Abstract
NHE5 is a brain-enriched Na+/H+ exchanger that dynamically shuttles between the plasma membrane and recycling endosomes, serving as a mechanism that acutely controls the local pH environment. In the current study we show that secretory carrier membrane proteins (SCAMPs), a group of tetraspanning integral membrane proteins that reside in multiple secretory and endocytic organelles, bind to NHE5 and co-localize predominantly in the recycling endosomes. In vitro protein-protein interaction assays revealed that NHE5 directly binds to the N- and C-terminal cytosolic extensions of SCAMP2. Heterologous expression of SCAMP2 but not SCAMP5 increased cell-surface abundance as well as transporter activity of NHE5 across the plasma membrane. Expression of a deletion mutant lacking the SCAMP2-specific N-terminal cytosolic domain, and a mini-gene encoding the N-terminal extension, reduced the transporter activity. Although both Arf6 and Rab11 positively regulate NHE5 cell-surface targeting and NHE5 activity across the plasma membrane, SCAMP2-mediated surface targeting of NHE5 was reversed by dominant-negative Arf6 but not by dominant-negative Rab11. Together, these results suggest that SCAMP2 regulates NHE5 transit through recycling endosomes and promotes its surface targeting in an Arf6-dependent manner.
Neurons and glial cells in the central and peripheral nervous systems are especially sensitive to perturbations of pH (1). Many voltage- and ligand-gated ion channels that control membrane excitability are sensitive to changes in cellular pH (1-3). Neurotransmitter release and uptake are also influenced by cellular and organellar pH (4, 5). Moreover, the intra- and extracellular pH of both neurons and glia are modulated in a highly transient and localized manner by neuronal activity (6, 7). Thus, neurons and glia require sophisticated mechanisms to finely tune ion and pH homeostasis to maintain their normal functions.
Na+/H+ exchangers (NHEs)3 were originally identified as a class of plasma membrane-bound ion transporters that exchange extracellular Na+ for intracellular H+, and thereby regulate cellular pH and volume. Since the discovery of NHE1 as the first mammalian NHE (8), eight additional isoforms (NHE2-9) that share 25-70% amino acid identity have been isolated in mammals (9, 10). NHE1-5 commonly exhibit transporter activity across the plasma membrane, whereas NHE6-9 are mostly found in organelle membranes and are believed to regulate organellar pH in most cell types at steady state (11). More recently, NHE10 was identified in human and mouse osteoclasts (12, 13). However, the cDNA encoding NHE10 shares only a low degree of sequence similarity with other known members of the NHE gene family, raising the possibility that this sodium-proton exchanger may belong to a separate gene family distantly related to NHE1-9 (see Ref. 9).
NHE gene family members contain 12 putative transmembrane domains at the N terminus followed by a C-terminal cytosolic extension that plays a role in regulation of the transporter activity by protein-protein interactions and phosphorylation. NHEs have been shown to regulate the pH environment of synaptic nerve terminals and to regulate the release of neurotransmitters from multiple neuronal populations (14-16). The importance of NHEs in brain function is further exemplified by the findings that spontaneous or directed mutations of the ubiquitously expressed NHE1 gene lead to the progression of epileptic seizures, ataxia, and increased mortality in mice (17, 18). The progression of the disease phenotype is associated with loss of specific neuron populations and increased neuronal excitability. However, NHE1-null mice appear to develop normally until 2 weeks after birth when symptoms begin to appear. Therefore, other mechanisms may compensate for the loss of NHE1 during early development and play a protective role in the surviving neurons after the onset of the disease phenotype.
NHE5 was identified as a unique member of the NHE gene family whose mRNA is expressed almost exclusively in the brain (19, 20), although more recent studies have suggested that NHE5 might be functional in other cell types such as sperm (21, 22) and osteosarcoma cells (23). Curiously, mutations found in several forms of congenital neurological disorders such as spinocerebellar ataxia type 4 (24-26) and autosomal dominant cerebellar ataxia (27-29) have been mapped to chromosome 16q22.1, a region containing NHE5. However, much remains unknown as to the molecular regulation of NHE5 and its role in brain function.
Very few if any proteins work in isolation. Therefore identification and characterization of binding proteins often reveal novel functions and regulation mechanisms of the protein of interest. To begin to elucidate the biological role of NHE5, we have started to explore NHE5-binding proteins. Previously, β-arrestins, multifunctional scaffold proteins that play a key role in desensitization of G-protein-coupled receptors, were shown to directly bind to NHE5 and promote its endocytosis (30). This study demonstrated that NHE5 trafficking between endosomes and the plasma membrane is regulated by protein-protein interactions with scaffold proteins. More recently, we demonstrated that receptor for activated C-kinase 1 (RACK1), a scaffold protein that links signaling molecules such as activated protein kinase C, integrins, and Src kinase (31), directly interacts with and activates NHE5 via integrin-dependent and independent pathways (32). These results further indicate that NHE5 is partly associated with focal adhesions and that its targeting to the specialized microdomain of the plasma membrane may be regulated by various signaling pathways.
Secretory carrier membrane proteins (SCAMPs) are a family of evolutionarily conserved tetra-spanning integral membrane proteins. SCAMPs are found in multiple organelles such as the Golgi apparatus, trans-Golgi network, recycling endosomes, synaptic vesicles, and the plasma membrane (33, 34) and have been shown to play a role in exocytosis (35-38) and endocytosis (39). Currently, five isoforms of SCAMP have been identified in mammals. The extended N terminus of SCAMP1-3 contain multiple Asn-Pro-Phe (NPF) repeats, which may allow these isoforms to participate in clathrin coat assembly and vesicle budding by binding to Eps15 homology (EH)-domain proteins (40, 41). Further, SCAMP2 was shown recently to bind to the small GTPase Arf6 (38), which is believed to participate in traffic between the recycling endosomes and the cell surface (42, 43). More recent studies have suggested that SCAMPs bind to organellar membrane type NHE7 (44) and the serotonin transporter SERT (45) and facilitate targeting of these integral membrane proteins to specific intracellular compartments. We show in the current study that SCAMP2 binds to NHE5, facilitates the cell-surface targeting of NHE5, and elevates Na+/H+ exchange activity at the plasma membrane, whereas expression of a SCAMP2 deletion mutant lacking the N-terminal domain containing the NPF repeats suppresses the effect. Further we show that this activity of SCAMP2 requires an active form of a small GTPase Arf6, but not Rab11. We propose a model in which SCAMPs bind to NHE5 in the endosomal compartment and control its cell-surface abundance via an Arf6-dependent pathway.
EXPERIMENTAL PROCEDURES
Antibodies—Mouse monoclonal anti-HA antibodies were obtained from Covance (Richmond, CA). Rabbit polyclonal anti-Myc (A-14), anti-HA (Y-11), and mouse monoclonal anti-SCAMP2 (8C10) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal anti-Rab11 and anti-Rab4 antibodies were obtained from Zymed Laboratories Inc. (South San Francisco, CA) and Stressgen (Victoria, British Columbia, Canada), respectively. Polyclonal antibodies against SCAMP1, -2, and -5 were purchased from Affinity BioReagents (Golden, CO). The purified mouse monoclonal antibody rho 1D4 against a 9-amino acid TETSQVAPA C-terminal epitope (46, 47) was obtained from the National Cell Culture Center (Minneapolis, MN). The antibody was coupled to CNBr-activated Sepharose beads as previously described (48). Affinity-purified anti-NHE5 rabbit polyclonal antibody raised against human NHE5 (G674-L896), which cross-reacts with rat NHE5 (32), was used for endogenous co-immunoprecipitation experiments. Goat anti-rabbit and goat anti-mouse horseradish-peroxidase fusion secondary antibodies were obtained from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). Alexa 647-coupled goat anti-mouse and Alexa 568 or Alexa 488-coupled goat anti-rabbit secondary antibodies were obtained from Invitrogen.
Mammalian Expression Constructs—First strand cDNA was synthesized from human brain RNA (Clontech, Mountain View, CA) by using random hexamers and SuperScript II reverse transcriptase (Invitrogen), and subjected to PCR using Pfu-Turbo (Stratagene, La Jolla, CA) to clone human Arf6 and Rab11. The following primers were used: Arf6 forward (5′-ATG GGG AAG GTG CTA TCC AAA ATC TTC GG-3′) and Arf6 reverse (5′-AGA TTT GTA GTT AGA GGT TAA CCA TGT G-3′); Rab11 forward (5′-ATG GGC ACC CGC GAC GAG TAC G-3′) and Rab11 reverse (5′-GAT GTT CTG ACA GCA CTG CAC CTT TGG-3′). The identities of the PCR fragments were verified by sequencing and subjected to a second round of PCR to introduce a Myc or HA tag at the extreme C terminus of the clone. The PCR fragment was ligated into mammalian expression vector pcDNA3, and the sequence of the Myc-tagged constructs was verified subsequently. Arf6T27N and Rab11S25N dominant-negative mutants were generated by using the QuikChange mutagenesis kit (Stratagene) using the HA-tagged Arf6/pcDNA3 or Myc-tagged Rab11/pcDNA3 as a template. The Myc-tagged human SCAMPs were described previously (44). The coding region for SCAMP2 and SCAMP5 was amplified by PCR and ligated into the pEGFP N1-vector in-frame to make GFP fusion constructs (SCAMP2GFP and SCAMP5GFP). GFP-tagged Arf6T27N (49) was a kind gift from Dr. Martin Schwartz (University of Virginia).
