Abstract
Glycogen storage disease type III (GSDIII) is a metabolic disorder characterized by a deficiency in the glycogen debranching enzyme, amylo-1,6-glucosidase,4-α-glucanotransferase (AGL). Patients with GSDIII commonly exhibit hypoglycemia, along with variable organ dysfunction of the liver, muscle or heart tissues. The AGL protein binds to glycogen through its C-terminal region, and possesses two separate domains for the transferase and glucosidase activities. Most causative mutations are nonsense, and how they affect the enzyme is not well understood. Here we investigated four rare missense mutations to determine the molecular basis of how they affect AGL function leading to GSDIII. The L620P mutant primarily abolishes transferase activity while the R1147G variant impairs glucosidase function. Interestingly, mutations in the carbohydrate-binding domain (CBD; G1448R and Y1445ins) are more severe in nature, leading to significant loss of all enzymatic activities and carbohydrate binding ability, as well as enhancing targeting for proteasomal degradation. This region (Y1445–G1448R) displays virtual identity across human and bacterial species, suggesting an important role that has been conserved throughout evolution. Our results clearly indicate that inactivation of either enzymatic activity is sufficient to cause GSDIII disease and suggest that the CBD of AGL plays a major role to coordinate its functions and regulation by the ubiquitin–proteasome system.
INTRODUCTION
Glycogen storage disease type III (GSDIII; OMIM 232400) is an autosomal recessive metabolic disorder resulting from a deficiency in amylo-1,6-glucosidase,4-α-glucanotransferase (AGL), the glycogen debranching enzyme (1,2). Afflicted individuals present with diverse symptoms. Clinically, this disorder is characterized by fasting hypoglycemia, growth retardation and hepatomegaly. However, more severe symptoms may also include liver cirrhosis, muscular weakness and cardiomyopathy.
AGL is expressed as a single protein containing two distinct active sites for transferase and glucosidase activities (3). During glycogenolysis, the removal of glucose-1-phosphate molecules from glycogen by phosphorylase is stalled when the enzyme encounters a branch point in glycogen four glucose residues away. To allow further glycogen degradation, the removal of this branch must be carried out by AGL. The 4-α-glucanotransferase (‘transferase’) activity relocates three glucose units of glycogen from one chain to another. This leaves one glucose unit at the branch point, which is subsequently released as glucose by the 1,6-glucosidase (‘glucosidase’) activity.
Studies using inhibitors that specifically target either activity have shown that both transferase and glucosidase function can occur independent of one another (4,5). Indeed, using yeast debranching enzyme as a model, the catalytic residues involved in both activities have been identified (6).
To date, over 50 AGL mutations have been discovered in GSDIII (1,2). The majority of these are nonsense mutations that lead to premature truncation of the protein. Among the missense variants, the G1448R mutation was the first identified (7), and is located within the carbohydrate-binding domain (CBD). We previously demonstrated that this mutant and a truncation mutant of the CBD (ΔCBD) are impaired in binding to carbohydrates and exhibit decreased stability owing to enhanced proteasomal-mediated degradation (8). Interestingly, an insertional mutation (Y1445ins) in close proximity to G1448R also occurs in the disease (9). However, whether any of these mutants are able to retain enzymatic activity in vitro was not investigated.
In this study, we characterize four rare missense mutations (L620P, R1147G, Y1445ins and G1448R) of GSDIII and determine that they alter diverse cellular functions. Detailed clinical features and genetic analyses will be described elsewhere. We discover mutants that impair either glucosidase or transferase activities without affecting the ability of AGL to bind to carbohydrates. In contrast, two mutants found in the glycogen-binding domain do exhibit enhanced proteasomal-mediated degradation, as well as significant abolishment of both activities. These results suggest that glycogen debranching requires the critical coordination of both enzymatic activities and protein stability of AGL, and that impairment of any one of these functions can lead to the development of GSDIII.
