Abstract
Restrictocin belongs to a family of site-specific ribonucleases that kill cells by inactivating the ribosome. The restrictocin–ribosome binding rate constant was observed to exceed 1010 M−1 s−1. We have developed a transient-complex theory to model the binding rates of protein–protein and protein–RNA complexes. The theory predicts the rate constant as ka = ka0 exp(−ΔGel*/kBT), where ka0 is the basal rate constant for reaching the transient complex, located at the outer boundary of the bound state, by random diffusion, and ΔGel* is the average electrostatic interaction free energy of the transient complex. Here, we applied the transient-complex theory to dissect the high restrictocin–ribosome binding rate constant. We found that the binding rate of restrictocin to the isolated sarcin/ricin loop is electrostatically enhanced by ≈300-fold, similar to results found in other protein–protein and protein–RNA complexes. The ribosome provides an additional 10,000-fold rate enhancement because of two synergistic mechanisms afforded by the distal regions of the ribosome. First, they provide additional electrostatic attraction with restrictocin. Second, they reposition the transient complex into a region where local electrostatic interactions of restrictocin with the sarcin/ricin loop are particularly favorable. Our calculations rationalize a host of experimental observations and identify a strategy for designing proteins that bind their targets with high speed.
Keywords: binding rate, electrostatic rate enhancement, transient complex
Ribotoxins such as α-sarcin are ribonucleases that kill cells by cleaving a specific nucleotide located in a universally conserved motif called the sarcin/ricin loop (SRL) in 23S–28S rRNA. The cleavage disrupts the binding of elongation factors to the ribosome, thereby halting protein synthesis and triggering apoptosis. Besides the sequence specificity of the substrate, another remarkable feature of ribotoxins is the record-setting rate constant, exceeding 1010 M−1 s−1 at low ionic strengths, for binding the ribosome target (1, 2). These two features are shared to a large extent by a related family, represented by ricin, known as ribosome-inactivating proteins (3). We have developed an approach, referred to as the transient-complex theory, for calculating absolute binding rate constants (4–7). In this work the theory was applied to dissect the contributions to the high ribosome-binding rate of restrictocin, a close homolog of α-sarcin. The results provide molecular bases for a host of experimental observations on the roles of sequence motifs, both on the toxin and on the SRL, and suggest a strategy for designing proteins that bind their targets with high speed.
The high speed with which ribotoxins bind the ribosome target is likely of biological importance. The toxins must compete against elongation factors for binding to the SRL. When different proteins compete for the same binding site, binding rate, not binding affinity, is the key determinant for which protein gets bound (5, 8). The binding rate constants of elongation factors for the SRL-binding site on the ribosome are ≈108 M−1 s−1 at an ionic strength ≈100 mM (9, 10), which is even somewhat higher than the binding rate constant of restrictocin at the same ionic strength (1). It can thus be suggested that ribotoxins need the high binding speed to compete against elongation factors effectively.
The high speed and high specificity with which ribotoxins recognize the ribosome target have been under intense investigation. A number of structural motifs have been implicated as contributing factors [Fig. 1 and supporting information (SI) Fig. S1]. However, regarding the molecular bases for their roles, several questions remain open. (i) Structural (11) and biochemical data (12–15) have implicated the importance of the interaction between loop 4 of the toxin with the bulged-G motif, formed by G4319, of the RNA (Fig. 1B). Does this interaction make the same contribution to kcat/Km whether the substrate is the intact ribosome or an SRL-containing oligonucleotide? If so, what explains the significant difference in kcat/Km between these two substrates [106 vs. 104 M−1 s−1 at ionic strength ≈100 mM (1)]? (ii) Comparison of the SRL RNA structure bound to restrictocin with the unbound counterpart (16) shows that the tetraloop, consisting of G4323–A4326, becomes unfolded and the bases flip out (Fig. S2). At what point along the binding pathway does this conformational change occur? (iii) Loop 1 of ribotoxins has been implicated in its specific ribosome recognition because an α-sarcin mutant with this loop deleted was found to exhibit ribonuclease activity against naked rRNA and synthetic substrates but lacked the specific ability of the wild-type protein to degrade rRNA in an intact ribosome (17). How does loop 1 contribute to the recognition of the ribosome target by ribotoxins?
Fig. 1.
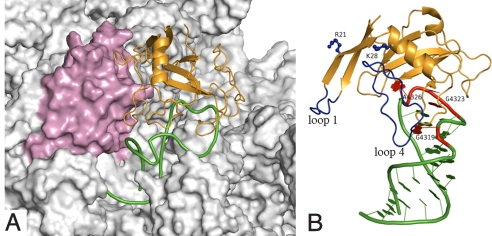
Interaction of restrictocin with the ribosome target. (A) Restrictocin is shown as orange ribbon; the SRL is shown as green ribbon; and ribosomal protein L14 is shown as pink surface. A full view of restrictocin bound to the whole 50S subunit of the ribosome is shown in Fig. S5A. (B) Restrictocin is shown as orange ribbon; its loop 1 and loop 4 are indicated in blue; and three residues, R21, K28, and K63 (hidden), studied by mutation, are shown as ball-and-stick. The SRL is shown as green ribbon; its tetraloop (G4323–A4326) and the bulged-G4319 are indicated in red. A view of B rotated by 180° around a vertical axis is shown in Fig. S1B.