Cell Culture and Transfection—AP-1 cells stably expressing NHE5 with a triple HA tag inserted after amino acid residue 36 (AP-1/NHE5HA cells) (50) were maintained in α-minimal essential medium with 10% fetal bovine serum, and PC12 and PC12 stably expressing 1D4-tagged NHE5 (PC12/NHE51D4) cells were maintained in RPMI supplemented with 5% fetal bovine serum. Transient transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. NHE51D4 was transfected to PC12 cells using the conventional calcium phosphate method (51), and cells stably expressing NHE51D4 were selected in selection media containing G418 (200 μg/ml). Approximately 20 independent clones were screened to test NHE51D4 expression by Western blot and immunofluorescence microscopy, and several independent clones expressing moderate levels of NHE5 were analyzed.
Expression and Purification of GST Fusion Proteins—For producing GST fusion proteins, PCR fragments corresponding to different regions of the SCAMP2 cytoplasmic domains were inserted into a pGEX-2T bacterial expression vector (Amersham Biosciences) in-frame with the N-terminal GST tag as described previously (44). Protein expression was induced by incubating transformed BL21 Escherichia coli cells with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 3 h. E. coli cells were collected by centrifugation and resuspended in lysis buffer containing 1% Triton X-100 and protease inhibitor mixture (Roche Diagnostics, Laval, Canada) in PBS. Cell lysates were then incubated for 30 min on ice and then sonicated four times for 30 s. After sonication, cell debris was cleared by centrifugation for 10 min at 16,000 × g at 4 °C. GST fusion proteins were purified by incubation with reduced form glutathione-Sepharose beads (Amersham Biosciences) at 4 °C.
GST Pulldown—A 35S-labeled NHE5 C-terminal domain (Gly491-Leu896) was produced by in vitro transcription-translation using the TnT-coupled reticulocyte lysate system (Promega, Madison, WI) according to the manufacturer's instructions. The 35S-labeled in vitro translated protein was diluted to 1 ml with cold PBS and then centrifuged at 16,000 × g for 5 min to remove insoluble materials. The supernatant was then further diluted to 6.2 ml in cold PBS plus protease inhibitor mixture (Roche Applied Science). 750 μl of this diluted solution was incubated with 2 μg of GST fusion protein immobilized to the reduced form glutathione-Sepharose beads for 90 min at room temperature. After extensive washing, 35S-labeled in vitro translated protein bound to the GST fusion protein was eluted with SDS sample buffer, resolved by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and bound 35S-labeled NHE5 C terminus was detected by phosphorimaging. Equal input of the different GST fusion proteins was confirmed by resolving these proteins on SDS-PAGE followed by visualization using Coomassie Blue protein stain.
Co-immunoprecipitation—AP-1/NHE5HA cells were transfected with SCAMP2Myc, SCAMP2ΔNPFMyc (deletion of amino acids 1-55), SCAMP2ΔCMyc, or SCAMP2-(1-154)Myc, and the cells were lysed in PBS containing 1% CHAPS and protease inhibitor mixture (Roche Applied Science) on ice for 30 min. Lysates were cleared by centrifugation at 16,000 × g for 10 min (two times) at 4 °C. Cell lysates were then incubated with anti-HA monoclonal antibody or pre-immune serum at 4 °C for 4 h, followed by overnight incubation with protein G-Sepharose beads (Amersham Biosciences). After extensive washing, eluted samples were resolved in SDS-PAGE, and the proteins present in the immunoprecipitate were detected by Western blot. To isolate membrane fractions, cells were resuspended in sonication buffer (250 mm sucrose, 10 mm HEPES-NaOH, pH 7.4, 1 mm EDTA, with protease inhibitor mixture (Roche Applied Science)) and homogenized by mild disruption through a 26.5-gauge needle. Insoluble cellular debris was removed by centrifugation at 800 × g for 10 min at 4 °C, and membrane fractions were isolated by ultracentrifugation at 100,000 × g at 4 °C. The membrane fraction was then solubilized in PBS containing 1% CHAPS and protease inhibitor mixture (Roche Applied Science), and debris was removed by centrifugation at 16,000 × g for 10 min (two times) at 4 °C. NHE51D4 was immunoprecipitated from the solubilized membrane fraction with 1D4 mouse monoclonal antibody coupled to Sepharose beads and, after washing seven times, bound endogenous SCAMPs were resolved by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and detected with anti-SCAMP1, -2, or -5 rabbit polyclonal antibodies. An association of endogenous SCAMP2 with endogenous NHE5 in brain tissues was assessed as follows. Rat brain was homogenized in sonication buffer using a glass homogenizer followed by mixing the homogenate with an equal volume of PBS containing 2% CHAPS and protease inhibitor mixture (Roche Applied Science). The lysate was cleared, as above, and then incubated with anti-NHE5 rabbit polyclonal antibody or pre-immune serum at 4 °C for 4 h, followed by overnight incubation with protein A-Sepharose beads (Amersham Biosciences). SCAMP2 found in the immunoprecipitate was detected by Western blot using an anti-SCAMP2 mouse monoclonal antibody.
Measurement of Cell-surface Expression and Internalization of NHE5—AP-1/NHE5HA cells were transiently transfected with SCAMP2Myc, SCAMP5Myc, Arf6HA, Rab11Myc (wild-type or dominant-negative), or empty vector using Lipofectamine 2000, rinsed with ice-cold PBS buffer containing 1 mm MgCl2 and 0.1 mm CaCl2, pH 8.0 (PBSCM), and membrane proteins exposed on the extracellular surface were then indiscriminately labeled with the membrane-impermeable biotinylation reagent N-hydroxysulfosuccinimydyl-SS-biotin (Pierce), 0.5 mg/ml, in PBSCM for 30 min at 4 °C. The labeling solution was then discarded, and unreacted biotinylation reagent was quenched with PBSCM containing 20 mm glycine twice for 7 min each. For internalization experiments, labeled cells were subjected to chase incubation at 37 °C in culture media. Cells were treated, or left untreated, with glutathione cleavage buffer (50 mm glutathione, 90 mm NaCl, 1 mm MgCl2, 0.1 mm CaCl2, 60 mm NaOH, 0.2% bovine serum albumin, pH 8.6) for 20 min (two times) at 4 °C and then solubilized in PBS containing 1% CHAPS plus protease inhibitor mixture (Roche Applied Science). Insoluble debris was removed from the lysate by centrifugation at 16,000 × g for 10 min two times at 4 °C. Protein concentration was determined using Bradford assay, and an equal amount of protein was collected from each sample for analysis. A small amount (5%) of lysate was removed and represents the total fraction; the remaining lysate was then incubated with NeutrAvidin-agarose beads (Pierce) overnight to extract biotinylated proteins. Following washing of the beads, biotinylated proteins were then eluted with SDS-sample buffer containing 100 mm dithiothreitol, resolved in SDS-PAGE, and detected by Western blotting. The intensity of the bands was analyzed by densitometry of films exposed in the linear range.
22Na+ Influx Assay—Sub-confluent AP-1/NHE5HA cells were plated into 24-well plates and transfected with SCAMP2Myc, SCAMP5Myc, or empty parental pcDNA3 vector. Transfection efficiency exceeded 50%, as determined by immunofluorescence microscopy. Forty-eight hours post-transfection, cells were acidified using the NH4Cl pre-pulse technique (50). In brief, cells were treated with ammonium choline solution (50 mm NH4Cl, 80 mm choline chloride, 1 mm MgCl2, 2 mm CaCl2, 5 mm glucose, 20 mm HEPES-Tris, pH 7.4) for 20 min at 37 °C followed by a rapid washout with isotonic choline chloride solution (130 mm choline chloride, 1 mm MgCl2, 2 mm CaCl2, 5 mm glucose, 20 mm HEPES-Tris, pH 7.4) to acutely acidify the cytosol. Assays were immediately initiated by adding radioactive 22Na+ (1 μCi/ml 22NaCl in choline chloride solution) to each well in the absence or presence of 1 mm amiloride. After 5 min, the influx of 22Na+ was terminated by rapidly washing each well three times with ice-cold NaCl-saline solution (130 mm NaCl, 5 mm KCl, 1 mm MgCl2, 2 mm CaCl2, 5 mm glucose, 20 mm HEPES-NaOH, pH 7.4). Cells were then lysed in 0.5 n NaOH to extract the radiolabel. Lysates were neutralized by the addition of an equal volume of 0.5 n HCl, and the radioactivity was counted by liquid scintillation spectroscopy. Influx values obtained in the presence of amiloride were subtracted from those in the absence of amiloride. The difference represents the “amiloride-sensitive” 22Na+ influx due to NHE. Each experiment was conducted in quadruplicate, and three independent experiments were performed.
pHi Measurements—AP-1/NHE5HA cells
were transfected with GFP-tagged SCAMP2, SCAMP5, SCAMP2ΔNPF,
SCAMP2ΔC, SCAMP2-(1-154), or empty parental pEGFP expression vector
(Clontech) or co-transfected with Rab11Myc or Arf6HA
(either wild-type or dominant-negative) and SCAMP2GFP or pEGFP
vector. Cells were then plated onto glass coverslips and grown for 48 h prior
to pHi measurements. Coverslips with cells attached were
mounted in a temperature-controlled recording chamber filled with NaCl-saline
solution, placed on the microscope stage, and GFP-expressing cells were
identified by viewing GFP fluorescence during excitation at 488 nm.
