Abstract
Type II topoisomerases are ubiquitous enzymes that play essential roles in a number of fundamental DNA processes. They regulate DNA under- and overwinding, and resolve knots and tangles in the genetic material by passing an intact double helix through a transient double-stranded break that they generate in a separate segment of DNA. Because type II topoisomerases generate DNA strand breaks as a requisite intermediate in their catalytic cycle, they have the potential to fragment the genome every time they function. Thus, while these enzymes are essential to the survival of proliferating cells, they also have significant genotoxic effects. This latter aspect of type II topoisomerase has been exploited for the development of several classes of anticancer drugs that are widely employed for the clinical treatment of human malignancies. However, considerable evidence indicates that these enzymes also trigger specific leukemic chromosomal translocations. In light of the impact, both positive and negative, of type II topoisomerases on human cells, it is important to understand how these enzymes function and how their actions can destablize the genome. This article discusses both aspects of human type II topoisomerases.
1. Introduction
1.1 DNA topology
Perhaps the most striking feature of DNA is the intertwining of the two complementary strands of the double helix [1]. Discovery of this characteristic led to the immediate recognition that biological processes such as replication would be severely affected by the topological state of the genetic material [2].
DNA is globally underwound (i.e., negatively supercoiled) in all species ranging from eubacteria to humans [3–6]. This underwinding makes it easier to separate complementary DNA strands from one another and greatly facilitates initiation and elongation of replication and transcription. Once the replication or transcription machinery begins to travel along the DNA template, however, deleterious effects of topology manifest themselves. Since helicases separate, but do not unwind the two strands of the double helix, fork movement results in acute overwinding (i.e., positive supercoiling) of the DNA ahead of the tracking systems [3,5–7]. In contrast to underwinding, overwinding dramatically increases the difficulty of separating duplex DNA into individual strands. Therefore, accumulation of positive supercoils presents a formidable block to replication, transcription, and other essential DNA processes [5,7–10].
In addition to issues related to DNA under/overwinding, nuclear processes such as recombination and replication generate knots and tangles in the genetic material. If knots accumulate in the genome, DNA tracking systems are unable to separate the two strands of the double helix [3,5–7,11]. Moreover, if tangled (i.e., catenated) daughter chromosomes are not resolved prior to cell division, cells will die of mitotic failure [7,12–16].
1.2 DNA topoisomerases
The topological state of DNA in the cell is modulated by enzymes known as topoisomerases [5,7,12–19]. These ubiquitous enzymes regulate DNA over- and underwinding, and remove knots and tangles from the genetic material by creating transient breaks in the sugar-phosphate backbone of the double-helix [5,7,12–19]. Topoisomerases maintain genomic integrity during this process by forming covalent attachments between active site tyrosyl residues and the terminal DNA phosphates that are generated during the cleavage reaction [5,12,14–20]. This covalent linkage is the hallmark characteristic of all DNA topoisomerases.
Cells encode two classes of topoisomerases that are distinguished by their catalytic mechanisms. Type I topoisomerases act by generating a transient single-stranded break in the double helix, followed by either a single-stranded DNA passage event or controlled rotation about the break [5,12,14,18,21,22]. As a result, these enzymes are able to alleviate torsional stress (i.e., remove superhelical twists) in duplex DNA. Type I topoisomerases are involved in all DNA processes that involve tracking systems and play important roles in maintaining genomic integrity [5,7,13,14,18,21,22].
Type II topoisomerases act by generating a transient double-stranded DNA break, followed by a double-stranded DNA passage event [14–16,19]. Consequently, these enzymes are able to remove superhelical twists from DNA and resolve knotted or tangled duplex molecules. Type II topoisomerases function in numerous DNA processes and are required for recombination, the separation of daughter chromosomes, and proper chromosome structure, condensation, and decondensation [5–7,12–16].
2. Topoisomerase II
2.1 Topoisomerase II isoforms
Whereas lower eukaryotes such as yeast and Drosophila encode only a single type II topoisomerase [23,24], vertebrate species express two discrete forms of the enzyme, topoisomerase IIα and IIβ [14,19,25,26]. These enzymes display a high degree of amino acid sequence identity (~70%), however, they differ in their protomer molecular masses (170 vs. 180 kDa, respectively) and are encoded by separate genes [7,14–16,19,25–30]. Topoisomerase IIα and IIβ can complement the loss of topoisomerase II in yeast [31–33], and with the exception of their abilities to discern DNA geometry (discussed later in this article), display similar enzymological characteristics [15,16,19,25,26,30]. Despite their similarities, the two enzymes have distinct patterns of expression and physiological functions in vertebrate cells [7,13,15,16,19,30].
Topoisomerase IIα is essential for the survival of actively growing cells. Enzyme concentrations are upregulated dramatically during periods of cell proliferation [34–36]. Furthermore, topoisomerase IIα levels increase over the cell cycle and peak in G2/M [36–38]. Topoisomerase IIα is found at replication forks and remains tightly associated with chromosomes during mitosis [13,39]. Thus, topoisomerase IIα is believed to be the isoform that functions in growth-dependent processes, such as DNA replication and chromosome segregation [7,13]. In contrast, expression of the β isoform is independent of proliferative status and the enzyme dissociates from chromosomes during mitosis [13,30,36,40]. Topoisomerase IIβ cannot compensate for the loss of topoisomerase IIα in mammalian cells [30,41,42] and its physiological functions have yet to be defined. Although topoisomerase IIβ appears to be dispensable at the cellular level, it is required for proper neural development in mice [43].
2.2 Topoisomerase II domain structure
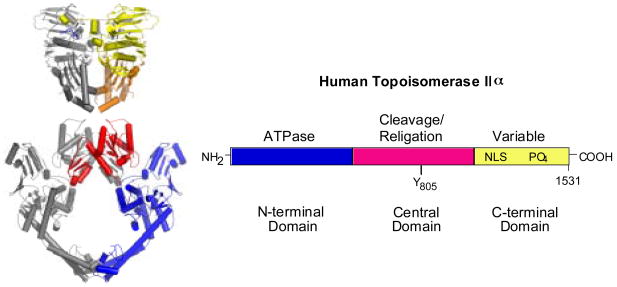
The primary structures of topoisomerase IIα and IIβ are very similar and can be divided into three domains based on sequence homology with the bacterial type II enzyme, DNA gyrase (Fig. 1) [5,13,15,19,30,44]. The N-terminal domain (first ~670 amino acids) of topoisomerase II is homologous to the B-subunit of DNA gyrase (GyrB). This portion of the enzyme contains the site of ATP binding and hydrolysis [14,19,45]. Crystal structures of this domain recently were solved for yeast topoisomerase II (Fig. 1) [46] and human topoisomerase IIα [47].
Fig. 1.
Structure of topoisomerase II. A ribbon diagram representing the crystal structure of a homodimer of yeast topoisomerase II is shown at left. The N-terminal domain is on the top (yellow and orange) and the central domain is on the bottom red and blue. At the present time, there is no structural information available for the C-terminal domain of any eukaryotic type II topoisomerase. The domain structure of human topoisomerase IIα is shown at right. The N-terminal domain is homologous to the B-subunit of DNA gyrase (GyrB) and contains the site of ATP binding and hydrolysis. The central domain is homologous to the A-subunit of DNA gyrase (GyrA) and contains the active site tyrosine (Y805) required for DNA cleavage and ligation. The C-terminal domain is highly variable among species and contains nuclear localization sequences (NLS) and sites of phosphorylation (PO4). Although the C-terminal domain was thought to contribute little to the enzymological activity of any type II topoisomerase, several recent studies suggest that this portion of the protein plays an important role in the recognition of DNA geometry.
The central domain (amino acids ~671–1200) of topoisomerase II is homologous to the A-subunit of DNA gyrase (GyrA) [14,19,48]. This portion of the enzyme contains the active site tyrosine (amino acid 805 for topoisomerase IIα and 821 for topoisomerase IIβ) required for DNA cleavage and ligation. A crystal structure for this domain in the absence of a DNA substrate was solved for yeast topoisomerase II (Fig. 1) [44].
The C-terminal domain (amino acids ~1201–1521 for topoisomerase IIα and ~1201–1621 for topoisomerase IIβ) is highly variable among species and between the two human isoforms. While it is dispensable for catalytic activity in vitro, this domain contains nuclear localization sequences [49–55] and sites of phosphorylation [49,56–58]. For many years, the C-terminal domain was thought to contribute little to the enzymatic activity of any type II topoisomerase. However, several recent studies suggest that this portion of the protein plays an intriguing and important role in the recognition of DNA geometry [59–65]. As such, it may impart unique attributes, such as the ability to supercoil DNA [59–61] or act in front of replication forks [62,64], to specific type II enzymes. Unfortunately, no structural information is available for the C-terminal domain of any eukaryotic type II enzyme at the present time.
2.3 Topoisomerase II catalytic cycle
Human topoisomerase IIα and IIβ function as homodimers and interconvert different topological forms of DNA by a “double-stranded DNA passage reaction” [15,19,44]. Briefly, these enzymes bind two separate segments of DNA, create a double-stranded break in one of the segments, translocate the second DNA segment through the cleaved nucleic acid “gate,” rejoin (i.e., ligate) the cleaved DNA, release the translocated segment through a gate in the protein, close the protein gate, and regain the ability to start a new round of catalysis [15,19,20,44,66–70]. The scissile bonds on the two strands of the double helix that are cut by topoisomerase II are staggered and are located across the major groove from one another. Thus, the enzyme generates cleaved DNA molecules that contain 4-base single-stranded ends at their 5′-termini. [71,72] During its cleavage event, topoisomerase II covalently attaches to these newly generated 5′-termini [48,71–73]. This covalent enzyme-cleaved DNA complex is known as the “cleavage complex.”
Topoisomerase II requires two cofactors in order to carry out its catalytic double-stranded DNA passage reaction. First, it needs a divalent cation for all steps beyond enzyme-DNA binding [67,71,72,74]. Magnesium(II) appears to be the divalent cation that the enzyme uses in vivo. Second, topoisomerase II uses the energy of adenosine triphosphate (ATP) to drive the overall DNA strand passage reaction [66,69,75,76]. While ATP is not required for either DNA cleavage or ligation, the binding of this nucleoside triphosphate triggers the translocation of DNA through the double-stranded nucleic acid gate [75,77]. ATP hydrolysis is necessary for enzyme recycling [66]. Normally, topoisomerase II binds two molecules of ATP [76]. Although ATP hydrolysis is not a prerequisite for the strand passage event, it appears that this step proceeds more rapidly if it is preceded by hydrolysis of one of the bound ATP molecules [77].
