Abstract
Background
Oxidative stress and excitotoxicity underlie the developmental neurotoxicity of numerous chemicals.
Objectives
We compared the effects of organophosphates (chlorpyrifos and diazinon), an organo-chlorine (dieldrin), and a metal [divalent nickel (Ni2+)] to determine how these mechanisms contribute to similar or dissimilar neurotoxic outcomes.
Methods
We used PC12 cells as a model of developing neurons and evaluated transcriptional profiles for genes for oxidative stress responses and glutamate receptors.
Results
Chlorpyrifos had a greater effect on oxidative-stress–related genes in differentiating cells compared with the undifferentiated state. Chlorpyrifos and diazinon showed significant concordance in their effects on glutathione-related genes, but they were negatively correlated for effects on catalase and superoxide dismutase isoforms and had no concordance for effects on ionotropic glutamate receptors. Surprisingly, the correlations were stronger between diazinon and dieldrin than between the two organophosphates. The effects of Ni2+ were the least similar for genes related to oxidative stress but had significant concordance with dieldrin for effects on glutamate receptors.
Conclusions
Our results point to underlying mechanisms by which different organophosphates produce disparate neurotoxic outcomes despite their shared property as cholinesterase inhibitors. Further, apparently unrelated neurotoxicants may produce similar outcomes because of convergence on oxidative stress and excitotoxicity. The combined use of cell cultures and microarrays points to specific end points that can distinguish similarities and disparities in the effects of diverse developmental neurotoxicants.
Keywords: chlorpyrifos, diazinon, dieldrin, excitotoxicity, glutamate receptors, glutathione, metal neurotoxicity, microarrays, nerve growth factor, neuronal development, neurotoxicity, nickel, organochlorine insecticides, oxidative stress, organophosphate insecticides, PC12 cells
A wide variety of toxicants elicit cell damage through their shared ability to produce oxidative stress (Gitto et al. 2002; Gupta 2004; Ohtsuka and Suzuki 2000; Olanow and Arendash 1994). The brain is especially vulnerable because it has a high rate of oxygen consumption, combined with a membrane lipid composition that is enriched in oxidizable polyunsaturated fatty acids (Gupta 2004). The developing brain is even more sensitive because it has lower reserves of protective enzymes and antioxidants (Gupta 2004) and has a higher ratio of neurons to glia, the cells that ordinarily protect neurons from oxidative molecules (Tanaka et al. 1999), while at the same time facing the increased metabolic demand associated with growth. Further, the fact that fetal arterial blood has substantially lower O2 concentrations means that the fetal brain is already hypoxic relative to that of a newborn or adult (Faber et al. 1985; Lagercrantz and Slotkin 1986), thus reducing the margin of safety for any agent that compromises oxidative metabolism. The combination of these factors explains why many environmental contaminants elicit oxidative stress within the developing brain (Henderson et al. 1999; Jett and Navoa 2000; Kern and Jones 2006; Mulholland et al. 2005; Pardo and Eberhart 2007; Sinha et al. 2006; Tata and Yamamoto 2007); indeed, this may provide a mechanism by which diverse compounds converge on common sets of neurodevelopmental disorders, such as autism (Kern and Jones 2006; Pardo and Eberhart 2007).
Exposure of the human population to organophosphate pesticides is ubiquitous (Barr et al. 2005; Bouvier et al. 2005; Casida and Quistad 2004), and these compounds are undergoing restriction because of their propensity to produce developmental neurotoxicity [U.S. Environmental Protection Agency (EPA) 2000, 2002, 2006]. Chlorpyrifos, diazinon, and other organophosphates produce oxidative stress in the developing brain, leading to shifts in expression and function of antioxidant genes, and accordingly, antioxidant therapy can offset some of the damage (Bagchi et al. 1995; Giordano et al. 2007; Jett and Navoa 2000; Qiao et al. 2005; Slotkin et al. 2007a; Slotkin and Seidler 2007). Nevertheless, it is increasingly clear that the various organophosphates do not produce the same patterns of neurodevelopmental damage or behavioral deficits, in part because they differ in other mechanisms that contribute to the net adverse outcomes (Pope 1999; Roegge et al. 2008; Slotkin 1999, 2004, 2005; Slotkin et al. 2006a, 2008a, 2008b, 2008c, 2009; Slotkin and Seidler 2007, 2008; Timofeeva et al. 2008a, 2008b). The participation of these additional mechanisms means that, although related compounds may produce similar degrees of oxidative stress, the cellular reactions to that stress may end up being substantially different, such that for the same degree of initial damage, the outcomes may be worse for particular agents. In the present study, we tested that hypothesis by examining the transcriptional responses to chlor pyrifos and diazinon for the various cellular targets involved in the response to oxidative stress: catalase (cat), the isoforms of superoxide dismutase (sod), glutathione synthase (gss), glutathione reductase (gsr), the family of glutathione peroxidases (gpx), the genes for the cytoplasmic (α, μ, ω, π, θ) glutathione S-transferases (gst), the microsomal and mitochondrial glutathione S-transferases (mgst, gst13-13), and the glutathione S-transferase yc2 subunit. In addition, we examined one of the major, indirect mechanisms by which organophosphates produce oxidative stress, namely, their actions on the function and expression of ionotropic glutamate receptors (Damodaran et al. 2006; Gupta 2004), which mediate excitotoxic cell death in the developing brain, including that evoked by hypoxia (Choi and Rothman 1990). We assessed expression of the AMPA receptor family (gria), δ-subunits (grid), kainate receptors (grik), NMDA (N-methyl D-aspartate) receptors (grin), and their associated glutamate-binding protein (grina). By way of contrast, we also assessed the metabotropic glutamate receptors (grm), which are not involved in excitotoxicity. For reference, Table 1 lists all genes tested, with their full names and GenBank accession numbers (National Center for Biotechnology Information 2008).