Subsequently, cells were loaded with BCECF by adding 2 μm BCECF
acetoxymethyl ester to the NaCl-saline solution for 10 min at room temperature
and were then superfused at 2 ml/min with NaCl-saline solution (without dye)
at 34 °C for the remainder of the experiment. BCECF-derived fluorescence
emission intensities during excitation at 488 nm and 452 nm were at least
20-fold higher than the original GFP fluorescence signal. The dual excitation
ratio method was used to estimate pHi employing a
fluorescence ratio-imaging system (Atto Biosciences, Rockville, MD); full
details of the methods employed have been presented previously
(52,
53). The
high-[K+]/nigericin technique was employed to convert
background-corrected BCECF emission intensity ratios into
pHi values. Intracellular acid loads were imposed by
exposing the cells for 2 min to
 -choline solution. The recovery of
pHi following an
-choline solution. The recovery of
pHi following an
 pre-pulse was fitted to a single
exponential function, and the first derivative of this function was used to
determine the rate of change of pHi
(dpHi/dt) at 0.05
pHi unit increments from the point of maximum
acidification (52,
53). Proton efflux was
calculated by multiplying the measured
dpHi/dt at a given
pHi value by the intrinsic intracellular buffering
capacity (βi) at the same pHi
value. We calculatedβi in AP-1/NHE5HA
cells by measuring the changes in pHi elicited by changing
the extracellular concentration of NH4Cl as described previously by
Roos and Boron (54) and
Boyarsky et al. (55).
Instantaneous proton efflux was then plotted against absolute
pHi values, and results from different experiments were
compared statistically (Student's unpaired two-tailed t test) at
corresponding values of pHi. To confirm the identity of
the acid-extrusion mechanism, the NHE inhibitor
5-(N-ethyl-N-isopropyl)amiloride (EIPA, 10 μm)
was added to the perfusion solution for 2.5 min during the
pHi recovery phase followed by a return to NaCl-saline
solution. The compositions of the NaCl-saline and
pre-pulse was fitted to a single
exponential function, and the first derivative of this function was used to
determine the rate of change of pHi
(dpHi/dt) at 0.05
pHi unit increments from the point of maximum
acidification (52,
53). Proton efflux was
calculated by multiplying the measured
dpHi/dt at a given
pHi value by the intrinsic intracellular buffering
capacity (βi) at the same pHi
value. We calculatedβi in AP-1/NHE5HA
cells by measuring the changes in pHi elicited by changing
the extracellular concentration of NH4Cl as described previously by
Roos and Boron (54) and
Boyarsky et al. (55).
Instantaneous proton efflux was then plotted against absolute
pHi values, and results from different experiments were
compared statistically (Student's unpaired two-tailed t test) at
corresponding values of pHi. To confirm the identity of
the acid-extrusion mechanism, the NHE inhibitor
5-(N-ethyl-N-isopropyl)amiloride (EIPA, 10 μm)
was added to the perfusion solution for 2.5 min during the
pHi recovery phase followed by a return to NaCl-saline
solution. The compositions of the NaCl-saline and
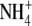 -choline solutions were the same as
those used for the 22Na+-influx assays.
-choline solutions were the same as
those used for the 22Na+-influx assays.
Immunofluorescence Microscopy—PC12/NHE51D4 cells grown on glass coverslips coated with poly-l-lysine and laminin (10 μg/ml, Sigma) were transfected with SCAMP2GFP or co-transfected with SCAMP2GFP and Arf6HA. 72 h post-transfection, cells were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature and permeabilized with 0.1% Triton X-100 in PBS for 5 min. Fixed cells were then treated with rabbit polyclonal anti-Rab4, anti-Rab11, or anti-HA antibodies and mouse monoclonal 1D4 antibodies followed by Alexa 647-conjugated goat anti-mouse IgG and Alexa 568-conjugated goat anti-rabbit IgG (Molecular Probes). To visualize recycling endosomes, AP-1/NHE5HA cells were serum-starved for 2 h and then incubated with Alexa 568-conjugated transferrin (25 μg/ml, Molecular Probes) for 30 min at 37 °C. Cells were rinsed with PBS, fixed with pre-chilled methanol at -20 °C for 5 min, permeabilized with 0.1% Triton X-100/PBS for 5 min, and internalized transferrin, NHE5HA, and endogenous SCAMP1 or SCAMP2 were visualized by immunofluorescence microscopy as described above, using anti-SCAMP1 or anti-SCAMP2 rabbit polyclonal and anti-HA monoclonal primary antibodies, followed by Alexa 488-conjugated goat anti-rabbit IgG and Alexa 647-conjugated goat anti-mouse IgG secondary antibodies. Prepared coverslips were then analyzed by triple immunofluorescence confocal microscopy.
RESULTS
SCAMPs Are Novel Binding Partners of NHE5—We showed previously that SCAMPs directly bind the cytosolic C-terminal extension of the organelle-enriched NHE7 isoform and govern its intracellular trafficking between the trans-Golgi network and recycling endosomes (44). NHE7 shuttles between the trans-Golgi network, plasma membrane, and recycling endosomes via the clathrin-dependent pathway (56). Similarly, NHE5 is internalized through the clathrin-dependent pathway and is predominantly associated with recycling endosomes following endocytosis (50). Thus, we reasoned that SCAMPs might also bind to NHE5 and regulate its targeting. To test this possibility, we first carried out co-immunoprecipitation using rat PC12 cells stably expressing 1D4-tagged NHE5 (PC12/NHE51D4). PC12 cells are widely used as a neuronal model system and endogenous expression of SCAMP1, -2, and -5 was observed by Western blot (Fig. 1A (44)). A membrane-enriched fraction was prepared from PC12 or PC12/NHE51D4 cells and subjected to immunoprecipitation using 1D4 antibody coupled to Sepharose beads. SCAMP1 and SCAMP2, but not SCAMP5, were readily detectable in the immunoprecipitate from PC12/NHE51D4 cells (Fig. 1A). Although equivalent levels of SCAMP expression were seen in lysates from both PC12 and PC12/NHE51D4 cells, SCAMPs were undetectable in the immunoprecipitated samples from untransfected PC12 cells.
FIGURE 1.
NHE5 interacts with SCAMPs. A, membrane fractions from control PC12 cells or PC12 cells stably expressing NHE51D4 (PC12/NHE51D4) were immunoprecipitated with 1D4 antibody conjugated to Sepharose beads. Bound endogenous SCAMP1, SCAMP2, and SCAMP5 found in the immunoprecipitate fraction (IP) were detected by Western blot using SCAMP-specific antibodies. Five percent of the membrane lysate (Lys.) was resolved as a positive control. B, 2.5, 5, and 10 μg of protein from rat brain lysate was probed by Western blot to assess the endogenous expression of NHE5 and SCAMP2 protein in brain tissue. C, NHE5 was immunoprecipitated from rat brain lysate using an anti-NHE5 antibody (IP) or pre-immune serum control (Con.), and bound endogenous SCAMP2 was detected by Western blot. One percent of the rat brain lysate (Lys.) was probed as a positive control. Western blots shown in A-C are representative of three independent experiments in each case.
Next we examined whether the SCAMP-NHE5 interaction was also found in brain tissue. Endogenous expression of NHE5 and SCAMP2 protein in rat brain was first confirmed by Western blot (Fig. 1B). Rat brain lysate was then subjected to immunoprecipitation using anti-NHE5 antibodies or pre-immune serum. A distinct band of ∼37 kDa in size corresponding to SCAMP2 was detected by Western blot in the lysate immunoprecipitated with anti-NHE5 antibody (IP, Fig. 1C), but not the lysate incubated with pre-immune serum (Con, Fig. 1C) suggesting the existence of a SCAMP2-NHE5 complex in brain.
Determination of the NHE5-binding Site of SCAMP2—The C-terminal cytosolic extension of NHE proteins serves as a major protein-protein interaction domain, and most of the previously identified NHE-binding proteins were shown to bind to this domain (10). SCAMPs contain possible protein-protein interaction interfaces in the N-terminal and C-terminal cytosolic extensions as well as the cytoplasmic loop between the second and third transmembrane domains (36, 57). To test whether these cytosolic domains of SCAMP2 and NHE5 directly interact, we performed in vitro GST pulldown protein-binding experiments. Immobilized GST alone or GST-SCAMP2 fusion proteins were incubated with in vitro transcribed/translated 35S-labeled NHE5 C terminus (amino acids 492-896). NHE5 bound to the immobilized GST fusion proteins was eluted, resolved in SDS-PAGE, and detected by phosphorimaging. Radiolabeled NHE5 protein exhibited a specific association with the GST-tagged SCAMP2 N terminus and C terminus but not the cytosolic loop or GST alone (Fig. 2A). Further GST pulldown experiments revealed strong interactions between NHE5 and GST-SCAMP2-(1-154), GST-SCAMP2-(45-154), GST-SCAMP2-(75-134), and GST-SCAMP2-(75-154), weaker interactions with GST-SCAMP2-(1-88) and GST-SCAMP2-(45-88) and no interaction with GST-SCAMP2-(75-117), GST-SCAMP2-(1-44), GST-SCAMP2-(134-154), or GST alone (Fig. 2, B and C). Together, these results indicate that the NHE5 C terminus binds to the cytosolic C terminus, and amino acids 45-75 and 117-134 within the cytosolic N terminus, of SCAMP2 (Fig. 2D).
FIGURE 2.
