Summary
DNA is subject to a multitude of oxidative damages generated by oxidizing agents from metabolism, from exogenous sources and by ionizing radiation. Guanine is particularly vulnerable to oxidation and the most common oxidative product, 8-oxoguanine (8-oxoG), is the most prevalent lesion observed in DNA molecules. 8-oxoG can form a normal Watson-Crick pair with cytosine (8-oxoG:C), but it can also form a stable Hoogsteen pair with adenine (8-oxoG:A) leading to a G:C →T:A transversion after replication. Fortunately, 8-oxoG is recognized and excised by either of two DNA glycosylases of the base excision repair (BER) pathway, formamidopyrimidine-DNA glycosylase (Fpg) and 8-oxoguanine DNA glycosylase (Ogg). While Clostridium acetobutylicum Ogg DNA glycosylase can specifically recognize and remove 8-oxoG it displays little preference for the base opposite the lesion, which is unusual for a member of the Ogg1 family. This paper describes the crystal structures of CacOgg in its apo-form and in complex with 8-oxo-2’-deoxyguanosine. A structural comparison between the apo and liganded forms of the enzyme reveals a structural reorganization of the C-terminal domain upon binding of 8-oxoG similar to that reported for hOGG1. A structural comparison of CacOgg with hOGG1 in complex with 8-oxoG containing DNA provides a structural rationale for the lack of opposite base specificity displayed by CacOgg.
Introduction
DNA is subject to a plethora of oxidative damages generated by radiation and oxidizing agents, which originate from an organism’s own metabolism and from exogenous sources. Guanine is particularly vulnerable to oxidation and the most common product, 7,8-dihydro-8-oxoguanine (8-oxoG), is the most prevalent oxidative lesion observed in DNA molecules.1 8-oxoG can form a normal Watson-Crick base pair with cytosine (8-oxoG:C), but can also form a stable Hoogsteen pair with adenine (8-oxoG:A) 2; 3 leading to a G:C →T:A transversion after replication. 2; 4 Fortunately, 8-oxoG is recognized and repaired by the base excision repair (BER) pathway 5; 6 via formamidopyrimidine-DNA glycosylase (Fpg) or 8-oxoguanine DNA glycosylase (Ogg). In humans, the repair of 8-oxoG is initiated by human OGG1 (hOGG1) 7; 8; 9; 10; 11; 12; 13 that excises 8-oxoG opposite C with the remainder of BER enzymes restoring the G:C base pair before replication. After replication has occurred and in the event of a newly formed 8-oxoG:A mispair, the DNA glycosylase hMYH can excise the opposite A giving a new chance to create an 8-oxoG:C substrate for hOGG114; 15. It is noteworthy that DNA polymerase β preferentially inserts a C opposite 8-oxoG relative to A.16 In eubacteria, 8-oxoG is usually removed by Fpg17; 18 although there is a significant number of bacterial species, including Clostridium, which instead use an Ogg1 homologue.19; 20 Gain of Ogg1 function seems to be associated with loss of a functional Fpg.19
Several Ogg glycosylases have been characterized and classified into three distinct families. The Ogg1 family encompasses the largest number of members including hOGG17; 8; 9; 10; 11; 12; 13 and Clostridium acetobutylicum Ogg (CacOgg).19 Enzymes of the Ogg2 family lack the N-terminal domain of Ogg1 family members and share a low sequence identity with hOGG1 (13–19%).21 Members of this family, mostly from archaea, show a reduced specificity for the base opposite the lesion compared to hOGG1.21; 22; 23 A third class of Ogg, AGOG (archaeal GO glycosylase),24; 25 was described and the crystal structure of one of its representatives, Pa-AGOG from Pyrobaculum aerophilum, was reported recently.26 Enzymes of this class efficiently remove 8-oxoG from single- and double-stranded DNA substrates and demonstrate no opposite base specificity. The crystal structure of Pa-AGOG revealed that the helix-hairpin-helix (HhH) differs from that found in other Ogg glycosylases.26
CacOgg differs in sequence from hOGG1 with which it shares only 28% amino acid identity. One striking difference between these two enzymes is the lack of opposite base specificity displayed by CacOgg.19 Several amino acids involved in opposite base recognition in hOGG1 are not conserved in CacOgg. This enzyme has two non-conserved amino acids substitutions compared to hOGG1: Met132 and Phe179 in CacOgg correspond to Arg154 and Tyr203, respectively, in hOGG1. Previous structural studies of hOGG1 have shown residue Arg154 to be crucial for the recognition of C opposite the lesion.27; 28 Tyr203 is involved in a stacking interaction with the cytosine leading to the bend observed in the DNA molecule when bound to the enzyme. Recent kinetic studies have demonstrated that mutating both Met132 and Phe179 in CacOgg to the corresponding Arg and Tyr in hOGG1 partially restores specificity for 8-oxoG:C.19 Also, unlike hOGG1 and EcoFpg, but similarly to Pa-AGOG, CacOgg can remove 8-oxoG from single stranded DNA.19; 21
Here we report the structural characterization of the first bacterial Ogg in both its apo and liganded forms. Our structures demonstrate, as seen with hOGG128, that the oxidized nucleoside is confined in a binding pocket which recognizes 8-oxoG specifically. We also show that CacOgg displays a substantial structural reorganization of the C-terminal domain upon binding of 8-oxo-2’-deoxyguanosine (8-oxodG). A model of a DNA duplex based on previous structural studies provides a rationale for the lack of specificity of CacOgg for the base opposite the lesion as compared to hOGG1.
Results
Crystallization and structure determination of CacOgg in its apo-form
Crystals of apo-CacOgg were obtained by the vapor diffusion method. Crystals grew rapidly at 4°C and streak seeding was necessary to obtain well diffracting crystals. Since phasing via molecular replacement failed and CacOgg contains 8 methionines for a total of 292 residues, we crystallized a selenomethionine derivative to obtain multiwavelength anomalous diffraction (MAD) phases. A single crystal was used to collect data at two different wavelengths (See Table 1 for diffraction statistics) at the APS synchrotron (Beam line 23-ID B). The resulting 2.3Å model was refined to an Rfree of 0.217 and Rcryst of 0.174 with good stereochemistry. The final model comprises residues 1–285. The last seven amino acids were not built in the model because they had poorly defined or non existent electron density certainly due to disorder.
Table 1.