Table 1.
Gene names and Genbank accession numbers.
| Gene symbol | Gene name | GenBank accession no. |
|---|---|---|
| cat | catalase | NM_012520 |
| gpx1 | glutathione peroxidase 1 | NM_030826 |
| gpx2 | glutathione peroxidase 2 | BQ196649 |
| gpx3 | glutathione peroxidase 3 | AI172411 |
| gpx4 | glutathione peroxidase 4 | NM_017165 |
| gpx6 | glutathione peroxidase 6 | NM_147165 |
| gria1 | glutamate receptor, ionotropic AMPA 1 | NM_031608 |
| gria2 | glutamate receptor, ionotropic AMPA 2 | NM_017261 |
| gria3 | glutamate receptor, ionotropic AMPA 3 | NM_032990 |
| gria4 | glutamate receptor, ionotropic AMPA 4 | NM_017263 |
| grid2 | glutamate receptor, ionotropic δ1 | NM_024379 |
| grik1 | glutamate receptor, ionotropic kainate 1 | AI111480 |
| grik2 | glutamate receptor, ionotropic kainate 2 | NM_019309 |
| grik3 | glutamate receptor, ionotropic kainate 3 | NM_181373 |
| grik4 | glutamate receptor, ionotropic kainate 4 | NM_012572 |
| grik5 | glutamate receptor, ionotropic kainate 5 | NM_017262 |
| grin1 | glutamate receptor, ionotropic N-methyl-D-aspartate 1 | NM_017010 |
| grin2a | glutamate receptor, ionotropic N-methyl-D-aspartate 2a | NM_012573 |
| grin2b | glutamate receptor, ionotropic N-methyl-D-aspartate 2b | NM_012574 |
| grin2d | glutamate receptor, ionotropic N-methyl-D-aspartate 2d | NM_022797 |
| grin3a | glutamate receptor, ionotropic N-methyl-D-aspartate 3a | AF061945 |
| grin3b | glutamate receptor, ionotropic N-methyl-D-aspartate 3b | NM_133308 |
| grina | glutamate receptor, ionotropic N-methyl-D-aspartate-associated protein | NM_153308 |
| grm1 | glutamate receptor, metabotropic 1 | NM_017011 |
| grm2 | glutamate receptor, metabotropic 2 | XM_343470 |
| grm3 | glutamate receptor, metabotropic 3 | M92076 |
| grm4 | glutamate receptor, metabotropic 4 | NM_022666 |
| grm5 | glutamate receptor, metabotropic 5 | NM_017012 |
| grm6 | glutamate receptor, metabotropic 6 | NM_022920 |
| grm7 | glutamate receptor, metabotropic 7 | NM_031040 |
| grm8 | glutamate receptor, metabotropic 8 | NM_022202 |
| gsr | glutathione reductase | NM_053906 |
| gss | glutathione synthetase | NM_012962 |
| gst13-13 | glutathione S-transferase, mitochondrial | NM_181371 |
| gsta2 | glutathione S-transferase, α 2 | BQ199390 |
| gsta4 | glutathione S-transferase, α 4 | XM_217195 |
| gstm1 | glutathione S-transferase, μ1 | NM_017014 |
| gstm2 | glutathione S-transferase, μ2 | NM_177426 |
| gstm3 | glutathione S-transferase, μ3 | NM_031154 |
| gstm4 | glutathione S-transferase, μ4 | NM_020540 |
| gstm5 | glutathione S-transferase, μ5 | NM_172038 |
| gstm6 | glutathione S-transferase, μ6 | XM_215682 |
| gsto1 | glutathione S-transferase, ω1 | NM_001007602 |
| gsto2 | glutathione S-transferase, ω2 | NM_001012071 |
| gstp2 | glutathione S-transferase, π2 | NM_138974 |
| gstt1 | glutathione S-transferase, θ1 | NM_053293 |
| gstt2 | glutathione S-transferase, θ2 | NM_012796 |
| gstt3 | glutathione S-transferase, θ3 | XM_574740 |
| mgst1 | microsomal glutathione S-transferase 1 | NM_134349 |
| mgst2 | microsomal glutathione S-transferase 2 | XM_215562 |
| mgst3 | microsomal glutathione S-transferase 3 | XM_213943 |
| sod1 | superoxide dismutase 1 | NM_017050 |
| sod2 | superoxide dismutase 2 | AI235842 |
| sod3 | superoxide dismutase 3 | NM_012880 |
| yc2 | glutathione S-transferase yc2 subunit | NM_001009920 |
In addition to comparing the effects of chlorpyrifos with those of diazinon, we evaluated two developmental neurotoxicants from different classes: dieldrin, an organochlorine pesticide, and divalent nickel (Ni2+). Both of these represent significant environmental concerns because like the organophosphates, they appear on the registry of Superfund chemicals (National Library of Medicine 2006). Although dieldrin acts primarily by blocking GABAA receptors (Brannen et al. 1998; Liu et al. 1998), it also elicits oxidative stress akin to that of the organophosphates (Kitazawa et al. 2001, 2003; Slotkin et al. 2007b) and produces fetal brain damage (Uzoukwu and Sleight 1972). Ni2+ is found in the fetal brain in concentrations up to 2 μg/g, similar to lead (Casey and Robinson 1978), and, like lead, interferes with the gating of calcium during neurodifferentiation (Benters et al. 1996; Nikodijevic and Guroff 1992). In contrast to the other agents, Ni2+ does not elicit oxidative stress in developing neuronal cells (Slotkin et al. 2007b), and in fact, it shifts cellular redox status toward reduction, likely because it can donate electrons to form higher valence states (Slotkin et al. 2007b).