The cytosolic C terminus of NHE5 interacts directly with SCAMP2. A, GST or GST fusion proteins containing either the cytoplasmic N terminus (amino acids 1-154), C terminus (amino acids 284-329), or the cytosolic loop (CL, amino acids 201-215) between the second and third trans-membrane domains of SCAMP2 were immobilized on reduced glutathione-Sepharose beads. The beads were then incubated with 35S-labeled in vitro transcribed/translated NHE5 C terminus (amino acids 492-896). After washing the beads, bound 35S-labeled NHE5 C terminus was eluted, resolved by SDS-PAGE, and detected by phosphorimaging. A small amount of the NHE5 C terminus input (3%), not subjected to pulldown assay, was also included as a control. B and C, two additional GST pulldown experiments were performed using GST fused to fragments of the SCAMP2 N terminus (amino acids 1-154, 1-88, 45-88, 45-154, 75-117, 75-134, and 75-154 in B, or amino acids 1-154, 1-44, and 134-154 in C) to determine the minimum NHE5-binding sites within the SCAMP2 N-terminal tail. Each pulldown experiment was performed three times; representative results are shown. D, a schematic representation showing the membrane topology of SCAMP2. NHE5-binding sites are highlighted with black rectangles, and the N-terminal NPF repeats are labeled with black circles. Numbers indicate amino acid residues.
Heterologous Expression of SCAMP2 Affects NHE5 Surface Localization, but Not NHE5 Internalization—SCAMPs have been suggested to play roles in both secretion (36) and endocytosis (39). We postulated that SCAMP2 might modulate the targeting of NHE5 between endosomes and the plasma membrane and thereby regulate transporter activity across the plasma membrane. To address the functional significance of the SCAMP-NHE5 interaction, we used Chinese hamster ovary AP-1 cells devoid of intrinsic NHE activity (58) stably expressing HA-tagged NHE5 (AP-1/NHE5HA, see “Experimental Procedures”). This is a widely used model system to measure the activity of different NHE isoforms in the absence of intrinsic NHE activity (30, 32, 50, 59). It was previously shown that NHE5 cycles between the plasma membrane and recycling endosomes (50). SCAMPs are also localized to the eukaryotic cell-surface recycling system (33). Therefore, we next tested whether NHE5 and SCAMPs co-localize in recycling endosomes. AP-1/NHE5HA cells grown on glass coverslips were incubated at 37 °C for 30 min in media containing fluorescently labeled transferrin to visualize recycling endosomes. In agreement with the previous study, NHE5HA was associated with internalized transferrin in a perinuclear location (Fig. 3). Some of the NHE5HA signal appeared dispersed. This is likely due to partial localization to the endoplasmic reticulum resulting from heterologous overexpression as noted earlier (50). As observed in the three color overlay picture, both SCAMP1 and SCAMP2 co-localized with NHE5 predominantly in the perinuclear location positive for fluorescently labeled transferrin (Fig. 3), suggesting that recycling endosomes are the site of the SCAMP-NHE5 interaction.
FIGURE 3.
The SCAMP·NHE5 complex is found in recycling endosomes. AP-1/NHE5HA cells were incubated with Alexa 568-conjugated transferrin at 37 °C for 30 min. Cells were then fixed, permeabilized and SCAMP1 or SCAMP2, and NHE5HA were visualized with anti-SCAMP and anti-HA antibodies, respectively, followed by fluorescently labeled secondary antibodies. Images were acquired using confocal microscopy. Shown are fluorescence images of NHE5HA, SCAMP1, or SCAMP2 and internalized transferrin (Tfn). Bars, 10 μm.
To investigate whether SCAMPs regulate the subcellular distribution of NHE5, we performed biotin-labeling and internalization assays to monitor the trafficking of NHE5. AP-1/NHE5HA cells were transiently transfected with either SCAMP2Myc or SCAMP5Myc, or empty pcDNA3 vector as a control. Cell-surface-exposed proteins were labeled with a membrane-impermeable protein-reactive biotinylation reagent containing a cleavable disulfide bond at 4 °C. Labeled cells were then incubated in culture media at 37 °C for 0-30 min to facilitate internalization of labeled proteins through endocytosis. Cells were then treated with reduced glutathione (cleavage buffer) to cleave the remaining surface biotin tags and allow for evaluation of the remaining internalized, biotinylated NHE5 population, or left untreated to assess the total surface-exposed and biotinylated NHE5 population. Biotinylated proteins were affinity-purified with avidin-coupled agarose beads and analyzed by SDS-PAGE and Western blot. NHE5 was efficiently biotinylated on the cell surface (Surface NHE5HA, Fig. 4, A and B). The cell-surface biotin tags of NHE5 were efficiently removed by incubation with cleavage buffer (time 0 in Fig. 4, A and B), whereas internalized NHE5 protected from cleavage after chase incubation was detectable by Western blot (time 15 and 30 min in Fig. 4, A and B). The rates of NHE5 endocytosis appeared to be unaffected by overexpression of either SCAMP2Myc or SCAMP5Myc as compared with vector-transfected controls (Fig. 4C). However, SCAMP2Myc, but not SCAMP5Myc, increased the surface abundance of NHE5HA by ∼50% relative to the control (p < 0.01, Fig. 4D). To further define whether SCAMP2 regulates the surface targeting of NHE5, we measured cell-surface Na+/H+ exchange activity in transfected AP-1/NHE5HA cells using the 22Na+-influx assay. Forced expression of SCAMP2 into these cells increased the amiloride-sensitive, acidic H+i-activated influx of 22Na+ typically by 50% or greater (p < 0.01), whereas expression of SCAMP5 caused only a slight increase that was not statistically significant (Fig. 4E).
FIGURE 4.
SCAMP2 controls NHE5 cell-surface abundance. A and B, AP-1/NHE5HA cells were transiently transfected with Myc-tagged SCAMP2 (SCAMP2Myc, A) or Myc-tagged SCAMP5 (SCAMP5Myc, B), or with empty pcDNA3 vector control. Transfected cells were incubated with a biotinylation reagent followed by chase incubation in the culture media for 0, 15, or 30 min (Chase) to permit endocytosis of labeled proteins. Following the chase period, surface biotin was removed by incubation with a cleavage reagent allowing visualization of internalized protein or left uncleaved (Cleavage: + or -). Cells were then lysed and biotinylated proteins were purified by incubation with avidin-conjugated agarose beads, resolved by SDS-PAGE, and surface-labeled and -internalized NHE5HA was detected by Western blotting using an anti-HA monoclonal antibody (Surface NHE5HA). A small amount of total lysate (5%) was analyzed as a loading control and probed for SCAMP2Myc or SCAMP5Myc and NHE5HA (Total NHE5HA, SCAMP2Myc, or SCAMP5Myc). The Western blots shown are representative of three independent experiments. C, the percentage of labeled NHE5HA internalized after 15 or 30 min of chase was calculated by comparing the signal in the cleaved samples (Cleavage: +) to the corresponding uncleaved samples (Cleavage: -). The amount of NHE5HA internalized at each time point in SCAMP2Myc- or SCAMP5Myc-transfected cells is expressed relative to control cells (pcDNA3) and are averaged from three independent experiments ± S.D. D, densitometric analysis of the biotinylated samples without chase (time 0 min), representing total surface-labeled protein, was used to measure the relative surface abundance of NHE5HA. Total surface NHE5HA in SCAMP2Myc- or SCAMP5Myc-transfected cells was compared directly to pcDNA3-transfected control cells from the same experiment and is expressed as a percentage relative to pcDNA3 transfected control. Values represent the averaged result from three independent experiments ± S.D. ** and NS, p < 0.01 and not significant, respectively (unpaired Student's t test). E, AP-1/NHE5HA cells were transfected with SCAMP2Myc, SCAMP5Myc or empty pcDNA3 vector, and NHE activity was measured by the amiloride-inhibitable 22Na+ influx technique (see “Experimental Procedures”). Results are expressed as a percentage relative to pcDNA3-transfected control. Data from a representative of three independent experiments are shown here ± S.D. Each experiment was performed in quadruplicate. ** and NS, p < 0.01 and not significant, respectively (unpaired Student's t test).
Heterologous Expression of SCAMPs Affects NHE5 Activity at the Cell Surface—The results of cell-surface biotinylation experiments and 22NaCl influx assays suggest the involvement of SCAMP2, but not SCAMP5, in controlling NHE5-surface expression. However, these assays involve measurements from a pool of transiently transfected cells, and the data could be affected by experimental conditions such as transfection efficiency. To address this possibility, pHi measurements were employed to assess Na+/H+ exchange activity in individual AP-1/NHE5HA cells. AP-1/NHE5HA cells were loaded with BCECF, the cytosol was acutely acidified by the ammonium pre-pulse technique, and rates of Na+-dependent pHi recovery were measured (see “Experimental Procedures”). pHi recovery was undetectable when cells were superfused with a Na+-free solution (not shown). Similarly, treatment with the NHE inhibitors EIPA (10 μm) (Fig. 5A) or amiloride (1 mm) (not shown, see Ref. 57) effectively blocked pHi recovery in a reversible manner. No recovery of pHi was observed in parental AP-1 cells following an intracellular acid load under HEPES-buffered conditions (not shown). Altogether, these results indicate that the recovery of pHi in AP-1/NHE5HA cells is mediated by NHE5. To determine NHE5-dependent proton efflux, we measured the intrinsic intracellular buffering capacity (βi) of AP-1/NHE5HA cells over the pHi range studied in the present experiments (see “Experimental Procedures”). Consistent with previous reports in AP-1 cell transfectants (50, 60), βi in AP-1/NHE5HA cells was 28.4 ± 4.6 mm/pH unit and was not significantly altered in cells transfected with GFP-tagged SCAMP2, SCAMP5, SCAMP2ΔC, SCAMP2-(1-154), or SCAMP2ΔNPF (see below).
FIGURE 5.