Summary of data collection and refinement statistics.
| Se-Peak | Se-Inflection | Complex with 8-oxoG | |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 0.9794 | 0.9796 | 1.5418 |
| Resolution (Å) | 20-2.2 (2.2–2.3)a | 20-2.3 (2.3–2.4) | 20-2.25 (2.3–2.25) |
| Space group | P41212 | P41212 | P4132 |
| Unit-cell parameters | |||
| a,b,c (Å) | 61.35, 61.35, 158.9 | 61.35, 61.35, 158.9 | 138.5, 138.5, 138.5 |
| Total reflection | 115697 (12730) | 105854 (12428) | 390143 (10371) |
| Unique reflection | 28397 (3384) | 24932 (2861) | 20809 (1192) |
| Completeness (%) | 96.6 (92.5) | 96.2 (92.3) | 94.1 (86.8) |
| I/σ(I) | 11.2 (6.0) | 11.4 (5.3) | 27.8 (3.1) |
| Rmerge (%) | 12.6 (28) | 13.3 (34.3) | 8.1 (53.3) |
| Redundancy | 4.1 (3.8) | 4.2 (4.3) | 18.8 (8.7) |
| Selenium sites | 8 | 8 | |
| Rcullis | 0.66 | 0.89 | |
| Phasing power | 1.72 | 0.69 | |
| Overall mean FOM b | 0.37/0.73 | ||
| Refinement | |||
| Rcryst (%) | 17.4 | 21.5 | |
| Rfree (%) | 21.7 | 25.4 | |
| Rmsd from ideal bond length (Å)/angles (°) | 0.005 / 1.2 | 0.006/1.2 | |
| Non-hydrogen atoms | |||
| All atoms | 2617 | 2561 | |
| Protein | 2364 | 2429 | |
| 8-oxoG | 20 | ||
| Water | 247 | 112 | |
| Heterogeneous atoms | 6 | 8 | |
| Average B factors (Å2) | 12.9 | 35.7 | |
| Wilson B-factor (Å2) | 7.2 | 30.4 | |
| 8-oxodG B factors (Å2) | - | 32.4 | |
| Ramachandran plot (%) | |||
| Most favored regions | 88.8 | 89.4 | |
| Allowed regions | 11.2 | 10.6 | |
High resolution shell is shown in parentheses.
Before and after density modification.
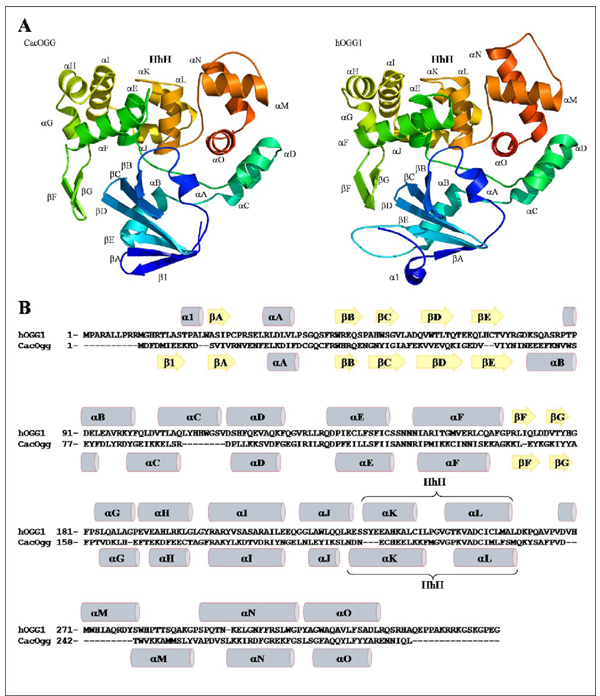
As illustrated in Figure 1, the overall fold of CacOgg is very similar to that of hOGG1. CacOgg is composed of three domains around the central HhH motif (αK-L, residues 206–232). The N-terminal domain (domain A) comprises a twisted antiparallel β-sheet composed of six β-strands (β1; βA–βE), a small one turn helix (αA) between βA and βB and a α-helix (αB) at the C-terminal part of this domain. Domain B (αE–αJ) is composed of six α-helices and two antiparallel β-strands (βF and βG) whereas domain C (αC–D and αM–O) comprises five α-helices. Both domains B and C are well conserved among the Ogg1 family. The central element of the Ogg1 proteins, the well conserved HhH motif (αK–L) is considered the fingerprint of DNA repair glycosylases of this superfamily.20 The catalytic lysine (Lys222) belongs to the second helix of the HhH motif (αL) while the other important strictly conserved catalytic residue (Asp241) belongs to αM.
Figure 1. Overall structure representation of CacOgg and unliganded hOGG1.
A) Ribbon diagram of CacOgg and unliganded hOGG1 (PDB ID 1KO9 34). Secondary structure elements are numbered according to their relative position in the sequence and the nomenclature follows that of the original hOGG1 structure 28. The helix-hairpin-helix (HhH) domain is composed of helices αK and αL in both proteins. Proteins are colored according to the amino acid sequence going from cold blue to warm red from N- to C-terminal. B) Sequence alignment of hOGG1 and CacOgg showing the secondary structures elements (grey cylinders for α-helices and yellow arrows for β-strands). All figures were prepared using PYMOL.48
Crystallization of CacOggK222Q in complex with 8-oxo-2’-deoxyguanosine and overall structure description
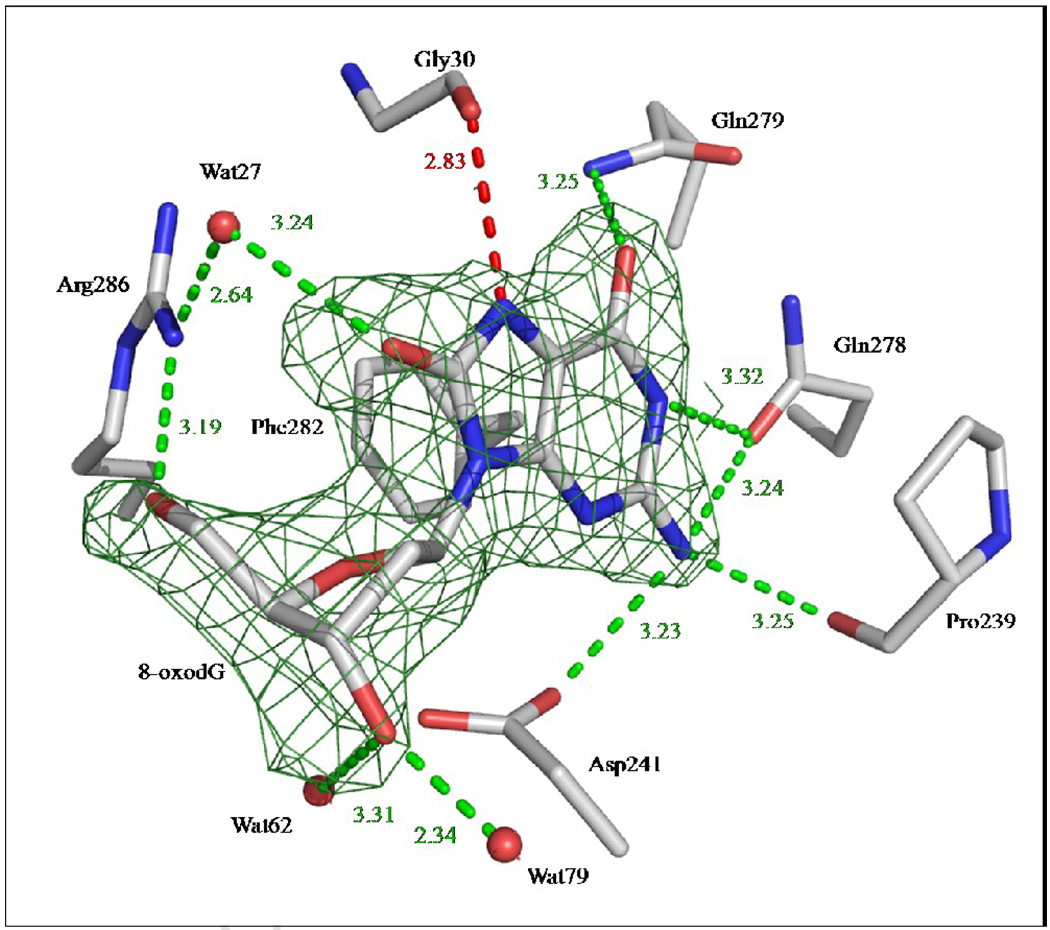
Soaking 8-oxodG into pre-formed apo-CacOgg crystals failed to achieve full occupancy of the ligand in the binding site. Attempts to co-crystallize wt-CacOgg with 8-oxodG were similarly unsuccessful. The catalytically inactive CacOggK222Q variant, on the other hand, readily yielded co-crystals with 8-oxo-deoxyguanosine. A complete 2.25Å data set was collected on our home X-ray source (See Table 1 for data collection statistics). Crystals of CacOggK222Q in complex with 8-oxo-deoxyguanosine belong to the cubic space group P4132 with unit-cell dimensions a=b=c=138.5Å, α=β=γ=90°. A molecular replacement solution (correlation factor of 0.71) was found using the atomic coordinates of apo-CacOgg as a search model. The resulting electron density map was well