Along the pathway to form a stereospecific native complex, two binding molecules first come into proximity with near-native orientation by translational and rotational diffusion, forming what we refer to as the transient complex (6). Conformational rearrangement and formation of short-range contacts then lead to the native complex. The location of the transient-complex ensemble, together with decomposition proposed here of contributions to the electrostatic interaction free energy of the transient complex, allows us to address the above questions. The calculations reproduce the observed high ribosome-binding rate constant of restrictocin, and through its dissection, they provide molecular insight for the restrictocin–ribosome system in particular and for achieving high binding speed in general.
Results and Discussion
The binding between the enzyme E, restrictocin, and the substrate S, either the intact ribosome or an SRL-containing oligonucleotide (referred to as the SRL RNA), and the subsequent cleavage can be described by the following kinetic scheme.
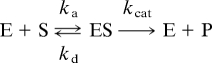 |
The overall catalytic efficiency is given by
Note that the binding rate constant ka, which is limited by the translational and rotational diffusional process that brings the enzyme and substrate into positional and orientational proximity, provides an upper boundary for kcat/Km. For catalytically “perfected” enzymes (i.e., those with kcat ≫ kd), kcat/Km approaches the diffusion-controlled limit ka. However, for the cleavage of either the SRL RNA or ribosome by restrictocin, experimental data (1, 2) indicate that kcat ≪ kd. In this situation
where Ka = ka/kd is the equilibrium constant for binding. We now present our calculation results for Ka and ka. The parallel results for the ribosome and the SRL RNA help dissect the contributions to the high specificity and high speed of restrictocin–ribosome recognition.
Electrostatic Contribution to kcat/Km.
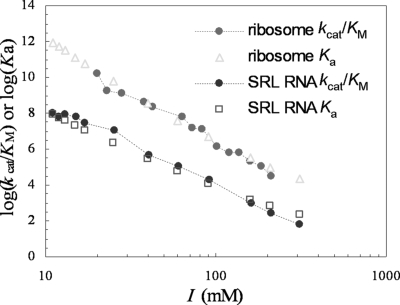
Experimental observation of strong dependence of kcat/Km on salt concentration (1, 2, 15) implicates significant electrostatic contribution to the binding of restrictocin with the ribosome. In Fig. 2, we display the calculated ionic-strength (I) dependences of the binding constants of restrictocin with the ribosome and with the SRL RNA. As the ionic strength increases from 10 to 310 mM, Ka decreases 7 × 107-fold and 6 × 105-fold, respectively, for the ribosome and the SRL RNA. The decreases can be attributed to salt screening of the electrostatic interactions with restrictocin. The stronger salt dependence of the ribosome can be attributed to the additional electrostatic interactions of its distal regions with restrictocin. At I = 60 mM, the electrostatic interaction free energies (ΔGel) of restrictocin with the ribosome and the SRL RNA are −6.7 and −4.9 kcal/mol, respectively. The contribution of the distal regions can be clearly seen by comparing the isoelectrostatic potential surfaces of the SRL RNA and the ribosome with the latter reaching a far greater distance (Fig. S3).
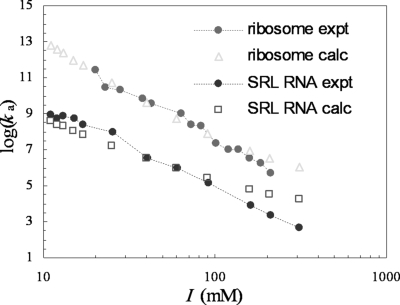
Fig. 2.
Comparison of ionic-strength dependences of experimental kcat/Km and calculated Ka.
Because kcat has very little dependence on salt (1), according to Eq. 3, Ka and kcat/Km are expected to exhibit the same dependence on ionic strength. Fig. 2 shows that the predicted salt effects agree very well with experimental results on kcat/Km for both the ribosome and the SRL RNA (1).
Effects of Mutations.
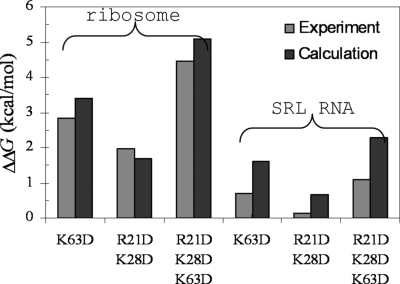
Korennykh et al. (1) studied the effects on kcat/Km of three basic residues, R21, K28, and K63, of restrictocin located away from the binding interface (Fig. 1B and Fig. S1); the effects were exclusively on Km. We found that the experimental results on the mutations can be reproduced well by their effects on ΔGel (Fig. 3). In particular, the triple mutation R21D/K28D/K63D was found to increase ΔGel (at I = 60 mM) by 5.1 and 2.3 kcal/mol, respectively, for the ribosome and the SRL RNA. The stronger effects of the mutations for binding with the ribosome than for the SRL RNA can again be attributed to the additional electrostatic interactions afforded by the distal regions of the ribosome.
