Abstract
It has previously been demonstrated that there are two distinct mechanisms for genetic resistance to human immunodeficiency virus type 1 (HIV-1) conferred by the CCR5Δ32 gene: the loss of wild-type CCR5 surface expression and the generation of CCR5Δ32 protein, which interacts with CXCR4. To analyse the protective effects of long-term expression of the CCR5Δ32 protein, recombinant lentiviral vectors were used to deliver the CCR5Δ32 gene into human cell lines and primary peripheral blood mononuclear cells that had been immortalized by human T-cell leukemia virus type 1. Blasticidin S-resistant cell lines expressing the lentivirus-encoded CCR5Δ32 showed a significant reduction in HIV-1 Env-mediated fusion assays. It was shown that CD4+ T lymphocytes expressing the lentivirus-encoded CCR5Δ32 gene were highly resistant to infection by a primary but not by a laboratory-adapted X4 strain, suggesting different infectivity requirements. In contrast to previous studies that analysed the CCR5Δ32 protective effects in a transient expression system, this study showed that long-term expression of CCR5Δ32 conferred resistance to HIV-1 despite cell-surface expression of the HIV co-receptors. The results suggest an additional unknown mechanism for generating the CCR5Δ32 resistance phenotype and support the hypothesis that the CCR5Δ32 protein acts as an HIV-suppressive factor by altering the stoichiometry of the molecules involved in HIV-1 entry. The lentiviral-CCR5Δ32 vectors offer a method of generating HIV-resistant cells by delivery of the CCR5Δ32 gene that may be useful for stem cell- or T-cell-based gene therapy for HIV-1 infection.
INTRODUCTION
Humans are inherently different in terms of their response to viral infections. Genetic factors are a major reason for the variables that can influence virus-host interactions and the diverse outcome of viral infections observed in different individuals. CCR5 is the major co-receptor used by macrophage-tropic (R5) human immunodeficiency virus type 1 (HIV-1) isolates (Alkhatib et al., 1996; Choe et al., 1996; Deng et al., 1996; Doranz et al., 1996; Dragic et al., 1996). Individuals who are homozygous for mutant alleles of CCR5 are highly resistant to HIV-1 infection (Braciak et al., 1996; Dean et al., 1996; Doranz et al., 1996; Dragic et al., 1996; Zimmerman et al., 1997). Individuals who are heterozygous for the mutant allele (CCR5+/-) are not protected against infection, but once infected their progression to AIDS is delayed (Braciak et al., 1996; Dean et al., 1996; Doranz et al., 1996; Meyer et al., 1997; Theodorou et al., 1997; Zimmerman et al., 1997), indicating that partial resistance can occur in the presence of a single copy of the mutant allele.
The mutant allele that renders homozygous individuals highly resistant to HIV-1 infection contains a 32 bp deletion that results in a truncated intracellular protein, designated CCR5Δ32. This frame-shift mutation introduces 31 new amino acid residues at the carboxyl terminus of the truncated protein that are not present in the wild-type CCR5 protein. Our previous work has suggested that HIV resistance in CCR5Δ32 homozygotes may result from the genetic loss of CCR5 on the cell surface as well as active downregulation of CXCR4 expression by the mutant CCR5Δ32 protein. There is some controversy over whether CCR5Δ32 protein expression negatively affects CCR5 expression by a sequestration-mediated mechanism (Venkatesan et al., 2002). We and others have demonstrated that CCR5Δ32 protein may form heterodimers with wild-type CCR5 and CXCR4, which are retained in the endoplasmic reticulum and result in reduced cell-surface expression of the HIV co-receptors (Agrawal et al., 2004; Benkirane et al., 1997; Chelli & Alizon, 2001). These findings suggested CCR5-CCR5Δ32 heterodimerization as a molecular mechanism for the slower progression to AIDS in individuals with a heterozygous genotype.
The effects of long-term expression of recombinant CCR5Δ32 protein in primary human CCR5+/+ lymphocytes have not been investigated. In this study, we addressed this question by constructing recombinant lentivirus vectors that carried the wild-type CCR5 or mutant CCR5Δ32 cDNA. The recombinant vectors were used to transduce established cell lines and human T-cell leukemia virus type 1 (HTLV-1)-immortalized peripheral blood mononuclear cell (PBMC) lines. Unlike our previous report demonstrating that transient expression of CCR5Δ32 conferred resistance to HIV (Agrawal et al., 2004), the present study showed that long-term stable expression of CCR5Δ32 in PBMC lines conferred resistance to HIV-1, despite detectable expression of the HIV co-receptors. These results have important implications for HIV pathogenesis and suggest a novel mechanism for generating the observed CCR5Δ32 resistance phenotype.
METHODS
Cells and viruses
HeLa and HEK-293 cells (ATCC) were cultured in Dulbecco’s minimal essential medium (Quality Biologicals) containing 10% fetal bovine serum (FBS; HyClone), 2 mM l-glutamine and antibiotics. Recombinant vaccinia virus stocks were prepared by standard procedures as described previously (Broder & Earl, 1999). The recombinant viruses used were: vCB-21R (pT7-lacZ), vTF7-3 (T7 RNA polymerase), vCB-3 (CD4), vCB-16 (Unc, a negative Env control with a mutation at the cleavage site, which therefore does not promote cell fusion), vCB-43 (Ba-L isolate) and vCB-41 (LAV isolate) and have been described previously (Alkhatib et al., 1996, 1997; Broder & Berger, 1995; Feng et al., 1996). All HIV-1 strains used in this study were obtained from the AIDS Reagent Program (NIAID, Rockville, MD, USA).
Donor PBMCs were either used as a total population or used to purify the CD4+ fraction by positive selection using microbeads coated with antibodies to CD4 (Mitenyi Biotec). PBMCs were activated with phytohaemagglutinin (PHA; 10 μg ml-1; Sigma Chemicals) and recombinant human interleukin (IL)-2 (100 U ml-1; AIDS Reagent Program) for 3 days before use.
Immortalization of CCR5+/+ PBMCs
PBMCs that had been stimulated previously were grown for 3 or more days and frozen in liquid nitrogen. The PBMCs were rethawed and cultured in RPMI 1640 with 15% FBS, 100 IU recombinant IL-2 (AIDS Reagent Program) ml-1, 5 mM l-glutamine, 100 U penicillin ml-1 and 100 μg streptomycin ml-1. After 24 h, infection of the PBMCs was carried out by co-culture of 5×106 cells in the same medium as above, with an equal number of gamma-irradiated (104 rad) human MT-2 cells (HTLV-1-transformed; AIDS Reagent Program) as described previously (Popovic et al., 1983). A single flask of irradiated MT-2 cells was cultured to ensure negative growth. Cells were cultured in a 37 °C incubator with 5% CO2, and the IL-2 was refreshed every 3-4 days. Viable cells were passaged as needed to minimize the effect of dead matter. After 2 weeks, the IL-2 concentration was decreased to 50 IU ml-1 and after about 8 weeks, it was decreased to 25 IU ml-1 and added only once on changing to fresh medium. The HTLV-1-transformed PBMCs stained positive for CD4, CD8, and CD25 (data not shown).
