures, we fused 5 Ig domains from ddFLN1–5 to the N terminus of the respective coiled coils (Fig. 1B). Adding ddFLN domains to protein structures does not alter their mechanical unfolding properties (32). The ddFLN1–5 domains serve as handles to anchor the protein between the cantilever tip and glass surface in the atomic force microscope. Unfolding of ddFLN1–5 provides a characteristic fingerprint in force-extension traces that allow identification of true single-molecule events. Mechanics of ddFLN1–5 have been extensively investigated before (33). Specifically, domain 4 (ddFLN4) unfolds via a mechanical unfolding intermediate, characterized by a double peak in the force-extension traces. We exploited this special unfolding behavior of the domain ddFLN4 to identify dimerized molecules that are anchored in the desired unzipping geometry (see Fig. 1C).
All curves that contain 2 ddFLN4 unfolding events show a coiled-coil unzipping event at low forces in the beginning of the force trace (framed in Fig. 1C for DmK construct). We find that the force signals of coiled-coil unzipping are close to the resolution limit of commercial AFM cantilevers. To increase force resolution in this regime, we recorded up to 30 forward and backward cycles on one single molecule. By averaging both forward and backward traces from up to 5 different measurements, we can further increase the force resolution to <1 pN. This allows detection of energetic differences, for example, due to point mutations down to 1 kBT (14).
Monte Carlo Simulation.
To model the mechanical unzipping experiment, we used a Monte Carlo simulation. A detailed description of the simulation is given in ref. 15. In brief, we define a complete energy landscape of the coiled coil Ecc(j) as a function of the number of open turns j. Each minimum of the energy landscape defines a state that the coiled coil can populate. As an example, the DmK-LZ10 energy landscape (see Fig. 3D) contains 3 states: fully closed [1], intermediate [2], and totally open [3]. A force tilts the energy landscape. Elastic contributions of stretching the unfolded polypeptide are included and lead to force-dependent motion of the position of the states as well as nonlinear dependence of barrier heights with force. The system can now switch between the states with a probability defined by the associated rates in the underlying kinetic scheme:
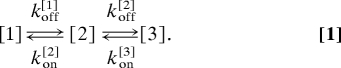 |
The rates kon/off[i](F) are associated to the transition state barriers of the underlying energy landscape by kon/off[i](F) = k0 exp(−ΔGon/off[i](F)/kBT). ΔGon[i](F) is defined as the free energy of activation separating state [i] from state [i − 1], and ΔGoff[i](F) is defined as the free energy of activation separating state [i] from state [i + 1]. We chose the preexponential factor as k0 = 107 1/s (34–36). At every time step a random variable determines whether a transition between the states in either forward or backward direction occurs. At low simulated pulling velocities, the probability to observe many alternating forward and backward transitions increases, as expected if thermodynamic equilibrium is approached (15). With the simulation we calculate multiple (n > 30) forward and backward force traces and average them with the same procedure used for the experimental data.
Cloning and Amino Acid Sequences.
All coiled-coil sequences were inserted in an pET28a(+) standard vector (Novagene) between NheI and XhoI restriction sites, directly after the ddFLN1–5 sequence that is described in ref. 33. The coiled-coil sequences we used were chosen to not contain repetitive DNA segments with codons optimised for expression in E. coli (Geneart). The complete amino acid sequence for DmK-LZ10 is AEEWKR R Y E K E K EKNARLKGKVEKLEIE L A R W R QLVGELEQKVEELLQ K N Y H L E Q E V A R L K Q LVGEC. The amino acid sequences of the LZ10 and LZ26 coiled coils are given in ref. 14, and the DmK sequence as well as local replacements, as for EKEK-LZ26 and HC-LZ25, are given in the text.
Supplementary Material
Acknowledgments.
We thank Manfred Schliwa, Elisabeth Wasner, Johann Jaud, Hendrik Dietz, and J. Christof M. Gebhard for stimulating discussions. Financial support of the German Excellence Initiative via the Nanosystems Initiative Munich and Deutsche Forschungsgemeinschaft SFB 486 is gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812620106/DCSupplemental.