Fig. 3.
Comparison of experimental and calculated effects of three mutations on the binding free energies of restrictocin with the ribosome and with the SRL RNA.
Calculation of Absolute Binding Rate Constant.
The results presented so far concern relative effects on the binding constant Ka. We now present results on the absolute binding rate constant ka for restrictocin binding to the ribosome and to the SRL RNA. The transient-complex theory predicts the rate constant as (4, 6, 7)
where ka0 is the basal rate constant for reaching the transient complex by random diffusion, ΔGel* is the average electrostatic interaction free energy of the transient complex, kB is the Boltzmann constant, and T is absolute temperature. Electrostatic attraction enhances the binding rate by increasing the probability of reaching the transient complex.
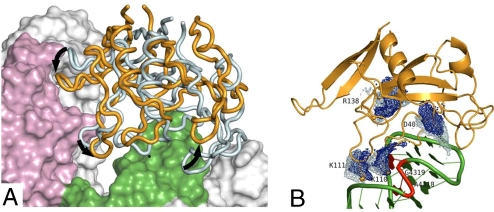
The transient complex has near-native separations and orientations between the two subunits but still misses a majority of the short-range interactions characterizing the native complex (4–7). It is located at the outer boundary of the bound-state energy well, which is dominated by short-range interactions (4). Fig. 4 presents the transient complexes for binding with the ribosome and with the SRL RNA. The two ensembles show distinct differences in relative separation and orientation between the subunits. By using the SRL RNA transient complex as reference, loop 1 and loop 4 of restrictocin in the ribosome transient complex move toward ribosomal protein L14 and the bulged-G, respectively; meanwhile the opposite end of restrictocin moves away from the ribosome. Overall the relative separation between the subunits in the ribosome transient complex is larger than in the SRL RNA counterpart: 4.5 ± 0.9 Å vs. 3.5 ± 1.0 Å (Fig. S4). The presence of the distal regions, in particular ribosomal protein L14, through short-range interactions, thus has a significant impact on the placement of the transient complex. Below it will be seen that the placement of the transient complex in turn has a significant impact on the strength of electrostatic interactions between the two subunits.
Fig. 4.
The transient-complex ensembles for the binding of restrictocin with the ribosome and with the SRL RNA. (A) Representative configurations of the transient complexes after superimposing the SRL. The restrictocin molecules in the transient complexes with the ribosome and with the SRL RNA are shown as orange and cyan tubes, respectively. The relative movement between the two restrictocin molecules is indicated by black arrows. The SRL is shown as green surface; ribosomal protein L14 is shown as pink surface. (B) Distributions of four atoms, each on a key restrictocin residue (K110, K111, R138, or D40), in the transient-complex ensembles, displayed on the structure of the native complex. The distributions are presented as isodensity surfaces, blue and cyan for the ribosome and the SRL RNA transient complexes, respectively. The atoms selected are Nζ, Cζ, and Cγ, respectively, for K, R, and D residues. The four residues and nucleotides A4318 and G4319 (shown in red) make the largest contributions to the difference between ΔGel*(rib_trun) and ΔGel*(SRL).
The basal rate constant, ka0, for reaching the transient complex by random diffusion, obtained by Brownian dynamics simulations, is 2.0 × 104 M−1 s−1 for binding with the ribosome and 4.4 × 104 M−1 s−1 for binding with the SRL RNA. These are within the ranges of values found for protein–protein and protein–RNA binding in previous studies (5–7). The 2-fold difference in ka0 can be accounted for by the fact that, although restrictocin and the SRL RNA have comparable translational diffusion constants (≈10 Å2/ns), the ribosome effectively has a translational diffusion constant of zero.
For the SRL RNA transient complex, with the SRL tetraloop in the unbound conformation (see below), the average electrostatic interaction free energy ΔGel* is reduced in magnitude by ≈50% relative to the counterpart ΔGel in the native complex. For example, at I = 25 mM, ΔGel* = −3.6 kcal/mol whereas ΔGel = −7.0 kcal/mol. According to Eq. 4, this value of ΔGel* corresponds to an electrostatic rate enhancement of ≈300-fold. The weakening of electrostatic attraction in the transient complex relative to that in the native complex and the magnitude of the rate enhancement are similar to what have been observed in other protein–protein and protein–RNA complexes (5–7). In contrast, ΔGel* for the ribosome transient complex (with the SRL tetraloop also in the unbound conformation) is nearly identical to ΔGel of the corresponding native complex. At I = 25 mM, ΔGel* = −9.1 kcal/mol whereas ΔGel = −9.8 kcal/mol. Correspondingly, the binding rate is enhanced electrostatically by 3 × 106-fold. The molecular basis of the dramatic 10,000-fold additional rate enhancement will be dissected below.