3. Topoisomerase II as a genotoxic enzyme
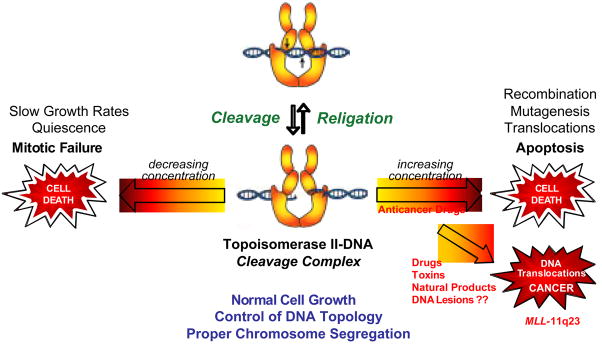
Topoisomerase II-DNA cleavage complexes are transient in nature and their cellular concentration is tightly regulated (Fig. 2). Cleavage complex formation is essential for topoisomerase II to perform is cellular functions [5,14–16,19]. If the level of topoisomerase II-DNA cleavage complexes falls too low (i.e., enzyme activity is lowered), cells are unable to undergo chromosome segregation and ultimately die of mitotic failure [7,12–16].
Fig. 2.
Topoisomerase II is an essential, but genotoxic enzyme. The formation of topoisomerase II-DNA cleavage complexes is required for the enzyme to perform its essential cellular functions. If the level of cleavage complexes falls too low (left arrow), cells are unable to undergo chromosome segregation and ultimately die of mitotic failure. If the level of cleavage complexes becomes too high (right arrow) the actions of DNA tracking systems can convert these transient complexes to permanent double-stranded breaks in the genetic material. The resulting strand breaks, as well as the inhibition of essential DNA processes, initiate multiple recombination/repair pathways and generate chromosome translocations and other DNA aberrations. If the DNA strand breaks overwhelm the cell, they trigger apoptotic pathways. This is the basis for the actions of several widely prescribed anticancer drugs. If the concentration of topoisomerase II-mediated DNA strand breaks is too low to overwhelm the cell, chromosomal translocations may be present in surviving populations and trigger the formation of leukemias that involve the MLL (mixed lineage leukemia) gene at chromosome band 11q23.
Although the cleavage complex is a requisite intermediate in the catalytic cycle of topoisomerase II, it also is potentially deleterious to the cell (Fig. 2). When a nucleic acid tracking system, such as a replication or transcription complex, attempts to traverse the cleavage complex, it converts this transient enzyme-DNA interaction to a permanent double-stranded break in the genetic material [15,16,78–81]. The resulting strand breaks, as well as the inhibition of essential DNA processes, initiate multiple recombination/repair pathways [15,16,80,82–84]. Accumulation of DNA breaks can lead to chromosome translocations and other DNA aberrations [82,84,85]. If the accumulation of breaks is overwhelming, they trigger apoptotic pathways and kill the cell [83]. If these DNA strand breaks do not result in cell death, chromosomal translocations may be present in surviving populations [84].
Increased levels of topoisomerase II-mediated DNA cleavage in humans is associated with translocations that involve the MLL (mixed lineage leukemia) gene at chromosome band 11q23 (Fig. 2) [84,86–94]. As discussed later in this article, these translocations often are linked to the initiation of specific types of acute leukemias. The mechanistic basis for the initiation of these leukemias has not been definitively elucidated, but it appears to be related to the function of the protein product of the MLL gene. MLL, which is the human homolog of the Drosophila trithorax and yeast Set1 proteins, is a histone methyltransferase that is involved in transcriptional regulation in hematopoietic cells [94–98]. Accumulating evidence suggests that the fusion of the MLL protein with other cellular partner proteins alters enzyme function and affects the differentiation of pluripotent hematopoietic stem cells or committed myeloid or lymphoid stem cells by deregulating the expression of the HOX gene [94–96,98,99].
4. Topoisomerase II poisons as anticancer agents and cellular toxins
Agents that increase levels of topoisomerase II-DNA cleavage complexes are known as “topoisomerase II poisons” because they convert this essential enzyme to a potent cellular toxin. Topoisomerase II poisons increase levels of enzyme-DNA cleavage complexes by two non-mutually exclusive mechanisms [15,16,79–81]. Some poisons act by inhibiting the ability of topoisomerase II to ligate the cleaved substrate [15,16,81,100]. These agents not only increase the level of cleavage complexes, but also increase the lifetime of these complexes. Other poisons have little effect on the rate of enzyme-mediated ligation and are believed to act primarily by enhancing the forward rate of cleavage complex formation [15,16,81,101]. The exact mechanism by which this second group of drugs increases levels of DNA cleavage is unknown. They may specifically act to enhance the forward rate of DNA scission. Alternatively, they may have some effect on the DNA binding/dissociation equilibrium, as the level of topoisomerase II-mediated DNA cleavage is proportional to the amount of enzyme bound.
Topoisomerase II poisons can be categorized into three broad classes: The first increases levels of enzyme-DNA cleavage complexes by interacting with topoisomerase II at the protein-DNA interface in a non-covalent manner; the second acts by covalently modifying the enzyme; and the third acts by covalently altering the structure of DNA. These will be discussed individually below.
4.1 Non-covalent topoisomerase II poisons
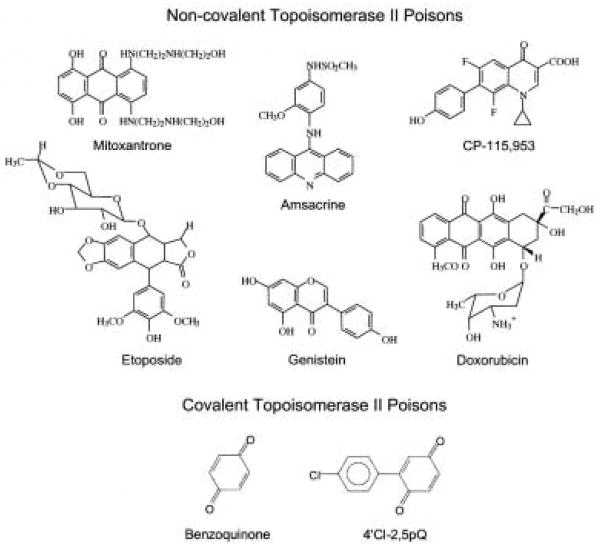
Topoisomerase IIα and IIβ are the targets of a diverse group of natural and synthetic compounds [15,16,19,79–81,102–104], some of which are depicted in Fig. 3. Although the compounds shown vary in their ring structures, all are potent topoisomerase II poisons in vitro and in human cells. Many of these agents are in wide clinical use as anticancer agents and represent some of the most successful chemotherapeutic drugs currently used for the treatment of human malignancies. On the basis of genetic and mutagenesis studies in yeast and cultured human cells, topoisomerase II is believed to be the cytotoxic target of drugs such as those shown in Fig. 3 [15,16,19,79–81,104]. Some of these drugs appear to favor either topoisomerase IIα or IIβ, however, no truly “isoform-specific” agents have been identified. The relative contribution of the two enzyme isoforms to the chemotherapeutic effects of drugs has yet to be resolved.
Fig. 3.
Structures of selected topoisomerase II poisons. Agents that act in a non-covalent fashion at the topoisomerase II-DNA interface are shown at the top. Quinones that act by covalently adducting the type II enzyme are shown at the bottom.
Half of all chemotherapy regimens include topoisomerase II-targeted drugs, and six such agents are approved for use in the USA [15,16,19,79–81,102–104]. Drugs such as etoposide and doxorubicin are front-line therapy for a variety of systemic cancers and solid tumors.
Bioflavonoids, such as genistein, are polyphenolic compounds that are constituents of many fruits, vegetables, legumes, and plant leaves [105–107]. They are an integral component of the human diet and are believed to provide a number of health benefits to adults, including chemoprevention leaves [105–112]. Bioflavonoids have a variety of effects on human cells. They represent the most abundant natural source of antioxidants, potently inhibit tyrosine kinases, and exhibit anti-proliferative and pro-apoptotic affects leaves [105–109,112–116]. However, they also display cytotoxic and genotoxic properties and many are potent topoisomerase II poisons [117–120]. Although the physiological actions of bioflavonoids are complex, the sensitivity of cells to genistein-induced toxicity has been correlated to the activity of the type II enzyme [119,121].
Quinolones, such as CP-115,953, are the only drugs that show high activity against eukaryotic and prokaryotic type II enzymes [101,122–125]. While this last drug class has not yet been exploited to treat cancer, quinolones such as ciprofloxacin and levofloxacin that target bacterial type II topoisomerases are the most active and broad-spectrum oral antibacterials in clinical use [124,126,127].
Non-covalent topoisomerase II poisons vary dramatically in their DNA binding properties. For example, etoposide is a non-intercalative compound that displays weak, if any, interaction with DNA in the absence of topoisomerase II [81,128]. Similarly, genistein and quinolones are also non-intercalative [101,123–125,129]. In contrast, amsacrine, doxorubicin, and mitoxantrone are all intercalative in nature, and the latter two compounds bind DNA with very high affinities [130–132]. It was originally thought that topoisomerase II-targeted drugs acted through interactions with DNA, “highjacking” the enzyme to sites of drug binding. We now believe that this is incorrect. All available evidence indicates that non-covalent topoisomerase II poisons act within the active site of the enzyme at the interface between the protein and DNA substrate [15,16,133–137]. Furthermore, mechanistic studies suggest that it is actually the interactions between topoisomerase II and these compounds that serve as the point of entry into the enzyme-DNA complex [135–138]. Although the non-intercalative/intercalative nature of compounds appears to have little effect on the action of drugs within the ternary topoisomerase II-DNA-drug complex, it has the potential to modulate the efficacy of these agents in a physiological setting. This issue is discussed in greater detail later in this article.
Despite the importance of topoisomerase II as a target for cancer chemotherapy, considerable evidence suggests that the enzyme also initiates chromosomal translocations that lead to specific types of leukemia [84,86,94,139–141]. For example, ~2–3% of patients treated with regimens that include etoposide ultimately develop acute myelocytic leukemia [84,86,94,139,140]. Recently, correlations between the rising use of mitoxantrone to treat breast cancer and the development of secondary leukemias have been reported [142]. Over 50% of these leukemias display translocations within an 8.3 kb breakpoint cluster region in the MLL gene at chromosomal band 11q23 [84,86,94,139,140,143]. DNA breakpoints found in these secondary leukemias are in close proximity to topoisomerase II cleavage sites [144–146]. It is notable that therapy-related leukemias with 11q23 rearrangements are unique to chemotherapeutic regimens that contain topoisomerase II-targeted drugs [84,86,94,139].
A high percentage of infant leukemias also display translocations involving chromosomal band 11q23 [147]. Even though genistein and other bioflavonoids appear to be chemopreventative in adults, the maternal consumption (during pregnancy) of foods that are naturally high in these topoisomerase II poisons increases the risk of developing these infant leukemias more than 3-fold [148]. Once again, chromosomal breakpoints in these leukemias are proximal to topoisomerase II cleavage sites [149].
