Abstract
The MCM2-7 (minichromosome maintenance) proteins are a family of evolutionarily highly conserved proteins. They are essential for DNA replication in yeast and are considered to function as DNA helicases. However, it has long been shown that there is an overabundance of the MCM2-7 proteins when compared with the number of DNA replication origins in chromatin. It has been suggested that the MCM2-7 proteins may function in other biological processes that require the unwinding of the DNA helix. In this report, we show that RNA polymerase II (Pol II)-mediated transcription is dependent on MCM5 and MCM2 proteins. Furthermore, the MCM2-7 proteins are co-localized with RNA Pol II on chromatins of constitutively transcribing genes, and MCM5 is required for transcription elongation of RNA Pol II. Finally, we demonstrate that the integrity of the MCM2-7 hexamer complex and the DNA helicase domain in MCM5 are essential for the process of transcription.
The MCM2-72 family is an evolutionarily conserved group of proteins that are essential for DNA replication (1–6). The MCM2-7 proteins bind to replication origins during G1 phase of the cell cycle along with members of the prereplication complex including origin recognition complex (ORC), cell division cycle 6 (Cdc6), and chromatin licensing and DNA replication factor 1 (Cdt1) (7–9), which is required for replication origin activation (reviewed in Refs. 2 and 3). Recruitment of MCM2-7 to replication origins is highly regulated and dependent on the interactions of Cdc6 and Cdt1 with origins (10). Once DNA replication is initiated, the MCM2-7 proteins appear to move along with the replication fork, further supporting their possible role as a replicative DNA helicase (8).
The molecular structure of the MCM2-7 proteins and biochemical analyses suggest that they function as a DNA helicase during DNA replication (11–13). The MCM2-7 proteins form several subcomplexes including MCM2/4/6/7, MCM4/6/7, MCM3/5, MCM4/7, and MCM2/3/5 (4, 11, 14–17). It has been suggested that different subcomplexes may be components or have a regulatory role in a stepwise assembly of the entire hexameric complex onto DNA (14, 18). A hexamer containing two trimers of MCM4/6/7 was shown to have helicase activity for DNA-DNA and DNA-RNA substrates in vitro (11, 14, 15, 19). Furthermore, in vivo data showed that all the subunits (MCM2-7) are required for DNA replication elongation and that the entire MCM2-7 complex has in vitro helicase activity (9, 12). Recent studies also showed that different members of the MCM2-7 family vary in their contributions to ATP hydrolysis and the MCM2-7 complex forms a toroidal structure containing a discontinuity between MCM2 and MCM5 that drives circularization of the complex (12, 13).
In addition to DNA replication, several lines of evidence suggest that MCM2-7 may also function in other cellular processes involving DNA. This possibility was first introduced based on the observation that MCM2-7 proteins are highly abundant in the cell and that the amount of these proteins exceeds the number of replication origins in yeast (1, 4–6). A potential role for the MCM2-7 proteins for transcription has been supported by a number of observations. Members of the MCM2-7 family have been shown to interact with components of the transcription machinery including RNA polymerase II (Pol II) and some transcription factors. Our previous studies showed that the MCM2-7 proteins are recruited by Stat1 for transcription activation in response to IFN-γ and that MCM5 is essential for Stat1-mediated transcription (20–22). The MCM2-7 proteins interact with the promoters of Stat1 target genes in response to IFN-γ, and they move along these genes with RNA Pol II during transcription (22), similar to their ability to move along with the DNA replication fork (8). MCM2 interacts with the carboxyl-terminal domain of RNA Pol II, and antibodies directed against MCM2 inhibit RNA Pol II transcription in Xenopus oocytes (23, 24).
In this report, the role of the MCM2-7 proteins in the process of cytokine-independent, RNA Pol II-mediated transcription of constitutively expressed genes was further analyzed. RNA interference techniques were utilized to knock down MCM2 and MCM5 followed by tritiated uridine incorporation assay and nuclear run-on assays to show that they are required for RNA Pol II-mediated transcription. Using chromatin immunoprecipitation (ChIP) assays, we demonstrate that members of the MCM2-7 proteins are present on the chromatins of constitutively transcribing genes along with RNA Pol II and that MCM5 is required for transcription elongation of RNA Pol II. Furthermore, using MCM5 mutants that are specifically deficient in the formation of MCM2-7 hexamer complex or DNA helicase domain, we demonstrate that the integrity of the MCM2-7 hexamer complex and the helicase domain of MCM5 are essential for the process of transcription.