RESULTS
Functional analysis of the AGL mutants
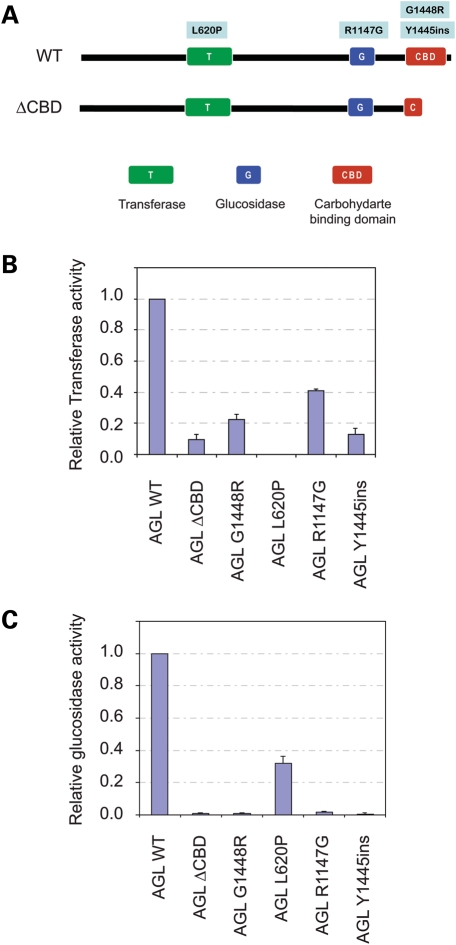
The molecular basis of how mutations in AGL disrupt enzymatic function is not well understood. In this study, we investigated four missense mutations of GSDIII (L620P, R1147G, Y1445ins and G1448R) that are located within the three domains of AGL (Fig. 1A). We expressed HA-tagged wild-type (WT), ΔCBD, and the missense variants of AGL to determine how the mutations affect its functions. Both glucosidase and transferase activities were assayed using fluorogenic substrates and the products analysed by high-performance liquid chromatography (10). Our results demonstrate that the L620P variant exhibits a complete loss of transferase activity while retaining approximately 35% of glucosidase activity compared with WT AGL (Fig. 1B and C). In contrast, the R1147G mutant produced negligible glucosidase activity while possessing 40% of transferase activity. Interestingly, CBD mutants (ΔCBD, Y1445ins and G1448R) display a severe loss (<20%) of both enzymatic functions. Similar results for glucosidase activity were obtained using glucose-β-cyclodextrin (GlcβCD) as a substrate (Supplementary Material, Fig. S1).
Figure 1.
Effect of amylo-1,6-glucosidase,4-α-glucanotransferase (AGL) mutations on enzymatic activities. (A) Schematic of wild-type AGL and the location of various mutants found in glycogen storage disease III. (B, C) Measurement of transferase and glucosidase activities. Cells expressing AGL wild-type or mutants were subjected to the indicated assays. Prior to the analysis, an aliquot of the cell lysate was used to determine AGL levels for subsequent normalization of activities (all are expressed relative to wild-type protein).
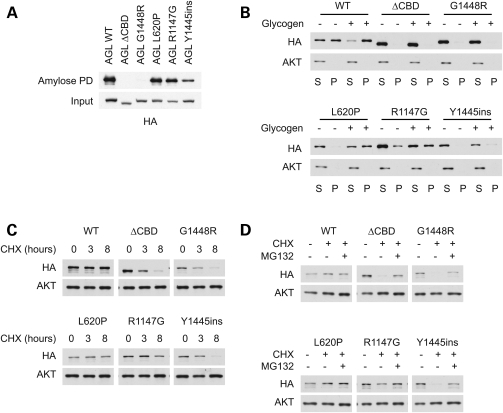
We previously demonstrated that both the G1448R variant and the ΔCBD mutants disrupt the ability of AGL to bind to carbohydrates (8). We therefore expressed the AGL proteins in cells, and subjected them to pull-down assays using amylose resin to evaluate carbohydrate-binding (Fig. 2A). Both AGL L620P and R1147G bind to amylose as well as AGL WT. In contrast, mutations in the CBD either abolish (ΔCBD and G1448R) or significantly impair (Y1445ins) its ability to bind to amylose.
Figure 2.