defined for the unique molecule per asymmetric unit. The CacOggK222Q/8-oxodG complex model was rebuilt and refined to a crystallographic R-factor of 0.215 (Rfree=0.254). The model does not lack any residues and residues Val18, Glu69, Asp84, Leu120 and Thr160 appear to adopt alternate conformations. A clear Fo-Fc electronic density (at 3σ) corresponding to 8-oxo-deoxyguanosine was observed in the binding site of the enzyme after molecular replacement. The map was well defined and unambiguous, allowing us to place the entire 8-oxodG molecule in the density. Figure 2 shows a simulated annealing omit map at 3σ around the 8-oxodG molecule confirming its presence in the active site.
Figure 2. Close-up view of CacOgg residues interacting with 8-oxodG.
Only residues involved in H-bonds and stacking interactions with 8-oxodG are depicted. Atoms are colored using standard CPK colors. H-bonds are represented by green dashed lines and distances are shown in Å. The red dashed line represents the H-bond between 8-oxodG N7 atom and main chain carbonyl of Gly30, which was shown to be essential for the recognition of 8-oxoG in hOGG128; 32. The simulated annealing omit map around 8-oxodG is shown in green and contoured at 3σ level.
Structural comparison of hOGG1 and CacOgg
The first secondary structure element differs between CacOgg and hOGG1. In CacOgg, the first secondary structure element is a βstrand (β1) while in hOGG1, a one turn α-helix (αA) is observed at this position. We cannot assume that this difference is significant because it might be a crystallization artefact in one or the other protein. Besides, the N-terminal domain is not known to interact with either DNA or the damaged base in hOGG1.
One major structural difference between CacOgg and hOGG1 is their size. CacOgg is made of 292 amino acids whereas hOGG1 is larger with 345 amino acids. The two proteins, however, share a very similar architecture. In order to accommodate the proper fold, hOGG1 harbors longer connecting loops than CacOgg. For example, the loop between βE and αB (see figure 1B) contains twelve residues in hOGG1 compared to only four residues in CacOgg. Also, a number of α-helices (αB, αF, αM, αN and αO) are longer in hOGG1 by at least one full turn, while the sizes of the β-strands are very similar.
CacOgg binding site and structural reorganization after substrate binding
As with other DNA glycosylases, CacOgg is presumed to bind its substrate in an extrahelical conformation.28; 29; 30 The damaged base is rotated out of the DNA helix and bound in a cavity lined with specific binding residues.31 The substrate binding cavity of CacOgg is located at the junction of the three domains and the helix-hairpin-helix motif. This deep binding cavity is composed of apolar and aromatic residues on the B-domain side while the opposite side of the cavity is formed of more hydrophilic residues of the C-domain. The conserved catalytic residues Lys222 and Asp241 are located near the entrance of the cavity. At this position, these residues can easily access the deoxyribose ring of a flipped 8-oxoG substrate, placing it in a position suitable for catalysis. A conserved glycine shown to play an important role in the discrimination between guanine and 8-oxoG in hOGG1 (Gly42) 28; 32 is located on loop αA–βB on the A-domain edge of the substrate binding site (Gly30 in CacOgg).
The structure of CacOggK222Q in complex with 8-oxodG revealed several interactions between the protein and its ligand. As seen in Figure 2, the deoxyribose interacts with Arg286 and two water molecules. The 8-oxoguanine moiety interacts with the protein via hydrophilic and hydrophobic interactions. The 8-oxygen atom of 8-oxodG hydrogen bonds with Arg286 via a water molecule. The other keto group, at position 6, is involved in a H-bond with Gln279. The 8-oxodG N1 and N2 atoms engage in hydrophilic interactions with Gln278, Asp241 and the main chain carbonyl of Pro239. Furthermore, stacking of Phe282 against the oxidized purine stabilizes the lesion in a position that facilitates the recognition of 8-oxoG by the enzyme (see below).