EXPERIMENTAL PROCEDURES
Cell Culture and Antibodies—2fTGH cells (I. Kerr, Cancer Research UK, London, UK) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% cosmic calf serum (Hyclone). 293T cells (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% bovine calf serum. Antibodies for Western blot analysis included: anti-MCM5 (Bethyl Laboratories), anti-Stat1 amino terminus, and anti-Stat3 (BD Biosciences). Antibodies used for ChIP assay were anti-Stat1 carboxyl terminus and anti-RNA Pol II (Santa Cruz Biotechnology) and anti-MCM5 (described in Ref. 21). Antibodies used in both Western blot analysis and ChIP assay were MCM2 and MCM3 (Bethyl Laboratories).
ChIP Assay—ChIP assays were performed as described previously (25) using 2.5 μg of each antibody (anti-Stat1C, anti-MCM2, anti-MCM3, anti-MCM5, anti-RNA Pol II). Primers for the ChIP assay were as follows: center of the GAPDH gene, 5′-GTATTCCCCCAGGTTTACAT-3′ and 5′-TTCTGTCTTCCACTCACTCC-3′; 5′ region of GAPDH, 5′-TCGTGTTCCCTAATCCCATC-3′ and 5′-ATAGCTCACGCCCTCTGC-3′; center of the E2F4 gene, 5′-AGATACCCTCTTGGCCATCC-3′ and 5′-GGATGCCCATCCTCCTAGAT-3′; center of the β-actin gene, 5′-AAGATGACCCAGGTGAGTGG-3′ and 5′-CCTGCAGAGTTCCAAAGGAG-3′; center of the β-tubulin gene, 5′-CTGGACCGCATCTCTGTGTA-3′ and 5′-GTTCACGAAAGGGGACAAAA-3′; 5′ region of β-actin, 5′-GTCTCTTGGCCAGCTGAATG-3′ and 5′-TCTTGCTGCCTGTTGATGAG-3′; 5′ region of EF4, 5′-TGCTAGACTTGGCCTCAACC-3′ and 5′-ACCACACCCCTCTTCACAAG-3′; 5′ region of β-tubulin, 5′-TCAAAACAATGGCTCGAAAA-3′ and 5′-AGCGAAAAATGGGGAGAAGT-3′; and GAPDH promoter region, 5′-ACGTAGCTCAGGCCTCAAGA-3′ and 5′-AAGAAGATGCGGCTGACTGT-3′.
RNAi—RNAi knockdown of MCM2 and MCM5 were performed with the RNAi human/mouse control kit from Qiagen. Two sets of MCM5 siRNAs were described previously (22), and MCM2 were from Qiagen (catalog number SI02653525). 293T cells were transfected with 0.25 μg of each MCM5 siRNA or 0.5 μg of MCM2 or negative control siRNA according to the manufacturer's instructions.
Tritium Incorporation Assay—Transcription efficiency was measured by tritium incorporation as described previously (26). A total of 1 × 106 cells were treated with 50 μm [5,6-H-3]uridine 40–50 Ci/mmol for 0, 20, and 120 min. Cells were washed once with 1× phosphate-buffered saline and resuspended in 500 μl of 10% trichloroacetic acid. Precipitated nucleotides were collected with glass microfiber filters, and counts per minute (cpm) were determined using a scintillation counter.
Nuclear Run-on Assay—A total of 1 × 106 cells were suspended in 4 ml of lysis buffer (10 mm Tris-Cl, pH 7.4, 10 mm NaCL, 3 mm MgCl2, 0.5% Nonidet P-40) and incubated on ice for 5 min. Cells were centrifuged at 1500 rpm for 5 min at 4 °C. Cells were again resuspended in 4 ml of lysis buffer and centrifuged. Cells were then resuspended in 200 μl of storage buffer (50 mm Tris-Cl, pH 8.3, 40% (v/v) glycerol, 5 mm MgCl2, 0.1 mm EDTA) and added to 200 μl of reaction buffer (10 mm Tris-Cl, pH 8.0, 5 mm MgCl2, 0.3 m KCl with 10 μl each of 100 mm ATP, CTP, GTP, 5 μl of 1 m dithiothreitol, 10 μl of [α-32P]UTP). Reactions were incubated at 30 °C for 30 min. RNA was extracted with TRIzol (Invitrogen) according to the manufacturer's instructions, and RNA was resuspended in 500 μl of diethylpyrocarbonate (DEPC) treated water. RNA was incubated at 65 °C for 10 min and incubated with slot-blot membranes in 25 ml of Church's buffer (1% bovine serum albumin, 0.5 m NaHPO4, 0.5 m NaH2PO4, 0.001 m EDTA, 7% SDS) at 65 °C overnight. Membranes contained 5 μg of the indicated plasmids and poly(A) RNA. Poly(A) RNA was isolated with the Promega poly(A) tract kit.
PI Staining and FACS—Cells were cultured as described in 10-cm dishes to 80–90% confluence and stained with PI (10 μg/ml PI in 38 mm sodium citrate, pH 7.0) as described previously (27). Cells were analyzed using FACSCalibur and CellQuest analysis software.