Although the involvement of topoisomerase II-mediated DNA cleavage in the development of leukemias with MLL translocations is widely accepted, the mechanism by which these translocations arise is controversial and poorly understood. The evidence discussed above suggests that breaks induced by topoisomerase II within the MLL gene play a direct role in the translocation process [94,147,150]. However, it also has been proposed that the breaks induced by topoisomerase II play an indirect role in the process and that breaks induced by apoptotic nucleases ultimately lead to the translocations [151–153] In support of this later theory, a major site of apoptotic cleavage is located in the breakpoint cluster region of the MLL gene [151–153]. Ultimately, the pathway(s) that converts topoisomerase II-mediated DNA cleavage into 11q23 chromosomal translocations is likely to be highly complex and may contain elements of both processes.
4.2 Covalent topoisomerase II poisons
Several recent studies indicate that selected sulfhydryl-reactive chemicals, especially quinones, are potent topoisomerase II poisons [154–157]. These compounds appear to act by covalently adducting to the enzyme. The best-characterized covalent topoisomerase II poisons are quinones (Fig. 3). Consequently, discussion will be confined to this class of compounds.
Quinones display the unusual property of acting as topoisomerase II poisons when incubated with the enzyme-DNA complex, but as inhibitors of enzyme activity when incubated with topoisomerase II in the absence of its nucleic acid substrate [155–161]. It has been proposed that this attribute is related to the ability of quinones to cross-link the N-terminal gate of the enzyme [157] [157]. This cross-linking traps DNA within the active site topoisomerase II, but blocks entry of nucleic acid molecules that are not already engaged with the enzyme.
Studies with quinones have focused on human topoisomerase IIα [155–161]. The few experiments carried out with topoisomerase IIβ suggest that these compounds are somewhat less reactive towards this isoform [156]. Quinones such as 1,4-benzoquinone (a major benzene metabolite), N–acetylbenzoquinone amine (the major toxic metabolite of acetaminophen), and polychlorinated biphenyl (PCB) quinones (PCB metabolites such as 4′Cl-2,5pQ) increase levels of DNA cleavage mediated by topoisomerase IIα in cultured human cells [155–157,160]. However, since quinones are reactive towards a number of proteins, it is not clear what role (if any) topoisomerase II plays in the cytotoxic or genotoxic effects of these compounds.
Benzene is carcinogenic in humans and causes primarily hematopoietic malignancies [162–164]. Several lines of evidence link benzene-induced leukemias to topoisomerase II. Occupational exposure to benzene has been associated with t(8;21), which that also are seen in patients that have been treated with topoisomerase II-targeted drugs [94,165–168]. In addition, there is also a case report of leukemia with t(4;11)(q21;q23) following exposure to benzol [169]. Finally, cigarette smoke is an environmental source of benzene, smoking during pregnancy is associated with chromosomal instability in fetal amniocytes, and the genomic region most affected by tobacco is 11q23 [170].
The mechanism by which benzene induces leukemias has not been fully elucidated. However, it is believed that benzene acts through a series of reactive phenolic and quinone-based metabolites [171]. The metabolite that is believed to be central to benzene-induced leukemias is 1,4-benzoquinone [171]. This compound is generated in high concentrations in the bone marrow by oxidation of hydroquinone by endogenous myeloperoxidase [166,171]. 1,4-Benzoquinone is reduced back to the less reactive hydroquinone by NAD(P)H:quinone oxidoreductase 1 (NQO1) [166,167,172]. It is notable that individuals who are heterozygous or homozygous for the C609T polymorphism of the NQO1 gene, which encodes an inactive form of NQO1, display an increased risk for leukemias with 11q23 chromosomal translocations [166,167,172]. The finding that 1,4-benzoquinone is a potent topoisomerase II poison in vitro and in cultured human cells [156, 190] provides a provocative biochemical link between the type II enzyme and these benzene-induced leukemias.
4.3 DNA lesions as topoisomerase II poisons
The remarkable ability of anticancer drugs to convert topoisomerase II to a toxic enzyme suggests that these agents take advantage of preexisting pathways and argues for the existence of endogenous topoisomerase II poisons. In this regard, a number of naturally occurring DNA lesions that result from endogenous or environmental stress increase levels of topoisomerase II-DNA cleavage complexes [173–179]. Both the α and β isoforms of the enzyme are affected similarly in vitro [177,179]. Furthermore, DNA damaging agents stimulate DNA cleavage mediated by topoisomerase IIα in cultured human cells (the cellular effects of DNA damage on topoisomerase IIβ have not been reported) [179].
The ability to distort the double helix appears to be a common feature among lesions that act as topoisomerase II poisons [174,176–179]. Abasic sites, which are the most common lesions in the cell, are among the strongest of poisons [173,174,176,177,179]. Abasic sites are generated by a variety of methods, including spontaneous hydrolysis, DNA reactive chemicals, ionizing radiation, and base excision repair pathways [180–182]. It is estimated that some human cells may contain in excess of 100,000 abasic sites per genome, even in the absence of environmental stress [182]. The presence of ~4 abasic sites randomly generated in a plasmid ~4,400 bp in size increases enzyme-mediated DNA cleavage by either topoisomerase IIα or IIβ to approximately the same extent as the presence of ~10,000 molecules of etoposide [177]. In addition to abasic sites, exocyclic DNA adducts, such as ethano-bases are strong topoisomerase II poisons [177,179]. These adducts are generated following peroxidation of cellular lipids or by exposure of cells to industrial chemicals such as vinyl chloride [183,184].
The position of the DNA damage relative to the scissile bond cleaved by the type II enzyme is critical to the actions of lesions as topoisomerase II poisons [173–179]. As discussed earlier, the two scissile bonds on the opposite strands of the double helix are separated by 4 base pairs. Lesions that occur within these 4 base pairs often stimulate topoisomerase II-mediated DNA cleavage. Conversely, lesions located immediately outside of the scissile bonds generally inhibit, or have little effect, on DNA cleavage.
The cellular consequences of interactions between type II topoisomerases and DNA lesions are not well understood. It may be that these interactions help to trigger the excision of chromosomal loops by topoisomerase II during the late stages of apoptosis or help to kill cells following oxidation by macrophages [185,186]. The ability of DNA lesions to poison topoisomerase II provides a fascinating teleological link between the actions of anticancer drugs and programmed cell death pathways.
5. Effects of DNA supercoiling on topoisomerase II-mediated DNA cleavage and the actions of anticancer drugs
As discussed above, topoisomerase II-DNA cleavage complexes are transient in nature and are converted to permanent DNA strand breaks in the cell when replication or transcription complexes (or other nucleic acid tracking systems) collide with the covalently attached enzyme [15,16,78,79]. It is these permanent DNA strand breaks that ultimately initiate the cytotoxic effects of topoisomerase II poisons [83].
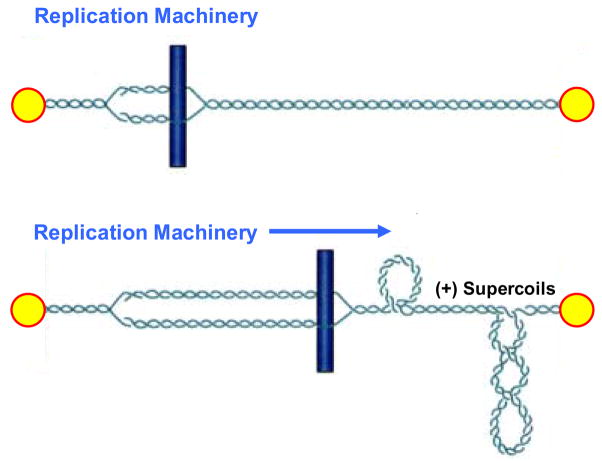
Globally, DNA in all eukaryotic cells is underwound (i.e., negatively supercoiled) [3–6]. This puts energy into the genetic material and makes it easier to separate the two strands of the double helix. In contrast to global underwinding, the movement of DNA tracking systems through the genetic material results in an acute overwinding (i.e., positive supercoiling) immediately ahead of replication or transcription enzymes (Fig. 4) [3,5–7]. Therefore, collisions with tracking systems, which are critical for the conversion of topoisomerase-DNA cleavage complexes to permanent strand breaks, most likely occur in overwound rather than underwound regions of the genome.
Fig. 4.
Generation of positive DNA supercoils (+SC) ahead of DNA tracking systems. The replication machinery is represented by a rod moving through the double helix. DNA ends are anchored to hypothetical immobile structures existing in the nucleus. Upon initiation of DNA replication, the two strands of duplex DNA are separated and the replication fork is formed (top). Movement of the replication machinery through the immobilized DNA template strands induces acute overwinding (i.e., positive supercoiling) ahead of the fork (bottom). Since collisions with DNA tracking systems (such as replication forks) are critical for the conversion of topoisomerase II-DNA cleavage complexes to permanent cytotoxic strand breaks, these collisions most likely occur in overwound regions of the genome.
Because of the universal nature of underwound DNA, negatively supercoiled substrates have been used for most previous studies that characterized the catalytic actions of type II topoisomerases and anticancer drugs that target these enzymes. Other studies have utilized linear nucleic acid substrates, especially for mapping sites of drug-induced DNA scission. Recent reports have begun to describe interactions between type II topoisomerases and positively supercoiled substrates [61,62,64,65,187,188]. Several unexpected findings have emerged from these experiments.
First, human topoisomerase IIα removes (i.e., relaxes) positive DNA supercoils >10–fold faster than it does negative supercoils [62]. Thus, the α isoform, which is involved in replicative processes, displays preferential activity with the DNA substrate that accumulates ahead of replication forks. In contrast, topoisomerase IIβ, which is not believed to play a role in DNA replication, relaxes positive and negative superhelical twists at similar rates [62]. On the basis of amino acid sequence comparisons between the two human topoisomerase II isoforms, as well as studies on bacterial and viral type II enzymes [59–61,65], it has been proposed that the ability to discern the geometry of DNA supercoils during relaxation resides in the C-terminal domain of human topoisomerase IIα.
Second, both topoisomerase IIα and IIβ maintain lower levels (~2– to 4–fold) of DNA cleavage complexes with positively supercoiled substrates [62,64]. This attribute decreases the probability that a collision with a replication fork (or other DNA tracking system) will result in the formation of a topoisomerase II-associated double-stranded chromosomal breaks and thus makes these enzymes “safer” participants in DNA processes. However, it also potentially renders them less lethal targets for anticancer drugs.