Lentivirus vector production and transduction
The genes for CCR5 and CCR5Δ32 were subcloned into the Gateway entry level plasmid pENTR 1A (Invitrogen). Following insertion into pENTR 1A, the genes were recombined into the Gateway adapted pL6 transfer plasmid using LR-Clonase (Invitrogen). The orientation of the cDNA fragments encoding either CCR5 or CCR5Δ32 was verified by restriction mapping to confirm their position downstream of the cytomegalovirus (CMV) promoter. The pL6 transfer plasmid contained the central polypurine tract and central termination sequence (cPPT) upstream of the CMV promoter and the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) downstream of the cloned transgene (CCR5 or CCR5Δ32).
Lentiviral vectors were produced by transient transfection of HEK-293 cells using a third-generation HIV-1 based system (Dull et al., 1998). Briefly, HEK-293 cells were seeded at 5×106 cells per 75 cm2 of flask surface area the day before transfection. The following day, the medium was changed 2-4 h prior to calcium phosphate transfection (Profection kit; Promega) with 13.2 μg of the transfer plasmid (L-Vector, L-CCR5 or L-CCR5Δ32; Fig. 1) and plasmids pMDLgpRRE (6.6 μg per 75 cm2), pRSV/Rev (3.3 μg per 75 cm2; both kindly provided by Cell Genesys) and pCMV-Ampho (4.6 μg per 75 cm2; Clontech). After refeeding the cells, vector-containing supernatants were harvested 48 h after transfection, filtered through a 0.45 μm filter and either frozen directly or concentrated with a Centricon 100 kDa molecular mass cut-off spin filter. Aliquots of 10-20 μl were stored at -80 °C until further use. For p24 titres, supernatants were diluted 1 : 10 000 and assayed by ELISA (p24 ELISA kit; Beckman Coulter) according to the manufacturer’s instructions. The HEK-293 and HeLa cell lines were transduced with the vectors and subjected to selection with 10 μg blasticidin S ml-1 (Invitrogen). Immortalized PBMCs isolated from different donors were transduced with the lentivirus vectors and subjected to selection with 3 μg blasticidin S ml-1.
Fig. 1.
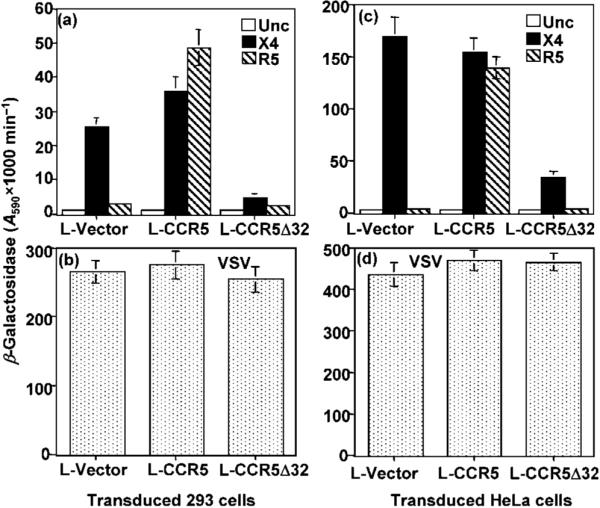
Gene delivery of CCR5 or CCR5Δ32 into HeLa and HEK-293 cell lines by recombinant lentiviral vectors. HeLa or HEK-293 cells were transduced with the indicated recombinant lentivirus (L-Vector, L-CCR5 or L-CCR5Δ32) and subjected to selection in the presence of 2 μg blasticidin S ml-1. Following six cycles of clonal expansion under drug selection, cells were co-infected with the vaccinia virus reporter vCB-21R (pT7-lacZ) and recombinant vaccinia virus vCB-3 (encoding human CD4). The infected cells were incubated overnight to allow expression of the recombinant proteins. Simultaneously, HeLa cells were infected with vaccinia virus recombinants encoding T7 RNA polymerase and one of the indicated HIV-1 Env proteins or VSV Env. The Env-expressing cells were mixed with HEK-293 cells (a, b) or HeLa cells (c, d) expressing the reporter gene and incubated for 2.5 h at 37 °C. The extent of cell fusion was evaluated by measuring β-galactosidase production. In (a) and (c), the X4 Env was from isolate LAV and the R5 Env was from isolate Ba-L.
Antisera against the first intracellular loop (ICL-1) of CCR5
A peptide corresponding to ICL-1 of CCR5 was used to obtain antisera that could recognize both the CCR5 and CCR5Δ32 proteins. The NCKRLKSMTDIY peptide was synthesized, conjugated to keyhole limpet haemocyanin and used to immunize rabbits. Antibodies from the IgG fraction were purified from the crude antiserum using protein G-Sepharose. The ELISA results demonstrated a specific reactivity at a dilution of 1 : 25 000, which was 10 times higher than that obtained with the pre-immune serum at the same dilution.
Western blot analysis and cell-surface staining
Expression of the CCR5 and CCR5Δ32 proteins was examined in CD4+ T lymphocytes isolated from the immortalized/lentivirus-transduced PBMCs. Cell lysates were prepared, fractionated by 12.5% SDS-PAGE and immunoblotted onto PVDF membrane (Millipore). After blocking, the membranes were reacted with the ICL-1 antibodies described above (diluted 1 : 500), washed and incubated with horseradish peroxidase-conjugated secondary antibodies. Protein bands were detected by the addition of substrate as described previously (Agrawal et al., 2007).
For surface expression, cells were washed twice in FACS buffer (supplemented with 0.5% FBS and 0.02% sodium azide), resuspended in 100 μl FACS buffer at 107 cells ml-1 and incubated with a 1 : 200 dilution of monoclonal antibodies (PharMingen) raised against the different co-receptors at 4 °C for 30 min. Cells were then washed twice, resuspended in 100 μl ice-cold FACS buffer in the presence of phycoerythrin-conjugated anti-mouse IgG (PharMingen) and incubated at 4 °C for 30 min. Finally, cells were washed twice, resuspended in 500 μl ice-cold FACS buffer and analysed in a FACScan flow cytometer (Becton Dickinson).
Env-mediated fusion
The basic features of the fusion assay were developed by using the HIV-1 Env-CD4 interaction of two different populations of cells: one expressing CD4 and the other expressing the HIV-1 envelope (Env) glycoprotein (Nussbaum et al., 1994). The PHA+IL-2-activated (non-immortalized) or immortalized PBMC samples were infected with vCB-21R (encoding LacZ under the control of the T7 promoter). HeLa cells co-infected with vTF7-3 (T7 RNA polymerase) and one of the HIV-1 Env proteins served as effector cells. After mixing the effector and target cell populations and incubation at 37 °C for 2.5 h, fusion specificity was measured by β-galactosidase production in a colorimetric lysate assay. To examine the effect of CCR5Δ32 expression on the lentivirus-transduced 293 and HeLa cell lines, the cells were co-infected with vCB-3 (CD4) and vTF7-3 (T7 RNA polymerase) and mixed with effector HeLa cells expressing the indicated HIV-1 Env and pT7-lacZ (LacZ under the control of the T7 promoter).