Because we wanted to compare the transcriptional responses inherent to each compound, we needed to avoid the confounds of pharmaco kinetic differences or effects on the maternal–fetal unit. Accordingly, we studied PC12 cells, a widely accepted in vitro model for neuronal development (Teng and Greene 1994) that has already been validated to reproduce the mechanisms and outcomes found after in vivo exposures of developing rats to organophosphates (Bagchi et al. 1995, 1996; Crumpton et al. 2000a, 2000b; Das and Barone 1999; Flaskos et al. 1994; Jameson et al. 2006, 2007; Li and Casida 1998; Nagata et al. 1997; Qiao et al. 2001, 2005; Slotkin et al. 2007a, 2007b, 2008d, 2008e; Song et al. 1998; Tuler et al. 1989). When nerve growth factor (NGF) is added to the culture medium, PC12 cells begin to differentiate, forming neuritic projections and acquiring electrical excitability and neuronal phenotypes (Fujita et al. 1989; Song et al. 1998; Teng and Greene 1994). The effects on neurodifferentiation in PC12 cells have been characterized previously for each of the four agents studied here (Qiao et al. 2001; Slotkin et al. 2007b; Slotkin and Seidler 2008, 2009), providing the necessary end points with which to interpret transcriptional responses. Finally, we compared effects in the undifferentiated and differentiating states for chlorpyrifos, as well as evaluating temporal responses for chlorpyrifos, diazinon, dieldrin, and Ni2+, so as to explore the role of critical developmental periods for vulnerability to oxidative stress and excitotoxicity.
Materials and Methods
Cell cultures
Because of the clonal instability of the PC12 cell line (Fujita et al. 1989), we performed the experiments on cells that had undergone fewer than five passages. As described previously (Qiao et al. 2003; Song et al. 1998), we seeded PC12 cells (American Type Culture Collection, 1721-CRL; obtained from the Duke Comprehensive Cancer Center (Durham, NC) onto poly-D-lysine–coated plates in RPMI-1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% inactivated horse serum (Sigma Chemical Co., St. Louis, MO), 5% inactivated fetal bovine serum (Sigma), and 50 μg/ mL penicillin streptomycin (Invitrogen). Incubations were carried out with 7.5% CO2 at 37°C, standard conditions for PC12 cells. To initiate neurodifferentiation (Jameson et al. 2006; Slotkin et al. 2007b; Teng and Greene 1994) 24 hr after seeding, the medium was changed to include 50 ng/mL of 2.5S murine NGF (Invitrogen). Along with the NGF, we added 30 μM of the test agent: chlorpyrifos (Chem Service, West Chester, PA), diazinon (Chem Service), dieldrin (Chem Service), or NiCl2 (Sigma). We chose this concentration from earlier studies that demonstrated adverse effects on differentiation of PC12 cells without outright cytotoxicity (Jameson et al. 2007; Qiao et al. 2001; Slotkin et al. 2007b, 2008d). Because of the limited water solubility of the three insecticides, we dissolved these agents in DMSO (final concentration, 0.1%), which was also added to the control cultures and to cultures containing NiCl2; this concentration of DMSO has no effect on PC12 cell growth or differentiation (Qiao et al. 2001, 2003; Song et al. 1998). We examined cultures 24 and 72 hr after commencing exposure, with five to eight independent cultures evaluated for each treatment at each time point. We used two time points so we could evaluate changes in gene expression regardless of whether the mRNA for a given gene has a rapid turnover (and hence can rise rapidly) or a slower turnover that would require a longer period to show corresponding increases or decreases. For chlorpyrifos, we evaluated the effects both on undifferentiated cells and during NGF-induced differentiation, whereas for the other agents, we studied only the effects during differentiation.
Microarray determinations
In the present study, we performed mRNA isolation, preparation of cDNA, conversion to cRNA incorporating cyanine-3 (reference RNA) or cyanine-5 (sample RNA), verification of RNA purity and quality, hybridization to the micro-arrays, washing, and scanning as described previously (Slotkin et al. 2007c, 2008d; Slotkin and Seidler 2007). The mRNA used for the reference standard was created by pooling aliquots from each of the samples in the study. Similarly, array normalizations and error detection were carried out using procedures described previously (Slotkin et al. 2007c, 2008d; Slotkin and Seidler 2007). We used Agilent Whole Rat Genome Arrays (Agilent Technologies, Palo Alto, CA), type G4131A for the studies of chlorpyrifos in undifferentiated and differentiating cells, and type G4131F for the studies of diazinon, dieldrin, and Ni2+ in differentiating cells. The two chips contain exactly the same sequences, but the latter has a lower detection threshold; however, all the genes reported here passed the quality control filters with both arrays.
For many of the genes, the arrays contain multiple probes for the same gene and/or replicates of the same probe in different locations on the chip, and we used these to verify the reliability of values and the validity of the measures on the chip. To avoid artificially inflating the number of positive findings, we limited each gene to a single set of values, selecting those obtained for the probe showing the smallest intragroup variance. We used the other values for that gene only to corroborate direction and magnitude of change. We also validated the readings on the arrays with duplicate arrays for selected samples (Slotkin et al. 2007c; Slotkin and Seidler 2007).
Statistical procedures
Because of the requirement to normalize the data across arrays and within each gene, the absolute values for a given gene are meaningless, so only the relative differences between treatments can be compared. Accordingly, we present results as means and SEs of the percent change from control values to allow for visual comparison of the effects across families of genes. However, statistical comparisons were based on the actual ratios (log-transformed, because the data are ratios) rather than the percent change.