The predicted absolute binding rate constants for the ribosome and for the SRL RNA are shown in Fig. 5 as functions of ionic strength. As the ionic strength increases from 10 to 310 mM, ka decreases from 1013 M−1 s−1 to 106 M−1 s−1 for the ribosome and from 5 × 108 M−1 s−1 to 2 × 104 M−1 s−1 for the SRL RNA. These decreases in ka are comparable with those presented above for the binding constant Ka. The similar effects of ionic strength on ka and Ka have been observed on many protein–protein complexes (8, 18, 19) and also on protein–RNA complexes (20, 21). This widely observed phenomenon has been explained within the transient-complex theory, in that the transient complex is structurally close to the native complex and therefore experiences salt screening to nearly the same extent (18, 19).
Fig. 5.
Comparison of calculated and experimental results for ka at different ionic strengths. Note that the experimental results for ka shown here are scaled from those shown in Fig. 2 for kcat/Km by a constant factor, kd/kcat (see Eq. 2).
According to Eq. 2, ka can be obtained from kcat/Km if kd and kcat are known. The data of Korennykh et al. (2) indicate kd ≈8 s−1 and kcat ≈1 s−1 for restrictocin cleaving the SRL RNA at I = 15 mM. Given that ionic strength has similar effects on ka and Ka, the dissociation constant kd is expected to be independent of ionic strength. In addition, as already noted, kcat is independent of ionic strength (1). Therefore, these values of kd and kcat can be used for all ionic strengths. Fig. 5 shows the results for ka obtained from the kcat/Km data of Korennykh et al. (1) for the SRL RNA along with kd/kcat = 8. It can be seen that the calculated values of ka, without any adjustable parameters, agree rather well with the experimental results. A small discrepancy at high ionic strengths perhaps can be attributed to a slight increase of kd with increasing ionic strength.
For restrictocin cleaving the ribosome, the results for ka obtained from the kcat/Km data of Korennykh et al. (1) along with kd/kcat = 16 also agree rather well with the calculated ka values. The 2-fold increase in kd/kcat is consistent with a 2-fold decrease in kcat when the substrate is changed from the SRL RNA to the ribosome (1).
It is of interest to note that, although kcat/Km for the SRL RNA falls below the diffusion-controlled limit ka by ≈8-fold, kcat is increased significantly when the 3′ oxygen of the scissile phosphate is replaced with sulfur, allowing restrictocin to reach the diffusion-controlled limit (2). The experimental value of kcat/Km for the substituted substrate is very close to our calculated value of ka and, as expected from theory, is inversely proportion to solution viscosity.
Base Flip.
The calculations of ka provide an opportunity to address the question of when the base flip of the SRL tetraloop (Fig. S2) occurs during the binding process. Specifically, does the base flip occur before or after forming the transient complex? This question can be addressed because the values of ΔGel* calculated on the transient complex with the SRL tetraloop adopting either the bound or unbound conformation are significantly different. As presented above, ΔGel* at I = 25 mM with the tetraloop in the unbound conformation is −3.6 kcal/mol for the SRL RNA and −9.1 kcal/mol for the ribosome. When the bound conformation is used, these values become −6.2 and −10.1 kcal/mol, respectively. Correspondingly, the calculated values of ka would increase from 2 × 107 M−1 s−1 and 5 × 1010 M−1 s−1 to 109 M−1 s−1 and 3 × 1011 M−1 s−1, respectively, for the SRL RNA and the ribosome. The latter predictions are an order of magnitude too high compared with the experimental results.
The effect of conformational changes on the binding rate constant critically depends on their time scales (22). Fast conformational changes (e.g., those on a subnanosecond time scale) are shown to have very little effect on the binding rate (22, 23). That our ka calculations implicate the unbound conformation for the SRL tetraloop in the transient complex suggests that, without interactions with restrictocin, the base flip can occur, if at all, only on a slow time scale (e.g., microseconds or longer). This suggestion is supported by molecular dynamics simulations of the SRL RNA, which show that the tetraloop has only limited flexibility on a 25-ns time scale (24). The apparent “induced fit” implicated here is reminiscent of the observations of a recent molecular dynamics study (25), in which binding to a protein partner was found to speed up the conformational transitions of another protein significantly.
Dissecting the Difference in ka Between Ribosome and SRL RNA.
What is the molecular basis for the dramatic 10,000-fold additional rate enhancement provided by the ribosome over the SRL RNA? The additional rate enhancement results from the difference in ΔGel* between the ribosome and the SRL RNA. These two quantities will now be denoted as ΔGel*(rib) and ΔGel*(SRL), respectively, and have values of −9.1 and −3.6 kcal/mol at I = 25 mM. The first obvious source for the difference between ΔGel*(rib) and ΔGel*(SRL) is the distal regions of the ribosome, which are seen above to make a significant contribution to ΔGel calculated on the native complex and can likewise make a significant contribution to ΔGel* calculated on the transient complex (see also Fig. S3). To find this contribution, we truncated the ribosome in its transient complex to just the nucleotides corresponding to the SRL RNA. The resulting ΔGel*, denoted as ΔGel*(rib_trun), was −6.8 kcal/mol. The difference between ΔGel*(rib) and ΔGel*(rib_trun) can be viewed as the contribution of the distal regions of the ribosome and amounts to −2.3 kcal/mol at I = 25 mM.