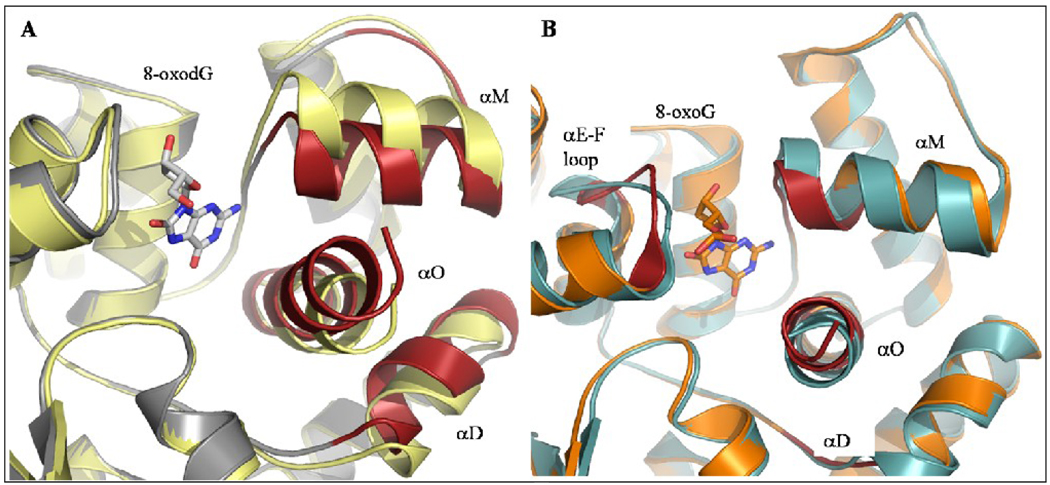
Superposition of apo-CacOgg onto the CacOggK222Q/8-oxodG complex revealed that domain C is flexible and undergoes a significant, contractive movement upon binding of the ligand (Figure 3A) even in the absence of DNA backbone. The calculated RMSD between the apo and the liganded forms for the three helices of domain C (αD, αM, and αO) is 1.77Å for backbone Cα atoms whereas the rest of the protein displays a RMSD of 0.58Å. The C-terminal helix αO engages in several interactions with 8-oxodG and these interactions can explain why we were able to build seven additional residues at the C-terminus end compared to the apo-CacOgg structure. Compared to hOGG1 (Figure 3B) the reorganization displayed by CacOgg is much wider and involves three of the four helices of domain C. The reorganization of αD, αM, and αO seems to be associated with the ligand binding and several residues of these helices contributes to its stabilization in the binding site. However, unlike CacOgg, a significant movement of the loop between helices αE and αF was observed in hOGG1. Because this loop interacts with DNA it is possible that a similar conformational change might be seen in the corresponding loop in CacOgg, but at this point the absence of DNA in our complex prevents us from observing such movement.
Figure 3. Superposition of apo- and liganded forms of CacOgg and hOGG1.
A) Ribbon diagram showing the closure of helices αD, αM and αO (in red) upon binding of 8-oxodG by CacOgg. B) Ribbon diagram showing the movement performed by helices αD, αM, αO and the αE–F loop (in red) upon binding of 8-oxoG containing DNA by hOGG1. Apo-CacOgg is shown in pale yellow, CacOggK22Q/8-oxodG is depicted in standard CPK colors, apo-hOGG1 (PDB ID 1K09 34) is depicted in cyan and hOGG1/DNA (1EBM28) is shown in orange. Secondary structure elements that undergo conformational change upon binding of the oxidized base are highlighted in red.
Discussion
8-oxoG binding site
The similarity between 8-oxoG and guanine makes the distinction between these two molecules a real challenge for 8-oxoguanine DNA glycosylases. At first glance, the presence of a keto moiety on the C8 atom of a guanine base might appear to be the major change between guanine and 8-oxoG, but earlier structural studies 28 of hOGG1 and the present study on CacOgg reveal that the C8 carbonyl group is completely devoid of any direct interactions with the protein. This result contrasts with what was described for Pa-AGOG. A H-bond was observed between the 8-oxo atom and the nitrogen of the indole group of Trp69 in the Pa-AGOG structure in complex with 8-oxodG.26 One consequence of the oxidation of the C8 atom of guanine is the electron delocalization of the double bond between the N7 and C8 atoms to the 8-oxo group and the protonation of N7. The N7 proton allows 8-oxoG to establish a crucial hydrophilic contact with the conserved glycine at position 30 (Gly42 in hOGG1) that cannot occur with G (See red dashed line on Figure 2).
The presence of this additional H-bond is not the only difference that allows Ogg to discriminate between 8-oxoG and G. Quantum calculations performed with 8-oxoG and G revealed an inversion of the dipole at positions C8 and N7. The charge carried by 8-oxoG complements perfectly the local charge on the active site of Ogg (Lys249 and Cys253 for hOGG133 and Lys222 and Cys226 for CacOgg). This dipole field creates an attractive force for 8-oxoG but also creates a repulsive force for G. These two elements combined (H-bond with protonated N7 and electrostatic attraction/repulsion) seem to be essential for the discrimination between G and 8-oxoG by Ogg glycosylases.
Structural reorganization of CacOgg upon 8-oxodG binding
The most dramatic movement of protein residues upon binding of 8-oxodG is observed for residue Phe282, which undergoes a wide reorganization of about 5Å from a distal position in the apo-CacOgg model to a proximal position (Figure 4) in the CacOggK222Q/8-oxodG complex. In this proximal position the ring of Phe282 stacks against the 6-membered ring of 8-oxodG. The stacking of 8-oxodG with Phe282 impedes the rotation of the oxidized base and constrains the N7 atom of 8-oxodG in a position suitable to form a H-bond with Gly30. A similar movement is observed in the homologous phenylalanine in hOGG1 (Phe319): When 8-oxoG is bound, the phenyl ring moves from a distal to a stacking position. 28; 34 The movement of helix αM towards 8-oxodG brings some residues in proximity to 8-oxodG: Asp241 makes a H-bond with N2 of 8-oxodG. Also, Gln278 moves forward slightly to form a H-bond with the N1 atom and N2 amine group of 8-oxodG. This structural reorganization is important for maintaining the damaged base in an extra-helical conformation at a distance from the catalytic residues (Asp241 and Lys222) that is suitable for glycosylase activity.
Figure 4. Close-up view of structural reorganization of CacOgg upon 8-oxodG binding.
Superposition of apo-CacOgg (pale yellow) and CacOggK222Q/8-oxodG complex (standard CPK colors) showing residues undergoing a wide reorganization upon binding of 8-oxodG. Residues Asp241 and Gln278 move closer to 8-oxodG. Phe282 moves from a distal to a proximal position. In the proximal position Phe282 stacks against the 6-membered ring of 8-oxoG.