Transfections—Transient transfections of 293T cells were performed with the Lipofectamine 2000 reagent according to the manufacturer's instructions (Invitrogen).
Western Blot Analysis—Western blot analysis was performed with the indicated antibodies on whole cell extracts separated by SDS-PAGE and analyzed by chemiluminescence (PerkinElmer Life Sciences).
Plasmids—The plasmids pMCM5WT, pMCM5KMD, and pMCM5RK were constructed as described previously (21). The plasmid pMCM5WT contains the wild-type hemagglutinin A (HA)-tagged MCM5 gene. The plasmid pMCM5KMD contains mutations in the ATP-binding site domain and the conserved helicase domain of MCM5 (described as KMDA4 in Ref. 21). The pMCM5RK construct contains mutations at residues Arg-732 and Lys-734 (described as R732D/K734D in Ref. 21). All MCM5 constructs were subcloned into RcCMV (Invitrogen) as described previously (21).
RESULTS
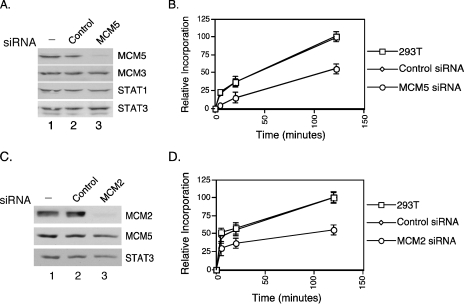
MCM5 and MCM2 Are Required for General Transcription—To determine whether MCM5 plays a role in general transcription, we utilized the RNAi knockdown technique that we have used in previous studies of Stat1 (22) to significantly reduce the amount of MCM5 in the cell but not eliminate it completely. 293T cells were transfected with a negative control siRNA or MCM5 siRNA for 4 days. Transfection with the MCM5 siRNA in 293T cells greatly reduced the level of MCM5 when compared with untransfected cells or those transfected with a negative control siRNA (Fig. 1A). MCM3 levels were unaffected by either siRNA; Stat1 and Stat3 are shown as loading controls (Fig. 1A). Cells containing the MCM5 siRNA, a negative control siRNA, and untransfected cells were analyzed by tritium incorporation assay for general transcription activity. Four days after siRNA transfection, tritiated uridine was added to the medium for 5, 20, and 120 min, and its incorporation into RNA was measured. There was an ∼50% decrease in tritium incorporation for the cells transfected with MCM5 siRNA when compared with untransfected cells or cells with the control siRNA (Fig. 1B).
FIGURE 1.
MCM5 and MCM2 are essential for general transcription. A, 293T cells were left untreated or transfected with either a negative control siRNA or MCM5 siRNA. Whole cell extracts were prepared after 4 days, and Western blot analysis was performed with anti-MCM5, anti-MCM3, anti-Stat1, and anti-Stat3 antibodies. B, 293T cells transfected as described in A were treated with 50 μm [5,6-H-3]uridine 40–50 Ci/mmol for 5, 20, and 120 min. Cells were washed once with 1× phosphate-buffered saline and resuspended in 500 μl of 10% trichloroacetic acid. The precipitated nucleotides were collected with glass microfiber filters, and cpm were determined with a scintillation counter. Data represent the averages and standard deviations (error bars) of three experiments. C, 293T cells were transfected with either a negative control siRNA or MCM2 siRNA and cultured for 3 days, and Western blot was performed with anti-MCM2, anti-MCM5, and anti-Stat3 antibodies. D, cells transfected as described in C were analyzed by tritium incorporation assays as described in B.
To test whether other members of the MCM2-7 hexamer complex are necessary for general transcription, we analyzed the effect of knockdown of MCM2 on transcription because MCM2 was co-purified with the RNA Pol II holoenzyme and has been shown to specifically interact with the carboxyl-terminal domain of RNA Pol II (23, 24). 293T cells were transiently transfected with an siRNA against MCM2 or a control siRNA. The MCM2 siRNA efficiently decreased levels of MCM2 protein when compared with untransfected (293T) cells and cells with a control siRNA (Fig. 1C, lane 3). The cellular levels of MCM5 and Stat3 were not significantly affected (Fig. 1C). Transcriptional activities in these cells were measured by incorporation of tritiated uridine. There was >50% decrease in tritium incorporation in cells containing the MCM2 siRNA when compared with untransfected cells (293T) and cells with negative control siRNA cells (Fig. 1D). All together, these results suggest that the MCM2-7 family plays an important role for transcription.