Given that ΔGel*(rib_trun) and ΔGel*(SRL) arise from the interactions of the same pair of molecules, it is surprising that they differ by −3.2 kcal/mol, which accounts for 60% of the difference between ΔGel*(rib) and ΔGel*(SRL). What distinguishes ΔGel*(rib_trun) and ΔGel*(SRL) is the relative separation and orientation between the interacting molecules, as defined by the respective transient-complex ensembles (Fig. 4). Apparently, the restrictocin molecule is positioned and oriented in the ribosome transient complex to have much more favorable electrostatic interactions than in the SRL RNA transient complex.
To gain further insight, we decomposed ΔGel*(rib_trun) − ΔGel*(SRL) into contributions of individual residues/nucleotides. Four restrictocin residues (K111, K110, D40, and R138) and two SRL nucleotides (A4318 and G4319) were found to top the list. Together, they contribute 55% of the total difference of −3.2 kcal/mol between ΔGel*(rib_trun) and ΔGel*(SRL). The energetic contributions of these residues/nucleotides can be easily explained by the structural differences between the two transient complexes. As noted above, relative to the SRL RNA transient complex, loop 4 of restrictocin in the ribosome transient complex moves toward the bulged-G whereas the opposite end of restrictocin moves away from the ribosome (Fig. 4B). Residues K110, K111, and R138 are located in loop 4 of restrictocin, and G4319 is the bulged-G. The closer placement between these two elements in the ribosome transient complex allows them to have stronger electrostatic attraction. Residue D40 is located at the opposite end of restrictocin; its farther placement from the ribosome reduces their electrostatic repulsion.
An important reason for the repositioning of the ribosome transient complex is the short-range interactions of restrictocin loop 1 and ribosomal protein L14 (Fig. 4A). This finding presents an explanation for the role of loop 1 in the specificity of restrictocin–ribosome recognition suggested in previous studies (17).
Mechanisms of Binding Rate Enhancement.
Two well-known ways for increasing the binding rate of a protein with a target are long-range electrostatic attraction, which increases the probability of reaching the transient complex, and nonspecific binding to the surface of the target, which reduces the dimensionality of the search space. Past work on electrostatic rate enhancement has focused on manipulating distal charges (6, 7, 26–31). The parallel results presented above on the binding of restrictocin with the ribosome and with the SRL RNA suggest a strategy for using electrostatic rate enhancement: By reshaping the binding interface, the transient complex can be placed into a region in configurational space where there is strong electrostatic attraction.
In conclusion, we found that the binding of restrictocin with the ribosome achieves a record rate constant, exceeding 1010 M−1 s−1, by two synergistic mechanisms afforded by the distal regions of the ribosome. They provide additional long-range electrostatic attraction and, through short-range interactions, reposition the transient complex into a region where electrostatic attraction between the primary recognition elements is particularly strong. The rigorous modeling of the large, highly charged ribosome–restrictocin complex demonstrates that the tools used and developed here will be useful for studying other challenging systems.
Methods
Structure Preparation.
The structure of the native complex, shown in Fig. 1, between restrictocin and the ribosome was built by combining the structures of the 50 S subunit of the Haloarcula marismortui ribosome [Protein Data Bank (PDB) ID code 1jj2 (32)] and of the complex between restrictocin and a 29-mer oligonucleotide containing the SRL sequence [PDB ID code 1jbt (11)]. Specifically, the base pair of A4321 and G4328 (rat rRNA numbering) of 1jbt was aligned with the counterpart in 1jj2, and the tetraloop (G4323–A4326) and restrictocin of 1jbt were transplanted to 1jj2, giving the restrictocin–ribosome complex. The structure was energy minimized with all crystal ions retained and hydrogens added. [The experimental study of Korennykh et al. (1) was done on the rat ribosome; that structure has not been determined, but the elements important for interactions with restrictocin are conserved.]
For comparison, we also studied the binding of restrictocin with a 27-mer oligonucleotide containing the SRL sequence, referred to as the SRL RNA. The structure of the restrictocin–SRL RNA complex was obtained by truncating the ribosome to just nucleotides 4311–4337 in the above prepared restrictocin–ribosome complex.
The SRL tetraloop assumes different conformations before and after binding restrictocin (11, 16) (Fig. S2). This conformational change, referred to as base flip, was specifically taken into consideration in our calculations (see below). The unbound and bound conformations of the tetraloop were taken from 1jj2 (the 50S subunit of the H. marismortui ribosome) and 1jbt (the complex of restrictocin with the SRL-containing 29-mer), respectively.
Mutations of three residues, R21, K28, and K63, on restrictocin into Asp were modeled individually by InsightII and energy minimized. All energy minimizations were done by running the AMBER program.
Electrostatic Calculations.
Electrostatic calculations were performed by the Adaptive Poisson–Boltzmann Solver (APBS version 0.5.1) (33), with AMBER charges (34) and Bondi radii (35). Because of the high charge densities of the systems studied here, the full, nonlinear Poisson–Boltzmann (PB) equation was solved. The solute dielectric constant was set to 4, and the solvent dielectric constant for solvent was set to 74, corresponding to the temperature of 310 K in the experimental study of Korennykh et al. (1). Atomic charges were mapped to grid points with the cubic B-spline discretization, with the chgm flag set to spl2. Following our previous studies on protein–protein and protein–RNA binding (6, 7, 36, 37), the dielectric boundary was specified as the van der Waals surface by setting the srfm flag to mol and srad to 0.