Effects of overexpression of SCAMPs on rates of pHi recovery from cytosolic acid loads. A, AP-1/NHE5HA cells grown on a glass coverslip were loaded with the pH-sensitive dye BCECF and acidified by exposure to 50 mm NH4Cl for 2 min followed by wash-out with Na+-containing HEPES-buffered saline solution. The recovery of intracellular pH (pHi) following the wash-out of NH4Cl was monitored in individual cells. Partway through the recovery phase of the experiment, cells were exposed to the NHE inhibitor EIPA (10 μm) for 2.5 min. The record is the mean of data obtained simultaneously from 17 cells on a single coverslip and is representative of four independent experiments. B, AP-1/NHE5HA cells were transfected with pEGFP, or GFP-tagged SCAMP2 or SCAMP5. pHi recoveries in the transfected cells were monitored 48 h following transfection. Records are means of data obtained simultaneously from 15, 8, and 8 cells transfected with pEGFP, GFP-tagged SCAMP2, or GFP-tagged SCAMP5, respectively, which exhibited similar peak acidifications. Each experiment was performed on a separate coverslip, and each record is representative of four to six independent experiments in each case. C, the pHi dependences of H+efflux in cells transfected with pEGFP, GFP-tagged SCAMP2, or GFP-tagged SCAMP5. Continuous lines represent the weighted non-linear least-squares regression fits to the data points (mean ± S.E.) indicated for each experimental condition. In each case, data points were obtained from at least four experiments of the type illustrated in B. D, AP-1/NHE5HA cells were transfected with Myc-tagged SCAMP2, SCAMP2ΔC, SCAMP2-(1-154), or SCAMP2ΔNPF. Transfected cells were lysed and subjected to immunoprecipitation with mouse anti-HA antibody (IP) or pre-immune serum (Con.). SCAMP2 constructs bound to the immunoprecipitate were detected by Western blotting using rabbit anti-Myc antibodies. Five percent volume of the cell lysate was analyzed as a positive control (Lys.). A nonspecific doublet of ∼24 kDa detected in both Con and IP lanes is indicated by asterisks. E, AP-1/NHE5HA cells were transfected with pEGFP, or GFP-tagged SCAMP2ΔC, SCAMP2-(1-154), or SCAMP2ΔNPF, and pHi measurements were conducted as in B. Records are means of data obtained simultaneously from 10, 7, 7, and 4 cells transfected with pEGFP or GFP-tagged SCAMP2ΔC, SCAMP2-(1-154), or SCAMP2ΔNPF, respectively, which exhibited similar peak acidifications. Each experiment was performed on a separate coverslip, and each record is representative of three to six independent experiments in each case. F, the pHi dependences of H+efflux in cells transfected with pEGFP, GFP-tagged SCAMP2ΔC, or GFP-tagged SCAMP2-(1-154). Continuous lines represent the weighted non-linear least-squares regression fits to the data points (mean ± S.E.) indicated for each experimental condition. In each case, data points were obtained from at least three experiments of the type illustrated in E. Cells transfected with GFP-tagged SCAMP2ΔNPF failed to exhibit measurable pHi recoveries (see E), and the recovery of pHi could not be fitted to a single exponential function to accurately determine dpHi/dt and thus proton efflux(see“Experimental Procedures”).
To define the role of SCAMPs on NHE5 activity across the plasma membrane, we expressed GFP or GFP-tagged SCAMP2 (SCAMP2GFP) or GFP-tagged SCAMP5 (SCAMP5GFP) into AP-1/NHE5HA cells and Na+-dependent, EIPA-sensitive pHi recovery following an ammonium pre-pulse was examined using single cell imaging. Cells transfected with SCAMP2GFP exhibited significantly faster (p < 0.05 at all absolute values of pHi) proton efflux than GFP-transfected controls (Fig. 5, B and C). In contrast, SCAMP5GFP expression failed to significantly affect (p > 0.05 at all absolute values of pHi) proton efflux, suggesting the specificity of SCAMP2GFP overexpression.
In vitro protein-protein interaction assays indicated that both the N- and C-terminal cytosolic extensions of SCAMP2 contribute to NHE5 binding (Fig. 2). To investigate the involvement of these domains of SCAMP2 in NHE5 targeting in the cell, we generated serial N-terminal and C-terminal deletion mutants. Because some of the mutants were either poorly expressed or exhibited cell toxicity during pHi measurements, the following three mutants were further characterized: SCAMP2ΔC, which lacks the cytosolic C-terminal tail, SCAMP2-(1-154), the soluble SCAMP2 N terminus alone, and SCAMP2ΔNPF lacking the N-terminal 55 amino acids containing multiple Asn-Pro-Phe (NPF) repeats. We first tested whether the mutants bind to NHE5 in a cellular context. When expressed in AP-1/NHE5HA cells, Myc-tagged SCAMP2 was co-immunoprecipitated with HA-tagged NHE5, while SCAMP2ΔC and SCAMP2-(1-154) showed little or no binding (Fig. 5D). In contrast, SCAMP2ΔNPF was co-immunoprecipitated with NHE5 as efficiently as full-length SCAMP2 (Fig. 5D). If SCAMP2 serves as a scaffold protein then some of the mutants lacking either NHE5 binding domains or binding motifs with other molecules might show dominant-negative effects by competing with intrinsic protein-protein interactions. To test this possibility, we transfected GFP-tagged SCAMP2 mutants and assessed their effects on NHE5 activity. AP1/NHE5HA cells transfected with SCAMP2ΔCGFP recovered from an imposed internal acid load (Fig. 5E); proton efflux values were not significantly different (p > 0.05 at all absolute values of pHi) to those observed in control (pEGFP-expressing) cells (Fig. 5F). In contrast, proton efflux in AP1/NHE5HA cells transfected with SCAMP2-(1-154)GFP was significantly slower than in GFP-transfected control cells (Fig. 5, E and F). Finally, cells transfected with SCAMP2ΔNPFGFP exhibited very little pHi recovery from imposed internal acid loads (Fig. 5E). Thus, the SCAMP2 N-terminal domain, and in particular the N-terminal NPF repeats, seem to play an important role in targeting NHE5 to the cell surface.
Small GTPases Arf6 and Rab11 Co-localize with NHE5 and SCAMP2 in Juxtanuclear Regions—The small GTPases Arf6 and Rab11 have been shown to facilitate the recycling of membrane proteins from recycling endosomes to the plasma membrane (42, 43, 61-63). Another small GTPase Rab4 also controls the recycling process of certain membrane proteins from early endosomes to the plasma membrane (43). We next asked whether these small GTPases are involved in SCAMP-mediated NHE5 recycling. NHE5 showed considerable co-localization with both transiently transfected SCAMP2GFP and Arf6HA in PC12/NHE51D4 cells in a perinuclear structure. Similarly, the localization of SCAMP2GFP and NHE5 coincided with that of endogenous Rab11 (white foci, Fig. 6). In contrast, the distribution of endogenous Rab4 was clearly distinct from that of NHE5 and SCAMP2.
FIGURE 6.
NHE5 and SCAMP2 co-localize with the small GTPases Arf6 and Rab11. PC12 cells stably expressing 1D4-tagged NHE5 (PC12/NHE51D4) grown on glass coverslips were transfected with GFP-tagged SCAMP2 (SCAMP2GFP) together with HA-tagged Arf6 (Arf6HA), and the localization of SCAMP2GFP, NHE51D4, and Arf6HA was assessed by immunofluorescence confocal microscopy. Alternatively, cells were transfected with SCAMP2GFP, and the localization of SCAMP2GFP, NHE51D4, and endogenous Rab11 or Rab4 was assessed. White foci in the merged images result from co-localization of the three proteins. Bars, 10 μm.
Arf6 and Rab11 Control the Cell-surface Abundance and Activity of NHE5—The immunofluorescence microscopic results showing that Arf6 and Rab11, but not Rab4, associate with NHE5 and SCAMP2 prompted us to test whether Arf6 and Rab11 influence the endosomeplasma membrane targeting of NHE5. AP-1/NHE5HA cells were transfected with wild-type or GTP-binding-deficient dominant-negative HA-tagged Arf6 (Arf6T27N) or Myc-tagged Rab11 (Rab11S25N), and changes in the surface abundance of NHE5 in transfected cells were then determined by surface biotin labeling followed by Western blot (see “Experimental Procedures”). Transfection with wild-type Arf6 or Rab11 each increased the cell-surface expression of NHE5 by ∼30% compared with vector-transfected control (p < 0.05, Fig. 7, A and B). Transfection of dominant-negative Arf6T27N or Rab11S25N caused no significant alterations in NHE5 surface abundance (p > 0.05), whereas co-expression of both Arf6T27N and Rab11S25N led to a significant (p < 0.05) reduction of ∼35% in surface NHE5 abundance (Fig. 7, A and B). To further define the role of Arf6 and Rab11 in NHE5 surface targeting, NHE5 activity across the plasma membrane was determined by single cell pHi measurements. AP-1/NHE5HA cells transfected with either wild-type Arf6 (Fig. 7, C and E) or wild-type Rab11 (Fig. 7, D and F) exhibited significantly faster proton efflux than GFP-transfected control cells (p < 0.05 at all absolute values of pHi in each case). Proton efflux values in Arf6T27N (Fig. 7, C and E)- or Rab11S25N (Fig. 7, D and F)-expressing cells were not significantly different (p > 0.05 at all absolute values of pHi) to those observed in control cells. Interestingly, however, co-expression of Arf6T27N and Rab11S25N significantly reduced proton efflux (p < 0.05 at all absolute values of pHi; Fig. 7, C and E). The results of the pHi-recovery assay and the biotin labeling assays suggest that NHE5 abundance and activity at the cell surface are regulated by both Arf6 and Rab11 GTPases.