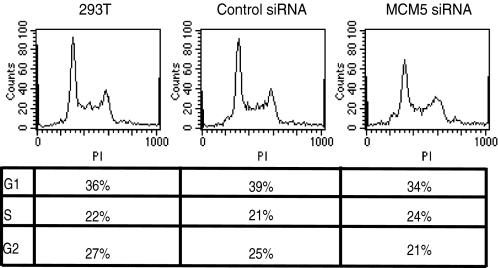
To ensure that the loss of transcription activity in the MCM5 knockdown cells is not due to cell death or cell cycle arrest, the cells were stained with PI and analyzed by FACS. Cells were growing normally and looked healthy at day four after siRNA transfection (data not shown), and FACS analyses of PI-stained cells showed that there was only a slight change in the distribution of cell population in the different phases of the cell cycle when compared with untransfected cells and cells transfected with a control siRNA (Fig. 2). These results indicate that a decrease in MCM5 protein levels resulted in a loss of general transcription activity and that the observed effect on transcription is not due to a disruption in DNA replication.
FIGURE 2.
Cell cycle analyses of cells with MCM5 knockdown by siRNA. 293T cells were transfected as described in the legend for Fig. 1. Cells were cultured for 4 days, and 1 × 106 cells were stained with propidium iodide (PI) and analyzed by FACs.
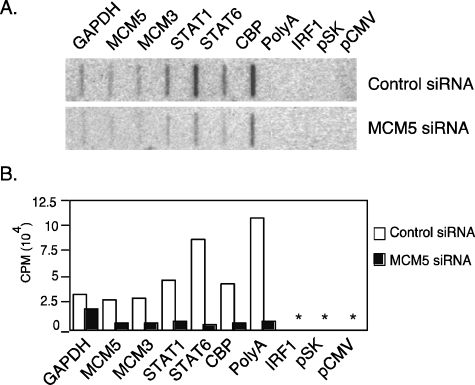
MCM5 Is Essential for RNA Pol II-mediated Transcription—The results obtained using tritiated uridine incorporation experiments suggest that MCM2 and MCM5 are required for general transcription. This assay does not distinguish RNA Pol II function from that of RNA Pol I and Pol III. To further demonstrate that MCM5 plays a role in RNA Pol II-dependent transcription and has a direct impact on transcription rate, nuclear run-on assays were performed on RNA Pol II genes. Plasmid DNA encoding RNA Pol II-dependent and constitutively expressed genes GAPDH, MCM5, MCM3, Stat1, Stat6, CREB-binding protein (CBP), and purified poly(A) mRNA were used to measure the mRNA transcription rate (Fig. 3). In cells transfected with the MCM5 siRNA, transcription for all these genes and the poly(A) mRNA was decreased when compared with the cells transfected with the control siRNA (Fig. 3, A and B). The IRF1 gene was used as a negative control because it is not constitutively expressed. Two plasmid vectors were also used as negative controls, and no signals were detected on these controls. Altogether, these results demonstrate that the MCM5 protein is required for RNA Pol II-dependent transcription of constitutively expressed genes.
FIGURE 3.
MCM5 is essential for RNA Pol II-mediated transcription. A, 293T cells transfected as in Fig. 1A were grown for 4 days, scraped, and washed with 1× phosphate-buffered saline. Nuclear run-on assays were performed with slot-blot membranes containing 5 μg of plasmid DNA or poly(A) RNA. CBP, CREB-binding protein. B, results from A were quantitated with a PhosphorImager (Amersham Biosciences). Results represent one of two independent experiments.
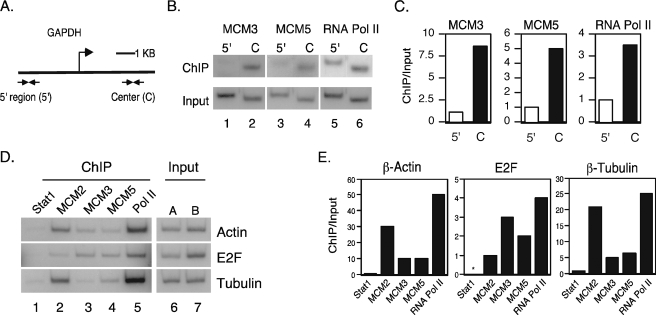
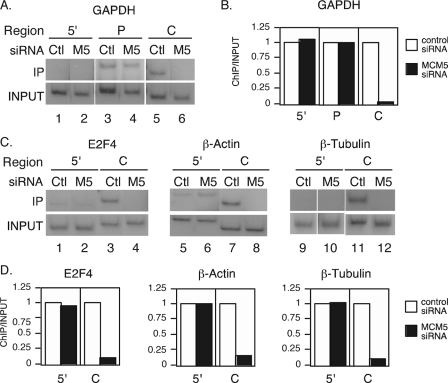
Members of the MCM2-7 Complex Specifically Co-localize with RNA Pol II in the Centers of Constitutively Transcribed Genes—We have previously demonstrated that members of the MCM2-7 family bound to the promoters of Stat1 target genes in response to IFN-γ and further move along with RNA Pol II for ligand-induced transcription (22). To further analyze the role of the MCM2-7 proteins for RNA Pol II-dependent transcription of constitutively expressed genes, we used ChIP assays to determine whether the MCM2-7 complex interacts with constitutively transcribed genes on the chromosome. Using PCR primers specific for the center or the 5′ intergenic region of the GAPDH gene (Fig. 4A), ChIP assays were performed with antibodies against MCM3, MCM5, and RNA Pol II. Both MCM3 and MCM5 were present at high levels in the center of the GAPDH gene (Fig. 4, B, lanes 2 and 4, and C) but at very low levels in the 5′ intergenic region (Fig. 4, B, lanes 1 and 3, and C). RNA Pol II was also present in the center of the GAPDH gene (Fig. 4, B, lane 6, and C), and a low level of RNA Pol II was also detected in the 5′ intergenic region (Fig. 4, B, lane 5, and C). These data demonstrate that there is a specific co-localization of MCM3 and MCM5 with RNA Pol II in the center of a constitutively transcribing gene on the chromosome.