Our design involved multiple planned comparisons of four agents at two time points, as well as the effects of one agent (chlorpyrifos) in undifferentiated versus differentiating states. It was therefore important to consider the false-positive rate and to protect against the increased probability of type 1 errors engendered by repeated testing of the same database. Accordingly, before looking at effects on individual genes, we performed a global analysis of variance (ANOVA) incorporating all the variables in a single comparison: treatment, time, and all genes. We then carried out lower-order ANOVAs on subdivisions of the data set as permitted by the interactions of treatment with the other variables. We evaluated differences for individual treatments for a specified gene at a single time point with Fisher’s protected least significant difference. However, for a given gene that showed no interaction of treatment with other variables (time, differentiation state), we report only the main treatment effect without subtesting effects at a single time point. We considered treatment effects significant at p < 0.05 (two-tailed, because we were interested in both increases and decreases in gene expression). Finally, concordance of patterns of effects between different agents was evaluated by linear regression analysis.
In addition to these parametric tests of the direction and magnitude of changes in gene expression, we evaluated the incidence of significant differences compared with the predicted false-positive rate, using Fisher’s exact test, applying a one-tailed criterion of p < 0.05, because only an increase above the false-positive rate would be predicted; at the criterion of p < 0.05, 1 gene of every 20 tested can be expected to show a difference at random. Finding a significant decrease in the incidence of detected differences relative to the false-positive rate would be biologically implausible and statistically meaningless.
Results
We compared the effects on undifferentiated versus differentiating cells with only one agent (chlorpyrifos), so we performed two sets of global statistical tests. For chlorpyrifos, the multivariate ANOVA incorporated the factors of treatment, differentiation state, time, and gene and identified interactions of treatment × gene (p < 0.0001), treatment × differentiation state × gene (p < 0.0001), and treatment × time × state × gene (p < 0.03). Because of the strong interaction with differentiation state, we then subdivided the results to isolate the effects on undifferentiated and differentiating cells and identified significant effects in both states: undifferentiated, treatment × gene (p < 0.0001); differentiating, treatment × time (p < 0.04), treatment × gene (p < 0.0001), and treatment × time × gene (p < 0.05). Chlorpyrifos exposure evoked significant changes in the expression of 40 of the total of 59 genes, compared with an expected false-positive rate of only 3 genes (p < 10−14), and the same was true for the separate analyses of undifferentiated cells (24 of 59 genes, p < 10−7) and differentiating cells (34 of 59 genes, p < 10−9); in addition, the incidence of changes in differentiating cells was significantly greater than in the undifferentiated state (p < 0.05). In light of the significant ANOVA interaction terms, we separated data according to pathway groups and then performed lower-order tests for each gene.
For the study of diazinon, dieldrin, and Ni2+ conducted in differentiating cells, global ANOVA (factors of treatment, gene, time) identified a main effect of treatment (p < 0.0001) and interactions of treatment × time (p < 0.02), treatment × gene (p < 0.0001), and treatment × time × gene (p < 0.0001). Of the 59 total genes, we found significant differences for 44 (p < 10−15 vs. the predicted false-positive rate). This was also true for each agent considered individually: diazinon, 32 genes, p < 10−10; dieldrin, 26 genes, p < 10−8; Ni2+, 27 genes, p < 10−8. In light of the interactions of treatment with the other variables, we divided the data into the separate treatments for presentation, grouping the genes by pathway and evaluating the effects on each gene.
Effects of chlorpyrifos in undifferentiated cells
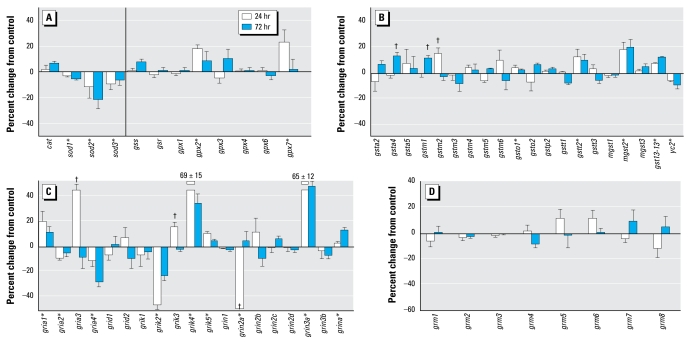
Exposure of undifferentiated PC12 cells to chlorpyrifos for 24 or 72 hr had significant but modest effects on genes mediating anti-oxidant responses and glutathione metabolism. Although cat expression was unaffected, all three sod subtypes showed small but significant down-regulation (Figure 1A). Neither gss nor gsr was affected, but two out of the six gpx genes showed significant up-regulation. The genes encoding the glutathione S-transferases likewise showed statistically significant changes in response to chlorpyrifos exposure, but the magnitude of effect did not exceed 20% (Figure 1B). In general, the main effect was up-regulation (gsta4, gstm1, gstm2, gsto1, gstt2, mgst2, gst13-13), with the exception of yc2, which showed a decrement.
Figure 1.
Effects of 30 μM chlorpyrifos exposure in undifferentiated PC12 cells. (A) Antioxidant genes. (B) Genes encoding the glutathione S-transferases. (C) Genes encoding the ionotropic glutamate receptors. (D) Genes encoding the metabotropic glutamate receptors. The vertical line in (A) separates genes encoding catalase and the superoxide dismutase isoforms from those involved in glutathione synthesis and redox status.
*Significant main treatment effect. †Treatment × time interaction and times for which treatment effects were present. Multivariate ANOVA (treatment, gene, time) indicates interactions of treatment × gene (p < 0.0001) and treatment × gene × time (p< 0.01).