FIGURE 7.
Arf6 and Rab11 up-regulate NHE5 cell-surface targeting and activity.
A and B, AP-1/NHE5HA cells transfected with empty
vector (pcDNA3), wild-type Arf6HA (Arf6WT),
Arf6T27NHA (Arf6T27N), Arf6T27NHA
plus Rab11S25NMyc
(Arf6T27N/Rab11S25N), wild-type
Rab11Myc (Rab11WT) or Rab11S25NMyc
(Rab11S25N) were subjected to surface labeling using a
protein reactive biotinylation reagent. Labeled proteins were isolated from
cell lysates by incubation with avidin-coupled agarose beads, and biotinylated
NHE5HA was analyzed by Western blot (Surface
NHE5HA). A five percent volume of the lysate, not subjected to
avidin-coupled beads, was analyzed by Western blot as a loading control
(Total NHE5HA, Rab11Myc, and
Arf6HA). A, representative Western blots.
B, surface NHE5HA from the different transfection
conditions was measured by densitometry and is expressed relative to
pcDNA3-transfected control ± S.D. Data are averaged from five
independent experiments, asterisks represent statistical
significance, p < 0.05 (Student's unpaired two-tailed t
test). C and D, AP-1/NHE5HA cells grown on glass
coverslips were transfected with pEGFP vector alone or together with wild-type
Arf6HA (Arf6WT), Arf6T27NHA
(Arf6T27N), Rab11S25NMyc plus
Arf6T27NHA
(Arf6T27N/Rab11S25N), wild-type
Rab11Myc (Rab11WT), or Rab11S25NMyc
(Rab11S25N). Transfected cells were loaded with BCECF, and
pHi recoveries from
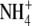 -induced internal acid loads were
examined 48 h following transfection. Records are means of data obtained
simultaneously from 12, 12, 19, 10, 7, and 20 cells transfected with pEGFP,
Arf6WT, Arf6T27N,
Arf6T27N/Rab11S25N, Rab11WT, or
Rab11S25N, respectively, which exhibited similar peak
acidifications. Each experiment was performed on a separate coverslip and each
record is representative of 4 independent experiments in each case. E
and F, the mean pHi-dependent proton efflux was
calculated based on four independent experiments ± S.E. The
pHi dependences of H+ efflux in cells
transfected with pEGFP, Arf6WT, Arf6T27N,
Arf6T27N/Rab11S25N, Rab11WT, or
Rab11S25N. Continuous lines represent the weighted
non-linear least-squares regression fits to the data points (mean ±
S.E.) indicated for each experimental condition. In each case, data points
were obtained from four experiments of the types illustrated in C and
D.
-induced internal acid loads were
examined 48 h following transfection. Records are means of data obtained
simultaneously from 12, 12, 19, 10, 7, and 20 cells transfected with pEGFP,
Arf6WT, Arf6T27N,
Arf6T27N/Rab11S25N, Rab11WT, or
Rab11S25N, respectively, which exhibited similar peak
acidifications. Each experiment was performed on a separate coverslip and each
record is representative of 4 independent experiments in each case. E
and F, the mean pHi-dependent proton efflux was
calculated based on four independent experiments ± S.E. The
pHi dependences of H+ efflux in cells
transfected with pEGFP, Arf6WT, Arf6T27N,
Arf6T27N/Rab11S25N, Rab11WT, or
Rab11S25N. Continuous lines represent the weighted
non-linear least-squares regression fits to the data points (mean ±
S.E.) indicated for each experimental condition. In each case, data points
were obtained from four experiments of the types illustrated in C and
D.
NHE5 Activation by SCAMP2 Is Arf6-dependent—To test the involvement of Arf6 and Rab11 in SCAMP2-mediated trafficking of NHE5, AP-1/NHE5HA cells were co-transfected with SCAMP2GFP and either dominant-negative Arf6T27N or Rab11S25N, and NHE5 activity was measured using the single cell pHi-recovery assay. Cells expressing exogenous SCAMP2 exhibited robust recoveries from the induced acid load. Arf6T27N significantly reduced the ability of concomitantly transfected SCAMP2 to up-regulate NHE5 activity (p < 0.05), whereas Rab11S25N did not influence the SCAMP2-mediated up-regulation of NHE5 (p > 0.05 (Fig. 8, A and B)). These results suggest that the activity of SCAMP2 in controlling NHE5 cell-surface targeting is Arf6-dependent and Rab11-independent.
FIGURE 8.
SCAMP2 facilitates cell-surface targeting of NHE5 via Arf6.
AP-1/NHE5HA cells were transfected with SCAMP2GFP
(SCAMP2), SCAMP2GFP plus Rab11S25NMyc
(Rab11S25N/SCAMP2), or SCAMP2GFP plus
Arf6T27NHA (Arf6T27N/SCAMP2). Transfected cells
were loaded with BCECF and examined 48 h following transfection. A,
pHi recoveries from
 -induced internal acid loads.
Records are means of data obtained simultaneously from 8, 5, and 5 cells
transfected with SCAMP2, Rab11S25N/SCAMP2, or
Arf6T27N/SCAMP2, respectively, which exhibited similar peak
acidifications. Each experiment was performed on a separate coverslip, and
each record is representative of three independent experiments in each case.
B, the pHi dependences of H+ efflux in
cells transfected with SCAMP2, Rab11S25N/SCAMP2, or
Arf6T27N/SCAMP2. Continuous lines represent the weighted
non-linear least-squares regression fits to the data points (mean ±
S.E.) indicated for each experimental condition. In each case, data points
were obtained from three experiments of the type illustrated in
A.
-induced internal acid loads.
Records are means of data obtained simultaneously from 8, 5, and 5 cells
transfected with SCAMP2, Rab11S25N/SCAMP2, or
Arf6T27N/SCAMP2, respectively, which exhibited similar peak
acidifications. Each experiment was performed on a separate coverslip, and
each record is representative of three independent experiments in each case.
B, the pHi dependences of H+ efflux in
cells transfected with SCAMP2, Rab11S25N/SCAMP2, or
Arf6T27N/SCAMP2. Continuous lines represent the weighted
non-linear least-squares regression fits to the data points (mean ±
S.E.) indicated for each experimental condition. In each case, data points
were obtained from three experiments of the type illustrated in
A.
DISCUSSION
Secretory carrier membrane proteins (SCAMPs) are a group of integral membrane proteins that cycle between multiple organelles and regulate membrane dynamics. In the current study, we have shown that SCAMP2 directly binds to NHE5 and facilitates its cell-surface targeting. SCAMP2 contains an N-terminal cytosolic extension, four transmembrane spans, and a C-terminal cytosolic tail (57). Using an in vitro protein binding assay, we have identified NHE5-binding sites within the cytosolic C terminus, and amino acids 45-75 and 117-134 within the cytosolic N terminus of SCAMP2. Further, we used a co-immunoprecipitation approach to show that NHE5 and SCAMP2 form a complex both in tissue culture cells and in brain tissue.
Exogenous expression of SCAMP2 increased both the cell-surface abundance and the ion-translocation activity of NHE5. The agreement between experiments examining cell-surface NHE5 abundance and NHE5 activity suggest the predominant action of SCAMP2 acts on membrane trafficking. Furthermore, SCAMP2 appeared to have no effect on the rates of endocytosis of NHE5 from the plasma membrane. Thus, SCAMP2 likely regulates the abundance of NHE5 at the cell surface by promoting its delivery from the perinuclear recycling endosomes. However, we cannot rule out the possibility that NHE5 ion-translocation activity may be partially regulated through interaction with SCAMP2 (see below). It is unlikely that the effect of SCAMP2 expression on NHE5 cell-surface targeting is an overexpression artifact, because expressing comparable levels of SCAMP5 or deletion mutants of SCAMP2 did not cause the same change. Interestingly, among the SCAMP2 deletion mutants tested, the N-terminal deletion mutant lacking the NPF repeats (SCAMP2ΔNPF) markedly suppressed NHE5 activity across the plasma membrane. Likewise, expression of a mini-gene encoding the N-terminal fragment of SCAMP2 (SCAMP2-(1-154)) caused a milder but significant decrease in NHE5 activity despite its weak binding affinity to NHE5. SCAMP2-(1-154) may compete with endogenous SCAMP2 for binding to other molecules such as soluble EH-domain proteins. In contrast, the ΔNPF mutant binds to NHE5 but may not be able to recruit necessary cytosolic factors to the NHE5·SCAMP2 complex. NPF repeats commonly interact with the EH-domain and regulate endocytosis and endocytic recycling (41, 64-66). Furthermore, intersectins, EH-domain-containing proteins that were reported to bind to the NPF repeats of SCAMP (39), regulate recycling of synaptic vesicles in Drosophila and Caenorhabditis elegans (67-70). Thus, we hypothesize that SCAMP2 recruits cytosolic EH-domain proteins to recycling endosomes via its N-terminal NPF repeats and promotes vesicle formation and the plasma membrane targeting of NHE5. This proposed model is in agreement with a previous report showing that newly formed transferrin-containing vesicles are SCAMP-deficient, which suggests that endocytosis of transferrin occurs independently of SCAMPs (33). The internalized transferrin-containing vesicles are fused with a pre-existing internal pool of SCAMP-positive membranes and then accumulate in the SCAMP-rich perinuclear region corresponding to the recycling endosomal compartment. Vesicles leaving this compartment, returning transferrin to the cell surface were again SCAMP-deficient, suggesting that the perinuclear recycling endosomes are a likely site of SCAMP function. Interestingly, SCAMP1 and SCAMP2 and to a lesser extent SCAMP3 showed considerably more overlap with trafficking transferrin than SCAMP4, which lacks a large part of the N-terminal cytosolic tail, including NPF repeats (33). Thus, these findings together with our own suggest that the N-terminal cytosolic extension of SCAMP2 is an important domain for cell-surface targeting through recycling endosomes.