Putative interactions of CacOgg with DNA and recognition of the base opposite the lesion
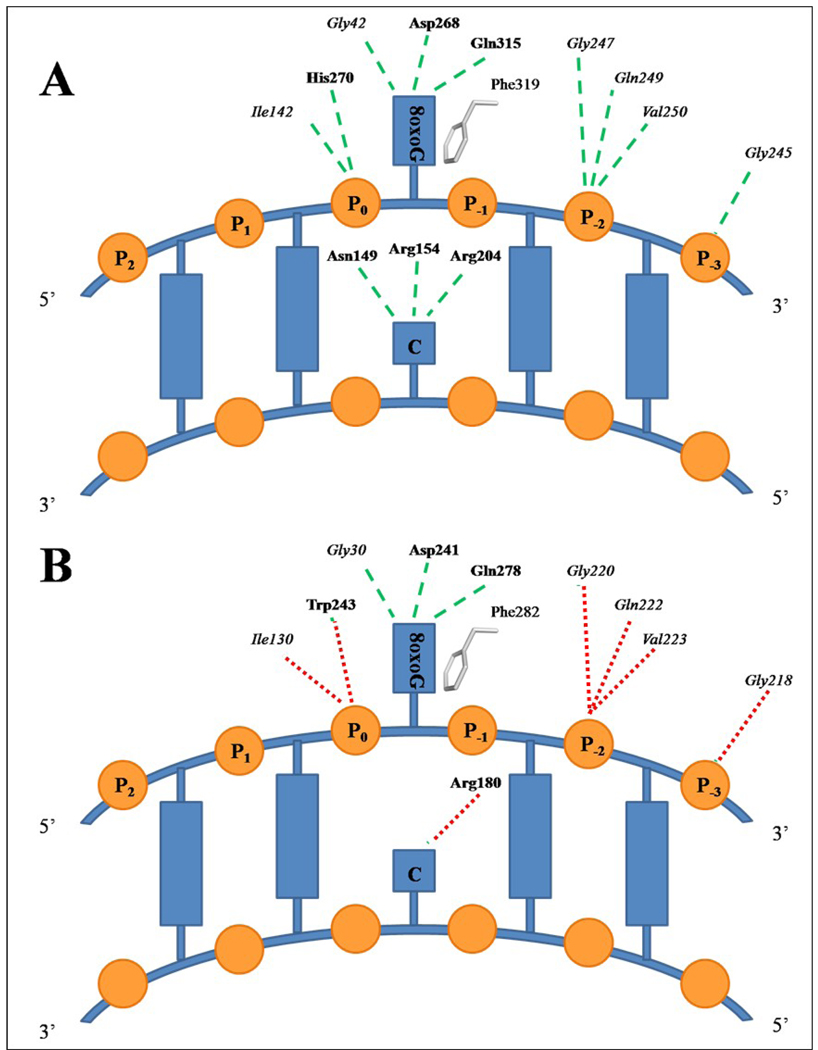
Similarly to hOGG1, CacOgg does not cleave oxidized pyrimidines but can efficiently remove 8-oxoG and 8-oxoA opposite C. In contrast to hOGG1, CacOgg can also remove 8-oxoG opposite A.19; 28 Both enzymes, however, are unable to excise the further oxidation product of 8-oxoG, guanidinohydantoin opposite C. 19; 35 hOGG1 and CacOgg share a very similar protein architecture (see Figure 1) and a structural alignment can be used to identify putative protein:DNA interactions in CacOgg. In the published structures of HhH-GPD enzymes in complex with DNA (hOGG128, MutY29, EndoIII36, and AlkA37), the DNA molecule is bent by about 60° at the site of the lesion. The DNA glycosylase mostly interacts with the minor groove of DNA along the interdomain region. In hOGG1 the DNA molecule is tightly bound by a complex network of hydrophilic interactions. Most of these interactions are made with the phosphate groups in close proximity to the lesion. As seen in Figure 5A, the phosphate (P0) 5’ to the lesion makes a H-bond with the main chain of Ile142 and the side chain Nε2 atom of His270. P−2 interacts with main chain atoms of Val250, Gly247, and Gln249 while P−3 interacts with Gly245. The 8-oxoG base is well stabilized by a combination of hydrophilic and hydrophobic interactions involving five residues. The main chain atoms of Gly42 and Pro266 and the side chains of Asp268 and Gln315 make H-bond with the 8-oxoG moiety while Phe319 stacks against the six membered ring of the 8-oxoG molecule. Few differences are expected for the binding of DNA by CacOgg since the majority of the interacting residues are very well conserved (Figure 5B). Only His270 is not conserved and is replaced by Trp243. In a manner similar to His270 in hOGG1, CacOgg Trp243 can make a H-bond between its Nε1 atom and one of the oxygen atoms of phosphate P0.
Figure 5. Schematic representation of enzyme-DNA interactions.
Schematic representation of A) DNA interactions with hOGG1 (PDB ID 1EBM 28) and B) putative interactions of DNA with CacOgg. Residues in bold are interacting with DNA via their side chain. Residues in italic interact with DNA via main chain atoms. Phe319 (hOGG1) and Phe282 (CacOgg) stack against the 6-membered ring of 8-oxoG. Green dashed lines indicate observed interactions while red dotted lines represent putative interactions.