Effect of amylo-1,6-glucosidase,4-α-glucanotransferase (AGL) mutations on protein stability. (A) Binding of AGL mutants to amylose. Cell lysates expressing HA-tagged AGL proteins were subjected to a pull-down assay using amylose resin. Mutations in the carbohydrate-binding domain (CBD) impair binding to amylose. (B) Ability of AGL proteins to bind to glycogen in a pelleting assay. Cell lysates were fractionated by high-speed ultracentrifugation (100 000g) into a supernatant and a pellet, in the presence or absence of glycogen. Mutations in the CBD domain prevent AGL from pelleting with glycogen. (C) Stability of AGL mutants. Cells expressing HA-tagged AGL proteins were treated with cycloheximide (CHX) to stop protein translation, and the stability of AGL proteins was monitored. (D) Decreased stability of CBD mutants is rescued by proteasomal inhibitors. Cells were treated with CHX for 8 h as in (C) in the presence or absence of MG-132.
To further assess the effect of the mutations on binding to carbohydrates, we tested the ability of AGL variants to bind to and pellet with glycogen after high speed (100 000g) ultracentrifugation (Fig. 2B). Compared with control lanes, addition of glycogen to AGL proteins significantly increased the percentage of AGL WT, L620P and R1147G proteins in the pelleted (P) fraction. In contrast, CBD mutants remained primarily in the supernatant (S) fraction regardless of whether glycogen was added.
Enhanced proteasomal targeting by carbohydrate-binding domain mutants
Our previous studies also suggested that the CBD of AGL is important for regulating its stability (8). We therefore expressed the AGL proteins in cells and then monitored their levels after treatment with the protein translation inhibitor cycloheximide (CHX). Our analysis revealed that the WT protein had a half-life of at least 8 h, whereas the mutants that disrupt glycogen binding (ΔCBD, Y1445ins and G1448R) exhibited a significantly shorter half-life of 3 h or less (Fig. 2C). In contrast, the stability of AGL L620P and R1147G were comparable with that of WT.
The rapid degradation of the CBD mutants strongly suggests the involvement of the ubiqutin–proteasome system. Treatment of cells with the proteasome inhibitor MG-132 had no effect on AGL WT, L620P or R1147G levels, consistent with their observed long half-life (Fig. 2D). In contrast, the levels of the CBD mutants were significantly stabilized upon proteasomal inhibition. Taken together, this suggests that mutations in the CBD alter the stability of AGL proteins.
Ubiquitination and aggresome formation of AGL mutants
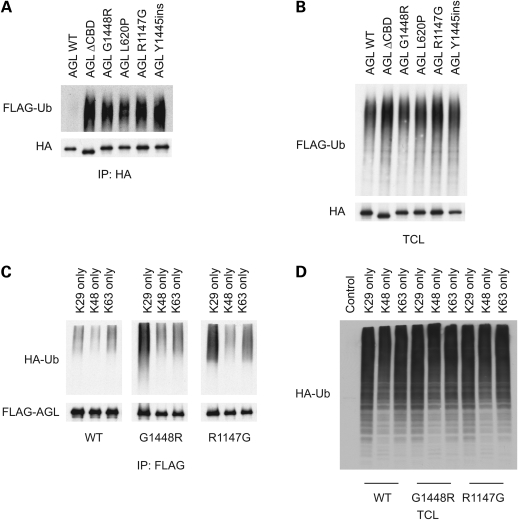
To further confirm the role of ubiquitination in the regulation of AGL, we co-expressed HA-AGL constructs together with FLAG-tagged ubiquitin (FLAG-UB) and assessed for the presence of high molecular-weight FLAG-UB conjugates in HA immunoprecipitates (Fig. 3A). In agreement with our previous model pointing to a role for the CBD in regulating AGL stability, we found that AGL ΔCBD, G1448R and Y1445ins exhibited enhanced ubiquitnation compared with AGL WT proteins. Unexpectedly, however, both AGL L620P and R1147G also displayed increased ubiquitination, despite no alteration in protein stability. As controls, we confirmed that similar expression of FLAG-UB conjugates was found in total cell lysates, thus excluding the possibility that AGL WT was not ubiquitinated because of different FLAG-UB expression. Since the AGL mutants display similar ubiquitination but differing stabilities, this suggests that additional signals are required to target AGL CBD mutants towards proteasomal degradation.