Third, the geometry of DNA supercoils differentially affects the efficacy of non-intercalative and intercalative topoisomerase II-targeted drugs [64]. In the case of non-intercalative drugs, such as etoposide, the supercoil geometry has relatively little effect on drug action. The relative increase in DNA cleavage mediated by topoisomerase IIα or IIβ is similar with positively or negatively supercoiled substrates [62,64]. However, overall levels of DNA scission are reduced in accordance with the lower baseline (i.e., no drug) levels of cleavage observed with overwound DNA [64]. The effects of supercoil geometry are considerably different with intercalative drugs such as amsacrine. Intercalative drugs produce the typical “bell-shaped” curve for the induction of topoisomerase II-mediated DNA cleavage with negatively supercoiled substrates, with scission dropping (sometimes precipitously) at high drug concentrations [64]. In contrast, levels of cleavage rise and remain high with positively supercoiled molecules [64].
The differential consequences of DNA geometry on the actions of intercalative drug probably result from the effects of intercalation on nucleic acid structure and drug binding. Since intercalative compounds locally underwind DNA, they induce compensatory unconstrained positive superhelical twists in distal regions of covalently closed circular molecules [130,189]. Thus, as the concentration of an intercalating agent increases, DNA that is topologically negatively supercoiled would appear to contain positive superhelical twists. Since baseline levels of DNA cleavage mediated by human type II topoisomerases are lower with positively supercoiled substrates [64], the apparent intercalation-induced change in DNA topology could diminish the ability of a compound to enhance cleavage with underwound substrates. In contrast, the apparent geometry of a positively supercoiled plasmid (which already is overwound) would not change substantially upon addition of an intercalative drug.
Although topoisomerase II-targeted anticancer drugs act at the enzyme-DNA interface [15,16,104,133–137], the accumulation of drugs in the double helix has the potential to inhibit enzyme binding or activity. Because the generation of positive superhelical twists by DNA intercalation induces torsional stress in the double helix [130,189], the ability of these molecules to absorb intercalative compounds is limited. Since overwound DNA is under positive torsional stress even in the absence of drugs, it cannot bind as many intercalative molecules as underwound DNA. Therefore, enzyme activity on positively supercoiled substrates is less likely to be inhibited by the accumulation of bound drug.
The differential influence of DNA geometry on the actions of intercalative anticancer drugs that target topoisomerase II allows these agents to maintain their efficacy ahead of tracking systems over a broad drug concentration range [64]. This aspect of drug activity may contribute to the clinical success of intercalative agents such as doxorubicin, mitoxantrone, and amsacrine.
6. Conclusions
Type II topoisomerases are ubiquitous enzymes that play essential roles in a number of critical DNA processes. In addition, they are the cytotoxic targets for a number of highly successful anticancer drugs. Despite the significance of topoisomerase IIα and IIβ to the survival of human cells and the efficacy of cancer chemotherapy, considerable evidence indicates that these enzymes have significant genotoxic effects and can trigger specific leukemic chromosomal translocations. In light of the profound impact, both good and bad, of type II topoisomerases on human cells, it is important to continue research on these fascinating enzymes. For example, recent work with positively supercoiled DNA has helped to elucidate why some topoisomerase II-targeted drugs may display unexpected activity in physiological settings. Future studies hopefully will generate even greater revelations.
Acknowledgments
Work in the senior author’s laboratory was supported by National Institutes of Health research grants GM33944 and GM53960. AKM was a trainee under National Institutes of Health grant 5 T32 CA09582.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Watson JD, Crick FHC. Molecular Structure of Nucleic Acids. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Watson JD, Crick FHC. Genetical Implications of the Structure of Deoxyribonucleic Acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 3.Cozzarelli NR, Wang JC, editors. DNA Topology and its Biological Effects. Cold Spring Harbor Laboratory Press: Cold Spring Harbor; 1990. [Google Scholar]
- 4.Kanaar R, Cozzarelli NR. Roles of supercoiled DNA structure in DNA transactions. Curr Opin Struct Biol. 1992;2:369–379. [Google Scholar]
- 5.Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 6.Schvartzman JB, Stasiak A. A topological view of the replicon. EMBO Rep. 2004;5:256–261. doi: 10.1038/sj.embor.7400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 8.Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. DNA topoisomerase activity is required as a swivel for DNA replication and for ribosomal RNA transcription. NCI Monographs. 1987;4:11–15. [PubMed] [Google Scholar]
- 9.Kim RA, Wang JC. Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. J Mol Biol. 1989;208:257–267. doi: 10.1016/0022-2836(89)90387-2. [DOI] [PubMed] [Google Scholar]
- 10.Peter BJ, Ullsperger C, Hiasa H, Marians KJ, Cozzarelli NR. The structure of supercoiled intermediates in DNA replication. Cell. 1998;94:819–827. doi: 10.1016/s0092-8674(00)81740-7. [DOI] [PubMed] [Google Scholar]
- 11.Deibler RW, Rahmati S, Zechiedrich EL. Topoisomerase IV, alone, unknots DNA in E. coli. Genes Dev. 2001;15:748–761. doi: 10.1101/gad.872301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osheroff N. DNA topoisomerases. Biochim Biophys Acta. 1998;1400:1–2. doi: 10.1016/s0167-4781(98)00123-7. [DOI] [PubMed] [Google Scholar]
- 13.Nitiss JL. Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim Biophys Acta. 1998;1400:63–81. doi: 10.1016/s0167-4781(98)00128-6. [DOI] [PubMed] [Google Scholar]
- 14.Champoux JJ. DNA topisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 15.Fortune JM, Osheroff N. Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog Nucleic Acid Res Mol Biol. 2000;64:221–253. doi: 10.1016/s0079-6603(00)64006-0. [DOI] [PubMed] [Google Scholar]
- 16.Wilstermann AM, Osheroff N. Stabilization of eukaryotic topoisomerase II-DNA cleavage complexes. Curr Top Med Chem. 2003;3:1349–1364. doi: 10.2174/1568026033452519. [DOI] [PubMed] [Google Scholar]
- 17.Sabourin M, Osheroff N. Topoisomerases Wiley Encyclopedia of Molecular Medicine. John Wiley & Sons, Inc; 2002. pp. 3192–3197. [Google Scholar]
- 18.Champoux JJ. DNA topoisomerases: type I, in: Encyclopedia of Biological Chemistry. Elsevier Inc; 2004. pp. 798–805. [Google Scholar]
- 19.Velez-Cruz R, Osheroff N. DNA topoisomerases: type II, in: Encyclopedia of Biological Chemistry. Elsevier Inc; 2004. pp. 806–811. [Google Scholar]
- 20.Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q Rev Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 21.Pommier Y, Pourquier P, Fan Y, Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim Biophys Acta. 1998;1400:83–106. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 22.Leppard JB, Champoux JJ. Human DNA topoisomerase I: relaxation, roles, and damage control. Chromosoma. 2005;114:75–85. doi: 10.1007/s00412-005-0345-5. [DOI] [PubMed] [Google Scholar]
- 23.Wyckoff E, Hsieh TS. Functional expression of a Drosophila gene in yeast: genetic complementation of DNA topoisomerase II. Proc Natl Acad Sci USA. 1988;85:6272–6276. doi: 10.1073/pnas.85.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goto T, Laipis P, Wang JC. The purification and characterization of DNA topoisomerases I and II of the yeast Saccharomyces cerevisiae. J Biol Chem. 1984;259:10422–10429. [PubMed] [Google Scholar]
- 25.Drake FH, Zimmerman JP, McCabe FL, Bartus HF, Per SR, Sullivan DM, Ross WE, Mattern MR, Johnson RK, Crooke ST, et al. Purification of topoisomerase II from amsacrine-resistant P388 leukemia cells. Evidence for two forms of the enzyme. J Biol Chem. 1987;262:16739–16747. [PubMed] [Google Scholar]
- 26.Drake FH, Hofmann GA, Bartus HF, Mattern MR, Crooke ST, Mirabelli CK. Biochemical and pharmacological properties of p170 and p180 forms of topoisomerase II. Biochemistry. 1989;28:8154–8160. doi: 10.1021/bi00446a029. [DOI] [PubMed] [Google Scholar]
- 27.Tsai-Pflugfelder M, Liu LF, Liu AA, Tewey KM, Whang-Peng J, Knutsen T, Huebner K, Croce CM, Wang JC. Cloning and sequencing of cDNA encoding human DNA topoisomerase II and localization of the gene to chromosome region 17q21–22. Proc Natl Acad Sci U S A. 1988;85:7177–7181. doi: 10.1073/pnas.85.19.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkins JR, Ayton P, Jones T, Davies SL, Simmons DL, Harris AL, Sheer D, Hickson ID. Isolation of cDNA clones encoding the beta isozyme of human DNA topoisomerase II and localisation of the gene to chromosome 3p24. Nucleic Acids Res. 1992;20:5587–5592. doi: 10.1093/nar/20.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan KB, Dorman TE, Falls KM, Chung TD, Mirabelli CK, Crooke ST, Mao J. Topoisomerase II alpha and topoisomerase II beta genes: characterization and mapping to human chromosomes 17 and 3, respectively. Cancer Res. 1992;52:231–234. [PubMed] [Google Scholar]