FIGURE 4.
Members of the MCM2-7 family co-localize with RNA Pol II at the loci of constitutively transcribed genes. A, map of the GAPDH gene and primers used for ChIP analysis against the 5′ region and the center (C) of the gene. B and C, 2fTGH cells were grown in 15-cm dishes to about 80–90% confluence. ChIP assays were performed using anti-MCM3, anti-MCM5, and anti-RNA Pol II antibodies. PCR reactions used primers against the 5′ region or the center of the GAPDH gene. C, results were quantitated using a PhosphorImager and expressed as ChIP/Input. D, ChIP assays were performed for 2fTGH cells with anti-Stat1, anti-MCM2, anti-MCM3, anti-MCM5, and anti-RNA Pol II antibodies. One 15-cm dish was used for the Stat1, MCM2, and MCM3 ChIP (lanes 1–3), and a second dish was used for the MCM5, RNA Pol II (lanes 4 and 5), and Stat1 ChIP. The result of one Stat1 IP (lane 1) is shown. Input A (lane 6) corresponds to the Stat1, MCM2, and MCM3 ChIP (lanes 1–3), and Input B (lane 7) corresponds to Mcm5 and RNA Pol II (lanes 4 and 5). Primers correspond to the centers of the β-actin, E2F4, and β-tubulin genes. E, signals in D were quantitated with a PhosphorImager, and values are expressed as ChIP/Input. Results represent at least three independent experiments.
To further demonstrate that the MCM2-7 complex interacts with other constitutively expressed genes in addition to GAPDH, ChIP assays were performed with antibodies against MCM2, MCM3, MCM5, and RNA Pol II, and primers were designed for the centers of the β-actin, E2F4, and β-tubulin genes. Stat1 was used as a negative control because it does not interact with constitutively expressed genes and only interacts with the promoters of specific target genes in response to IFN-γ stimulation (22). MCM2, MCM3, MCM5, and RNA Pol II were all detected in the center of these genes, whereas Stat1 was not (Fig. 4, D and E). These results further indicate that the MCM2-7 complex is involved in RNA Pol II-mediated transcription.
MCM5 Is Required for Transcription Elongation of RNA Pol II—To determine whether the MCM2-7 proteins are required for the presence of RNA Pol II in the centers of constitutively expressed genes, ChIP analysis was performed with antibodies against RNA Pol II in the MCM5 RNAi knockdown cells. RNA Pol II was present in the promoter region and center of the GAPDH gene in cells transfected with a negative control siRNA (Fig. 5, A, lanes 3 and 5, and B). In contrast, RNA Pol II was only detected in the promoter region in the MCM5 knockdown cells (Fig. 5, A, lane 4, and B), and there was almost no RNA Pol II in the center of the gene (Fig. 5, A, lane 6, and B). Very low levels of RNA Pol II were detected in the 5′ intergenic region in both cell types (Fig. 5, A, lanes 1 and 2, and B). Together, these results indicate that MCM5 is required for transcription elongation of RNA Pol II but not required for the recruitment of RNA Pol II to the promoter.
FIGURE 5.
MCM5 is required for transcription elongation of RNA Pol II. RNAi-mediated MCM5 knockdown was performed in 293T cells as described in the legend for Fig. 1, and ChIP analysis was performed with RNA Pol II antibody as described in the legend for Fig. 4. A, primers against the 5′ intergenic region (5′), promoter region (P), and center (C) of the GAPDH gene were used for ChIP analysis in both control (Ctl) and MCM5 siRNA-transfected (M5) cells. C, ChIP was performed with primers against the 5′ regions (5′) and centers (C) of E2F4, β-actin, and β-tubulin. B and D, signals from A and C were quantitated with a PhosphorImager and shown as ChIP/INPUT with results from control siRNA cells set as 1.