HIV-1 infection of CD4+ T lymphocytes
The CD4+ T lymphocytes isolated from immortalized/transduced PBMCs were infected with the primary X4 strain (AIDS Reagent Program), the laboratory-adapted IIIB strain or the R5 Ba-L isolate. The HIV-1 virus was adsorbed for 2 h, washed three times with PBS and maintained in RPMI 1640 supplemented with 10% FBS, and 25 IU IL-2 ml-1. Culture fluid (50 μl) was harvested after cell resuspension every 3 days and replaced with fresh medium. The amount of p24 antigen in the cell-containing supernatants was measured using an ELISA kit purchased from the National Cancer Institute (Frederick, MD, USA).
RESULTS
Construction of lentivirus vectors carrying the CCR5 or CCR5Δ32 gene
The CCR5 or CCR5Δ32 gene was inserted into the pL6 transfer vector downstream of the CMV promoter sequences that control expression of the inserted gene. The pL6 vector also encodes blasticidin S resistance under the control of the EM7 promoter. The pL6 plasmid had been modified previously to contain the cPPT tract upstream of the CMV promoter and the WPRE element downstream of the simian virus 40 promoter-driven blasticidin S resistance gene. The cPPT tract increases transduction of non-dividing cells, whilst the WPRE element increases expression levels of the transgene (Barry et al., 2001).
The CCR5 and CCR5Δ32 genes delivered by the lentiviral vectors show the expected biological activity in established cell lines
To test the transduction efficiency of the constructed lentiviral vectors (L-CCR5, L-CCR5Δ32 and L-Vector), HeLa or HEK-293 cells were transduced with the lentiviral vectors and cultured in the presence of 10 μg blasticidin S ml-1. The transduction efficiency, assessed by expression of green fluorescent protein (GFP), was ∼50% in HeLa and HEK-293 cell lines (data not shown). The transduced cells were examined for the acquisition of CCR5 co-receptor activity using a vaccinia virus-based Env-mediated fusion assay. The transduced cells expressing vaccinia virus-encoded T7-lacZ reporter were mixed with Env-expressing HeLa cells that co-expressed vaccinia virus-encoded T7 RNA polymerase. The results demonstrated that L-CCR5-transduced HEK-293 (Fig. 1a) or HeLa (Fig. 1c) cells showed efficient Env-mediated fusion for X4 and R5 Env. In contrast, the L-vector-transduced cells showed efficient Env-mediated fusion with X4 but not R5 Env, whereas the L-CCR5Δ32-transduced cells showed a significant reduction in X4 Env-mediated fusion (Fig. 1a, c). All transduced cell populations showed efficient vesicular stomatitis virus (VSV) Env-induced fusion indicating the competence of the cells to undergo cell fusion (Fig. 1b, d). Expression of CCR5 or CCR5Δ32 protein was confirmed by FACS analysis and Western blotting (data not shown). Thus, these results demonstrated efficient gene delivery by the lentiviral vectors with the expected function of the transduced genes; the L-CCR5-transduced cells acquired efficient CCR5 co-receptor activity, whilst L-CCR5Δ32 transduced cells showed a significant reduction in X4 fusion with HeLa and HEK-293 cells.
Immortalized PBMCs show the expected HIV-1 Env-mediated fusion
We first utilized PHA+IL-2-activated PBMCs to perform the gene delivery experiments and examine transduction efficiency in primary cells. After 3 days in culture, we transduced the activated PBMCs with the lentivirus vectors and subjected them to blasticidin S selection. The transduced PBMCs did not survive long enough to select blasticidin S-resistant clones: the cells died after 3 weeks in the presence of the drug. Therefore, we decided to immortalize the CCR5+/+ PBMCs before transduction.
All of the HTLV-1-transformed lines expressed HTLV antigens as demonstrated by RT-PCR analysis of expressed HTLV-1 mRNA (data not shown). The transformed PBMC lines stained positive for CD4, CD8 and CD25 antigens (data not shown). To determine whether the HTLV-1-mediated transformation of CCR5+/+ PBMCs affected their ability to fuse with the different HIV-1 Env proteins, we compared them with Env-mediated fusion with the PHA+IL-2-activated counterparts (Fig. 2a). The immortalized PBMCs produced efficient Env fusion signals mediated by the X4 and R5 Env proteins (Fig. 2b). The Env-mediated fusion signals of the three donors were significantly higher than those obtained before immortalization (Fig. 2b). The higher Env fusion signals in the three donors correlated with the detection of higher levels of CCR5 and CXCR4 in CD4+ T cells isolated from the immortalized PBMC lines (Table 1). These results demonstrated that the immortalized CCR5+/+ PBMCs retained the ability to fuse efficiently with HIV-1 Env-expressing cells.
Fig. 2.
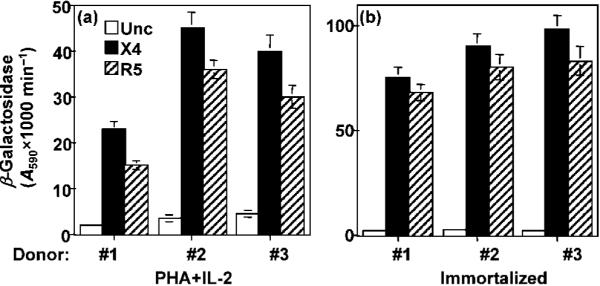
HIV-1 Env-mediated fusion activity of immortalized CCR5+/+ PBMCs. The immortalized PBMC lines and their original PHA+IL-2-activated counterparts were infected with a vaccina virus recombinant encoding the reporter gene under the control of the T7 promoter (pT7-lacZ). The infected cell samples were mixed with an equal number of HeLa cells that had been infected with vaccinia virus recombinants encoding T7 RNA polymerase and one of the indicated HIV-1 Env proteins. Following incubation for 2.5 h, the levels of β-galactosidase produced as a result of cell fusion with PHA+IL-2-activated cells (a) or with their HTLV-immortalized counterparts (b) were determined. The X4 Env was from isolate LAV and the R5 Env was from isolate Ba-L.
Table 1. Cell-surface expression of CCR5 and CXCR4 in CD4+ T lymphocytes isolated from immortalized PBMCs.
Results are shown as mean fluorescence values. Numbers in parentheses indicate the mean values obtained with cells stained with the matched isotype control. IMM, Immortalized
| Receptor | Donor #1 |
Donor #2 |
Donor #3 |
|||
|---|---|---|---|---|---|---|
| IMM | PHA+IL-2* | IMM | PHA+IL-2* | IMM | PHA+IL-2* | |
| CCR5 | 63 (2.6) | 28 (2.5) | 78 (2.7) | 21.6 (3.2) | 38 (3.4) | 24 (3.9) |
| CXCR4 | 255 (2.6) | 185 (3.7) | 242 (3.5) | 169 (4.6) | 389 (3.9) | 268 (3.7) |
Unstimulated PBMCs from each donor were frozen in liquid nitrogen until the immortalized cells from the three donors were ready. Once the immortalized lines were established, the PBMC samples were thawed, stimulated with PHA and IL-2, and used to purify CD4+ T cells as described in Methods.