Each APBS calculation started with a coarse grid with dimensions of 161 × 193 × 193 covering a volume of 299.4 Å × 379.2 Å × 372.7 Å around the solute molecule, with the “single Debye–Hückel” boundary condition. A central volume of 196.1 Å × 224.1 Å × 239.3 Å was then divided into 4 × 3 × 3 partitions, with a default value of 0.1 for the overlap parameter ofrac (i.e., every partition was enlarged in each direction by 10% of the initial length). Centered on each partition, an intermediate grid with dimensions of 161 × 193 × 193 and spacings of 0.83 Å × 1.0 Å × 0.98 Å was used to solve the PB equation for the second time. Finally, each partition was discretized into a grid with dimensions of 161 × 193 × 193 and spacings of 0.37 Å × 0.50 Å × 0.50 Å to obtain the electrostatic free energy of the solute molecule.
The electrostatic contribution to the binding free energy of restrictocin with the ribosome was calculated as (6, 7, 36, 37)
where Gel is the total electrostatic free energy (Coulomb plus solvation) of a solute molecule. Here, “complex” refers to the restrictocin–ribosome native complex, as prepared above. The APBS program gave overflow error when the solute included the whole 50S subunit of the ribosome. We took advantage of the fact that only the region of the ribosome close to the binding interface with restrictocin makes significant contributions to ΔGel and hence calculated ΔGel on a truncated ribosome. The truncated region was contained in a cubic box centered at the Cβ atom of restrictocin residue S46, located in the binding interface; residues of protein chains and nucleotides of RNA chains not fully inside the cube were truncated (Fig. S5A). To determine the appropriate value of the side length, bcut, of the cube, ΔGel was calculated at increasing bcut until a plateau was reached. As Fig. S5B shows, the value of ΔGel was essentially constant between bcut = 100 Å and bcut = 140 Å. The region of the 50S subunit inside the box with bcut = 120 Å was used in all of the calculations reported here.
The corresponding calculations for binding with the SRL RNA were carried out in a similar manner, except here no truncation was necessary.
Salt Effects.
Salt effects on binding were modeled by the dependence of ΔGel on salt concentration. In the experimental studies of Korennykh et al. (1), salt effects were reported by the slope n = –dln(kcat/Km)/dln[KCl]. However, it is important to recognize that the solutions in these studies contained 10 mM Tris buffer, which also contributes to the ionic strength. To model the combined effects, in the APBS calculations of ΔGel we included a 1:1 salt at the total concentration of the Tris buffer and the added KCl. The ion exclusion radius was 2 Å. The correction for the Tris buffer was especially important at low [KCl].
The calculation of ΔGel outlined above assumed the bound conformation for the ribosome (or the SRL RNA) even when restrictocin was absent. To obtain the full salt dependence of the binding free energy −kBTlnKa, we also accounted for salt effects on the conformational change of the SRL tetraloop. Specifically, the changes in the electrostatic solvation free energy with ionic strength were calculated for the ribosome (or the SRL RNA) in both the bound and unbound conformations. Their difference was then added to the salt dependence of ΔGel.
Mutational Effects.
The effect of a charge mutation on the binding affinity was predicted as
where the two terms on the right side denote ΔGel after and before the mutation, respectively. Note that the base flip of the SRL tetraloop does not affect ΔΔGel, because its contributions are canceled when the difference in Eq. 6 is taken.
Generation of Transient-Complex Ensemble and Calculation of ΔGel*.
The procedure for generating the transient complex was described (6, 7). Briefly, after mapping the energy landscape around the native complex in the 6-dimensional space of relative translation and rotation, the transient complex was identified with the outer boundary of the bound-state energy well (4). Relative translation was represented by a displacement vector r. Relative rotation was represented by a body-fixed unit vector and a rotation angle, χ, around this vector. In the native complex, r = 0 and χ = 0.
The short-range interaction energy around the native complex was represented by the total number, Nc, of contacts between two lists of representative atoms across the binding interface. The value of Nc decreases when the two subunits move away from the native complex; along the way the range of values available to χ shows a sharp increase. The value of Nc at the onset of this sharp increase, denoted as Nc*, defined the transient complex. The transient-complex ensemble consisted of all of the configurations with this Nc value. In the native complexes of the ribosome and the SRL RNA with restrictocin, the values of Nc were 42 and 31, respectively. From 1.7 × 106 configurations for the ribosome complex and 1.7 × 107 configurations for the SRL RNA complex, the values of Nc* were determined to be 15 and 17, respectively.
As in previous studies (5–7), 100 configurations from the transient-complex ensemble were randomly selected to calculate ΔGel*. For each configuration, the procedure was just as described for calculating ΔGel on the native complex. The results were then averaged to yield ΔGel*. Separate calculations of ΔGel* were carried out with the SRL tetraloop taking either the unbound or bound conformation.
Decomposition of Electrostatic Solvation Free Energy.