In contrast, we identified widespread and robust effects of chlorpyrifos on the gene family for ionotropic glutamate receptors (Figure 1C). We found consistent increases for gria1, grik4, grik5, grin3a, and grina and persistent decreases for gria2, gria4, and grik2. In addition, some genes showed transient effects, with increases (gria3, grik3) or decreases (grin2a) after 24 hr of exposure that waned by 72 hr. We did not see these effects for the metabotropic glutamate receptors, none of which showed significant effects of chlorpyrifos in undifferentiated cells (Figure 1D).
Antioxidant genes in differentiating cells
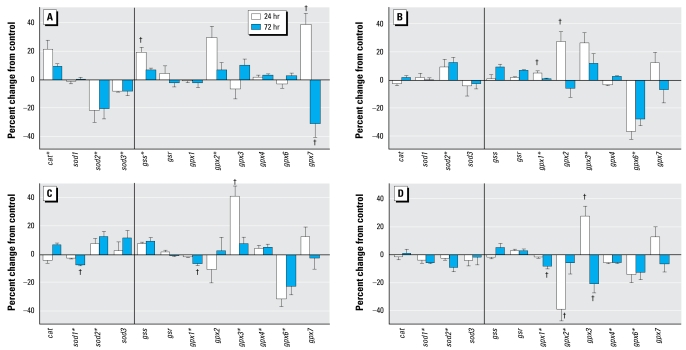
Compared with the effects on undifferentiated PC12 cells, chlorpyrifos exposure during differentiation elicited a greater overall response for the genes involved in antioxidant activity (Figure 2A). Across all these genes, the absolute magnitude of effect was doubled (net 12% change in differentiating cells vs. 6% in undifferentiated cells, p < 0.02). Chlorpyrifos elicited significant up-regulation of cat and transient up-regulation of gss, effects that were not seen in the undifferentiated state, as well as eliciting much larger initial increases in gpx2 and gpx7; for the latter, we found a significant subsequent rebound suppression at 72 hr. Two additional changes resembled those seen in the undifferentiated cells, namely, down-regulation of sod2 and sod3.
Figure 2.
Effects of 30 μM chlorpyrifos (A), diazinon (B), dieldrin (C), and Ni2+ (D) exposure on expression of antioxidant genes in differentiating PC12 cells. The vertical lines separate genes encoding catalase and the superoxide dismutase isoforms from those involved in glutathione synthesis and redox status.
*Significant main treatment effect. †Treatment × time interaction and times for which treatment effects were present. Multivariate ANOVA (all treatments, all genes, time) indicates a significant main effect of treatment (p < 0.02) and interactions of treatment × gene ( p < 0.0001) and treatment × gene × time (p < 0.006).
The response to diazinon in differentiating PC12 cells was distinctly different from that evoked by chlorpyrifos (Figure 2B). Diazinon failed to alter cat, gss, or gpx7 expression; enhanced sod2 instead of suppressing it; and evoked additional changes not seen with chlorpyrifos, namely, increases in gpx1 and gpx3 and decreases in gpx6. The only point of clear overlap between the two agents was for gpx2, which showed the same transient elevation for diazinon and for chlorpyrifos.
Dieldrin evoked some of the same changes as did diazinon, including up-regulation of sod2 and gpx3 and down-regulation of gpx6 (Figure 2C). It differed in that dieldrin reduced sod1, gpx1, and gpx4, leaving gpx2 unaffected. Exposure to Ni2+ produced a different response pattern, with small but significant reductions in sod1, sod2, gpx1, and gpx6 and a larger transient decrease in gpx2 (Figure 2D). The only major point of consonance was for gpx3, which showed the same type of transient increase as seen with diazinon and dieldrin, except that with Ni2+, it was followed by a decrease at 72 hr.
Glutathione S-transferases in differentiating cells
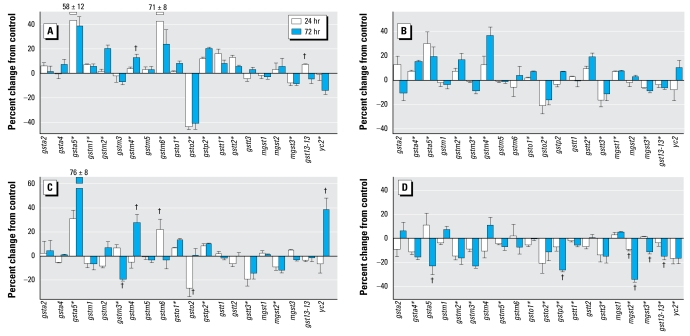
Again, the genes encoding the various glutathione S-transferases showed much greater responses to chlorpyrifos in differentiating PC12 cells (Figure 3A) than in undifferentiated cells, with double the absolute magnitude of change (net 12% in differentiating cells vs. 6% in undifferentiated cells, p < 0.03). Indeed, we found significant changes in expression for 13 of the 18 genes in this group, with increases (10 genes) predominating over decreases (3 genes). The largest changes were seen for gsta5 (increase), gstm6 (increase), and gsto2 (decrease). We found smaller increases for gstm1, gstm2, gstm4, gsto1, gstp2, gstt1, gstt2, and gst13-13; minor but significant decreases were confined to mgst3 and yc2.
Figure 3.
Effects of 30 μM chlorpyrifos (A), diazinon (B), dieldrin (C), and Ni2+ (D) exposure on expression of genes encoding the glutathione S-transferases in differentiating PC12 cells.
*Significant main treatment effect. †Treatment × time interaction and times for which treatment effects were present. Multivariate ANOVA (all treatments, all genes, time) indicates a significant main effect of treatment (p < 0.0001) and interactions of treatment × time ( p < 0.001), treatment × gene (p< 0.0001), and treatment × gene × time (p < 0.0001).