ChIP experiments were also performed for the E2F4, β-actin, and β-tubulin genes in the MCM5 knockdown cells. Similar to the results observed for GAPDH, RNA Pol II was localized to the centers of these genes only in cells transfected with a negative control siRNA (Fig. 5, C, lanes 3, 7, and 11, and D). Very low levels of RNA Pol II were detected in the centers of these genes in the MCM5 RNAi knockdown (Fig. 5, C, lanes 4, 8, and 12, and D). The 5′ intergenic regions are shown as a negative control. These results further demonstrate that MCM5 is required for transcription elongation of RNA Pol II.
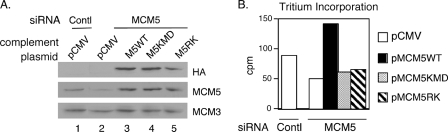
The Intact MCM2-7 Hexamer and Helicase Activity Are Required for Transcription—Several studies suggest that the MCM2-7 hexamer complex functions as a DNA helicase during DNA replication (11–13). However, the functional role of the MCM2-7 complex in the process of transcription is not as well defined. To determine whether the intact MCM2-7 hexamer complex and the helicase activity of each member are required for transcription, we set up a knockdown complementation procedure with MCM5 as an example because of the availability of many MCM5 mutants generated from our previous transcriptional studies of Stat1 (21). 293T cells were first transfected with the MCM5 siRNA or control siRNA for 3 days to allow sufficient knockdown. The cells were then divided and further transfected with the following expression plasmids: vector, MCM5 wild type, MCM5KMD (mutations in both the ATP-binding site and the helicase domain), and MCM5RK (a mutant defective in forming the hexamer) (21). The exogenous MCM5 was tagged with hemagglutinin A (HA) (Fig. 6) to allow for detection of expression by Western blot analyses. The twice-transfected cells were further cultured for 24 h and then harvested and analyzed by Western blot analyses and tritium incorporation analyses. As shown in Fig. 6, the expression of wild-type MCM5 in MCM5 knockdown cells (Fig. 6A, lane 3) rescued the transcription activity (Fig. 6B), whereas MCM5 mutants defective in either helicase activity (Fig. 6A, lane 4) or hexamer formation (Fig. 6A, lane 5) could not rescue the general transcription activity (Fig. 6B). Due to the high expression level of exogenous MCM5 from the RcCMV plasmids, the MCM5 siRNA at day 4 after transfection did not inhibit the expression of exogenous MCM5. Furthermore, a wild-type MCM5 construct with a modified sequence at the siRNA target site also rescued transcription efficiency similar to the unmodified wild-type construct (data not shown). These results demonstrate that the intact MCM2-7 hexamer and helicase activity are required for transcription.
FIGURE 6.
Requirement of the MCM2-7 hexamer and helicase activity for transcription. A, 293T cells were transfected with either MCM5 siRNA or a negative control siRNA (Contl) and grown for 3 days. Cells were then transfected with pCMV, pMCM5WT, pMCM5KMD, or pMCM5RK and grown for 24 h. Whole cell extracts were prepared from a portion of the cells and analyzed by Western blot analysis. HA, hemagglutinin A (HA). B, the remaining cells were treated with 50 mm [5,6-H-3]uridine 40–50 Ci/mmol for 2 h followed by trichloroacetic acid precipitation and analysis of tritium incorporation. Results represent one of three independent experiments.
DISCUSSION
It has long been shown that there is an overabundance of the MCM2-7 proteins when compared with the number of DNA replication origins in chromatin (1, 4–6). It has been suggested that the MCM2-7 proteins may function in other biological processes that require the unwinding of the DNA helix, such as DNA recombination, DNA repair, and transcription (3, 28). Recent studies showed that the excess of MCM2-7 proteins protects human cells from replicative stress by licensing backup origins of replication (29, 30). Our previous studies demonstrated that one specific transcriptional event, the cytokine-induced transcription activation mediated by a transcription factor called Stat1, requires one member of the MCM2-7 family and that the whole MCM2-7 complex was inducibly bound to the Stat1 target gene promoters (22). Together with the earlier findings of a specific interaction between MCM2 and the carboxyl-terminal domain of RNA Pol II (23, 24), we set out to test the hypothesis that the MCM2-7 proteins are essential for RNA Pol II-dependent transcription of constitutively expressed genes.