The small GTPases Arf6 and Rab11 have both been implicated as master regulators of membrane traffic from recycling endosomes to the cell surface (43). Overexpression of either Arf6 or Rab11 significantly enhanced NHE5 abundance and activity at the cell surface, whereas expression of dominant-negative Arf6 and Rab11 alone had very little effect on NHE5 activity. Co-expression of both dominant-negative GTPases caused a substantial decrease in NHE5 activity and cell-surface abundance. When concomitantly expressed with SCAMP2, Arf6T27N but not Rab11S25N impaired the SCAMP2-mediated NHE5 translocation to the plasma membrane. These results indicate that, although both Arf6 and Rab11 participate in controlling the membrane traffic of NHE5, SCAMP2-mediated trafficking is Arf6-dependent and Rab11-independent. Thus we propose that NHE5 accesses the cell surface from the recycling endosomes via at least two distinct pathways: a Rab11-dependent pathway and an Arf6/SCAMP2 pathway. It was previously shown that Arf6 binds to SCAMP2 and regulates fusion pore formation during dense-core vesicle exocytosis in PC12 cells (38). It is tempting to hypothesize that the SCAMP2-Arf6 complex targets NHE5 to vesicular docking sites on the plasma membrane and that the locally elevated NHE5 activity controls secretion of dense core vesicles.
We found previously that NHE5 was able to bind to integrin β1 subunits and could localize to focal adhesion complexes. We further showed that NHE5 could be activated by stimulating the integrin signaling pathway, in a process that required the receptor for activated C-kinase 1 (32). Integrin β1 has also been shown to traffic between the cell surface and the recycling endosomes, which was found to be sensitive to extracellular stimuli in a process termed “regulated recycling.” Moreover, the regulated traffic of integrin β1 was found to require the activity of both Rab11 and Arf6 (71). Because NHE5 is able to bind to integrin and traffics through recycling endosomes in a pathway that also involves Arf6 and Rab11, NHE5 may follow a similar regulated recycling pathway. Thus, it is tempting to speculate that SCAMP2 promotes the targeting of NHE5 from endomembrane stores to the sites of nascent focal adhesion complexes following activation of the integrin signaling pathway. Activated NHE5 would then be in a position to create a specific ionic environment favorable for focal adhesion formation and downstream signaling.
Interestingly, heterologous expression of SCAMP2 not only increased proton efflux at a given absolute pHi but also shifted the pHi dependence of NHE5-dependent acid extrusion in an alkaline direction (e.g. Fig. 5C). In addition to a H+ transport site, many NHE isoforms are believed to contain a second H+ binding site with positive cooperative binding characteristics, which mediates the allosteric H+ activation of transport activity, thus forming the basis of the pH set-point concept (72-75). However, NHE5 does not exhibit a greater than first order dependence on H+i concentration, suggesting the presence of only a single internal H+ binding site (59, 76). Taken together, these considerations suggest the possibility that SCAMP2 might regulate NHE5 activation by binding to NHE5 and changing its conformation into a form possessing a higher affinity for intracellular protons, as recently proposed for the activation of NHE1 by mitogens (77).
In summary, we have identified SCAMPs as novel NHE5-interacting proteins. We propose a model in which SCAMP2 binds to NHE5 in recycling endosomes and promotes its cell-surface targeting. This process is Arf6-dependent and Rab11-independent, and the N-terminal 54 amino acids of SCAMP2 containing the NPF repeats represents a crucial domain. Hence, regulation of NHE5-trafficking behavior may serve as a major mechanism in controlling NHE5 activity across the plasma membrane.
Acknowledgments
We acknowledge Dr. Paulo Lin for technical assistance and helpful discussions. We thank Dr. Martin Schwartz (University of Virginia) for the GFP-tagged Arf6T27N construct, and Dr. Roger Brownsey (University of British Columbia) for kindly providing rat brain.
This work was supported in part by Canadian Institutes of Health Research Operating Grants MOP144919 (to M. N.) and MOP77616 (to J. C.), and by Michael Smith Foundation for Health Research (MSFHR) establishment grants (to M. N.).
Footnotes
The abbreviations used are: NHE, Na+/H+ exchanger; SCAMP, secretory carrier membrane protein; EH, Eps15 homology; GFP, green fluorescent protein; EGFP, enhanced GFP; PBS, phosphate-buffered saline; GST, glutathione S-transferase; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; EIPA, 5-(N-ethyl-N-isopropyl)amiloride; BCECF/AM, 2′,7′-bis(2-carboxyethyl)-5(6)-carboxyfluorescein acetoxymethyl ester.
References
- 1.Chesler, M. (2003) Physiol. Rev. 83 1183-1221 [DOI] [PubMed] [Google Scholar]
- 2.DeCoursey, T. E. (2003) Physiol. Rev. 83 475-579 [DOI] [PubMed] [Google Scholar]
- 3.Traynelis, S. F., and Cull-Candy, S. G. (1990) Nature 345 347-350 [DOI] [PubMed] [Google Scholar]
- 4.Edwards, R. H. (2007) Neuron 55 835-858 [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y. H., Wu, M. L., and Fu, W. M. (1998) J. Neurosci. 18 2982-2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeVries, S. H. (2001) Neuron 32 1107-1117 [DOI] [PubMed] [Google Scholar]
- 7.Krishtal, O. A., Osipchuk, Y. V., Shelest, T. N., and Smirnoff, S. V. (1987) Brain Res. 436 352-356 [DOI] [PubMed] [Google Scholar]
- 8.Sardet, C., Franchi, A., and Pouysségur, J. (1989) Cell 56 271-280 [DOI] [PubMed] [Google Scholar]
- 9.Brett, C. L., Donowitz, M., and Rao, R. (2005) Am. J. Physiol. 288 C223-C239 [DOI] [PubMed] [Google Scholar]
- 10.Orlowski, J., and Grinstein, S. (2004) Pflügers Arch. 447 549-565 [DOI] [PubMed] [Google Scholar]
- 11.Orlowski, J., and Grinstein, S. (2007) Curr. Opin. Cell Biol. 19 483-492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Battaglino, R. A., Pham, L., Morse, L. R., Vokes, M., Sharma, A., Odgren, P. R., Yang, M., Sasaki, H., and Stashenko, P. (2008) Bone 42 180-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, S. H., Kim, T., Park, E.-S., Yang, S., Jeong, D., Choi, Y., and Rho, J. (2008) Biochem. Biophys. Res. Commun. 369 320-326 [DOI] [PubMed] [Google Scholar]
- 14.Rocha, M. A., Crockett, D. P., Wong, L. Y., Richardson, J. R., and Sonsalla, P. K. (2008) J. Neurochem. 106 231-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jang, I. S., Brodwick, M. S., Wang, Z. M., Jeong, H. J., Choi, B. J., and Akaike, N. (2006) J. Neurochem. 99 1224-1236 [DOI] [PubMed] [Google Scholar]
- 16.Trudeau, L. E., Parpura, V., and Haydon, P. G. (1999) J. Neurophysiol. 81 2627-2635 [DOI] [PubMed] [Google Scholar]
- 17.Bell, S. M., Schreiner, C. M., Schultheis, P. J., Miller, M. L., Evans, R. L., Vorhees, C. V., Shull, G. E., and Scott, W. J. (1999) Am. J. Physiol. 276 C788-C795 [DOI] [PubMed] [Google Scholar]
- 18.Cox, G. A., Lutz, C. M., Yang, C. L., Biemesderfer, D., Bronson, R. T., Fu, A., Aronson, P. S., Noebels, J. L., and Frankel, W. N. (1997) Cell 91 139-148 [DOI] [PubMed] [Google Scholar]
- 19.Attaphitaya, S., Park, K., and Melvin, J. E. (1999) J. Biol. Chem. 274 4383-4388 [DOI] [PubMed] [Google Scholar]
- 20.Baird, N. R., Orlowski, J., Szabo, E. Z., Zaun, H. C., Schultheis, P. J., Menon, A. G., and Shull, G. E. (1999) J. Biol. Chem. 274 4377-4382 [DOI] [PubMed] [Google Scholar]
- 21.Wang, D., King, S. M., Quill, T. A., Doolittle, L. K., and Garbers, D. L. (2003) Nature Cell Biol. 5 1117-1122 [DOI] [PubMed] [Google Scholar]
- 22.Woo, A. L., James, P. F., and Lingrel, J. B. (2002) Mol. Reprod. Dev. 62 348-356 [DOI] [PubMed] [Google Scholar]