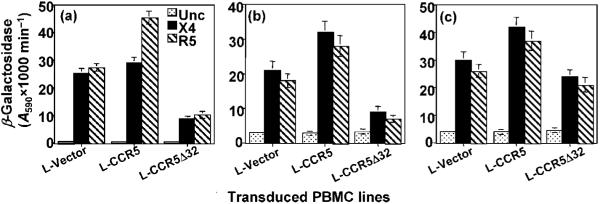
HIV-1 Env-mediated fusion with PBMC lines transduced with the lentivirus vectors
The transduction efficiency (assessed by GFP expression) of the PBMC lines was at best ∼20% and was different for each donor (data not shown). We performed blasticidin S selection to enrich the cell populations that expressed the recombinant proteins. We evaluated the HIV-1 Env-mediated fusion profile of the PBMC lines transduced with the lentiviral vectors. Enhanced Env-mediated fusion was consistently observed with CCR5-transduced PBMC lines compared with those transduced with the vector alone (Fig. 3a-c). In contrast, the PBMC lines transduced with L-CCR5Δ32 consistently showed a significant reduction in the Env fusion signals with cells derived from donors #1 and #2 (Fig. 3a, b) but a less significant reduction with cells derived from donor #3 (Fig. 3c). All transduced PBMC lines were competent for VSV Env-mediated fusion (data not shown). The results demonstrated that the transduced PBMC lines were competent in the HIV-1 Env-mediated fusion assay and showed the expected phenotype resulting from gene delivery of CCR5Δ32; the CCR5Δ32-transduced lines showed significantly lower levels of X4 fusion.
Fig. 3.
HIV-1 Env-mediated fusion with immortalized CCR5+/+ PBMC lines transduced with lentivirus vectors encoding either CCR5 or CCR5Δ32. PBMC lines transduced with the indicated lentivirus vectors were infected with a vaccinia virus recombinant encoding pT7-lacZ. The effect of the CCR5Δ32 protective effect in donor #1 (a), donor #2 (b) and donor #3 (c) was analysed by mixing the pT7-lacZ-expressing PBMC lines with HeLa cells expressing the indicated HIV-1 Env and T7 RNA polymerase and determining the amount of β-galactosidase resulting from cell fusion. VSV Env fusion was used as a positive control to indicate the competence of the PBMC lines in cell fusion (not shown). In all experiments the X4 Env was from isolate LAV and the R5 Env was from isolate Ba-L.
Analysis of co-receptor expression in CD4+ T lymphocytes isolated from the transduced PBMC lines
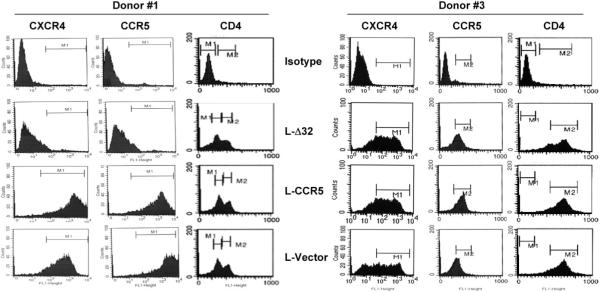
FACS analysis was performed to monitor CCR5 and CXCR4 expression in CD4+ T lymphocytes isolated from the immortalized PBMC lines before (Table 1) and after (Fig. 4 and Table 2) lentivirus transduction. Efficient HIV co-receptor expression was observed in the immortalized/transduced PBMC lines 6 months after selection in the presence of blasticidin S (Fig. 4 and Table 2). Fig. 4 shows the FACS data performed on the PBMC lines developed from donors #1 and #3. The CD4 staining did not seem to be affected by long-term expression of either CCR5Δ32 or CCR5; however, the CD4 expression index seemed to be more efficient in donor #3 (Fig. 4). The FACS analysis demonstrated efficient cell-surface expression of CCR5 and CXCR4 in the vector and CCR5-transduced PBMCs, but significantly reduced surface expression of the co-receptors in the CCR5Δ32-transduced PBMCs (Table 2). This reduction in co-receptor surface expression was more pronounced in transduced PBMC lines from donors #1 and #2 and less significant in donor #3 (Table 2). We consistently observed increased cell-surface expression levels of CCR5 in PBMCs transduced with L-CCR5 and reduced levels of CCR5 in PBMCs transduced with L-CCR5Δ32 (Table 2).
Fig. 4.
FACS analysis of CCR5, CXCR4 and CD4 in CD4+ T lymphocytes isolated from the indicated lentivirus-transduced PBMC lines developed from donors #1 and #3. The staining procedure is outlined in Methods. This analysis was performed at a different time from the experiment shown in Table 2. Isotype, isotype-matched control; L-Δ32, PBMC line transduced with a lentivirus encoding the CCR5Δ32 protein; L-CCR5, PBMC line transduced with a lentivirus encoding the CCR5 protein; L-Vector, PBMC line transduced with an empty lentivirus vector.
Table 2. Cell-surface expression of CCR5 and CXCR4 in CD4+ T lymphocytes isolated from lentivirus-transduced PBMC lines.
Transduced/immortalized PBMCs from the three donors were cultured for 6 months in the presence of blasticidin S and then used to purify CD4+ T lymphocytes. The CD4+ T cells were analysed for receptor/co-receptor cell-surface expression. Results are shown as mean fluorescence values. Numbers in parentheses indicate the mean values obtained with cells stained with the matched isotype control
| Receptor | Transduced lentivirus |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Donor #1 |
Donor #2 |
Donor #3 |
|||||||
| Vector | CCR5 | CCR5Δ32 | Vector | CCR5 | CCR5Δ32 | Vector | CCR5 | CCR5Δ32 | |
| CCR5 | 78 (1.8) | 142 (2.1) | 27 (2.3) | 110 (2.6) | 176 (2.3) | 34 (2.2) | 74 (3.8) | 178 (2.1) | 63 (2.2) |
| CXCR4 | 225 (1.9) | 252 (2.2) | 75 (2.2) | 222 (3.1) | 232 (2.7) | 81 (3.5) | 419 (3.9) | 435 (3.4) | 387 (3.8) |
To verify the expression of the lentivirus-encoded genes, we performed RT-PCR and Western blot analysis of the CCR5 and CCR5Δ32 proteins in the three donors. The RT-PCR analysis indicated efficient expression of the CCR5 transcripts in CD4+ T cells isolated from all three PBMC lines (Fig. 5a). The primers were designed to amplify an internal fragment spanning the 32 bp deletion. As expected, the CCR5Δ32-specific transcript was only detected in the PBMC samples transduced with the L-CCR5Δ32 recombinant lentivirus (Fig. 5a). Amplification of β-actin mRNA was used as an internal control (Fig. 5b).