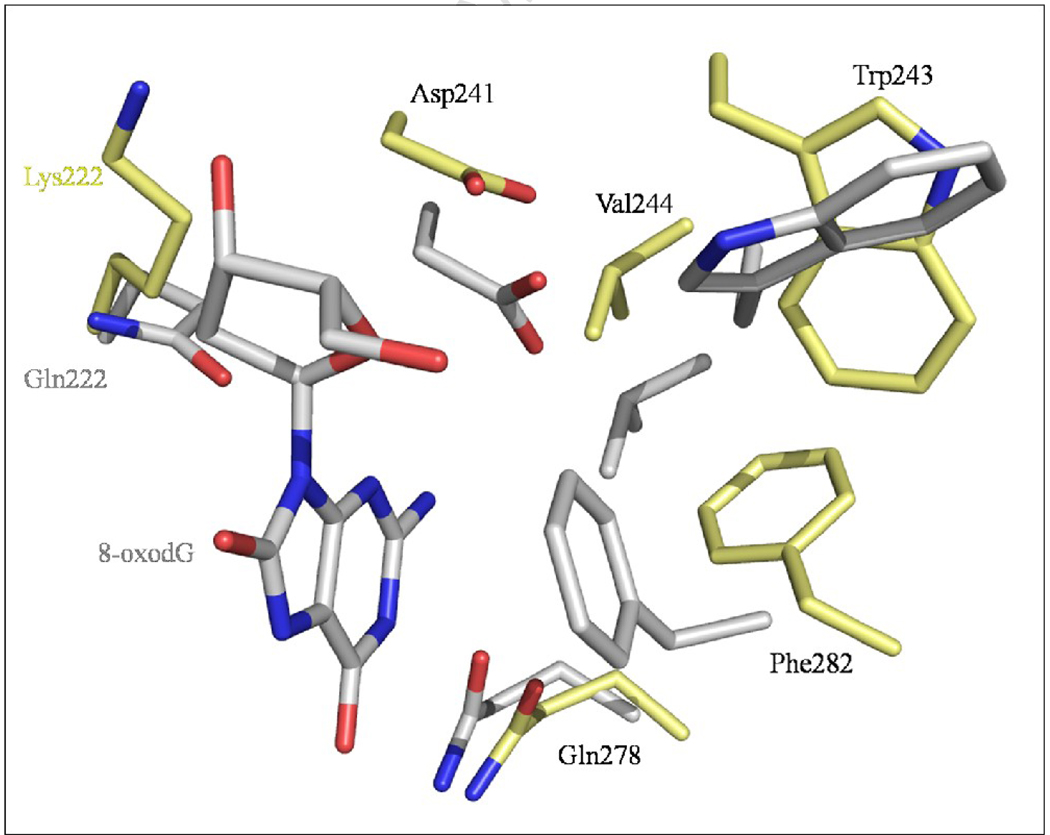
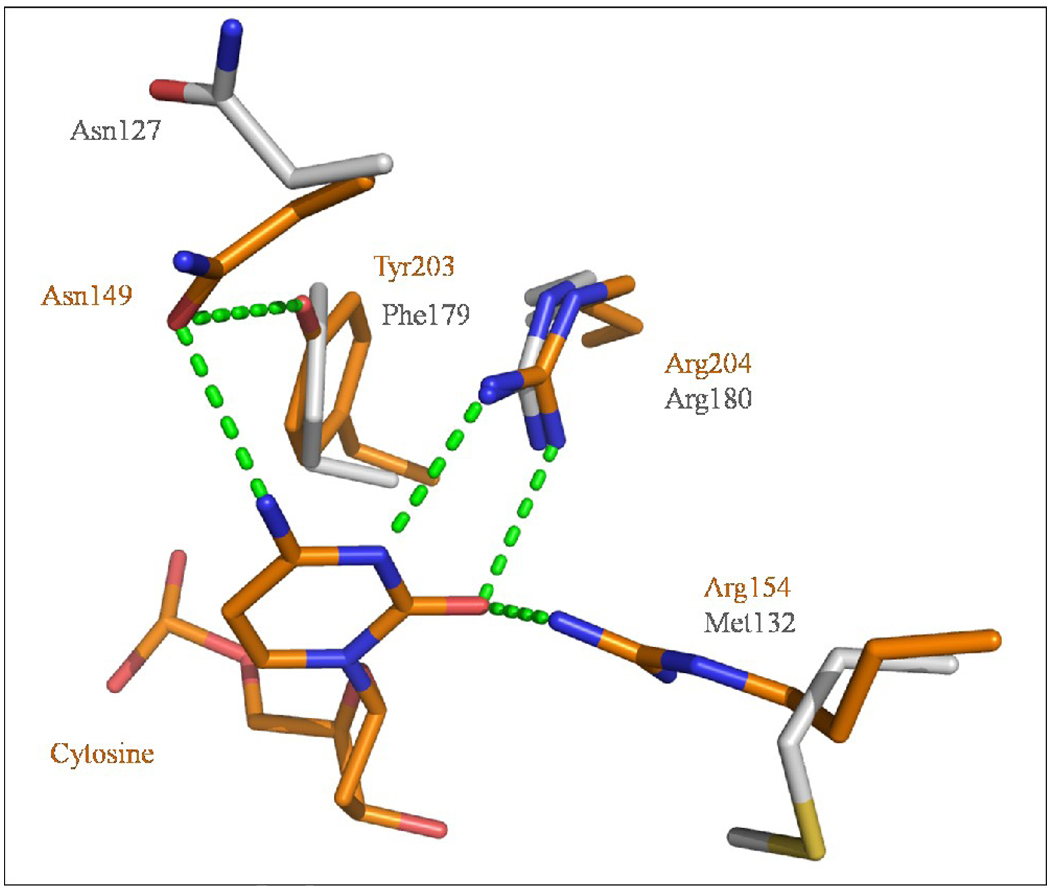
The most notable difference between these two Oggs is their opposite base specificity. hOGG1 removes 8-oxoG opposite C but not paired to G or A whereas CacOgg efficiently removes 8-oxoG opposite each of the four bases.19; 28; 35 A structural comparison with hOGG1 helps shed light on CacOgg’s lack of selectivity. In the hOGG1 model, three very important residues interact with the cytosine via their side chains: Asn149, Arg154, and Arg204 form a network of five H-bond that specify C. In CacOgg three of these H-bonds are lost: Arg154 is replaced by a methionine (Met132), which is unable to form a H-bond with the keto group of C. Although Asn127 is conserved it does not adopt the same conformation in our structures as its counterpart in hOGG1 probably because the absence of a DNA backbone in our model. The remaining two H-bonds originate from Arg180, which forms two H-bonds with N3 and O4 (see Figure 6). Adenine is the second most favored opposite base for CacOgg.19 This can be explained by the nature of adenine in which N1/N6 can mimic N3/N4 in C and can interact in a similar way with Asn127 and Arg180. Another interesting interaction is observed in hOGG1, which can explain the increased specificity for the opposite base for this enzyme compared to CacOgg. As seen in Figure 6, the hydroxyl group of residue Tyr203 makes a H-bond with the Oδ1 atom of Asn149, which is also involved in a H-bond with the 8-oxoG N4 amine of C. This strong network contributes to stabilize Asn149 in a position which facilitates a tight interaction with the N4 amine of the C opposite the lesion. Because Tyr is replaced by a Phe (Phe179) in CacOgg, the H-bond is lost. In fact, mutating Phe179 and Met132 into the corresponding residues in hOGG1 (Tyr and Arg) yields a CacOgg protein variant with an increased specificity for 8-oxoG:C. 19
Figure 6. Residues implicated in recognition of the base opposite the lesion.
Superposition of residues interacting with the C opposite the lesion in the hOGG1-DNA complex (PDB ID 1EBM 28) with corresponding residues in CacOgg. Residues belonging to CacOgg are depicted in standard CPK colors while residues from hOGG1 are colored in orange. H-bonds are shown as green dashed lines. Residue numbers are highlighted in grey for CacOgg and orange for hOGG1.