References
- 1.Hackney DD, Stock MF, Moore J, Patterson RA. Modulation of kinesin half-site ADP release and kinetic processivity by a spacer between the head groups. Biochemistry. 2003;42:12011–12018. doi: 10.1021/bi0349118. [DOI] [PubMed] [Google Scholar]
- 2.Rosenfeld SS, Fordyce PM, Jefferson GM, King PH, Block SM. Stepping and stretching. How kinesin uses internal strain to walk processively. J Biol Chem. 2003;278:18550–18556. doi: 10.1074/jbc.M300849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crevel IM, et al. What kinesin does at roadblocks: The coordination mechanism for molecular walking. EMBO J. 2004;23:23–32. doi: 10.1038/sj.emboj.7600042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hahlen K, et al. Feedback of the kinesin-1 neck-linker position on the catalytic site. J Biol Chem. 2006;281:18868–18877. doi: 10.1074/jbc.M508019200. [DOI] [PubMed] [Google Scholar]
- 5.Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asbury CL, Fehr AN, Block SM. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stock MF, et al. Formation of the compact confomer of kinesin requires a COOH-terminal heavy chain domain and inhibits microtubule-stimulated ATPase activity. J Biol Chem. 1999;274:14617–14623. doi: 10.1074/jbc.274.21.14617. [DOI] [PubMed] [Google Scholar]
- 8.Adio S, Reth J, Bathe F, Woehlke G. Review: Regulation mechanisms of Kinesin-1. J Muscle Res Cell Motil. 2006;27:153–160. doi: 10.1007/s10974-005-9054-1. [DOI] [PubMed] [Google Scholar]
- 9.Tripet B, Vale RD, Hodges RS. Demonstration of coiled-coil interactions within the kinesin neck region using synthetic peptides. Implications for motor activity. J Biol Chem. 1997;272:8946–8956. doi: 10.1074/jbc.272.14.8946. [DOI] [PubMed] [Google Scholar]
- 10.Morii H, Takenawa T, Arisaka F, Shimizu T. Identification of kinesin neck region as a stable alpha-helical coiled coil and its thermodynamic characterization. Biochemistry. 1997;36:1933–1942. doi: 10.1021/bi962392l. [DOI] [PubMed] [Google Scholar]
- 11.Kirchner J, Woehlke G, Schliwa M. Universal and unique features of kinesin motors: Insights from a comparison of fungal and animal conventional kinesins. Biol Chem. 1999;380:915–921. doi: 10.1515/BC.1999.113. [DOI] [PubMed] [Google Scholar]
- 12.Tripet B, Hodges RS. Helix capping interactions stabilize the N-terminus of the kinesin neck coiled-coil. J Struct Biol. 2002;137:220–235. doi: 10.1006/jsbi.2002.4475. [DOI] [PubMed] [Google Scholar]
- 13.Thormahlen M, Marx A, Sack S, Mandelkow E. The coiled-coil helix in the neck of kinesin. J Struct Biol. 1998;122:30–41. doi: 10.1006/jsbi.1998.3986. [DOI] [PubMed] [Google Scholar]
- 14.Bornschlögl T, Rief M. Single molecule unzipping of coiled coils: Sequence resolved stability profiles. Phys Rev Lett. 2006;96:118102. doi: 10.1103/PhysRevLett.96.118102. [DOI] [PubMed] [Google Scholar]
- 15.Bornschlögl T, Rief M. Single-molecule dynamics of mechanical coiled-coil unzipping. Langmuir. 2008;24:1338–1342. doi: 10.1021/la7023567. [DOI] [PubMed] [Google Scholar]
- 16.Zimm BH, Bragg JK. Theory of the phase transition between helix and random coil in polypeptide chains. J Chem Phys. 1959;31:526. [Google Scholar]
- 17.Zitzewitz JA, Ibarra-Molero B, Fishel DR, Terry KL, Matthews CR. Preformed secondary structure drives the association reaction of GCN4–p1, a model coiled-coil system. J Mol Biol. 2000;296:1105–1116. doi: 10.1006/jmbi.2000.3507. [DOI] [PubMed] [Google Scholar]