Fig. 5.
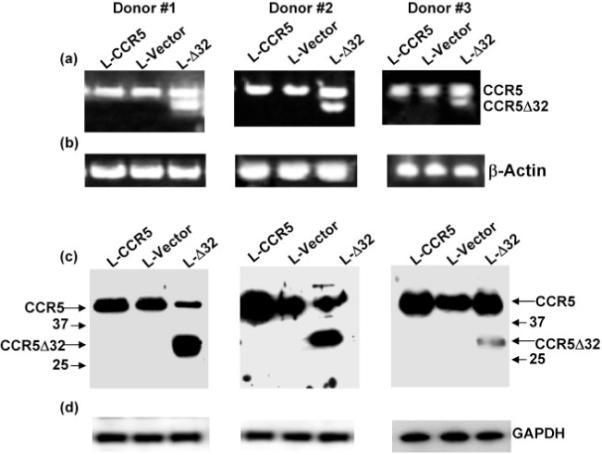
Transgene expression in CD4+ T lymphocytes isolated from PBMC lines transduced with lentivirus vectors encoding CCR5 or CCR5Δ32. Total RNA purified from the CD4+ T lymphocytes was used as template to amplify a fragment spanning the 32 bp deletion (a). A similar reaction was carried out in parallel using β-actin-specific primers as an internal control (b). Cell lysates of the indicated CD4+ T lymphocytes were fractionated by 12% SDS-PAGE and transferred to membrane. The blots were probed with polyclonal antibodies to ICL-1 of CCR5 (c) or with antibodies to GAPDH as an internal control (d). The arrows indicate the positions of the CCR5 and CCR5Δ32 protein bands. The numbers 37 and 25 indicate the molecular masses (kDa) of the marker proteins.
As CCR5Δ32 is a truncated CCR5 protein that is not expressed at the cell surface, we performed Western blotting to verify the intracellular expression of CCR5Δ32. We utilized polyclonal antibodies against ICL-1 of CCR5 to demonstrate expression of the CCR5 and CCR5Δ32 proteins in PBMCs transduced with L-CCR5Δ32. As expected, CCR5 was detectable in all transduced samples from the three donors (Fig. 5c). In contrast, the presence of both CCR5 and CCR5Δ32 protein bands was only detected in PBMCs transduced with L-CCR5Δ32 (Fig. 5c). The CD4+ T cells expressing lentivirus-encoded CCR5Δ32 showed abundant expression of intracellular CCR5Δ32 protein in donors #1 and #2 and a faint band in donor #3 (Fig. 5c). The blots were also analysed for GAPDH expression to confirm that equivalent amounts of cell lysates were loaded on the gels (Fig. 5d). These results confirmed the expression of CCR5Δ32 and CCR5 in the transduced PBMC lines.
Kinetics of HIV-1 productive infection of CD4+ T lymphocytes isolated from transduced PBMC lines
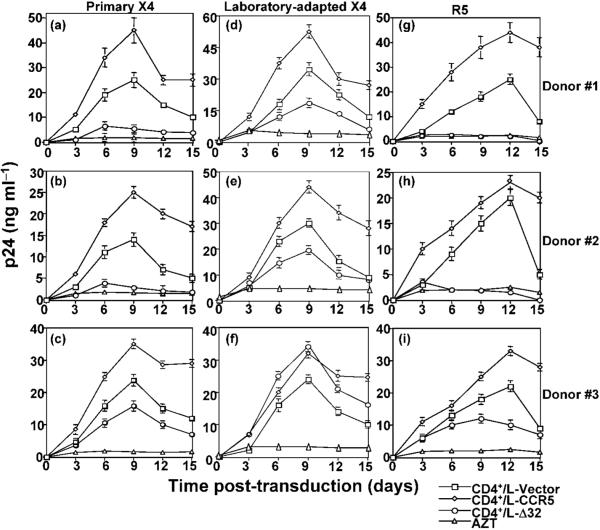
To determine whether the observed CCR5Δ32 effects (i.e. reduced co-receptor expression and Env fusion) could be demonstrated over a longer period of time than that required to perform the Env-mediated fusion assay, purified CD4+ T cells from the three PBMC lines were infected with the primary X4 strain (AIDS Reagent Program), the laboratory-adapted X4 strain IIIB or the R5 Ba-L isolate. We have demonstrated previously that CCR5Δ32 protein interacts with CCR5 and CXCR4 producing a resistant phenotype (Agrawal et al., 2004).
We reasoned that stable expression of the recombinant CCR5Δ32 protein should result in resistance that could be monitored during the course of productive infection. Productive infection by X4 (Fig. 6a-f) and R5 (Fig. 6g-i) isolates was consistently observed in CD4+ T cells isolated from either L-Vector- or L-CCR5-transduced PBMC lines. In contrast, efficient resistance to primary X4 (Fig. 6a-c) and R5 (Fig. 6g-i) was consistently observed during the course of infection of CD4+ T cells isolated from PBMC lines transduced with L-CCR5Δ32 (donors #1 and #2). We observed lower levels of productive infection with the CD4+ T cells isolated from donor #3, but these cells did not show the resistance we observed with donors #1 and #2.
Fig. 6.
Resistance of CD4+ T lymphocytes isolated from PBMC lines to R5 and primary X4 but not to laboratory-adapted X4. CD4+ T lymphocytes were isolated from immortalized/transduced PBMCs using magnetic beads, washed several times in complete medium and infected with primary X4 (a-c), the laboratory-adapted X4 IIIB (d-f) or R5 Ba-L (g-i). Infections were performed using 10 ng p24 ml-1 of the indicated HIV-1 isolate. Culture fluids were harvested every 3 days and replaced with fresh medium. The amount of p24 antigen in the cell-containing supernatants was measured by ELISA. As a negative control for HIV infection, azidothymidine (AZT) was added at a final concentration of 10 μM to infected cells and maintained throughout the course of infection; AZT is a nucleoside analogue antiviral drug that inhibits the replication of retroviruses such as HIV by blocking the enzyme reverse transcriptase.
The CD4+ T cells isolated from PBMC lines transduced with L-CCR5Δ32 showed either lower rates (Fig. 6d, e) or higher rates (Fig. 6f) of productive infection with the laboratory-adapted X4 IIIB. These results demonstrated that resistance to HIV-1 productive infection can be induced by gene delivery of CCR5Δ32 to CD4+ T lymphocytes and suggest that resistance to X4 infection might depend on the HIV-1 isolate used.