Concluding remarks
We describe here the first crystal structures of an atypical Ogg1 enzyme cloned from a bacterium, CacOgg in both apo- and liganded forms. These structures allowed us to identify the interactions made by the glycosylase with 8-oxodG. A superposition of the two structures revealed a structural reorganization of helices in the C-terminal domain upon binding of 8-oxodG reminiscent of that seen in hOGG1. Furthermore, our structural work provides a structural rationale as to why CacOgg is less specific for the base opposite the lesion than its human homologue. It may appear aberrant for an anaerobic bacterium like Clostridium acetobutylicum to encode an enzyme able to restore DNA after damage from reactive oxygen species, but it is noteworthy that most anaerobic bacteria can tolerate brief exposure to oxygen. 19
Materials and Methods
Recombinant CacOgg expression and purification
Recombinant CacOgg (wild-type and K222Q mutant) were expressed and purified essentially as described.19 Briefly, CacOgg wild-type and K222Q mutant were expressed in ER2566 fpg- E. coli co-transfected with a pLysS RIR vector.38 After overnight expression with IPTG at 16°C, bacterial pellets were sonicated in lysis buffer (50 mM Tris pH 8.0, 500 mM NaCl, 1 mM EDTA, 1 mM PMSF). The centrifuged lysate was loaded on a chitin column and eluted using 50 mM DTT. Pooled fractions of CacOgg were loaded on a Q column and eluted with a salt gradient. The selenomethionyl variant was engineered by inhibiting the methionine biosynthesis pathway.39 The resulting protein was purified using the same purification protocol as for the wild type enzyme. The purified protein was dialyzed in crystallization buffer (20 mM Tris-HCl pH 8.5, 100 mM NaCl, 1 mM DTT and 2 % (v/v) glycerol) then concentrated to 6 mg/ml for apo-enzyme. CacOggK222Q was concentrated to 15 mg/ml and 8-oxo-2’deoxyguanosine (Berry and Associates Inc.) was added to a final concentration of 7 mM.
Crystallization of the recombinant CacOgg in its apo-form and in complex with 8-oxodG
Crystals of apo-CacOgg were obtained by hanging-drop vapor diffusion at 4°C. Crystals were obtained in drops of a 1:1 ratio of purified protein and well solution (14% (w/v) PEG-4000 and 0.1 M Tris-HCl pH 8.5) but the resulting crystals were of poor quality and not suitable for X-ray diffraction. In order to improve the size and the shape of apo-CacOgg crystals, streak seeding was performed using the same crystallization condition. This procedure allowed us to obtain crystals with dimensions suitable (250 × 80 × 80 µm3) for X-ray diffraction experiments. First crystal hits of CacOggK222Q in complex with 8-oxodG were obtained in condition D4 (20% (v/v) isopropanol, 0.1M NaCitrate pH 5.6, 20% (w/v) PEG-4000) and D5 (10% isopropanol 0.1M HEPES pH 7.5, 20% (w/v) PEG-4000) of Crystal Screen HT (Hampton Research). After minor optimization of precipitant and pH, crystals of suitable size (120 × 120 × 120 µm3) were obtained in 10–20% (v/v) isopropanol, 0.1M NaCitrate pH 5.5, 12% (w/v) PEG-4000.
X-ray analysis and structure determination of apo-CacOgg and CacOggK222Q in complex with 8-oxodG.
All X-ray diffraction experiments were done at 100K after crystals were allowed to equilibrate for 1–2 minutes in a cryoprotectant solution consisting of the crystallization buffer supplemented with 20% (v/v) ethylene glycol or glycerol. Since molecular replacement attempts failed to produce a clear solution, we collected a MAD data set at two wavelengths corresponding to the peak and inflection of the K edge of selenium (See Table 1 for data collection statistics) at beamline 23-ID B at the Argonne Advanced Photon Source for the selenomethionyl variant of apo-CacOgg. The diffraction images were integrated using XDS 40 and merged/scaled with CCP4. 41 The program SOLVE 42 identified seven of the eight selenium sites. AutoSHARP 43 was then used for refinement of the selenium parameters. The space group was judged to be P41212 and not the enantiomorphic P43212, based on the map quality and continuity. The phasing information was then used in ARP/wARP 44 for density modification and iterative model building. The refinement procedure was performed with CNS. 45 The initial model issued from ARP/wARP was submitted to a cycle of simulated annealing at 3000 K followed by energy minimization and B-factor refinement cycle. Afterwards, the model was refined by simple energy minimization followed by isotropic B-factors refinement (restrained and individual) and corrected by manual rebuilding using O. 46 Missing parts of the model, glycerol and water molecules were progressively added during the refinement procedure. Finally, the quality of the model was verified with PROCHECK. 47 There are no residues in the disallowed region of the Ramachandran plot.
X-ray diffraction images of CacOggK222Q-8-oxoG crystals were recorded on our laboratory MAR345 (MAR Research) detector mounted on a Rigaku RU-200 rotating anode-generator equipped with Xenocs focusing mirrors. Diffraction data were indexed using XDS40 and scaled with XSCALE. The structure of CacOggK222Q was solved by molecular replacement with MOLREP software from the CCP4 suite 41 using the coordinates of apo CacOgg as a model. A very clear Fo-Fc electron density map corresponding to 8-oxodG was observed in the putative substrate binding site immediately after the molecular replacement. A calculated simulated annealing omit map is shown in Figure 2. Refinement procedure was performed as stated above for the apo- enzyme. There are no residues in the disallowed region of the Ramachandran plot.
Protein Data Bank accession codes
Atomic coordinates and structure factor amplitudes have been deposited with the Protein Data Bank and are available under the following accession codes: 3F0Z for CacOgg in its apo-form; 3F10 for CacOggK222Q in complex with 8-oxo-2’-deoxyguanosine.
Acknowledgements
This work is dedicated to the memory of Wendy Cooper. We thank Alicia S. Holmes for her help with protein purification and Dr. Pierre Aller and Karl Zahn for collecting the MAD data set at the APS synchrotron. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357. This research was supported by National Institutes of Health grants R01CA33657 and P01CA098993 awarded by the National Cancer Institute.