The Coulomb part of the electrostatic ΔGel* consists of contributions of individual pairs of atoms across the binding interface. However, the solvation part calculated by solving the nonlinear PB equation is not decomposable into contributions of individual atoms. However, we showed that the solvation free energy of the nonlinear PB equation can be reproduced well by a generalized Born (GB) method (38, 39). The GB solvation free energy consists of one-body and pair contributions (40). We used the results of our GB method for the solvation part of ΔGel* for its decomposition. The GB results were scaled (41) so the total solvation part matched that calculated by the nonlinear PB equation.
The decomposition was applied to analyze the difference between ΔGel*(rib_trun) and ΔGel*(SRL), which are caused by the interactions of the same pair of molecules in two different transient-complex ensembles. The one-body contributions of ΔGel* represent the changes in solvation free energy of individual atoms upon binding. They did not play any role in the difference between ΔGel*(rib_trun) and ΔGel*(SRL) because their sums were virtually identical in the two transient complexes. The pair contributions (Coulomb plus solvation) of ΔGel* arise from interactions across the binding interface. We assigned each pair contribution evenly to the partner atoms. These assigned values were then accumulated to the residue/nucleotide level. The difference in the accumulated values for each residue/nucleotide between the two transient complexes was reported as its contribution to the difference between ΔGel*(rib_trun) and ΔGel*(SRL).
Calculation of Basal Binding Rate by Force-Free Brownian Dynamics Simulations.
The basal binding rate constant was obtained from Brownian dynamics simulations as described in ref. 6. The translational diffusion constants of restrictocin and the SRL RNA were assigned values of 10.6 and 12.4 Å2/ns, respectively; the ribosome was assumed to be immobile. Trajectories of restrictocin (1.8 × 105 and 2.0 × 105) were launched to calculate ka0 for binding with the ribosome and the SRL RNA, respectively.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grant GM058187.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900291106/DCSupplemental.
References
- 1.Korennykh AV, Piccirilli JA, Correll CC. The electrostatic character of the ribosomal surface enables extraordinarily rapid target location by ribotoxins. Nat Struct Mol Biol. 2006;13:436–443. doi: 10.1038/nsmb1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korennykh AV, Plantinga MJ, Correll CC, Piccirilli JA. Linkage between substrate recognition and catalysis during cleavage of sarcin/ricin loop RNA by restrictocin. Biochemistry. 2007;46:12744–12756. doi: 10.1021/bi700931y. [DOI] [PubMed] [Google Scholar]
- 3.Korennykh AV, Correll CC, Piccirilli JA. Evidence for the importance of electrostatics in the function of two distinct families of ribosome inactivating toxins. RNA. 2007;13:1391–1396. doi: 10.1261/rna.619707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alsallaq R, Zhou H-X. Energy landscape and transition state of protein–protein association. Biophys J. 2007;92:1486–1502. doi: 10.1529/biophysj.106.096024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alsallaq R, Zhou H-X. Prediction of protein–protein association rates from a transition-state theory. Structure. 2007;15:215–224. doi: 10.1016/j.str.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Alsallaq R, Zhou H-X. Electrostatic rate enhancement and transient complex of protein–protein association. Proteins. 2008;71:320–335. doi: 10.1002/prot.21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin S, Zhou H-X. Prediction of salt and mutational effects on the association rate of U1A protein and U1 small nuclear RNA stem/loop II. J Phys Chem B. 2008;112:5955–5960. doi: 10.1021/jp075919k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schreiber G, Haran G, Zhou H-X. Fundamental aspects of protein–protein association kinetics. Chem Rev. 2009;109:839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005;24:4316–4323. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X, Gerczei T, Glover LT, Correll CC. Crystal structures of restrictocin–inhibitor complexes with implications for RNA recognition and base flipping. Nat Struct Biol. 2001;8:968–973. doi: 10.1038/nsb1101-968. [DOI] [PubMed] [Google Scholar]
- 12.Gluck A, Wool IG. Determination of the 28S ribosomal RNA identity element (G4319) for α-sarcin and the relationship of recognition to the selection of the catalytic site. J Mol Biol. 1996;256:838–848. doi: 10.1006/jmbi.1996.0130. [DOI] [PubMed] [Google Scholar]
- 13.Kao R, Davies J. Molecular dissection of mitogillin reveals that the fungal ribotoxins are a family of natural genetically engineered ribonucleases. J Biol Chem. 1999;274:12576–12582. doi: 10.1074/jbc.274.18.12576. [DOI] [PubMed] [Google Scholar]
- 14.Dey P, Tripathi M, Batra JK. Involvement of loops L2 and L4 of ribonucleolytic toxin restrictocin in its functional activity. Protein Pept Lett. 2007;14:125–129. doi: 10.2174/092986607779816131. [DOI] [PubMed] [Google Scholar]