DISCUSSION
This study was designed to examine the effects of long-term expression of the CCR5Δ32 gene and the resistance phenotype of cells selected to express the CCR5Δ32 protein. Several lines of experimental evidence were provided to demonstrate the functional transfer of the protective effect of the CCR5Δ32 gene. First, cell lines expressing the CCR5Δ32 protein consistently displayed reduced Env fusion signals mediated by both R5 and X4 Env proteins. Secondly, CD4+ T lymphocytes isolated from PBMC lines expressing CCR5Δ32 but not CCR5 showed a reduction in surface expression of the major HIV co-receptors. Finally, the lentivirus-encoded CCR5Δ32 protein conferred resistance to productive HIV-1 infection of CD4+ T cells isolated from two HTLV-transformed PBMC lines. Interestingly, the observed CCR5Δ32 effects occurred despite detectable cell-surface expression of the HIV co-receptors. This is a critical finding that suggests another novel mechanism for CCR5Δ32 activity different from that reported previously by us (Agrawal et al., 2004). The results of the current study and our recent report (Agrawal et al., 2007) suggest a potential role for other host factors in generating the CCR5Δ32 protective phenotype.
The ability of HTLV-1 to transform CD4+ T lymphocytes was utilized to immortalize CCR5+/+ PBMCs isolated from three different donors. HTLV-1 can infect primary human cells upon co-cultivation with the virus-producing tumour MT-2 cell line. Immortalized CD4+ T lymphocytes closely resemble the cells present in leukaemia patients and can be derived from peripheral blood, cord blood and thymocytes infected with HTLV-1 (Markham et al., 1983; Miyoshi et al., 1981; Popovic et al., 1983; Yamamoto et al., 1982). Previous studies have demonstrated that the HTLV-1-transformed culture is typically a CD4+ CD25+ T-lymphocyte population (Collins et al., 1996; Popovic et al., 1983). In this study, the CD4+ T lymphocytes isolated from the immortalized PBMC lines were CD25+ and expressed the HIV co-receptors. The HTLV-transformed cells were mostly CD4+/CD8+ and retained their ability to fuse efficiently with HIV-1 Env-expressing cells. These transformed cells may not represent either naïve or memory cells, which are the main target for the X4 and R5 viruses, respectively, in vivo. The purpose of this study was to analyse the biological effects resulting from long-term expression of the CCR5Δ32 and CCR5 proteins in CD4+ T lymphocytes. Although our isolation method utilized the total CD4+ T-cell population, it will be interesting to examine the CCR5Δ32 protective effect in naïve and memory T-cell populations.
Efficient resistance to HIV-1 infection was accomplished in two out of three PBMC lines. The CD4+ T cells purified from the third PBMC line (donor #3) expressed lower amounts of CCR5Δ32. The relative CCR5Δ32 protein levels were significantly higher in samples #1 and #2. These results suggest that the CCR5Δ32 gene product is sufficient to generate an HIV-1-resistant phenotype in two out of three donors. We cannot, however, exclude the role of other host factors in establishing the resistance phenotype. We have consistently observed that the resistance of PBMC lines (donors #1 and #2) was always associated with detectable levels of the HIV co-receptors. It is possible that other host factors affecting the translation or the stability of the CCR5Δ32 protein may have contributed to the observed lack of resistance in cells from donor #3. We have reported previously that expression levels and the stability of the CCR5Δ32 protein are critical in conferring resistance to HIV-1 infection (Agrawal et al., 2007). Recent studies have demonstrated that host factors other than CCR5 influence the inhibition of HIV-1 infection of human lymphocytes by CCR5 ligands (Ketas et al., 2007).
There is abundant evidence for heterodimerization of chemokine receptors and the creation of new signalling pathways (reviewed by Mellado et al., 2006). Previous studies have demonstrated that a monoclonal antibody to CCR2 induces heterodimerization of CCR2 with CXCR4 and CCR5, creating a resistant phenotype to R5 and X4 infection (Rodríguez-Frade et al., 2004). Other studies have demonstrated that CXCR4 heterodimerizes with CCR2, resulting in a cross-inhibition of the functional response to their specific ligands (Sohy et al., 2007). It is reasonable to assume that CXCR4 in CCR5-/- cells may heterodimerize with other G protein-coupled receptors (GPCRs) producing a dominant-negative phenotype that might explain the resistance to HIV-1. Recent studies have supported this hypothesis by demonstrating that δ-opioid heterodimerizes with CXCR4 and results in suppression of signalling, promoting a dominant-negative effect (Pello et al., 2008). We hypothesize that the HIV co-receptor expression levels may contribute significantly to either favourable or unfavourable physiological conditions for CXCR4 to form heterodimers with other GPCR molecules. Therefore, it is possible that the ability to heterodimerize with other GPCRs might depend on the HIV co-receptor levels. Donor variability in co-receptor expression levels has been reported previously (Lee et al., 1999). It is possible that the donor variability influences how the HIV co-receptors heterodimerize with other GPCRs and respond differently to the CCR5Δ32 protective effects. The PBMC lines generated here will provide us with an excellent opportunity to examine these possibilities.
The resistance of the CD4+ T cells purified from PBMC lines to a primary X4 but not to a laboratory-adapted X4 may suggest different infectivity requirements. We have observed this previously with PBMCs isolated from individuals who are homozygous for CCR5Δ32 (Agrawal et al., 2007). Although the CD4+ T cells purified from donor #3 were not resistant to infection, they showed reduced p24 values by primary but not by laboratory-adapted X4. Previous studies have demonstrated differences in CD4 dependence for infectivity of laboratory-adapted and primary HIV-1 isolates (Kabat et al., 1994). They showed that laboratory-adapted HIV-1 isolates infected all HeLa/CD4 cell clones with equal efficiency regardless of the levels of CD4, whereas primary HIV-1 isolates infected these clones in direct proportion to cellular CD4 expression. The different CD4 levels in the different donors might explain the observed differences in X4 infectivity; however, it is possible that other host-cell factors such as co-receptor density and/or the CD4 : co-receptor ratio play a role in conferring the observed resistance phenotype. To examine whether it is a general phenomenon, more experiments are needed to confirm the observed different infectivity requirements using a number of HIV-1 isolates.
We have reported previously that adenovirus-mediated delivery of CCR5Δ32 gene expression resulted in protection against the laboratory-adapted X4 strain IIIB (Agrawal et al., 2004). This study showed that PBMC lines expressing the lentivirus-encoded CCR5Δ32 protein were susceptible to IIIB infection. The different expression levels of CCR5Δ32 in the two systems may explain this discrepancy. It is possible that the CCR5Δ32 protein levels produced by the transducing lentivirus were insufficient to downmodulate CXCR4 expression to levels that would confer resistance to IIIB infection. The approach taken in this study is different from the previously described transient adenovirus 5 expression system (Agrawal et al., 2004) in that it utilized lentivirus-transduced PBMC lines with long-term expression of the CCR5Δ32 protein. It is possible that higher expression levels of the CCR5Δ32 protein were accomplished in the adenovirus vector system. The advantage of the lentivirus expression system is the long-term expression of the protein, which may have more physiological relevance to the effect of CCR5Δ32.