Figure 3.
Ubiquitination of amylo-1,6-glucosidase,4-α-glucanotransferase (AGL) mutants. (A) Mutant AGL proteins exhibit increased ubiquitination. Cells expressing HA-tagged AGL proteins, in addition with FLAG-UB were lysed in denaturing immunoprecipitation buffer, and lysates were immunoprecipitated with anti-HA antibodies. Immunoprecipitates were immunoblotted with the indicated antibodies. (B) Total cell lysates of the samples from (A) were probed for the indicated antibodies. (C) Mutant AGL proteins exhibit ubiquitination primarily through K29-mediated linkages. Cells were transfected as in (A) except HA-UB mutants (K29, K48, K63 only) were used. (D) Total cell lysates of the samples from (C) were probed for the indicated antibodies.
In addition to protein degradation, ubiquitination serves as a signal for other processes, such as DNA repair, protein trafficking and cell signaling (11,12). Moreover, polyubiquitin can occur through linkages of several lysine (K) residues, the most common ones being K29, K48 and K63. Therefore, we were interested in determining the type of ubiquitin linkage that occurs with the mutant AGL proteins. We repeated the above experiment except that we utilized HA-UB mutants, containing only K29, K48 or K63 residues and assessed for ubiquitin conjugates. As shown in Figure 3C, the primary type of ubiquitin linkage observed was K29-mediated. This is interesting, considering proteasomal-mediated degradation are primarily mediated by K48 linkages. Again, this is consistent with the fact that the non-CBD mutants do not display altered stability, and that additional signals involving the CBD are required for proteasomal targeting.
Previous results have indicated that mutation of the catalytic residues in the transferase or glucosidase domains do not affect the activity of the other, suggesting that these enzymatic activities can occur separately from one another (4–6). Since our results (Fig. 1) have indicated that the non-CBD mutants (L620P and R1147G) display diminished function of one enzymatic activity and complete loss of the other, it is possible that these AGL mutants are misfolded. It is known that misfolded proteins can undergo either the proteasomal pathway or a chaperone-mediated refolding route. In cells where the capacities of these processes are impaired, these misfolded proteins can be temporarily stored in compartments termed the ‘aggresome’ (13). Indeed, we have previously shown that this is the case for the ΔCBD and G1448R mutants (8).
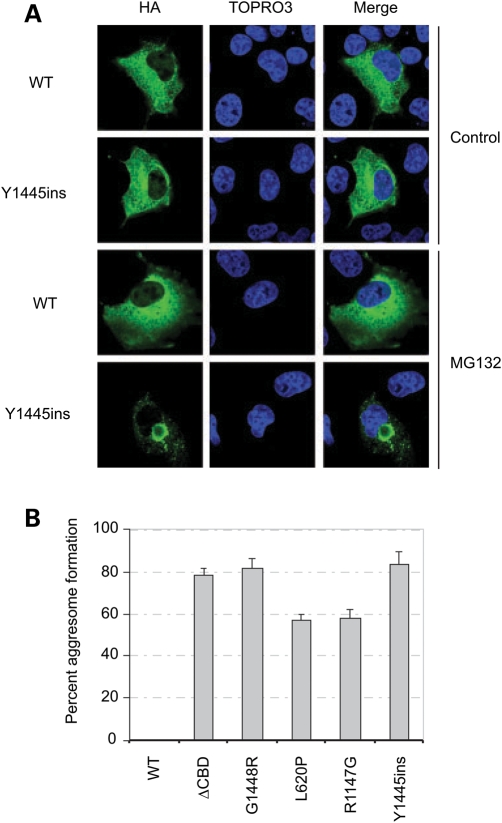
To determine whether the same occurs for the other AGL mutants, we transfected cells and treated them in the presence or absence of MG-132. As shown, the Y1445ins mutant formed aggresomes, whereas the WT protein did not (Fig. 4A). Upon quantification of the other mutants, we see that they all formed aggresomes (Fig. 4B), consistent with them all being ubiquitinated.