Abbreviations
- 8-oxoG
8-oxoguanine
- 8-oxodG
8-oxo-2’-deoxy-guanosine
- Ogg
8-oxoguanine DNA glycosylase
- BER
base excision repair
- CacOgg
Clostridium acetobutylicum Ogg
- HhH
helix-hairpin-helix
- hOGG1
human OGG1
- PEG
polyethylene glycol
- RMSD
root mean square deviation
- ROS
reactive oxygen species
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 2.Kuchino Y, Mori F, Kasai H, Inoue H, Iwai S, Miura K, Ohtsuka E, Nishimura S. Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature. 1987;327:77–79. doi: 10.1038/327077a0. [DOI] [PubMed] [Google Scholar]
- 3.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 4.Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 5.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 7.Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kodama T, Takao M, Yasui A, Yamamoto K, Asano M. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- 8.Arai K, Morishita K, Shinmura K, Kohno T, Kim SR, Nohmi T, Taniwaki M, Ohwada S, Yokota J. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene. 1997;14:2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- 9.Rosenquist TA, Zharkov DO, Grollman AP. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjørås M, Luna L, Johnsen B, Hoff E, Haug T, Rognes T, Seeberg E. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. Embo J. 1997;16:6314–6322. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roldan-Arjona T, Wei YF, Carter KC, Klungland A, Anselmino C, Wang RP, Augustus M, Lindahl T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagashima M, Sasaki A, Morishita K, Takenoshita S, Nagamachi Y, Kasai H, Yokota J. Presence of human cellular protein(s) that specifically binds and cleaves 8-hydroxyguanine containing DNA. Mutat Res. 1997;383:49–59. doi: 10.1016/s0921-8777(96)00045-6. [DOI] [PubMed] [Google Scholar]
- 13.Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu Y, Lu AL. Differential DNA recognition and glycosylase activity of the native human MutY homolog (hMYH) and recombinant hMYH expressed in bacteria. Nucleic Acids Res. 2001;29:2666–2674. doi: 10.1093/nar/29.12.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slupska MM, Luther WM, Chiang JH, Yang H, Miller JH. Functional expression of hMYH, a human homolog of the Escherichia coli MutY protein. J Bacteriol. 1999;181:6210–6213. doi: 10.1128/jb.181.19.6210-6213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krahn JM, Beard WA, Miller H, Grollman AP, Wilson SH. Structure of DNA polymerase beta with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure. 2003;11:121–127. doi: 10.1016/s0969-2126(02)00930-9. [DOI] [PubMed] [Google Scholar]
- 17.Chung MH, Kasai H, Jones DS, Inoue H, Ishikawa H, Ohtsuka E, Nishimura S. An endonuclease activity of Escherichia coli that specifically removes 8-hydroxyguanine residues from DNA. Mutat Res. 1991;254:1–12. doi: 10.1016/0921-8777(91)90035-n. [DOI] [PubMed] [Google Scholar]
- 18.Tchou J, Kasai H, Shibutani S, Chung MH, Laval J, Grollman AP, Nishimura S. 8-oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc Natl Acad Sci U S A. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robey-Bond SM, Barrantes-Reynolds R, Bond JP, Wallace SS, Bandaru V. Clostridium acetobutylicum 8-oxoguanine DNA glycosylase (Ogg) differs from eukaryotic Oggs with respect to opposite base discrimination. Biochemistry. 2008;47:7626–7636. doi: 10.1021/bi800162e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denver DR, Swenson SL, Lynch M. An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol Biol Evol. 2003;20:1603–1611. doi: 10.1093/molbev/msg177. [DOI] [PubMed] [Google Scholar]
- 21.Gogos A, Clarke ND. Characterization of an 8-oxoguanine DNA glycosylase from Methanococcus jannaschii. J Biol Chem. 1999;274:30447–30450. doi: 10.1074/jbc.274.43.30447. [DOI] [PubMed] [Google Scholar]
- 22.Chung JH, Suh MJ, Park YI, Tainer JA, Han YS. Repair activities of 8-oxoguanine DNA glycosylase from Archaeoglobus fulgidus, a hyperthermophilic archaeon. Mutat Res. 2001;486:99–111. doi: 10.1016/s0921-8777(01)00081-7. [DOI] [PubMed] [Google Scholar]
- 23.Im EK, Hong CH, Back JH, Han YS, Chung JH. Functional identification of an 8-oxoguanine specific endonuclease from Thermotoga maritima. J Biochem Mol Biol. 2005;38:676–682. doi: 10.5483/bmbrep.2005.38.6.676. [DOI] [PubMed] [Google Scholar]
- 24.Gondichatnahalli L, Sartori A, Hunziker P, Kostrewa D, Prota A, Jiricny J, Winkler F. Pa-AGOG, a novel 8-oxoguanine DNA glycosylase from the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Faseb Journal. 2004;18:C177–C177. [Google Scholar]
- 25.Sartori AA, Lingaraju GM, Hunziker P, Winkler FK, Jiricny J. Pa-AGOG, the founding member of a new family of archaeal 8-oxoguanine DNA-glycosylases. Nucleic Acids Res. 2004;32:6531–6539. doi: 10.1093/nar/gkh995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lingaraju GM, Sartori AA, Kostrewa D, Prota AE, Jiricny J, Winkler FK. A DNA glycosylase from Pyrobaculum aerophilum with an 8-oxoguanine binding mode and a noncanonical helix-hairpin-helix structure. Structure. 2005;13:87–98. doi: 10.1016/j.str.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Audebert M, Radicella JP, Dizdaroglu M. Effect of single mutations in the OGG1 gene found in human tumors on the substrate specificity of the Ogg1 protein. Nucleic Acids Res. 2000;28:2672–2678. doi: 10.1093/nar/28.14.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 29.Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- 30.Labahn J, Scharer OD, Long A, EzazNikpay K, Verdine GL, Ellenberger TE. Structural basis for the excision repair of alkylation-damaged DNA. Cell. 1996;86:321–329. doi: 10.1016/s0092-8674(00)80103-8. [DOI] [PubMed] [Google Scholar]
- 31.Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 32.Radom CT, Banerjee A, Verdine GL. Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations. J Biol Chem. 2007;282:9182–9194. doi: 10.1074/jbc.M608989200. [DOI] [PubMed] [Google Scholar]
- 33.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 34.Bjørås M, Seeberg E, Luna L, Pearl LH, Barrett TE. Reciprocal "flipping" underlies substrate recognition and catalytic activation by the human 8-oxo-guanine DNA glycosylase. J Mol Biol. 2002;317:171–177. doi: 10.1006/jmbi.2002.5400. [DOI] [PubMed] [Google Scholar]
- 35.Leipold MD, Workman H, Muller JG, Burrows CJ, David SS. Recognition and removal of oxidized guanines in duplex DNA by the base excision repair enzymes hOGG1, yOGG1, and yOGG2. Biochemistry. 2003;42:11373–11381. doi: 10.1021/bi034951b. [DOI] [PubMed] [Google Scholar]
- 36.Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. Embo J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hollis T, Ichikawa Y, Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bandaru V, Blaisdell JO, Wallace SS. Oxidative DNA glycosylases: recipes from cloning to characterization. Methods Enzymol. 2006;408:15–33. doi: 10.1016/S0076-6879(06)08002-5. [DOI] [PubMed] [Google Scholar]
- 39.Doublié S. In: Macromolecular Crystallography Protocols. S D, editor. Vol. 1. Totowa: Humana Press; 2007. pp. 91–108. [Google Scholar]
- 40.Kabsch W. Automatic Processing of Rotation Diffraction Data from Crystals of Initially Unknown Symmetry and Cell Constants. Journal of Applied Crystallography. 1993;26:795–800. [Google Scholar]
- 41.Bailey S. The Ccp4 Suite -Programs for Protein Crystallography. Acta Crystallographica Section D-Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 42.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallographica Section D-Biological Crystallography. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vonrhein C, Blanc E, Roversi P, Bricogne G. In: Macromolecular Crystallography Protocols. S D, editor. Vol. 1. Totowa: Humana Press; 2007. pp. 215–230. [Google Scholar]
- 44.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 45.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D-Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 46.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved Methods for Building Protein Models in Electron-Density Maps and the Location of Errors in These Models. Acta Crystallographica Section A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 47.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 48.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: 2008. http://www.pymol.org. [Google Scholar]