- 30.Austin CA, Marsh KL. Eukaryotic DNA topoisomerase IIβ. Bioessays. 1998;20:215–226. doi: 10.1002/(SICI)1521-1878(199803)20:3<215::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 31.Wasserman RA, Austin CA, Fisher LM, Wang JC. Use of yeast in the study of anticancer drugs targeting DNA topoisomerases: expression of a functional recombinant human DNA topoisomerase II alpha in yeast. Cancer Res. 1993;53:3591–3596. [PubMed] [Google Scholar]
- 32.Jensen S, Redwood CS, Jenkins JR, Andersen AH, Hickson ID. Human DNA topoisomerases II alpha and II beta can functionally substitute for yeast TOP2 in chromosome segregation and recombination. Mol Gen Genet. 1996;252:79–86. [PubMed] [Google Scholar]
- 33.Meczes EL, Marsh KL, Fisher LM, Rogers MP, Austin CA. Complementation of temperature-sensitive topoisomerase II mutations in Saccharomyces cerevisiae by a human TOP2 beta construct allows the study of topoisomerase II beta inhibitors in yeast. Cancer Chemother Pharmacol. 1997;39:367–375. doi: 10.1007/s002800050585. [DOI] [PubMed] [Google Scholar]
- 34.Heck MM, Earnshaw WC. Topoisomerase II: a specific marker for cell proliferation. J Cell Biol. 1986;103:2569–2581. doi: 10.1083/jcb.103.6.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsiang YH, Wu HY, Liu LF. Proliferation-dependent regulation of DNA topoisomerase II in cultured human cells. Cancer Res. 1988;48:3230–3235. [PubMed] [Google Scholar]
- 36.Woessner RD, Mattern MR, Mirabelli CK, Johnson RK, Drake FH. Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 1991;2:209–214. [PubMed] [Google Scholar]
- 37.Heck MM, Hittelman WN, Earnshaw WC. Differential expression of DNA topoisomerases I and II during the eukaryotic cell cycle. Proc Natl Acad Sci U S A. 1988;85:1086–1090. doi: 10.1073/pnas.85.4.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura K, Saijo M, Ui M, Enomoto T. Growth state- and cell cycle-dependent fluctuation in the expression of two forms of DNA topoisomerase II and possible specific modification of the higher molecular weight form in the M phase. J Biol Chem. 1994;269:1173–1176. [PubMed] [Google Scholar]
- 39.Bauman ME, Holden JA, Brown KA, Harker WG, Perkins SL. Differential immunohistochemical staining for DNA topoisomerase II alpha and beta in human tissues and for DNA topoisomerase II beta in non-Hodgkin’s lymphomas. Mod Pathol. 1997;10:168–175. [PubMed] [Google Scholar]
- 40.Isaacs RJ, Davies SL, Sandri MI, Redwood C, Wells NJ, Hickson ID. Physiological regulation of eukaryotic topoisomerase II. Biochim Biophys Acta. 1998;1400:121–137. doi: 10.1016/s0167-4781(98)00131-6. [DOI] [PubMed] [Google Scholar]
- 41.Dereuddre S, Delaporte C, Jacquemin-Sablon A. Role of topoisomerase II beta in the resistance of 9-OH-ellipticine-resistant Chinese hamster fibroblasts to topoisomerase II inhibitors. Cancer Res. 1997;57:4301–4308. [PubMed] [Google Scholar]
- 42.Grue P, Grasser A, Sehested M, Jensen PB, Uhse A, Straub T, Ness W, Boege F. Essential mitotic functions of DNA topoisomerase IIα are not adopted by topoisomerase IIβ in human H69 cells. J Biol Chem. 1998;273:33660–33666. doi: 10.1074/jbc.273.50.33660. [DOI] [PubMed] [Google Scholar]
- 43.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIb and neural development. Science. 2000;287:131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 44.Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 45.Berger JM, Fass D, Wang JC, Harrison SC. Structural similarities between topoisomerases that cleave one or both DNA strands. Proc Natl Acad Sci USA. 1998;95:7876–7881. doi: 10.1073/pnas.95.14.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Classen S, Olland S, Berger JM. Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187. Proc Natl Acad Sci U S A. 2003;100:10629–10634. doi: 10.1073/pnas.1832879100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei H, Ruthenburg AJ, Bechis SK, Verdine GL. Nucleotide-dependent domain movement in the ATPase domain of a human type IIA DNA topoisomerase. J Biol Chem. 2005;280:37041–37047. doi: 10.1074/jbc.M506520200. [DOI] [PubMed] [Google Scholar]
- 48.Worland ST, Wang JC. Inducible overexpression, purification, and active site mapping of DNA topoisomerase II from the yeast Saccharomyces cerevisiae. J Biol Chem. 1989;264:4412–4416. [PubMed] [Google Scholar]
- 49.Shiozaki K, Yanagida M. Functional dissection of the phosphorylated termini of fission yeast DNA topoisomerase II. J Cell Biol. 1992;119:1023–1036. doi: 10.1083/jcb.119.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crenshaw DG, Hsieh T. Function of the hydrophilic carboxyl terminus of type II DNA topoisomerase from Drosophila melanogaster. I. In vitro studies. J Biol Chem. 1993;268:21328–21334. [PubMed] [Google Scholar]
- 51.Mirski SE, Cole SP. Cytoplasmic localization of a mutant M(r) 160,000 topoisomerase II alpha is associated with the loss of putative bipartite nuclear localization signals in a drug-resistant human lung cancer cell line. Cancer Res. 1995;55:2129–2134. [PubMed] [Google Scholar]
- 52.Wessel I, Jensen PB, Falck J, Mirski SE, Cole SP, Sehested M. Loss of amino acids 1490Lys-Ser-Lys1492 in the COOH-terminal region of topoisomerase IIalpha in human small cell lung cancer cells selected for resistance to etoposide results in an extranuclear enzyme localization. Cancer Res. 1997;57:4451–4454. [PubMed] [Google Scholar]
- 53.Adachi N, Miyaike M, Kato S, Kanamaru R, Koyama H, Kikuchi A. Cellular distribution of mammalian DNA topoisomerase II is determined by its catalytically dispensable C-terminal domain. Nucleic Acids Res. 1997;25:3135–3142. doi: 10.1093/nar/25.15.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mirski SE, Gerlach JH, Cummings HJ, Zirngibl R, Greer PA, Cole SP. Bipartite nuclear localization signals in the C terminus of human topoisomerase II alpha. Exp Cell Res. 1997;237:452–455. doi: 10.1006/excr.1997.3805. [DOI] [PubMed] [Google Scholar]
- 55.Cowell IG, Willmore E, Chalton D, Marsh KL, Jazrawi E, Fisher LM, Austin CA. Nuclear distribution of human DNA topoisomerase IIbeta: a nuclear targeting signal resides in the 116-residue C-terminal tail. Exp Cell Res. 1998;243:232–240. doi: 10.1006/excr.1998.4150. [DOI] [PubMed] [Google Scholar]
- 56.DeVore RF, Corbett AH, Osheroff N. Phosphorylation of topoisomerase II by casein kinase II and protein kinase C: effects on enzyme-mediated DNA cleavage/religation and sensitivity to the antineoplastic drugs etoposide and 4′-(9-acridinylamino)methane-sulfon-m-anisidide. Cancer Res. 1992;52:2156–2161. [PubMed] [Google Scholar]
- 57.Cardenas ME, Dang Q, Glover CV, Gasser SM. Casein kinase II phosphorylates the eukaryote-specific C-terminal domain of topoisomerase II in vivo. EMBO J. 1992;11:1785–1796. doi: 10.1002/j.1460-2075.1992.tb05230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wells NJ, Addison CM, Fry AM, Ganapathi R, Hickson ID. Serine 1524 is a major site of phosphorylation on human topoisomerase II alpha protein in vivo and is a substrate for casein kinase II in vitro. J Biol Chem. 1994;269:29746–29751. [PubMed] [Google Scholar]
- 59.Corbett KD, Shultzaberger RK, Berger JM. The C-terminal domain of DNA gyrase A adopts a DNA-bending beta-pinwheel fold. Proc Natl Acad Sci USA. 2004;101:7293–7298. doi: 10.1073/pnas.0401595101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsieh TJ, Farh L, Huang WM, Chan NL. Structure of the topoisomerase IV C-terminal domain: a broken beta-propeller implies a role as geometry facilitator in catalysis. J Biol Chem. 2004;279:55587–55593. doi: 10.1074/jbc.M408934200. [DOI] [PubMed] [Google Scholar]
- 61.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J Mol Biol. 2005;351:545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 62.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase IIalpha rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J Biol Chem. 2005;280:39337–39345. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 63.Dickey JS, Osheroff N. Impact of the C-terminal domain of topoisomerase IIa on the DNA cleavage activity of the human enzyme. Biochemistry. 2005;44:11546–11554. doi: 10.1021/bi050811l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McClendon AK, Osheroff N. The Geometry of DNA Supercoils Modulates Topoisomerase-mediated DNA Cleavage and Enzyme Response to Anticancer Drugs. Biochemistry. 2006;45:3040–3050. doi: 10.1021/bi051987q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McClendon AK, Dickey JS, Osheroff N. Ability of Viral Topoisomerase II To Discern the Handedness of Supercoiled DNA: Bimodal Recognition of DNA Geometry by Type II Enzymes. Biochemistry. 2006;45:11674–11680. doi: 10.1021/bi0520838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Osheroff N. Eukaryotic topoisomerase II. Characterization of enzyme turnover. J Biol Chem. 1986;261:9944–9950. [PubMed] [Google Scholar]
- 67.Osheroff N. Role of the divalent cation in topoisomerase II mediated reactions. Biochemistry. 1987;26:6402–6406. doi: 10.1021/bi00394a015. [DOI] [PubMed] [Google Scholar]
- 68.Roca J, Wang JC. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell. 1992;71:833–840. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 69.Lindsley JE, Wang JC. On the coupling between ATP usage and DNA transport by yeast DNA topoisomerase II. J Biol Chem. 1993;268:8096–8104. [PubMed] [Google Scholar]
- 70.Wilstermann AM, Osheroff N. Positioning the 3′-DNA terminus for topoisomerase II-mediated religation. J Biol Chem. 2001;276:17727–17731. doi: 10.1074/jbc.M100197200. [DOI] [PubMed] [Google Scholar]