Utilizing the siRNA technique to knock down the MCM proteins without causing cell death (22) (Fig. 2), we have generated an experimental condition in which one of the MCM2-7 proteins is transiently knocked down, allowing us to study the role of MCM2-7 in transcription without affecting DNA replication and cell cycle progression. Other studies have also shown that efficient knockdown of MCM5 did not affect cell cycle progression or cell proliferation (29), although varying the concentration of MCM siRNAs and multiple rounds of siRNA transfection could affect proliferation and cell cycle progression (29, 30). Our initial experiments utilizing RNAi-mediated knockdown of MCM2 and MCM5 and tritiated uridine incorporation assay determined that the MCM2-7 complex is required for transcription in general. The nuclear run-on assay using RNA Pol II-dependent constitutively transcribing genes and poly(A) RNA specifically demonstrate that MCM5 is indeed necessary for RNA Pol II-mediated transcription. Although the detailed analyses presented in this report focused on the role of MCM2-7, specifically in RNA Pol II-mediated transcription, they did not rule out a possible role for MCM2-7 in RNA Pol I and Pol III-mediated transcription. Preliminary experiments using nuclear run-on assays with the 18S rRNA and 5S rRNA genes in MCM5 knockdown cells suggest that MCM5 may also be required for efficient transcription of these genes.3 Further detailed studies are needed to examine the role of MCM2-7 in transcription mediated by Pol I and Pol III. It is likely that the MCM2-7 proteins may be an essential component of the transcription machinery in general. Therefore, the abundance of these MCM2-7 proteins allows them to participate in two fundamental cellular processes: DNA replication and RNA transcription.
Because of the conserved helicase domains present in each member of the MCM2-7 family, they have been assumed to function as DNA helicases for unwinding of the DNA helix during replication (3, 18). It is not clear whether the hexamer ring detected by electronic microscopy (16, 31) contains all six members (heterohexamer) or only a few members (dimer of trimers or trimer of dimers, etc.). MCM2-7 proteins can form a heterohexamer complex and subcomplexes such as MCM4/6/7 and MCM3/5 (4, 11, 32, 33). It has been suggested that these subcomplexes are components in the assembly of the hexameric structure or that they may have regulatory activities (18). For our studies of the involvement of MCM2-7 in transcription, we developed a procedure to introduce mutants defective in formation of the heterohexamer or helicase domain (21) in MCM5 knockdown cells to assess the function of the hexamer helicase in transcription in vivo. Our results using mutations in MCM5 suggest that it is likely that each member of the MCM2-7 family is necessary to form the hexamer on chromatin and contribute to the helicase function for transcription.
The RNA Pol II holoenzyme contains DNA helicase components such as the TFIIH complex required for transcription initiation (34, 35). However, the TFIIH complex has not been shown to move with the RNA Pol II during transcription elongation. Chromatin remodeling complexes have been implicated in transcription elongation by RNA Pol II, yet no direct interaction with RNA Pol II has been demonstrated with these factors (36). Our findings of the co-presence of MCM2-7 proteins with the RNA Pol II in the central portions of transcribing genes (22) (Fig. 4) strongly suggest that it is the MCM2-7 hexamer that provides the necessary DNA helicase activity during transcription elongation. Furthermore, we demonstrate that, at least one of the MCM2-7 proteins, MCM5, is required for transcription elongation of RNA Pol II (Fig. 5). It is conceivable that through the specific interaction between MCM2 and the carboxyl-terminal domain of RNA Pol II (23, 24), the whole MCM2-7 hexamer complex travels with RNA Pol II to unwind the DNA helix during transcription elongation.
Acknowledgments
We thank Olivia Lou for technical advice regarding FACS analysis and Selena Chen-Kiang for use of the slot-blot apparatus. We thank all the laboratory members for their help.
This work was supported, in whole or in part, by National Institutes of Health Grant GM61652 (to J. J. Z.).
Footnotes
The abbreviations used are: MCM, minichromosome maintenance protein; RNA Pol II, RNA polymerase II; IFN, interferon; ChIP, chromatin immunoprecipitation; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; RNAi, RNA interference; siRNA, small interfering RNA; FACS, fluorescence-activated cell sorter; CREB, cAMP-response element-binding protein; Stat1, signal transducer and activator of transcription 1; PI, propidium iodide.