Figure 4.
Formation of amylo-1,6-glucosidase,4-α-glucanotransferase (AGL) mutants. (A) AGL mutants form aggresomes. Cells were transfected with HA-tagged AGL constructs for 12 h, and then 10 µm MG-132 was included during the media change and cells were allowed to continue for an additional 12 h before immunofluorescence analysis. AGL proteins were immunostained with HA-antibodies and the nuclei were visualized with TOPRO3.
DISCUSSION
To date, only a handful of point mutants of AGL have been identified in GSDIII (1,2). We have fortuitously analyzed four mutants that are found in the three major domains of AGL. The L620P mutant located in the transferase domain specifically abolishes transferase activity while retaining significant glucosidase function. On the other hand, the R1147G mutant in the glucosidase domain exhibits the reverse phenotype. Moreover, the selective loss of glucosidase or transferase activity in these cases may provide the first molecular basis for the specific GSDIIIc and GSDIIId subtypes, respectively (14,15).
Interestingly, the CBD mutants (Y1445ins and G1448R) are more severe in nature, leading to significant abolishment of both enzymatic activities, loss of carbohydrate-binding ability as well as enhanced targeting for proteasomal degradation. This suggests that this region/domain may have additional crucial functions. We would like to present this as a new subtype (GSDIIIe). In this case, mutations in the CBD prevent AGL from binding to glycogen, leading to complete loss of both transferase and glucosidase activity.
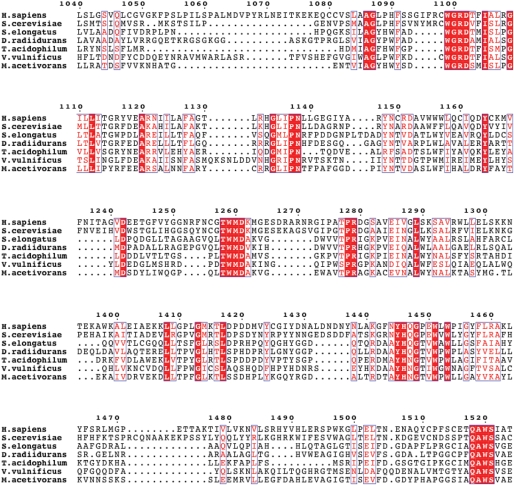
Sequence alignment of the mammalian AGL proteins reveals over 80% identity throughout the protein, and thus is not entirely informative (data not shown). We then decided to perform a sequence alignment between human, yeast and bacterial species to obtain further insight (Supplementary Material, Fig. S2). Limited homology exists within the first 1000 amino acids of human AGL compared with other species. Both L620 and R1147 are conserved in humans and yeast but not the bacterial species. However, considerable homology (and identity in some regions) is seen in the remaining region, primarily encompassing the CBD (Fig. 5). For example, in the region where the two CBD mutants are found (1445-YHQ/NG-1448), virtual identity has been conserved in these diverse organisms. This strongly suggests that this region serves an important purpose that has been conserved throughout evolution. Theoretical modeling analysis coupled with the Protein Homology/analogY Recognition Engine (16) indicates that this stretch of amino acids is embedded within the structure, and that the Y1445ins or G1448R mutations likely disrupt and open-up the structure (data not shown). Future structure–function analysis will shed more light on the importance of this region and may help us understand the regulation of AGL as well as other glycogen-associated enzymes and their dysregulation in their respective genetic diseases.
Figure 5.
Conservation of the carbohydrate-binding region of AGL. A sequence alignment showing regions of high similarity within the CBD of AGL. The species and the accession numbers for the sequences are as follows: Homo sapiens (P35573), Saccharomyces cerevisiae (Q06625), Synechococcus elongatus PCC 7942 (Q8KPV1), Deinococcus radiidurans (Q9RXW4), Thermoplasma acidophilum (Q9HL89), Vibrio vulnificus (Q8D4R0) and Methanosarcina acetivorans (Q8TMM8).