- 18.Moran LB, Schneider JP, Kentsis A, Reddy GA, Sosnick TR. Transition state heterogeneity in GCN4 coiled coil folding studied by using multisite mutations and crosslinking. Proc Natl Acad Sci USA. 1999;96:10699–10704. doi: 10.1073/pnas.96.19.10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 20.Izrailev S, Stepaniants S, Balsera M, Oono Y, Schulten K. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys J. 1997;72:1568–1581. doi: 10.1016/S0006-3495(97)78804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tripet B, Wagschal K, Lavigne P, Mant CT, Hodges RS. Effects of side-chain characteristics on stability and oligomerization state of a de novo-designed model coiled-coil: 20 amino acid substitutions in position “d”. J Mol Biol. 2000;300:377–402. doi: 10.1006/jmbi.2000.3866. [DOI] [PubMed] [Google Scholar]
- 22.Friedman DS, Vale RD. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat Cell Biol. 1999;1:293–297. doi: 10.1038/13008. [DOI] [PubMed] [Google Scholar]
- 23.Cai D, Hoppe AD, Swanson JA, Verhey KJ. Kinesin-1 structural organization and conformational changes revealed by FRET stoichiometry in live cells. J Cell Biol. 2007;176:51–63. doi: 10.1083/jcb.200605097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozielski F, et al. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 1997;91:985–994. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- 25.Romberg L, Pierce DW, Vale RD. Role of the kinesin neck region in processive microtubule-based motility. J Cell Biol. 1998;140:1407–1416. doi: 10.1083/jcb.140.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 27.Tomishige M, Vale RD. Controlling kinesin by reversible disulfide cross-linking. Identifying the motility-producing conformational change. J Cell Biol. 2000;151:1081–1092. doi: 10.1083/jcb.151.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyeon C, Onuchic JN. Internal strain regulates the nucleotide binding site of the kinesin leading head. Proc Natl Acad Sci USA. 2007;104:2175–2180. doi: 10.1073/pnas.0610939104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter NJ, Cross RA. Mechanics of the kinesin step. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 30.Tomishige M, Stuurman N, Vale RD. Single-molecule observations of neck linker conformational changes in the kinesin motor protein. Nat Struct Mol Biol. 2006;13:887–894. doi: 10.1038/nsmb1151. [DOI] [PubMed] [Google Scholar]
- 31.Mori T, Vale RD, Tomishige M. How kinesin waits between steps. Nature. 2007;450:750–754. doi: 10.1038/nature06346. [DOI] [PubMed] [Google Scholar]
- 32.Bertz M, Rief M. Mechanical unfoldons as building blocks of maltose-binding protein. J Mol Biol. 2008;378:447–458. doi: 10.1016/j.jmb.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 33.Schwaiger I, Kardinal A, Schleicher M, Noegel AA, Rief M. A mechanical unfolding intermediate in an actin-crosslinking protein. Nat Struct Mol Biol. 2004;11:81–85. doi: 10.1038/nsmb705. [DOI] [PubMed] [Google Scholar]
- 34.Lapidus LJ, Eaton WA, Hofrichter J. Measuring the rate of intramolecular contact formation in polypeptides. Proc Natl Acad Sci USA. 2000;97:7220–7225. doi: 10.1073/pnas.97.13.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang WY, Gruebele M. Folding at the speed limit. Nature. 2003;423:193–197. doi: 10.1038/nature01609. [DOI] [PubMed] [Google Scholar]
- 36.Fierz B, et al. Loop formation in unfolded polypeptide chains on the picoseconds to microseconds time scale. Proc Natl Acad Sci USA. 2007;104:2163–2168. doi: 10.1073/pnas.0611087104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.