- 71.Liu LF. DNA topoisomerases--enzymes that catalyse the breaking and rejoining of DNA. [Review] CRC Crit Rev Biochem. 1983;15:1–24. doi: 10.3109/10409238309102799. [DOI] [PubMed] [Google Scholar]
- 72.Sander M, Hsieh T. Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J Biol Chem. 1983;258:8421–8428. [PubMed] [Google Scholar]
- 73.Zechiedrich EL, Christiansen K, Andersen AH, Westergaard O, Osheroff N. Double-stranded DNA cleavage/religation reaction of eukaryotic topoisomerase II: evidence for a nicked DNA intermediate. Biochemistry. 1989;28:6229–6236. doi: 10.1021/bi00441a014. [DOI] [PubMed] [Google Scholar]
- 74.Osheroff N, Zechiedrich EL. Calcium-promoted DNA cleavage by eukaryotic topoisomerase II: trapping the covalent enzyme-DNA complex in an active form. Biochemistry. 1987;26:4303–4309. doi: 10.1021/bi00388a018. [DOI] [PubMed] [Google Scholar]
- 75.Osheroff N, Shelton ER, Brutlag DL. DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA. J Biol Chem. 1983;258:9536–9543. [PubMed] [Google Scholar]
- 76.Lindsley JE, Wang JC. Proteolysis patterns of epitopically labeled yeast DNA topoisomerase II suggest an allosteric transition in the enzyme induced by ATP binding. Proc Natl Acad Sci USA. 1991;88:10485–10489. doi: 10.1073/pnas.88.23.10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baird CL, Harkins TT, Morris SK, Lindsley JE. Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc Natl Acad Sci U S A. 1999;96:13685–13690. doi: 10.1073/pnas.96.24.13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu LF, D’Arpa P. Topoisomerase-targeting antitumor drugs: mechanisms of cytotoxicity and resistance. Important Adv Oncol. 1992:79–89. [PubMed] [Google Scholar]
- 79.Li TK, Liu LF. Tumor cell death induced by topoisomerase-targeting drugs. Annu Rev Pharmacol Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- 80.Walker JV, Nitiss JL. DNA topoisomerase II as a target for cancer chemotherapy. Cancer Invest. 2002;20:570–589. doi: 10.1081/cnv-120002156. [DOI] [PubMed] [Google Scholar]
- 81.Baldwin EL, Osheroff N. Etoposide, topoisomerase II and cancer. Curr Med Chem Anti-Cancer Agents. 2005;5:363–372. doi: 10.2174/1568011054222364. [DOI] [PubMed] [Google Scholar]
- 82.Baguley BC, Ferguson LR. Mutagenic properties of topoisomerase-targeted drugs. Biochim Biophys Acta. 1998;1400:213–222. doi: 10.1016/s0167-4781(98)00137-7. [DOI] [PubMed] [Google Scholar]
- 83.Kaufmann SH. Cell death induced by topoisomerase-targeted drugs: more questions than answers. Biochim Biophys Acta. 1998;1400:195–211. doi: 10.1016/s0167-4781(98)00136-5. [DOI] [PubMed] [Google Scholar]
- 84.Felix CA. Secondary leukemias induced by topoisomerase-targeted drugs. Biochim Biophys Acta. 1998;1400:233–255. doi: 10.1016/s0167-4781(98)00139-0. [DOI] [PubMed] [Google Scholar]
- 85.Burden DA, Osheroff N. Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim Biophys Acta. 1998;1400:139–154. doi: 10.1016/s0167-4781(98)00132-8. [DOI] [PubMed] [Google Scholar]
- 86.Felix CA. Leukemias related to treatment with DNA topoisomerase II inhibitors. Med Pediatr Oncol. 2001;36:525–535. doi: 10.1002/mpo.1125. [DOI] [PubMed] [Google Scholar]
- 87.DeVore R, Whitlock J, Hainsworth JD, Johnson DH. Therapy-related acute nonlymphocytic leukemia with monocytic features and rearrangement of chromosome 11q. Ann Intern Med. 1989;110:740–742. doi: 10.7326/0003-4819-110-9-740. [DOI] [PubMed] [Google Scholar]
- 88.Pedersen-Bjergaard J, Philip P. Balanced translocations involving chromosome bands 11q23 and 21q22 are highly characteristic of myelodysplasia and leukemia following therapy with cytostatic agents targeting at DNA-topoisomerase II. Blood. 1991;78:1147–1148. [PubMed] [Google Scholar]
- 89.Pedersen-Bjergaard J. Radiotherapy- and chemotherapy-induced myelodysplasia and acute myeloid leukemia. A review. Leuk Res. 1992;16:61–65. doi: 10.1016/0145-2126(92)90102-d. [DOI] [PubMed] [Google Scholar]
- 90.Felix CA, Winick NJ, Negrini M, Bowman WP, Croce CM, Lange BJ. Common region of ALL-1 gene disrupted in epipodophyllotoxin-related secondary acute myeloid leukemia. Cancer Res. 1993;53:2954–2956. [PubMed] [Google Scholar]
- 91.Felix CA, Lange BJ, Hosler MR, Fertala J, Bjornsti MA. Chromosome band 11q23 translocation breakpoints are DNA topoisomerase II cleavage sites. Cancer Res. 1995;55:4287–4292. [PubMed] [Google Scholar]
- 92.Broeker PL, Super HG, Thirman MJ, Pomykala H, Yonebayashi Y, Tanabe S, Zeleznik-Le N, Rowley JD. Distribution of 11q23 breakpoints within the MLL breakpoint cluster region in de novo acute leukemia and in treatment-related acute myeloid leukemia: correlation with scaffold attachment regions and topoisomerase II consensus binding sites. Blood. 1996;87:1912–1922. [PubMed] [Google Scholar]
- 93.Sung PA, Libura J, Richardson C. Etoposide and illegitimate DNA double-strand break repair in the generation of MLL translocations: new insights and new questions. DNA Repair (Amst) 2006;5:1109–1118. doi: 10.1016/j.dnarep.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 94.Felix CA, Kolaris CP, Osheroff N. Topoisomerase II and the etiology of chromosomal translocations. DNA Repair (Amst) 2006;5:1093–1108. doi: 10.1016/j.dnarep.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 95.Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10:500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 96.Hess JL. Mechanisms of transformation by MLL. Crit Rev Eukaryot Gene Expr. 2004;14:235–254. doi: 10.1615/critreveukaryotgeneexpr.v14.i4.10. [DOI] [PubMed] [Google Scholar]
- 97.Tenney K, Shilatifard A. A COMPASS in the voyage of defining the role of trithorax/MLL-containing complexes: linking leukemogensis to covalent modifications of chromatin. J Cell Biochem. 2005;95:429–436. doi: 10.1002/jcb.20421. [DOI] [PubMed] [Google Scholar]
- 98.Lappin TR, Grier DG, Thompson A, Halliday HL. HOX genes: seductive science, mysterious mechanisms. Ulster Med J. 2006;75:23–31. [PMC free article] [PubMed] [Google Scholar]
- 99.Li ZY, Liu DP, Liang CC. New insight into the molecular mechanisms of MLL-associated leukemia. Leukemia. 2005;19:183–190. doi: 10.1038/sj.leu.2403602. [DOI] [PubMed] [Google Scholar]
- 100.Osheroff N. Effect of antineoplastic agents on the DNA cleavage/religation reaction of eukaryotic topoisomerase II: inhibition of DNA religation by etoposide. Biochemistry. 1989;28:6157–6160. doi: 10.1021/bi00441a005. [DOI] [PubMed] [Google Scholar]
- 101.Robinson MJ, Martin BA, Gootz TD, McGuirk PR, Moynihan M, Sutcliffe JA, Osheroff N. Effects of quinolone derivatives on eukaryotic topoisomerase II. A novel mechanism for enhancement of enzyme-mediated DNA cleavage. J Biol Chem. 1991;266:14585–14592. [PubMed] [Google Scholar]
- 102.Hande KR. Clinical applications of anticancer drugs targeted to topoisomerase II. Biochim Biophys Acta. 1998;1400:173–184. doi: 10.1016/s0167-4781(98)00134-1. [DOI] [PubMed] [Google Scholar]
- 103.Hande KR. Etoposide: four decades of development of a topoisomerase II inhibitor. Eur J Cancer. 1998;34:1514–1521. doi: 10.1016/s0959-8049(98)00228-7. [DOI] [PubMed] [Google Scholar]
- 104.Pommier Y, Marchand C. Interfacial inhibitors of protein-nucleic acid interactions. Curr Med Chem Anti-Cancer Agents. 2005;5:421–429. doi: 10.2174/1568011054222337. [DOI] [PubMed] [Google Scholar]
- 105.Kurzer MS, Xu X. Dietary phytoestrogens. Annu Rev Nutr. 1997;17:353–381. doi: 10.1146/annurev.nutr.17.1.353. [DOI] [PubMed] [Google Scholar]
- 106.Scalbert A, Williamson G. Dietary intake and bioavailability of polyphenols. J Nutr. 2000;130:2073S–2085S. doi: 10.1093/jn/130.8.2073S. [DOI] [PubMed] [Google Scholar]
- 107.Yao LH, Jiang YM, Shi J, Tomas-Barberan FA, Datta N, Singanusong R, Chen SS. Flavonoids in food and their health benefits. Plant Foods Hum Nutr. 2004;59:113–122. doi: 10.1007/s11130-004-0049-7. [DOI] [PubMed] [Google Scholar]
- 108.Kanadaswami C, Lee LT, Lee PP, Hwang JJ, Ke FC, Huang YT, Lee MT. The antitumor activities of flavonoids. In Vivo. 2005;19:895–909. [PubMed] [Google Scholar]
- 109.Siddiqui IA, Adhami VM, Saleem M, Mukhtar H. Beneficial effects of tea and its polyphenols against prostate cancer. Mol Nutr Food Res. 2006;50:130–143. doi: 10.1002/mnfr.200500113. [DOI] [PubMed] [Google Scholar]
- 110.Barnes S, Peterson TG, Coward L. Rationale for the use of genistein-containing soy matrices in chemoprevention trials for breast and prostate cancer. J Cell Biochem Suppl. 1995;22:181–187. doi: 10.1002/jcb.240590823. [DOI] [PubMed] [Google Scholar]
- 111.Stoll BA. Eating to beat breast cancer: potential role for soy supplements. Ann Oncol. 1997;8:223–225. doi: 10.1023/a:1008237505645. [DOI] [PubMed] [Google Scholar]
- 112.Lamartiniere CA. Protection against breast cancer with genistein: a component of soy. Am J Clin Nutr. 2000;71:1705S–1707S. doi: 10.1093/ajcn/71.6.1705S. discussion 1708S–1709S. [DOI] [PubMed] [Google Scholar]
- 113.Dragsted LO. Antioxidant actions of polyphenols in humans. Int J Vitam Nutr Res. 2003;73:112–119. doi: 10.1024/0300-9831.73.2.112. [DOI] [PubMed] [Google Scholar]
- 114.Sang S, Hou Z, Lambert JD, Yang CS. Redox properties of tea polyphenols and related biological activities. Antioxid Redox Signal. 2005;7:1704–1714. doi: 10.1089/ars.2005.7.1704. [DOI] [PubMed] [Google Scholar]
- 115.Adlercreutz H, Markkanen H, Watanabe S. Plasma concentrations of phyto-oestrogens in Japanese men. Lancet. 1993;342:1209–1210. doi: 10.1016/0140-6736(93)92188-y. [DOI] [PubMed] [Google Scholar]