For this set of genes, diazinon exposure altered expression in a pattern somewhat similar to that of chlorpyrifos, albeit with much smaller overall effects on the genes that showed the biggest changes with chlorpyrifos (Figure 3B). Like chlorpyrifos, diazinon evoked up-regulation of gsta5, gstm2, gstm4, gsto1, and gstt2 and down-regulation of gsto2 and mgst3. In addition, diazinon enhanced gsta4 and suppressed gstm3, gstt3, and gst13-13, effects that were not seen with chlorpyrifos, while failing to affect many of the genes that were significantly altered by chlorpyrifos (gstm1, gstm6, gstp2, gstt1, yc2). With dieldrin exposure (Figure 3C), we again saw features shared by chlorpyrifos and/or diazinon, namely, robust up-regulation of gsta5; lesser stimulation of gstm4, gstm6, gsto1, and gstp2; and down-regulation of gstm3, gsto2, and gstt3. We also found features unique to dieldrin: a decrease in mgst2 and an increase in yc2 expression. Although exposure to Ni2+ (Figure 3D) produced some changes in gene expression resembling those of the other three agents (e.g., decreased gsto2 expression), the overall response pattern was distinctly different, with inhibition predominating over stimulation (main treatment effect, p < 0.0001). We found significant decreases for all but 6 of the 20 genes, and no individual gene showed a significant increase.
Glutamate receptors in differentiating cells
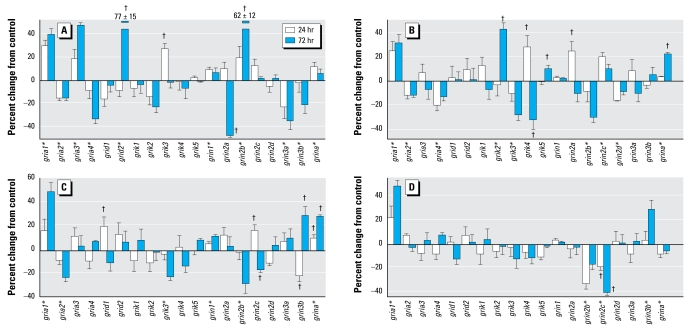
Unlike the situation with the genes involved in oxidative stress, chlorpyrifos exposure in differentiating cells showed approximately the same net absolute response for glutamate receptor genes as in undifferentiated cells (16% change for differentiating cells vs. 12% change for undifferentiated cells; not significant); this was also true even if the comparison was restricted to ionotropic glutamate receptors (18% vs. 16%, respectively), the sub group that participates in organophosphate-induced neuro toxicity (Chebabo et al. 1999; Damodaran et al. 2006; Dekundy et al. 2007; Gupta 2004). However, we found significant differences in the response of specific genes in the differentiating versus undifferentiated states (treatment × state × gene, p < 0.004) and in the time course of effect (treatment × state × gene × time, p < 0.006). Just as in undifferentiated cells, chlorpyrifos exposure during differentiation elicited significant up-regulation of gria1, gria3, grik3, and grina and down-regulation of gria2, gria4, and grin2a (Figure 4A). However, we also found key differences: increases in grid2, grin1, and grin2b and decreases in grin3b restricted to differentiating cells (Figure 4A); increases in grik4 and grik5, and decreases in grik2 restricted to undifferentiated cells (Figure 1C); increases in grin3a in undifferentiated cells (Figure 1C) but decreases for the same gene in differentiating cells (Figure 4A); and differences between undifferentiated (Figure 1C) and differentiating cells (Figure 4A) in the time course of effect for gria3 and grin2a.
Figure 4.
Effects of 30 μM chlorpyrifos (A), diazinon (B), dieldrin (C), and Ni2+ (D) exposure on expression of genes encoding the ionotropic glutamate receptors in differentiating PC12 cells.
*Significant main treatment effect. †Treatment × time interaction and times for which treatment effects were present. Multivariate ANOVA (all treatments, all genes, time) indicates interactions of treatment × time (p < 0.01), treatment × gene ( p < 0.0001), and treatment × gene × time (p < 0.0003).
Like chlorpyrifos, diazinon exposure altered the expression for most of the genes encoding ionotropic glutamate receptors (Figure 4B). However, the patterns of effects showed major disparities between the two organophosphates, with similar changes for only four genes (gria1, gria2, gria4, grina) and dissimilar effects for 11 genes (gria3, grid2, grik2, grik3, grik4, grik5, grin2a, grin2b, grin2d, grin3a, grin3b), even to the extent of changes in the opposite direction. Dieldrin elicited more modest changes, but in general, the pattern was closer to that of diazinon, sharing similar changes for gria1, gria2, grik3, grin2b, grin2c, and grina, as well as a lack of effect on gria3, grid2, grik1, and grin3a (Figure 4C); the two differed for gria4, grid1, grik2, grik4, and grin2a. Although exposure to Ni2+ elicited the same increase in gria1 seen with the other three agents, in general it affected a much smaller repertoire of genes, with only five significant changes among the total of 19 (Figure 4D). Like diazinon or dieldrin, Ni2+ reduced the expression of grin2b, an effect opposite that of chlorpyrifos, but Ni2+ also affected grin2c (decrease), grina (decrease), and grin3b (increase) in a manner distinct from one or more of the other agents.