M. Snyder and J. J. Zhang, unpublished data.
References
- 1.Donovan, S., Harwood, J., Drury, L. S., and Diffley, J. F. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 5611–5616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tye, B. K. (1999) Annu. Rev. Biochem. 68 649–686 [DOI] [PubMed] [Google Scholar]
- 3.Forsburg, S. L. (2004) Microbiol. Mol. Biol. Rev. 68 109–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lei, M., Kawasaki, Y., and Tye, B. K. (1996) Mol. Cell. Biol. 16 5081–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raghuraman, M. K., Winzeler, E. A., Collingwood, D., Hunt, S., Wodicka, L., Conway, A., Lockhart, D. J., Davis, R. W., Brewer, B. J., and Fangman, W. L. (2001) Science 294 115–121 [DOI] [PubMed] [Google Scholar]
- 6.Wyrick, J. J., Aparicio, J. G., Chen, T., Barnett, J. D., Jennings, E. G., Young, R. A., Bell, S. P., and Aparicio, O. M. (2001) Science 294 2357–2360 [DOI] [PubMed] [Google Scholar]
- 7.Tanaka, T., Knapp, D., and Nasmyth, K. (1997) Cell 90 649–660 [DOI] [PubMed] [Google Scholar]
- 8.Aparicio, O. M., Weinstein, D. M., and Bell, S. P. (1997) Cell 91 59–69 [DOI] [PubMed] [Google Scholar]
- 9.Labib, K., Tercero, J. A., and Diffley, J. F. (2000) Science 288 1643–1647 [DOI] [PubMed] [Google Scholar]
- 10.Tsuyama, T., Tada, S., Watanabe, S., Seki, M., and Enomoto, T. (2005) Nucleic Acids Res. 33 765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishimi, Y. (1997) J. Biol. Chem. 272 24508–24513 [DOI] [PubMed] [Google Scholar]
- 12.Bochman, M. L., and Schwacha, A. (2008) Mol. Cell 31 287–293 [DOI] [PubMed] [Google Scholar]
- 13.Bochman, M. L., Bell, S. P., and Schwacha, A. (2008) Mol. Cell. Biol. 28 5865–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, J. K., and Hurwitz, J. (2000) J. Biol. Chem. 275 18871–18878 [DOI] [PubMed] [Google Scholar]
- 15.Prokhorova, T. A., and Blow, J. J. (2000) J. Biol. Chem. 275 2491–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yabuta, N., Kajimura, N., Mayanagi, K., Sato, M., Gotow, T., Uchiyama, Y., Ishimi, Y., and Nojima, H. (2003) Genes Cells 8 413–421 [DOI] [PubMed] [Google Scholar]
- 17.Kanter, D. M., Bruck, I., and Kaplan, D. L. (2008) J. Biol. Chem. 283 31172–31182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tye, B. K., and Sawyer, S. (2000) J. Biol. Chem. 275 34833–34836 [DOI] [PubMed] [Google Scholar]
- 19.Shin, J. H., and Kelman, Z. (2006) J. Biol. Chem. 281 26914–26921 [DOI] [PubMed] [Google Scholar]
- 20.Zhang, J. J., Zhao, Y., Chait, B. T., Lathem, W. W., Ritzi, M., Knippers, R., and Darnell, J. E., Jr. (1998) EMBO J. 17 6963–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DaFonseca, C. J., Shu, F., and Zhang, J. J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 3034–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snyder, M., He, W., and Zhang, J. J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 14539–14544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yankulov, K., Todorov, I., Romanowski, P., Licatalosi, D., Cilli, K., McCracken, S., Laskey, R., and Bentley, D. L. (1999) Mol. Cell. Biol. 19 6154–6163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holland, L., Gauthier, L., Bell-Rogers, P., and Yankulov, K. (2002) Eur. J. Biochem. 269 5192–5202 [DOI] [PubMed] [Google Scholar]
- 25.Yang, E., Henriksen, M. A., Schaefer, O., Zakharova, N., and Darnell, J. E., Jr. (2002) J. Biol. Chem. 277 13455–13462 [DOI] [PubMed] [Google Scholar]
- 26.Ausubel, F. M., Brent, R, Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. (eds) (1997) Current Protocols in Molecular Biology, pp. 18.7.14–18.7.15, John Wiley & Sons, Inc., New York
- 27.Crissman, H. A., and Steinkamp, J. A. (1973) J. Cell Biol. 59 766–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newlon, C. S. (1997) Cell 91 717–720 [DOI] [PubMed] [Google Scholar]
- 29.Ge, X. Q., Jackson, D. A., and Blow, J. J. (2007) Genes Dev. 21 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ibarra, A., Schwob, E., and Mendez, J. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 8956–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato, M., Gotow, T., You, Z., Komamura-Kohno, Y., Uchiyama, Y., Yabuta, N., Nojima, H., and Ishimi, Y. (2000) J. Mol. Biol. 300 421–431 [DOI] [PubMed] [Google Scholar]
- 32.Holthoff, H. P., Baack, M., Richter, A., Ritzi, M., and Knippers, R. (1998) J. Biol. Chem. 273 7320–7325 [DOI] [PubMed] [Google Scholar]
- 33.Thommes, P., Kubota, Y., Takisawa, H., and Blow, J. J. (1997) EMBO J. 16 3312–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guzder, S. N., Sung, P., Bailly, V., Prakash, L., and Prakash, S. (1994) Nature 369 578–581 [DOI] [PubMed] [Google Scholar]
- 35.Moreland, R. J., Tirode, F., Yan, Q., Conaway, J. W., Egly, J. M., and Conaway, R. C. (1999) J. Biol. Chem. 274 22127–22130 [DOI] [PubMed] [Google Scholar]
- 36.Eisen, A., and Lucchesi, J. C. (1998) BioEssays 20 634–641 [DOI] [PubMed] [Google Scholar]