- 116.Sarkar FH, Adsule S, Padhye S, Kulkarni S, Li Y. The role of genistein and synthetic derivatives of isoflavone in cancer prevention and therapy. Mini Rev Med Chem. 2006;6:401–407. doi: 10.2174/138955706776361439. [DOI] [PubMed] [Google Scholar]
- 117.Strick R, Strissel PL, Borgers S, Smith SL, Rowley JD. Dietary bioflavonoids induce cleavage in the MLL gene and may contribute to infant leukemia. Proc Natl Acad Sci U S A. 2000;97:4790–4795. doi: 10.1073/pnas.070061297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Austin CA, Patel S, Ono K, Nakane H, Fisher LM. Site-specific DNA cleavage by mammalian DNA topoisomerase II induced by novel flavone and catechin derivatives. Biochem J. 1992;282(Pt 3):883–889. doi: 10.1042/bj2820883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Markovits J, Junqua S, Goldwasser F, Venuat AM, Luccioni C, Beaumatin J, Saucier JM, Bernheim A, Jacquemin-Sablon A. Genistein resistance in human leukaemic CCRF-CEM cells: selection of a diploid cell line with reduced DNA topoisomerase II beta isoform. Biochem Pharmacol. 1995;50:177–186. doi: 10.1016/0006-2952(95)00131-i. [DOI] [PubMed] [Google Scholar]
- 120.Bandele OJ, Osheroff N. Bioflavonoids as poisons of human topoisomerase IIalpha and IIbeta. Biochemistry. 2007;46:6097–6108. doi: 10.1021/bi7000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lopez-Lazaro M, Willmore E, Austin CA. Cells Lacking DNA Topoisomerase IIbeta are Resistant to Genistein. J Nat Prod. 2007;70:763–767. doi: 10.1021/np060609z. [DOI] [PubMed] [Google Scholar]
- 122.Elsea SH, McGuirk PR, Gootz TD, Moynihan M, Osheroff N. Drug features that contribute to the activity of quinolones against mammalian topoisomerase II and cultured cells: correlation between enhancement of enzyme-mediated DNA cleavage in vitro and cytotoxic potential. Antimicrob Agents Chemother. 1993;37:2179–2186. doi: 10.1128/aac.37.10.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Spitzner JR, Chung IK, Gootz TD, McGuirk PR, Muller MT. Analysis of eukaryotic topoisomerase II cleavage sites in the presence of the quinolone CP-115,953 reveals drug-dependent and -independent recognition elements. Mol Pharmacol. 1995;48:238–249. [PubMed] [Google Scholar]
- 124.Anderson VE, Osheroff N. Type II topoisomerases as targets for quinolone antibacterials: turning Dr. Jekyll into Mr. Hyde. Curr Pharm Des. 2001;7:337–353. doi: 10.2174/1381612013398013. [DOI] [PubMed] [Google Scholar]
- 125.Bromberg KD, Burgin AB, Osheroff N. Quinolone action against human topoisomerase IIa: stimulation of enzyme-mediated double-stranded DNA cleavage. Biochemistry. 2003;42:3393–3398. doi: 10.1021/bi027383t. [DOI] [PubMed] [Google Scholar]
- 126.Hooper DC, Wolfson JS. Fluoroquinolone antimicrobial agents. N Engl J Med. 1991;324:384–394. doi: 10.1056/NEJM199102073240606. [DOI] [PubMed] [Google Scholar]
- 127.Hooper DC, Wolfson JS, editors. Qunolone Antimicrobial Agents. American Society for Microbiology: Washington, D.C; 1993. [Google Scholar]
- 128.Chow KC, Macdonald TL, Ross WE. DNA binding by epipodophyllotoxins and N-acyl anthracyclines: implications for mechanism of topoisomerase II inhibition. Mol Pharmacol. 1988;34:467–473. [PubMed] [Google Scholar]
- 129.Capranico G, Palumbo M, Tinelli S, Zunino F. Unique sequence specificity of topoisomerase II DNA cleavage stimulation and DNA binding mode of streptonigrin. J Biol Chem. 1994;269:25004–25009. [PubMed] [Google Scholar]
- 130.Waring MJ. Drugs and DNA: uncoiling of the DNA double helix as evidence of intercalation. Humangenetik. 1970;9:234–236. doi: 10.1007/BF00279229. [DOI] [PubMed] [Google Scholar]
- 131.Graves DE. Targeting DNA through-covalent interactions of reversible binding drugs. Methods Enzymol. 2001;340:377–395. doi: 10.1016/s0076-6879(01)40432-0. [DOI] [PubMed] [Google Scholar]
- 132.Graves DE. Drug-DNA interactions. Methods Mol Biol. 2001;95:161–169. doi: 10.1385/1-59259-057-8:161. [DOI] [PubMed] [Google Scholar]
- 133.Beck WT, Danks MK, Wolverton JS, Kim R, Chen M. Drug resistance associated with altered DNA topoisomerase II. Adv Enzyme Reg. 1993;33:113–127. doi: 10.1016/0065-2571(93)90012-3. [DOI] [PubMed] [Google Scholar]
- 134.Vassetzky YS, Alghisi GC, Gasser SM. DNA topoisomerase II mutations and resistance to anti-tumor drugs. Bioessays. 1995;17:767–774. doi: 10.1002/bies.950170906. [DOI] [PubMed] [Google Scholar]
- 135.Burden DA, Kingma PS, Froelich-Ammon SJ, Bjornsti MA, Patchan MW, Thompson RB, Osheroff N. Topoisomerase II-etoposide interactions direct the formation of drug-induced enzyme-DNA cleavage complexes. J Biol Chem. 1996;271:29238–29244. doi: 10.1074/jbc.271.46.29238. [DOI] [PubMed] [Google Scholar]
- 136.Kingma PS, Burden DA, Osheroff N. Binding of etoposide to topoisomerase II in the absence of DNA: decreased affinity as a mechanism of drug resistance. Biochemistry. 1999;38:3457–3461. doi: 10.1021/bi982855i. [DOI] [PubMed] [Google Scholar]
- 137.Leroy D, Kajava AV, Frei C, Gasser SM. Analysis of etoposide binding to subdomains of human DNA topoisomerase II alpha in the absence of DNA. Biochemistry. 2001;40:1624–1634. doi: 10.1021/bi0019141. [DOI] [PubMed] [Google Scholar]
- 138.Wilstermann AM, Bender RP, Godfrey M, Choi S, Anklin C, Berkowitz DB, Osheroff N, Graves DE. Topoisomerase II-drug interaction domains: identification of substituents on etoposide that interact with the enzyme. Biochemistry. 2007 doi: 10.1021/bi700272u. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Smith MA, Rubinstein L, Anderson JR, Arthur D, Catalano PJ, Freidlin B, Heyn R, Khayat A, Krailo M, Land VJ, Miser J, Shuster J, Vena D. Secondary leukemia or myelodysplastic syndrome after treatment with epipodophyllotoxins. J Clin Oncol. 1999;17:569–577. doi: 10.1200/JCO.1999.17.2.569. [DOI] [PubMed] [Google Scholar]
- 140.Andersen MK, Christiansen DH, Jensen BA, Ernst P, Hauge G, Pedersen-Bjergaard J. Therapy-related acute lymphoblastic leukaemia with MLL rearrangements following DNA topoisomerase II inhibitors, an increasing problem: report on two new cases and review of the literature since 1992. Br J Haematol. 2001;114:539–543. doi: 10.1046/j.1365-2141.2001.03000.x. [DOI] [PubMed] [Google Scholar]
- 141.Libura J, Slater DJ, Felix CA, Richardson C. Therapy-related acute myeloid leukemia-like MLL rearrangements are induced by etoposide in primary human CD34+ cells and remain stable after clonal expansion. Blood. 2005;105:2124–2131. doi: 10.1182/blood-2004-07-2683. [DOI] [PubMed] [Google Scholar]
- 142.Mistry AR, Felix CA, Whitmarsh RJ, Mason A, Reiter A, Cassinat B, Parry A, Walz C, Wiemels JL, Segal MR, Ades L, Blair IA, Osheroff N, Peniket AJ, Lafage-Pochitaloff M, Cross NC, Chomienne C, Solomon E, Fenaux P, Grimwade D. DNA topoisomerase II in therapy-related acute promyelocytic leukemia. N Engl J Med. 2005;352:1529–1538. doi: 10.1056/NEJMoa042715. [DOI] [PubMed] [Google Scholar]
- 143.Rowley JD. 1993 Robert R. deVilliers Lecture. Chromosome translocations: dangerous liaisons. Leukemia. 1994;8(Suppl 1):S1–6. [PubMed] [Google Scholar]
- 144.Felix CA, Lange BJ, Hosler MR, Fertala J, Bjornsti MA. Chromosome band 11q23 translocation breakpoints are DNA topoisomerase II cleavage sites. Cancer Res. 1995;55:4287–4492. [PubMed] [Google Scholar]
- 145.Lovett BD, Lo Nigro L, Rappaport EF, Blair IA, Osheroff N, Zheng N, Megonigal MD, Williams WR, Nowell PC, Felix CA. Near-precise interchromosomal recombination and functional DNA topoisomerase II cleavage sites at MLL and AF-4 genomic breakpoints in treatment-related acute lymphoblastic leukemia with t(4;11) translocation. Proc Natl Acad Sci U S A. 2001;98:9802–9807. doi: 10.1073/pnas.171309898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lovett BD, Strumberg D, Blair IA, Pang S, Burden DA, Megonigal MD, Rappaport EF, Rebbeck TR, Osheroff N, Pommier YG, Felix CA. Etoposide metabolites enhance DNA topoisomerase II cleavage near leukemia-associated MLL translocation breakpoints. Biochemistry. 2001;40:1159–1170. doi: 10.1021/bi002361x. [DOI] [PubMed] [Google Scholar]
- 147.Felix CA, Lange BJ. Leukemia in infants. Oncologist. 1999;4:225–240. [PubMed] [Google Scholar]
- 148.Spector LG, Xie Y, Robison LL, Heerema NA, Hilden JM, Lange B, Felix CA, Davies SM, Slavin J, Potter JD, Blair CK, Reaman GH, Ross JA. Maternal diet and infant leukemia: the DNA topoisomerase II inhibitor hypothesis: a report from the children’s oncology group. Cancer Epidemiol Biomarkers Prev. 2005;14:651–655. doi: 10.1158/1055-9965.EPI-04-0602. [DOI] [PubMed] [Google Scholar]
- 149.Slater DJ, Hilgenfeld E, Rappaport EF, Shah N, Meek RG, Williams WR, Lovett BD, Osheroff N, Autar RS, Ried T, Felix CA. MLL-SEPTIN6 fusion recurs in novel translocation of chromosomes 3, X, and 11 in infant acute myelomonocytic leukaemia and in t(X;11) in infant acute myeloid leukaemia, and MLL genomic breakpoint in complex MLL-SEPTIN6 rearrangement is a DNA topoisomerase II cleavage site. Oncogene. 2002;21:4706–4714. doi: 10.1038/sj.onc.1205572. [DOI] [PubMed] [Google Scholar]
- 150.Rowley JD. The critical role of chromosome translocations in human leukemias. Ann Rev Genet. 1998;32:495–519. doi: 10.1146/annurev.genet.32.1.495. [DOI] [PubMed] [Google Scholar]
- 151.Betti CJ, Villalobos MJ, Diaz MO, Vaughan AT. Apoptotic triggers initiate translocations within the MLL gene involving the nonhomologous end joining repair system. Cancer Res. 2001;61:4550–4555. [PubMed] [Google Scholar]