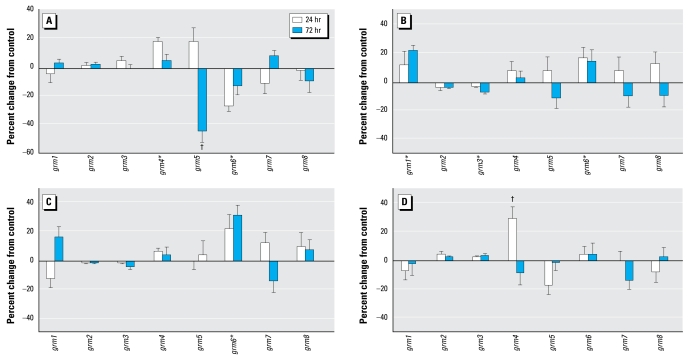
Although chlorpyrifos exposure in undifferentiated PC12 cells did not alter the expression of genes encoding the metabotropic glutamate receptors, differentiating cells were more sensitive, displaying changes for three of the eight subtypes (Figure 5A): increased grm4 and decreased grm5 and grm6. The effects of diazinon were totally different, with increases in grm1 and grm6 and a small but significant decrease in grm3 (Figure 5B). Dieldrin produced even less of an effect, restricted to an increase in grm6 (Figure 5C). Likewise, Ni2+ evoked only a transient change in grm4 expression (Figure 5D). Taken across all four agents, the incidence of changes in metabotropic glutamate receptor genes (25%) was much lower than for ionotropic receptors (50%, p< 0.01 by Fisher’s exact test).
Figure 5.
Effects of 30 μM chlorpyrifos (A), diazinon (B), dieldrin (C), and Ni2+ (D) exposure on expression of genes encoding the metabotropic glutamate receptors in differentiating PC12 cells.
*Significant main treatment effect. †Treatment × time interaction and times for which treatment effects were present. Multivariate ANOVA (all treatments, all genes, time) indicates interactions of treatment × time (p < 0.05) and treatment × gene ( p < 0.02).
Discussion
In earlier work with the PC12 cell model, we showed that lipid peroxidation evoked by chlorpyrifos was enhanced by coexposure to NGF (Qiao et al. 2005), consistent with increased sensitivity to oxidative stress during neuro differentiation. We confirmed this by studies conducted in vivo with chlorpyrifos administered to neonatal rats, which likewise evoked greater lipid peroxidation in vulnerable brain regions during peak periods of axonogenesis and synapto genesis (Slotkin et al. 2005). In the present study, we found much larger and widespread transcriptional changes elicited by chlorpyrifos exposure in differentiating PC12 cells compared with undifferentiated cells, for genes involved in the oxidative stress response (cat, sod, gss, gsr, gpx, gst, mgst, yc2; including all subtypes); the correspondence of the changes in gene expression with the functional end point of lipid peroxidation thus serves as a validation of this approach. Further, genes encoding the glutamate receptors did not show a corresponding net increase in chlorpyrifos effects during differentiation, demonstrating the specificity for those elements directly involved in mustering anti oxidant defenses; we found selective differences for individual receptor genes that depended on differentiation state, but the overall magnitude of effect was no greater in differentiating cells compared with undifferentiated cells, unlike the situation for anti-oxidant genes. Indeed, for undifferentiated cells, the effect of chlorpyrifos on ionotropic glutamate receptor genes was far more robust than for the genes involved in oxidative stress; further, this was not true for metabotropic glutamate receptors, which are not involved in excitotoxicity. Our results thus suggest a greater role for excitotoxicity than for oxidative stress in the undifferentiated state (i.e., earlier stages of neurodevelopment) but an increasing role for oxidative stress as cells undergo differentiation (later stages of neurodevelopment). The comparative effects of chlorpyrifos in the two states thus reinforce the idea that both oxidative stress and excitotoxicity are likely contributors that define the critical windows in which specific neuronal populations in the developing brain are most vulnerable to different aspects of chlorpyrifos neurotoxicity (Garcia et al. 2005; Slotkin 1999, 2004, 2005).
In differentiating cells, both chlorpyrifos and diazinon produced widespread changes in the expression of genes involved in oxidative stress. Although we found a significant correlation between the effects on these genes for the two organophosphates (Table 2), the relatively modest correlation coefficient (r = 0.40) points to substantial differences, as well. In fact, the effects on the primary anti oxidant genes—cat and the sod subtypes—were totally dissimilar, as evidenced by a strong negative correlation between chlorpyrifos and diazinon (Table 2); this reflected not only an opposite direction of change for sod2 but also a substantially greater overall effect of chlorpyrifos across this set of genes (average change 11% for chlor pyrifos vs. 4% for diazinon, p < 0.02). These results point to the likelihood that chlorpyrifos either produces a greater degree of oxidative stress or, through its other mechanisms of action, exacerbates the net effect of oxidative stress. In contrast, our results for the glutathione-related genes suggest a similar outcome from the two agents, evidenced by a significant positive correlation (Table 2); the larger number of glutathione-related genes also accounts for the overall concordance between chlorpyrifos and diazinon across all the oxidative-stress–related genes and points to why examination of subdivisions is important to delineate different mechanisms and potential outcomes. Indeed, despite the positive correlation for the glutathione-related genes, individual components in that set displayed markedly different effects of the two agents (e.g., gpx6 and gstm6).
Table 2.
Concordance between test agents.