MATERIALS AND METHODS
Reagents and plasmids
Protein A/G agarose and antibodies against HA were from Santa Cruz Biotechnology (Santa Cruz). Anti-FLAG antibodies and anti-FLAG (M2) agarose beads were from Sigma (St Louis, MO, USA). The proteasomal inhibitor MG132 was purchased from EMD Biosciences (San Diego, CA, USA). Constructs for FLAG-Ubiquitin, HA-AGL WT, ΔCBD and G1448R mutants have been previously described (8). The additional missense mutations (L620P, R1147G and Y1445ins) were generated using the Stratagene Quickchange site-directed mutagenesis kit (La Jolla, CA, USA). All constructs were verified by DNA sequencing.
Cell culture and transfection
CV-1 origin, SV40 cells were grown in Dulbecco's Modified Eagle's Medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (Invitrogen). CV-1 origin, SV40 cells were transfected with FUGENE 6 (Roche) according to the manufacturer's instructions.
Preparation of cell lysates and immunoblotting
For isolation of total cell lysates, cells were washed twice in ice-cold phosphate-buffered saline, and then scraped in lysis buffer [50 mm Tris pH 7.5, 150 mm NaCl, 1% Triton X-100, 1 mm Na3VO4, 10 mm NaF and Complete ethylene diamine tetraacetic acid (EDTA)-free protease inhibitor]. The lysates were rocked end-over-end for 15 min at 4°C and then clarified by centrifugation at 14 000g for 10 min.
Glycogen-pelleting experiments were performed as previously described with minor modifications (17). Briefly, cells lysates were obtained as above and then incubated in the presence or absence of 5 mg/ml glycogen at 4°C for 30 min. After ultracentifugation at 100 000g for 90 min, supernatant (S) and pellet (P) fractions were obtained and the pellet was solubilized in lysis buffer by rotating end-over-end for 15 min. The lysates were then cleared one more time at 14 000g to remove unwanted debris.
Denaturing immunoprecipitations were performed as previously described with minor modifications (8,18). Briefly, cells were lysed in denaturing buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton, 1% SDS, 1 mm Na3VO4, 10 mm NaF and Complete EDTA-free protease inhibitor) and boiled for 10 min. Lysates were diluted 1:10 with the same buffer without SDS and incubated with the appropriate antibody/Protein A/G agarose beads for 2 h rotating at 4°C. Immunoprecipitates were extensively washed in lysis buffer (without SDS) and proteins were eluted at 95°C in SDS loading buffer, separated by SDS–PAGE and transferred to nitrocellulose.
Confocal fluorescence microscopy
Immunofluorescence studies were performed as previously described (19). Cells were grown on glass coverslips in 6- or 12-well dishes. Following the fixation with 10% formalin for 20 min, cells were permeabilized with 0.5% Triton X-100 for 5 min and then blocked with 1% bovine serum albumin, 1% ovalbumin and 2% goat serum for 1 h. For some experiments, methanol fixation at −20°C for 10 min was performed. Coverslips were incubated with primary and Alexa Fluor secondary antibodies in blocking solution and TOPRO3 (Invitrogen), and mounted on glass slides with Vectashield (Vector Laboratories). Cells were imaged using confocal fluorescence microscope (Olympus IX SLA). Images were then imported into Photoshop (Adobe Systems, Inc.) for processing.
Determination of glucosidase and transferase activities using fluorogenic substrates
The glucosidase and transferase activities of AGL proteins were determined using fluorogenic substrates as follows. A Wakosil-II 5C18 HG column (4.6 × 150 cm) was purchased from Wako Pure Chemicals (Osaka, Japan). Glcα1-4Glcα1-4Glcα1-4Glcα1-4(Glcα1-4Glcα1-4Glcα1-4Glcα1-6)Glcα1-4Glcα1-4Glcα1-4GlcPA (B4/84) and Glcα1-4Glcα1-4Glcα1-4Glcα1-4(Glcα1-6)Glcα1-4Glcα1-4GlcPA (B3/71) were prepared as described previously (10,20).