- 152.Betti CJ, Villalobos MJ, Diaz MO, Vaughan AT. Apoptotic stimuli initiate MLL-AF9 translocations that are transcribed in cells capable of division. Cancer Res. 2003;63:1377–1381. [PubMed] [Google Scholar]
- 153.Vaughan AT, Betti CJ, Villalobos MJ, Premkumar K, Cline E, Jiang Q, Diaz MO. Surviving apoptosis: a possible mechanism of benzene-induced leukemia. Chem Biol Interact. 2005;153–154:179–185. doi: 10.1016/j.cbi.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 154.Wang H, Mao Y, Chen AY, Zhou N, LaVoie EJ, Liu LF. Stimulation of topoisomerase II-mediated DNA damage via a mechanism involving protein thiolation. Biochemistry. 2001;40:3316–3323. doi: 10.1021/bi002786j. [DOI] [PubMed] [Google Scholar]
- 155.Bender RP, Lindsey RH, Jr, Burden DA, Osheroff N. N-acetyl-p-benzoquinone imine, the toxic metabolite of acetaminophen, is a topoisomerase II poison. Biochemistry. 2004;43:3731–3739. doi: 10.1021/bi036107r. [DOI] [PubMed] [Google Scholar]
- 156.Lindsey RH, Jr, Bromberg KD, Felix CA, Osheroff N. 1,4-Benzoquinone is a topoisomerase II poison. Biochemistry. 2004;43:7563–7574. doi: 10.1021/bi049756r. [DOI] [PubMed] [Google Scholar]
- 157.Bender RP, Lehmler HJ, Robertson LW, Ludewig G, Osheroff N. Polychlorinated biphenyl quinone metabolites poison human topoisomerase IIalpha: altering enzyme function by blocking the N-terminal protein gate. Biochemistry. 2006;45:10140–10152. doi: 10.1021/bi0524666. [DOI] [PubMed] [Google Scholar]
- 158.Chen H, Eastmond DA. Topoisomerase inhibition by phenolic metabolites: a potential mechanism for benzene’s clastogenic effects. Carcinogenesis. 1995;16:2301–2307. doi: 10.1093/carcin/16.10.2301. [DOI] [PubMed] [Google Scholar]
- 159.Baker RK, Kurz EU, Pyatt DW, Irons RD, Kroll DJ. Benzene metabolites antagonize etoposide-stabilized cleavable complexes of DNA topoisomerase IIalpha. Blood. 2001;98:830–833. doi: 10.1182/blood.v98.3.830. [DOI] [PubMed] [Google Scholar]
- 160.Lindsey RH, Jr, Bender RP, Osheroff N. Effects of benzene metabolites on DNA cleavage mediated by human topoisomerase II alpha: 1,4-hydroquinone is a topoisomerase II poison. Chem Res Toxicol. 2005;18:761–770. doi: 10.1021/tx049659z. [DOI] [PubMed] [Google Scholar]
- 161.Eastmond DA, Mondrala ST, Hasegawa L. Topoisomerase II inhibition by myeloperoxidase-activated hydroquinone: a potential mechanism underlying the genotoxic and carcinogenic effects of benzene. Chem Biol Interact. 2005;153–154:207–216. doi: 10.1016/j.cbi.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 162.Snyder R, Kalf GF. A perspective on benzene leukemogenesis. Crit Rev Toxicol. 1994;24:177–209. doi: 10.3109/10408449409021605. [DOI] [PubMed] [Google Scholar]
- 163.Hayes RB, Yin SN, Dosemeci M, Li GL, Wacholder S, Travis LB, Li CY, Rothman N, Hoover RN, Linet MS. Benzene and the dose-related incidence of hematologic neoplasms in China. Chinese Academy of Preventive Medicine--National Cancer Institute Benzene Study Group. J Natl Cancer Inst. 1997;89:1065–1071. doi: 10.1093/jnci/89.14.1065. [DOI] [PubMed] [Google Scholar]
- 164.Irons RD. Molecular models of benzene leukemogenesis. J Toxicol Environ Health A. 2000;61:391–397. doi: 10.1080/00984100050166415. [DOI] [PubMed] [Google Scholar]
- 165.Smith MT, Zhang L, Wang Y, Hayes RB, Li G, Wiemels J, Dosemeci M, Titenko-Holland N, Xi L, Kolachana P, Yin S, Rothman N. Increased translocations and aneusomy in chromosomes 8 and 21 among workers exposed to benzene. Cancer Res. 1998;58:2176–2181. [PubMed] [Google Scholar]
- 166.Smith MT. Benzene, NQO1, and genetic susceptibility to cancer. Proc Natl Acad Sci U S A. 1999;96:7624–7626. doi: 10.1073/pnas.96.14.7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Smith MT, Wang Y, Skibola CF, Slater DJ, Lo Nigro L, Nowell PC, Lange BJ, Felix CA. Low NAD(P)H:quinone oxidoreductase activity is associated with increased risk of leukemia with MLL translocations in infants and children. Blood. 2002;100:4590–4593. doi: 10.1182/blood-2001-12-0264. [DOI] [PubMed] [Google Scholar]
- 168.Kim SY, Choi JK, Cho YH, Chung EJ, Paek D, Chung HW. Chromosomal aberrations in workers exposed to low levels of benzene: association with genetic polymorphisms. Pharmacogenetics. 2004;14:453–463. doi: 10.1097/01.fpc.0000114751.08559.7b. [DOI] [PubMed] [Google Scholar]
- 169.Sole F, Caballin MR, Coll MD, Woessner S, Egozcue J. Acute lymphoblastic leukemia with t(4;11) in a patient previously exposed to a carcinogen. Cancer Genet Cytogenet. 1990;49:133–136. doi: 10.1016/0165-4608(90)90174-9. [DOI] [PubMed] [Google Scholar]
- 170.de la Chica RA, Ribas I, Giraldo J, Egozcue J, Fuster C. Chromosomal instability in amniocytes from fetuses of mothers who smoke. JAMA. 2005;293:1212–1222. doi: 10.1001/jama.293.10.1212. [DOI] [PubMed] [Google Scholar]
- 171.Snyder R, Hedli CC. An overview of benzene metabolism. Environ Health Perspect. 1996;104(Suppl 6):1165–1171. doi: 10.1289/ehp.961041165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Larson RA, Wang Y, Banerjee M, Wiemels J, Hartford C, Le Beau MM, Smith MT. Prevalence of the inactivating 609C-->T polymorphism in the NAD(P)H:quinone oxidoreductase (NQO1) gene in patients with primary and therapy-related myeloid leukemia. Blood. 1999;94:803–807. [PubMed] [Google Scholar]
- 173.Kingma PS, Greider CA, Osheroff N. Spontaneous DNA lesions poison human topoisomerase IIα and stimulate cleavage proximal to leukemic 11q23 chromosomal breakpoints. Biochemistry. 1997;36:5934–5939. doi: 10.1021/bi970507v. [DOI] [PubMed] [Google Scholar]
- 174.Kingma PS, Osheroff N. The response of eukaryotic topoisomerases to DNA damage. Biochim Biophys Acta. 1998;1400:223–232. doi: 10.1016/s0167-4781(98)00138-9. [DOI] [PubMed] [Google Scholar]
- 175.Cline SD, Osheroff N. Cytosine arabinoside lesions are position-specific topoisomerase II poisons and stimulate DNA cleavage mediated by the human type II enzymes. J Biol Chem. 1999;274:29740–29743. doi: 10.1074/jbc.274.42.29740. [DOI] [PubMed] [Google Scholar]
- 176.Cline SD, Jones WR, Stone MP, Osheroff N. DNA abasic lesions in a different light: solution structure of an endogenous topoisomerase II poison. Biochemistry. 1999;38:15500–15507. doi: 10.1021/bi991750s. [DOI] [PubMed] [Google Scholar]
- 177.Sabourin M, Osheroff N. Sensitivity of human type II topoisomerases to DNA damage: stimulation of enzyme-mediated DNA cleavage by abasic, oxidized and alkylated lesions. Nucleic Acids Res. 2000;28:1947–1954. doi: 10.1093/nar/28.9.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Khan QA, Kohlhagen G, Marshall R, Austin CA, Kalena GP, Kroth H, Sayer JM, Jerina DM, Pommier Y. Position-specific trapping of topoisomerase II by benzo[a]pyrene diol epoxide adducts: implications for interactions with intercalating anticancer agents. Proc Natl Acad Sci U S A. 2003;100:12498–12503. doi: 10.1073/pnas.2032456100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Velez-Cruz R, Riggins JN, Daniels JS, Cai H, Guengerich FP, Marnett LJ, Osheroff N. Exocyclic DNA lesions stimulate DNA cleavage mediated by human topoisomerase II alpha in vitro and in cultured cells. Biochemistry. 2005;44:3972–3981. doi: 10.1021/bi0478289. [DOI] [PubMed] [Google Scholar]
- 180.Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 181.Lindahl T, Karlstrom O. Heat-induced depyrimidination of deoxyribonucleic acid in neutral solution. Biochemistry. 1973;12:5151–5154. doi: 10.1021/bi00749a020. [DOI] [PubMed] [Google Scholar]
- 182.Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. [PubMed] [Google Scholar]
- 183.Swenberg JA, Fedtke N, Ciroussel F, Barbin A, Bartsch H. Etheno adducts formed in DNA of vinyl chloride-exposed rats are highly persistent in liver. Carcinogenesis. 1992;13:727–729. doi: 10.1093/carcin/13.4.727. [DOI] [PubMed] [Google Scholar]
- 184.Lee SH, Oe T, Blair IA. 4,5-Epoxy-2(E)-decenal-induced formation of 1,N(6)-etheno-2′-deoxyadenosine and 1,N(2)-etheno-2′-deoxyguanosine adducts. Chem Res Toxicol. 2002;15:300–304. doi: 10.1021/tx010147j. [DOI] [PubMed] [Google Scholar]
- 185.Solovyan VT, Bezvenyuk ZA, Salminen A, Austin CA, Courtney MJ. The role of topoisomerase II in the excision of DNA loop domains during apoptosis. J Biol Chem. 2002;277:21458–21467. doi: 10.1074/jbc.M110621200. [DOI] [PubMed] [Google Scholar]
- 186.Belyaev IY. DNA loop organization and DNA fragmentation during radiation-induced apoptosis in human lymphocytes. Radiats Biol Radioecol. 2005;45:541–548. [PubMed] [Google Scholar]
- 187.Crisona NJ, Strick TR, Bensimon D, Croquette V, Cozzarelli NR. Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 2000;14:2881–2892. doi: 10.1101/gad.838900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Stone MD, Bryant Z, Crisona NJ, Smith SB, Vologodskii A, Bustamante C, Cozzarelli NR. Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases. Proc Natl Acad Sci USA. 2003;100:8654–8659. doi: 10.1073/pnas.1133178100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Waring M. Variation of the supercoils in closed circular DNA by binding of antibiotics and drugs: evidence for molecular models involving intercalation. J Mol Biol. 1970;54:247–279. doi: 10.1016/0022-2836(70)90429-8. [DOI] [PubMed] [Google Scholar]
- 190.Eastmond DA, Schuler M, Frantz C, Chen H, Parks R, Wang L, Hawegawa L. Characterization and mechanisms of chromosomal alterations induced by benzene in mice and humans. Res Rep Health Eff Inst. 2001;103:1–80. [PubMed] [Google Scholar]