The results described in this study suggest that resistance to HIV-1 can occur despite detectable expression of the co-receptors. Previous studies have demonstrated that individuals heterozygous for CCR5Δ32 mutation show slow disease progression to AIDS and are considered partially protected against HIV-1 infection (reviewed by O’Brien & Moore, 2000). The partial resistance to HIV-1 was associated with reduced surface levels of CCR5 (Bleul et al., 1997; Hladik et al., 2005; Venkatesan et al., 2002). The present study demonstrated that expression of CCR5 and CXCR4 was downmodulated but not abolished in CD4+ T cells purified from PBMC lines expressing the lentivirus-encoded CCR5Δ32 protein. This suggests that blocking HIV-1 infection by entry inhibitors may not require the complete elimination of HIV co-receptor expression. The results strengthen the hypothesis that the CCR5Δ32 protein may alter the optimal stoichiometry of the receptor/co-receptor molecules required for HIV-1 entry. It has been demonstrated previously that efficient HIV-1 infection requires an optimal number of co-receptor molecules on the target cell (Kuhmann et al., 2000). Previous studies have also demonstrated an association between CD4 and co-receptor molecules (Xiao et al., 2000). It is possible that an optimal CD4: co-receptor ratio is required for efficient HIV-1 infection.
A number of methods to disrupt HIV co-receptor expression have been described previously. These methods involve vector-delivered genetic disruption mechanisms that target the HIV co-receptors, such as RNA interference, ribozymes, zinc fingers, intrakines and intrabodies (reviewed by Swan & Torbett, 2006; Wolkowicz & Nolan, 2005). Although these methods have shown promising results in vitro, their in vivo application remains controversial. The advantage of the CCR5Δ32 approach is its natural expression in individuals who possess the 32 bp deletion in the CCR5 gene (Agrawal et al., 2004, 2007). Individuals homozygous for CCR5Δ32 do not suffer any known immunological defects as a result of missing CCR5 expression (reviewed by Alkhatib & Berger, 2007).
Haematopoietic stem cells (HSCs) are postulated to have the capacity for self-renewal and the ability to give rise to lineage-restricted progeny, which in turn have the capacity to proliferate and differentiate into mature blood cells (reviewed by Broxmeyer et al., 2006). Therefore, HSCs might be the ideal target for lentivirus gene transfer. A relatively small number of genetically modified HSCs can potentially give rise to large numbers of differentiated haematopoietic cells containing and expressing the CCR5Δ32 gene for long periods of time. Recent studies have described a humanized mouse system that is dependent on the differentiation of HSCs (Sun et al., 2007). Intrarectal inoculation of the humanized mice resulted in systemic infection with depletion of CD4+ T cells in gut-associated lymphoid tissue. This model may provide an excellent opportunity to test whether differentiation of HSCs that have been modified to express CCR5Δ32 would produce a protective effect.
ACKNOWLEDGEMENTS
This study was supported by NIH grant no. A152019-01 to G. A. Q. J. was supported by a scholarship from the Chinese Scholarship Council, Beijin, China. We thank Zainab VanHorn-Ali for excellent technical help, Chinghai Kao for critical comments and Bashar Alkhatib for editing the manuscript. Human PBMC samples were obtained under Indiana University exempt IRB study #4. The Indiana University Vector Production Facility is an NIH designated National Gene Vector Laboratory (U42 RR11148) and this work was supported, in part, by a Core Centers of Excellence in Molecular Hematology (CCEMH) grant (PHS P50 DK49218). K. C. is supported in part by the Indiana Genomics Initiative (INGEN) created through a grant from Lilly Endowment, Inc.
REFERENCES
- Agrawal L, Lu X, Qingwen J, VanHorn-Ali Z, Nicolescue V, McDermott D, Murphy PM, Alkhatib G. Role for CCR5Δ32 protein in resistance to R5, R5X4, and X4 human immunodeficiency virus type 1 in primary CD4+ cells. J Virol. 2004;78:2277–2287. doi: 10.1128/JVI.78.5.2277-2287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal L, Jin Q, Altenburg J, Meyer L, Tubiana R, Theodorou I, Alkhatib G. CCR5Δ32 protein expression and stability are critical for resistance to human immunodeficiency virus type 1 in vivo. J Virol. 2007;81:8041–8049. doi: 10.1128/JVI.00068-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhatib G, Berger EA. HIV coreceptors: from discovery and designation to new paradigms and promise. Eur J Med Res. 2007;12:375–384. [PubMed] [Google Scholar]
- Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- Alkhatib G, Liao F, Berger EA, Farber JM, Peden KW. A new SIV co-receptor, STRL33. Nature. 1997;388:238. doi: 10.1038/40789. [DOI] [PubMed] [Google Scholar]
- Barry SC, Harder B, Brzezinski M, Flint LY, Seppen J, Osborne WR. Lentivirus vectors encoding both central polypurine tract and posttranscriptional regulatory element provide enhanced transduction and transgene expression. Hum Gene Ther. 2001;12:1103–1108. doi: 10.1089/104303401750214311. [DOI] [PubMed] [Google Scholar]
- Benkirane M, Jin DY, Chun RF, Koup RA, Jeang KT. Mechanism of transdominant inhibition of CCR5-mediated HIV-1 infection by ccr5Δ32. J Biol Chem. 1997;272:30603–30606. doi: 10.1074/jbc.272.49.30603. [DOI] [PubMed] [Google Scholar]
- Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci U S A. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braciak TA, Bacon K, Xing Z, Torry DJ, Graham FL, Schall TJ, Richards CD, Croitoru K, Gauldie J. Overexpression of RANTES using a recombinant adenovirus vector induces the tissue-directed recruitment of monocytes to the lung. J Immunol. 1996;157:5076–5084. [PubMed] [Google Scholar]
- Broder CC, Berger EA. Fusogenic selectivity of the envelope glycoprotein is a major determinant of human immunodeficiency virus type 1 tropism for CD4+ T-cell lines vs. primary macrophages. Proc Natl Acad Sci U S A. 1995;92:9004–9008. doi: 10.1073/pnas.92.19.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder CC, Earl PL. Recombinant vaccinia viruses. Design, generation, and isolation. Mol Biotechnol. 1999;13:223–245. doi: 10.1385/MB:13:3:223. [DOI] [PubMed] [Google Scholar]
- Broxmeyer HE, Srour E, Orschell C, Ingram DA, Cooper S, Plett PA, Mead LE, Yoder MC. Cord blood stem and progenitor cells. Methods Enzymol. 2006;419:439–473. doi: 10.1016/S0076-6879(06)19018-7. [DOI] [PubMed] [Google Scholar]
- Chelli M, Alizon M. Determinants of the trans-dominant negative effect of truncated forms of the CCR5 chemokine receptor. J Biol Chem. 2001;276:46975–46982. doi: 10.1074/jbc.M106432200. [DOI] [PubMed] [Google Scholar]
- Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. other authors. [DOI] [PubMed] [Google Scholar]
- Collins ND, Newbound GC, Ratner L, Lairmore MD. In vitro CD4+ lymphocyte transformation and infection in a rabbit model with a molecular clone of human T-cell lymphotrophic virus type 1. J Virol. 1996;70:7241–7246. doi: 10.1128/jvi.70.10.7241-7246.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, Goedert JJ, Buchbinder SP, Vittinghoff E. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. other authors. [DOI] [PubMed] [Google Scholar]
- Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. other authors. [DOI] [PubMed] [Google Scholar]
- Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- Dragic T, Litwin V, Allaway GP, Martin SR, Huang YX, Nagashima KA, Cayanan C, Maddon PJ, Koup RA. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. other authors. [DOI] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- Hladik F, Liu H, Speelmon E, Livingston-Rosanoff D, Wilson S, Sakchalathorn P, Hwangbo Y, Greene B, Zhu T, McElrath MJ. Combined effect of CCR5-Δ32 heterozygosity and the CCR5 promoter polymorphism 22459 A/G on CCR5 expression and resistance to human immunodeficiency virus type 1 transmission. J Virol. 2005;79:11677–11684. doi: 10.1128/JVI.79.18.11677-11684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat D, Kozak SL, Wehrly K, Chesebro B. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J Virol. 1994;68:2570–2577. doi: 10.1128/jvi.68.4.2570-2577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketas TJ, Kuhmann SE, Palmer A, Zurita J, He W, Ahuja SK, Klasse PJ, Moore JP. Cell surface expression of CCR5 and other host factors influence the inhibition of HIV-1 infection of human lymphocytes by CCR5 ligands. Virology. 2007;364:281–290. doi: 10.1016/j.virol.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhmann SE, Platt EJ, Kozak SL, Kabat D. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J Virol. 2000;74:7005–7015. doi: 10.1128/jvi.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci U S A. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham PD, Salahuddin SZ, Kalyanaraman VS, Popovic M, Sarin P, Gallo RC. Infection and transformation of fresh human umbilical cord blood cells by multiple sources of human T-cell leukemia-lymphoma virus (HTLV) Int J Cancer. 1983;31:413–420. doi: 10.1002/ijc.2910310404. [DOI] [PubMed] [Google Scholar]
- Mellado M, Serrano A, Martínez C, Rodríguez-Frade JM. G protein-coupled receptor dimerization and signaling. Methods Mol Biol. 2006;332:141–157. doi: 10.1385/1-59745-048-0:141. [DOI] [PubMed] [Google Scholar]
- Meyer L, Magierowska M, Hubert JB, Rouzioux C, Deveau C, Sanson F, Debre P, Delfraissy JF, Theodorou I, The SEROCO Study Group Early protective effect of CCR-5Δ32 heterozygosity on HIV-1 disease progression: relationship with viral load. AIDS. 1997;11:F73–F78. doi: 10.1097/00002030-199711000-00001. [DOI] [PubMed] [Google Scholar]
- Miyoshi I, Kubonishi I, Yoshimoto S, Akagi T, Ohtsuki Y, Shiraishi Y, Nagata K, Hinuma Y. Type C virus particles in a cord T-cell line derived by co-cultivating normal human cord leukocytes and human leukaemic T cells. Nature. 1981;294:770–771. doi: 10.1038/294770a0. [DOI] [PubMed] [Google Scholar]
- Nussbaum O, Broder CC, Berger EA. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien SJ, Moore JP. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunol Rev. 2000;177:99–111. doi: 10.1034/j.1600-065x.2000.17710.x. [DOI] [PubMed] [Google Scholar]
- Pello OM, Martínez-Muñoz L, Parrillas V, Serrano A, Rodríguez-Frade JM, Toro MJ, Lucas P, Monterrubio M, Martínez AC, Mellado M. Ligand stabilization of CXCR4/δ-opioid receptor heterodimers reveals a mechanism for immune response regulation. Eur J Immunol. 2008;38:537–549. doi: 10.1002/eji.200737630. [DOI] [PubMed] [Google Scholar]
- Popovic M, Lange-Wantzin G, Sarin PS, Mann D, Gallo RC. Transformation of human umbilical cord blood T cells by human T-cell leukemia/lymphoma virus. Proc Natl Acad Sci U S A. 1983;80:5402–5406. doi: 10.1073/pnas.80.17.5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Frade JM, del Real G, Serrano A, Hernanz-Falcón P, Soriano SF, Vila-Coro AJ, de Ana AM, Lucas P, Prieto I. Blocking HIV-1 infection via CCR5 and CXCR4 receptors by acting in trans on the CCR2 chemokine receptor. EMBO J. 2004;23:66–76. doi: 10.1038/sj.emboj.7600020. other authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohy D, Parmentier M, Springael JY. Allosteric transinhibition by specific antagonists in CCR2/CXCR4 heterodimers. J Biol Chem. 2007;282:30062–30069. doi: 10.1074/jbc.M705302200. [DOI] [PubMed] [Google Scholar]
- Sun Z, Denton PW, Estes JD, Othieno FA, Wei BL, Wege AK, Melkus MW, Padgett-Thomas A, Zupancic M. Intrarectal transmission, systemic infection, and CD4+ T cell depletion in humanized mice infected with HIV-1. J Exp Med. 2007;204:705–714. doi: 10.1084/jem.20062411. other authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan CH, Torbett BE. Can gene delivery close the door to HIV-1 entry after escape? J Med Primatol. 2006;35:236–247. doi: 10.1111/j.1600-0684.2006.00172.x. [DOI] [PubMed] [Google Scholar]
- Theodorou I, Meyer L, Magierowska M, Katlama C, Rouzioux C. HIV-1 infection in an individual homozygous for CCR5Δ32. Lancet. 1997;349:1219–1220. [PubMed] [Google Scholar]
- Venkatesan S, Petrovic A, Van Ryk DI, Locati M, Weissman D, Murphy PM. Reduced cell surface expression of CCR5 in CCR5Δ32 heterozygotes is mediated by gene dosage, rather than by receptor sequestration. J Biol Chem. 2002;277:2287–2301. doi: 10.1074/jbc.M108321200. [DOI] [PubMed] [Google Scholar]
- Wolkowicz R, Nolan GP. Gene therapy progress and prospects: novel gene therapy approaches for AIDS. Gene Ther. 2005;12:467–476. doi: 10.1038/sj.gt.3302488. [DOI] [PubMed] [Google Scholar]
- Xiao X, Kinter A, Broder CC, Dimitrov DS. Interactions of CCR5 and CXCR4 with CD4 and gp120 in human blood monocyte-derived dendritic cells. Exp Mol Pathol. 2000;68:133–138. doi: 10.1006/exmp.1999.2300. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Okada M, Koyanagi Y, Kannagi M, Hinuma Y. Transformation of human leukocytes by cocultivation with an adult T cell leukemia virus producer cell line. Science. 1982;217:737–739. doi: 10.1126/science.6980467. [DOI] [PubMed] [Google Scholar]
- Zimmerman PA, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, Combadiere C, Weissman D, Cohen O, Rubbert A. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol Med. 1997;3:23–36. other authors. [PMC free article] [PubMed] [Google Scholar]