Aliquots of cell homogenates were centrifuged and 5 µl of the resulting supernatants were dissolved to 195 µl of 5 mm sodium maleate buffer (pH 6.0) containing 0.005% gelatin, 0.5 mm EDTA and 1.0 mm β-mercaptoethanol and the solutions were used as AGL preparations.
4-α-Glucanotransferase assay was carried out using B4/84 and maltohexaose as the donor substrate and the acceptor substrate, respectively (21). 4-α-Glucanotransferase removes maltotriosyl residue from the branch of B4/84 to transfer to the non-reducing-end glucosyl residue of maltohexaose. Thus, the products are maltononaose, the transfer product and fluorogenic B4/81. Some of B4/84 liberated is further hydrolyzed to PA-maltooctaose (G8PA) when glycogen debranching enzyme (GDE) preparation has an amylo-α-1,6-glucosidase activity. Thus, the activity of 4-α-glucanotransferase was calculated from the total amounts of the fluorogenic products, 65-O-α-glucosyl-PA maltooctaose and G8PA.
A mixture of B4/84 (3 µmol) and maltohexaose (1.0 mm) was incubated at 37°C for 5 min with 5 µl of each GDE preparation in 30 µl of 50 mm sodium maleate buffer (pH 6.0) containing 0.05% gelatin, 5 mm EDTA and 10 mm β-mercaptoethanol. To stop the enzymatic reaction, 200 µl of 0.1 m acetic acid was added to the reaction mixture, and the mixture was heated at 100°C for 5 min. The fluorogenic products, B4/81 and G8PA, in the enzymatic reaction mixtures were separated and quantified by high-performance liquid chromatography (HPLC).
Amylo-α-1,6-Glucosidase assay was done using B3/71 as the substrate. Amylo-α-1,6-glucosidase hydrolyzes B3/71 to glucose and PA-maltoheptaose (G7PA) (21). The activity was calculated from the amount of G7PA. B3/71 (3 µm) was incubated at 37°C for 5 min with 5 µl of each GDE preparation in 30 µl of 50 mm sodium maleate buffer (pH 6.0) containing 0.05% gelatin, 5 mm EDTA and 10 mm β-mercaptoethanol. To the enzymatic reaction mixture, 200 µl of 0.1 m acetic acid was added and the mixture was heated at 100°C for 5 min. G7PA was separated and quantified by HPLC.
HPLC: the column used was a Wakosil-II 5C18 HG column (4.6 × 150 mm), and the elution buffer was 0.05 m ammonium acetate buffer of pH 4.3 containing 0.1% 1-butanol at a flow rate of 1.3 ml/min. The elution was monitored by measuring the fluorescence (excitation wavelength, 320 nm; emission wavelength, 400 nm).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (5R01DK060597-07 to A.R.S); and the American Heart Association (0930045N to A.C).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Junyu Xiao for help with ClustalW and ESPrint alignment software, and molecular modeling techniques.
Conflict of Interest statement. The authors have no conflicts of interest to declare.
REFERENCES
- 1.Shen J.J., Chen Y.T. Molecular characterization of glycogen storage disease type III. Curr. Mol. Med. 2002;2:167–175. doi: 10.2174/1566524024605752. [DOI] [PubMed] [Google Scholar]
- 2.Lucchiari S., Santoro D., Pagliarani S., Comi G.P. Clinical, biochemical and genetic features of glycogen debranching enzyme deficiency. Acta Myol. 2007;26:72–74. [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor C., Cox A.J., Kernohan J.C., Cohen P. Debranching enzyme from rabbit skeletal muscle. Purification, properties and physiological role. Eur. J. Biochem. 1975;51:105–115. doi: 10.1111/j.1432-1033.1975.tb03911.x. [DOI] [PubMed] [Google Scholar]
- 4.Gillard B.K., Nelson T.E. Amylo-1,6-glucosidase/4-alpha-glucanotransferase: use of reversible substrate model inhibitors to study the binding and active sites of rabbit muscle debranching enzyme. Biochemistry. 1977;16:3978–3987. doi: 10.1021/bi00637a007. [DOI] [PubMed] [Google Scholar]
- 5.Liu W., Madsen N.B., Braun C., Withers S.G. Reassessment of the catalytic mechanism of glycogen debranching enzyme. Biochemistry. 1991;30:1419–1424. doi: 10.1021/bi00219a036. [DOI] [PubMed] [Google Scholar]