- 15.Plantinga MJ, Korennykh AV, Piccirilli JA, Correll CC. Electrostatic interactions guide the active site face of a structure-specific ribonuclease to its RNA substrate. Biochemistry. 2008;47:8912–8918. doi: 10.1021/bi800592g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Correll CC, et al. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc Natl Acad Sci USA. 1998;95:13436–13441. doi: 10.1073/pnas.95.23.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Ortega L, et al. Deletion of the NH2-terminal β-hairpin of the ribotoxin α-sarcin produces a nontoxic but active ribonuclease. J Biol Chem. 2002;277:18632–18639. doi: 10.1074/jbc.M200922200. [DOI] [PubMed] [Google Scholar]
- 18.Zhou H-X. Disparate ionic-strength dependencies of on and off rates in protein–protein association. Biopolymers. 2001;59:427–433. doi: 10.1002/1097-0282(200111)59:6<427::AID-BIP1047>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 19.Zhou H-X. Association and dissociation kinetics of colicin E3 and immunity protein 3: Convergence of theory and experiment. Protein Sci. 2003;12:2379–2382. doi: 10.1110/ps.03216203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Law MJ, et al. The role of positively charged amino acids and electrostatic interactions in the complex of U1A protein and U1 hairpin II RNA. Nucleic Acids Res. 2006;34:275–285. doi: 10.1093/nar/gkj436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auweter SD, et al. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou H-X, Wlodek ST, McCammon JA. Conformation gating as a mechanism for enzyme specificity. Proc Natl Acad Sci USA. 1998;95:9280–9283. doi: 10.1073/pnas.95.16.9280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou H-X. Theory of the diffusion-influenced substrate binding rate to a buried and gated active site. J Chem Phys. 1998;108:8146–8154. [Google Scholar]
- 24.Spackova N, Sponer J. Molecular dynamics simulations of sarcin–ricin rRNA motif. Nucleic Acids Res. 2006;34:697–708. doi: 10.1093/nar/gkj470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bui JM, McCammon JA. Protein complex formation by acetylcholinesterase and the neurotoxin fasciculin-2 appears to involve an induced-fit mechanism. Proc Natl Acad Sci USA. 2006;103:15451–15456. doi: 10.1073/pnas.0605355103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharp K, Fine R, Honig B. Computer simulations of the diffusion of a substrate to an active site of an enzyme. Science. 1987;236:1460–1463. doi: 10.1126/science.3589666. [DOI] [PubMed] [Google Scholar]
- 27.Schreiber G, Fersht AR. Rapid, electrostatically assisted association of proteins. Nat Struct Biol. 1996;3:427–431. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- 28.Gabdoulline RR, Wade RC. Simulation of the diffusional association of barnase and barstar. Biophys J. 1997;72:1917–1929. doi: 10.1016/S0006-3495(97)78838-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vijayakumar M, et al. Electrostatic enhancement of diffusion-controlled protein–protein association: Comparison of theory and experiment on barnase and barstar. J Mol Biol. 1998;278:1015–1024. doi: 10.1006/jmbi.1998.1747. [DOI] [PubMed] [Google Scholar]
- 30.Elcock AH, Gabdoulline RR, Wade RC, McCammon JA. Computer simulation of protein–protein association kinetics: Acetylcholinesterase-fasciculin. J Mol Biol. 1999;291:149–162. doi: 10.1006/jmbi.1999.2919. [DOI] [PubMed] [Google Scholar]
- 31.Kiel C, Selzer T, Shaul Y, Schreiber G, Herrmann C. Electrostatically optimized Ras-binding Ral guanine dissociation stimulator mutants increase the rate of association by stabilizing the encounter complex. Proc Natl Acad Sci USA. 2004;101:9223–9228. doi: 10.1073/pnas.0401160101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: A new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cornell WD, et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1996;117:5179–5197. [Google Scholar]
- 35.Bondi A. Van der Waals volumes and radii. J Phys Chem. 1964;68:441–451. [Google Scholar]
- 36.Dong F, Vijayakumar M, Zhou H-X. Comparison of calculation and experiment implicates significant electrostatic contributions to the binding stability of barnase and barstar. Biophys J. 2003;85:49–60. doi: 10.1016/S0006-3495(03)74453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin S, Zhou H-X. Do electrostatic interactions destabilize protein-nucleic acid binding? Biopolymers. 2007;86:112–118. doi: 10.1002/bip.20708. [DOI] [PubMed] [Google Scholar]
- 38.Tjong H, Zhou H-X. GBr6: A parameterization-free, accurate, analytical generalized Born method. J Phys Chem B. 2007;111:3055–3061. doi: 10.1021/jp066284c. [DOI] [PubMed] [Google Scholar]
- 39.Tjong H, Zhou H-X. GBr6NL: A generalized Born method for accurately reproducing solvation energy of the nonlinear Poisson–Boltzmann equation. J Chem Phys. 2007;126:195102. doi: 10.1063/1.2735322. [DOI] [PubMed] [Google Scholar]
- 40.Still A, Tempczyk WC, Hawley RC, Hendrikson R. Semianalytical treatment of solvation for molecular mechanics and dynamics. J Am Chem Soc. 1990;112:6127–6129. [Google Scholar]
- 41.Tjong H, Zhou H-X. Accurate calculations of binding, folding, and transfer free energies by a scaled generalized Born method. J Chem Theory Comput. 2008;4:1733–1744. doi: 10.1021/ct8001656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.