- 23.Porcelli, A. M., Scotlandi, K., Strammiello, R., Gislimberti, G., Baldini, N., and Rugolo, M. (2002) Biochim. Biophys. Acta 1542 125-138 [DOI] [PubMed] [Google Scholar]
- 24.Flanigan, K., Gardner, K., Alderson, K., Galster, B., Otterud, B., Leppert, M. F., Kaplan, C., and Ptacek, L. J. (1996) Am. J. Hum. Genet. 59 392-399 [PMC free article] [PubMed] [Google Scholar]
- 25.Hellenbroich, Y., Pawlack, H., Rub, U., Schwinger, E., and Zuhlke, C. (2005) J. Neurol. 252 1472-1475 [DOI] [PubMed] [Google Scholar]
- 26.Hellenbroich, Y., Bubel, S., Pawlack, H., Opitz, S., Vieregge, P., Schwinger, E., and Zuhlke, C. (2003) J. Neurol. 250 668-671 [DOI] [PubMed] [Google Scholar]
- 27.Nagaoka, U., Takashima, M., Ishikawa, K., Yoshizawa, K., Yoshizawa, T., Ishikawa, M., Yamawaki, T., Shoji, S., and Mizusawa, H. (2000) Neurology 54 1971-1975 [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa, K., and Mizusawa, H. (2006) Neuropathology 26 352-360 [DOI] [PubMed] [Google Scholar]
- 29.Owada, K., Ishikawa, K., Toru, S., Ishida, G., Gomyoda, M., Tao, O., Noguchi, Y., Kitamura, K., Kondo, I., Noguchi, E., Arinami, T., and Mizusawa, H. (2005) Neurology 65 629-632 [DOI] [PubMed] [Google Scholar]
- 30.Szabo, E. Z., Numata, M., Lukashova, V., Iannuzzi, P., and Orlowski, J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 2790-2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sklan, E. H., Podoly, E., and Soreq, H. (2006) Prog. Neurobiol. 78 117-134 [DOI] [PubMed] [Google Scholar]
- 32.Onishi, I., Lin, P. J., Diering, G. H., Williams, W. P., and Numata, M. (2007) Cell. Signal. 19 194-203 [DOI] [PubMed] [Google Scholar]
- 33.Castle, A., and Castle, D. (2005) J. Cell Sci. 118 3769-3780 [DOI] [PubMed] [Google Scholar]
- 34.Fernandez-Chacon, R., and Sudhof, T. C. (2000) J. Neurosci. 20 7941-7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez-Chacon, R., Alvarez de Toledo, G., Hammer, R. E., and Sudhof, T. C. (1999) J. Biol. Chem. 274 32551-32554 [DOI] [PubMed] [Google Scholar]
- 36.Guo, Z., Liu, L., Cafiso, D., and Castle, D. (2002) J. Biol. Chem. 277 35357-35363 [DOI] [PubMed] [Google Scholar]
- 37.Liu, L., Guo, Z., Tieu, Q., Castle, A., and Castle, D. (2002) Mol. Biol. Cell 13 4266-4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, L., Liao, H., Castle, A., Zhang, J., Casanova, J., Szabo, G., and Castle, D. (2005) Mol. Biol. Cell 16 4463-4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez-Chacon, R., Achiriloaie, M., Janz, R., Albanesi, J. P., and Sudhof, T. C. (2000) J. Biol. Chem. 275 12752-12756 [DOI] [PubMed] [Google Scholar]
- 40.Polo, S., Confalonieri, S., Salcini, A. E., and Di Fiore, P. P. (2003) Sci. STKE 2003, re17. [DOI] [PubMed]
- 41.Montesinos, M. L., Castellano-Munoz, M., Garcia-Junco-Clemente, P., and Fernandez-Chacon, R. (2005) Brain Res. Brain Res. Rev. 49 416-428 [DOI] [PubMed] [Google Scholar]
- 42.D'Souza-Schorey, C., and Chavrier, P. (2006) Nat. Rev. Mol. Cell Biol. 7 347-358 [DOI] [PubMed] [Google Scholar]
- 43.Jones, M. C., Caswell, P. T., and Norman, J. C. (2006) Curr. Opin. Cell Biol. 18 549-557 [DOI] [PubMed] [Google Scholar]
- 44.Lin, P. J., Williams, W. P., Luu, Y., Molday, R. S., Orlowski, J., and Numata, M. (2005) J. Cell Sci. 118 1885-1897 [DOI] [PubMed] [Google Scholar]
- 45.Muller, H. K., Wiborg, O., and Haase, J. (2006) J. Biol. Chem. 281 28901-28909 [DOI] [PubMed] [Google Scholar]
- 46.Hodges, R. S., Heaton, R. J., Parker, J. M., Molday, L., and Molday, R. S. (1988) J. Biol. Chem. 263 11768-11775 [PubMed] [Google Scholar]
- 47.MacKenzie, D., and Molday, R. S. (1982) J. Biol. Chem. 257 7100-7105 [PubMed] [Google Scholar]
- 48.Oprian, D. D., Molday, R. S., Kaufman, R. J., and Khorana, H. G. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 8874-8878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balasubramanian, N., Scott, D. W., Castle, J. D., Casanova, J. E., and Schwartz, M. A. (2007) Nat. Cell Biol. 9 1381-1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szaszi, K., Paulsen, A., Szabo, E. Z., Numata, M., Grinstein, S., and Orlowski, J. (2002) J. Biol. Chem. 277 42623-42632 [DOI] [PubMed] [Google Scholar]
- 51.Chen, C., and Okayama, H. (1987) Mol. Cell. Biol. 7 2745-2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baxter, K. A., and Church, J. (1996) J. Physiol. 493 457-470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, G. A. M., Brett, C. L., and Church, J. (1998) J. Physiol. 512 487-505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roos, A., and Boron, W. F. (1981) Physiol. Rev. 61 296-434 [DOI] [PubMed] [Google Scholar]
- 55.Boyarsky, G., Ganz, M. B., Sterzel, R. B., and Boron, W. F. (1988) Am. J. Physiol. 255 C844-C856 [DOI] [PubMed] [Google Scholar]
- 56.Lin, P. J., Williams, W. P., Kobiljski, J., and Numata, M. (2007) Cell. Signal. 19 978-988 [DOI] [PubMed] [Google Scholar]
- 57.Hubbard, C., Singleton, D., Rauch, M., Jayasinghe, S., Cafiso, D., and Castle, D. (2000) Mol. Biol. Cell 11 2933-2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rotin, D., and Grinstein, S. (1989) Am. J. Physiol. 257 C1158-C1165 [DOI] [PubMed] [Google Scholar]
- 59.Szabo, E. Z., Numata, M., Shull, G. E., and Orlowski, J. (2000) J. Biol. Chem. 275 6302-6307 [DOI] [PubMed] [Google Scholar]
- 60.Bianchini, L., Kapus, A., Lukacs, G., Wasan, S., Wakabayashi, S., Pouysségur, J., Yu, F. H., Orlowski, J., and Grinstein, S. (1995) Am. J. Physiol. 269 C998-C1007 [DOI] [PubMed] [Google Scholar]
- 61.Schlierf, B., Fey, G. H., Hauber, J., Hocke, G. M., and Rosorius, O. (2000) Exp. Cell Res. 259 257-265 [DOI] [PubMed] [Google Scholar]
- 62.Ren, M., Xu, G., Zeng, J., De Lemos-Chiarandini, C., Adesnik, M., and Sabatini, D. D. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 6187-6192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Donaldson, J. G. (2003) J. Biol. Chem. 278 41573-41576 [DOI] [PubMed] [Google Scholar]
- 64.Naslavsky, N., and Caplan, S. (2005) J. Cell Sci. 118 4093-4101 [DOI] [PubMed] [Google Scholar]
- 65.Naslavsky, N., Boehm, M., Backlund, P. S., Jr, and Caplan, S. (2004) Mol. Biol. Cell 15 2410-2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi, A., Pant, S., Balklava, Z., Chen, C. C., Figueroa, V., and Grant, B. D. (2007) Curr. Biol. 17 1913-1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koh, T. W., Verstreken, P., and Bellen, H. J. (2004) Neuron 43 193-205 [DOI] [PubMed] [Google Scholar]
- 68.Marie, B., Sweeney, S. T., Poskanzer, K. E., Roos, J., Kelly, R. B., and Davis, G. W. (2004) Neuron 43 207-219 [DOI] [PubMed] [Google Scholar]
- 69.Rose, S., Malabarba, M. G., Krag, C., Schultz, A., Tsushima, H., Di Fiore, P. P., and Salcini, A. E. (2007) Mol. Biol. Cell 18 5091-5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, W., Bouhours, M., Gracheva, E. O., Liao, E. H., Xu, K., Sengar, A. S., Xin, X., Roder, J., Boone, C., Richmond, J. E., Zhen, M., and Egan, S. E. (2008) Traffic 9 742-754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Powelka, A. M., Sun, J., Li, J., Gao, M., Shaw, L. M., Sonnenberg, A., and Hsu, V. W. (2004) Traffic 5 20-36 [DOI] [PubMed] [Google Scholar]
- 72.Aronson, P. S., Nee, J., and Suhm, M. A. (1982) Nature 299 161-163 [DOI] [PubMed] [Google Scholar]
- 73.Otsu, K., Kinsella, J. L., Koh, E., and Froehlich, J. P. (1992) J. Biol. Chem. 267 8089-8096 [PubMed] [Google Scholar]
- 74.Wakabayashi, S., Hisamitsu, T., Pang, T., and Shigekawa, M. (2003) J. Biol. Chem. 278 11828-11835 [DOI] [PubMed] [Google Scholar]
- 75.Wakabayashi, S., Shigekawa, M., and Pouyssegur, J. (1997) Physiol. Rev. 77 51-74 [DOI] [PubMed] [Google Scholar]
- 76.Attaphitaya, S., Nehrke, K., and Melvin, J. E. (2001) Am. J. Physiol. 281 C1146-C1157 [DOI] [PubMed] [Google Scholar]
- 77.Lacroix, J., Mallorie, P., Maehrel, C., and Counillon, L. (2004) EMBO Rep. 5 91-96 [DOI] [PMC free article] [PubMed] [Google Scholar]