- 6.Nakayama A., Yamamoto K., Tabata S. Identification of the catalytic residues of bifunctional glycogen debranching enzyme. J. Biol. Chem. 2001;276:28824–28828. doi: 10.1074/jbc.M102192200. [DOI] [PubMed] [Google Scholar]
- 7.Okubo M., Kanda F., Horinishi A., Takahashi K., Okuda S., Chihara K., Murase T. Glycogen storage disease type IIIa: first report of a causative missense mutation (G1448R) of the glycogen debranching enzyme gene found in a homozygous patient. Hum. Mutat. 1999;14:542–543. doi: 10.1002/(SICI)1098-1004(199912)14:6<542::AID-HUMU15>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 8.Cheng A., Zhang M., Gentry M.S., Worby C.A., Dixon J.E., Saltiel A.R. A role for AGL ubiquitination in the glycogen storage disorders of Lafora and Cori's disease. Genes Dev. 2007;21:2399–2409. doi: 10.1101/gad.1553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okubo M., Horinishi A., Takeuchi M., Suzuki Y., Sakura N., Hasegawa Y., Igarashi T., Goto K., Tahara H., Uchimoto S., et al. Heterogeneous mutations in the glycogen-debranching enzyme gene are responsible for glycogen storage disease type IIIa in Japan. Hum. Genet. 2000;106:108–115. doi: 10.1007/s004390051017. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe Y., Makino Y., Omichi K. Fluorogenic substrates of glycogen debranching enzyme for assaying debranching activity. Anal. Biochem. 2005;340:279–286. doi: 10.1016/j.ab.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 11.Kirkin V., Dikic I. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr. Opin. Cell Biol. 2007;19:199–205. doi: 10.1016/j.ceb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Ravid T., Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopito R.R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 14.Ding J.H., de Barsy T., Brown B.I., Coleman R.A., Chen Y.T. Immunoblot analyses of glycogen debranching enzyme in different subtypes of glycogen storage disease type III. J. Pediatr. 1990;116:95–100. doi: 10.1016/s0022-3476(05)81652-x. [DOI] [PubMed] [Google Scholar]
- 15.Van Hoof F., Hers H.G. The subgroups of type 3 glycogenosis. Eur. J. Biochem. 1967;2:265–270. doi: 10.1111/j.1432-1033.1967.tb00134.x. [DOI] [PubMed] [Google Scholar]
- 16.Bennett-Lovsey R.M., Herbert A.D., Sternberg M.J., Kelley L.A. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 17.Wang J., Stuckey J.A., Wishart M.J., Dixon J.E. A unique carbohydrate binding domain targets the Lafora disease phosphatase to glycogen. J. Biol. Chem. 2002;277:2377–2380. doi: 10.1074/jbc.C100686200. [DOI] [PubMed] [Google Scholar]
- 18.Gentry M.S., Worby C.A., Dixon J.E. Insights into Lafora disease: malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc. Natl Acad. Sci. USA. 2005;102:8501–8506. doi: 10.1073/pnas.0503285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang M., Liu J., Cheng A., Deyoung S.M., Chen X., Dold L.H., Saltiel A.R. CAP interacts with cytoskeletal proteins and regulates adhesion-mediated ERK activation and motility. EMBO J. 2006;25:5284–5293. doi: 10.1038/sj.emboj.7601406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto E., Makino Y., Omichi K. Active site mapping of amylo-alpha-1,6-glucosidase in porcine liver glycogen debranching enzyme using fluorogenic 6-O-alpha-glucosyl-maltooligosaccharides. J. Biochem. 2007;141:627–634. doi: 10.1093/jb/mvm065. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe Y., Makino Y., Omichi K. Donor substrate specificity of 4-alpha-glucanotransferase of porcine liver glycogen debranching enzyme and complementary action to glycogen phosphorylase on debranching. J. Biochem. 2008;143:435–440. doi: 10.1093/jb/mvm240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.