| Chlorpyrifos vs. diazinon | Chlorpyrifos vs. dieldrin | Chlorpyrifos vs. Ni2+ | Diazinon vs. dieldrin | Diazinon vs. Ni2+ | Dieldrin vs Ni2+ | |
|---|---|---|---|---|---|---|
| All oxidative stress- and glutathione-related genesa | r = 0.40 | r = 0.37 | r = 0.11 | r = 0.62 | r = 0.20 | r = 0.30 |
| p < 0.001 | p < 0.003 | NS | p < 0.0001 | NS | p < 0.02 | |
| cat, sod | r = −0.66 | r = −0.62 | r = 0.53 | r = 0.51 | r = −0.52 | r = −0.08 |
| p < 0.05 | p < 0.05 | NS | NS | NS | NS | |
| All glutathione-related genes | r = 0.46 | r = 0.43 | r = 0.13 | r = 0.63 | r = 0.22 | r = 0.31 |
| p < 0.0004 | p < 0.002 | NS | p < 0.0001 | NS | p < 0.03 | |
| All glutamate receptor genes | r = 0.02 | r = 0.02 | r = 0.03 | r = 0.58 | r = 0.23 | r = 0.56 |
| NS | NS | NS | p < 0.0001 | NS | p < 0.0001 | |
| Ionotropic glutamate receptors | r = 0.05 | r = 0.06 | r = 0.02 | r = 0.59 | r = 0.25 | r = 0.60 |
| NS | NS | NS | p < 0.0001 | NS | p < 0.0001 | |
| Metabotropic glutamate receptors | r = 0.09 | r = −0.37 | r = −0.02 | r = 0.60 | r = 0.05 | r = 0.31 |
| NS | NS | NS | p < 0.02 | NS | NS | |
| All genes | r = 0.20 | r = 0.18 | r = 0.04 | r = 0.60 | r = 0.21 | r = 0.42 |
| p < 0.04 | p < 0.05 | NS | p < 0.0001 | p < 0.03 | p < 0.0001 | |
NS, not significant.
All genes in the cat, sod, gss, gsr, gpx, and gst families.
The much more widespread and robust effects of chlorpyrifos on expression of ion-otropic glutamate receptors compared with metabotropic receptors in differentiating PC12 cells reflects the specific involvement of these receptors in the excitotoxicity associated with organophosphate-induced neuronal injury (Damodaran et al. 2006; Gupta 2004) and are similar to results reported previously for effects on the developing rat brain after neonatal chlorpyrifos exposure (Slotkin and Seidler 2007). Here, although diazinon likewise produced widespread effects on ionotropic glutamate receptors, the overall pattern was totally distinct from that of chlorpyrifos, evidenced by a lack of concordance whether considering each receptor class separately or together (Table 2). Again, this is consistent with major differences between the two agents observed for expression of the same receptor genes after in vivo exposures (Slotkin and Seidler 2007). One likely reason is that chlor pyrifos may interact directly with ion channel receptors that gate calcium entry (Katz et al. 1997; Smulders et al. 2004). Because this mechanism obviously reflects actions unrelated to the shared property of cholinesterase inhibition, the structural differences between chlorpyrifos and diazinon could clearly contribute to dissimilar effects on ionotropic glutamate receptor function, leading to disparities in receptor expression.
Perhaps the most surprising result is the close similarity between the outcomes of exposure to dieldrin and those of diazinon. We found high concordance between these two agents across the oxidative-stress–related genes as well as the glutamate receptor genes, resulting in a substantially higher correlation across all genes than for chlorpyrifos and diazinon, the two organophosphates (Table 2). This was also reflected in the lesser correlations between chlorpyrifos and dieldrin, which were just as robust as those between chlorpyrifos and diazinon (Table 2). In our earlier work with PC12 cells, we similarly found convergent outcomes for diazinon and dieldrin on lipid peroxidation and on differentiation end points, including indices of neurite formation and neurotransmitter phenotype (Slotkin et al. 2007b; Slotkin and Seidler 2008, 2009). Our present findings can thus guide future studies of in vivo dieldrin exposure to confirm the prediction that this agent will produce developmental neurotoxicity akin to that of diazinon.
Ni2+ does not evoke oxidative stress (Slotkin et al. 2007b), and accordingly, this agent elicited changes in gene expression that were distinct from those of the organophosphates. We found no significant concordance in the outcomes for chlorpyrifos compared with Ni2+ and only a weak overall correlation for diazinon and Ni2+ that did not achieve significance for any subset of genes (Table 2). The difference between Ni2+ and the pesticides is best illustrated by comparing effects on the glutathione S-transferases, where the metal produced widespread down-regulation compared with the predominance of up-regulation for three pesticides. Further, the fact that all four agents (including Ni2+) down-regulated gsto2 indicates that this particular transcriptional change is probably not involved in the response to oxidative stress per se. We found a significant overall concordance for dieldrin and Ni2+, primarily reflecting their shared actions on glutamate receptors. Interestingly, although we found concordance between dieldrin and Ni2+ and between diazinon and dieldrin for the ionotropic glutamate receptors, the correlation was much poorer between dieldrin and Ni2+; this points out that the concordance patterns between any two agents can involve sets of genes different from those generating the concordance between one of those agents and a third compound. In any case, the unexpected similarities between apparently unrelated neurotoxicants in their effects on ionotropic glutamate receptors points to future experiments on the potential for convergent underlying mechanisms and outcomes involving excitotoxicity.
The limitations and advantages of the combination of in vitro model and planned comparison approaches to micro array data have been detailed in earlier work (Slotkin et al. 2007c, 2008d, 2008e; Slotkin and Seidler 2007, 2008) but are worth repeating in relation to the present study. We used cells from a transformed cell line, which, unlike primary neurons in culture, maintain their ability to divide, an important consideration when, as here, the neurotoxicants target the cell cycle as part of their injury pattern. Nevertheless, transformed cells are inherently less sensitive to toxicant injury than are developing neurons in vivo. Further, cell culture treatments involve much shorter durations than with environmental exposures extending throughout brain development. We considered both of these factors in our selection of the 30 μM test concentrations. In the case of the organophosphates, this is approximately an order of magnitude higher than the levels in newborn babies after nonsymptomatic environmental exposures in agricultural communities (Ostrea et al. 2002); however, the cultures contain high concentrations of serum proteins, so < 10% of the nominal concentration is actually available to diffuse into the cells (Qiao et al. 2001). The most important proof of relevance, however, is that for chlorpyrifos and diazinon, parallel outcomes have been identified between in vivo exposures and the PC12 model (Bagchi et al. 1995, 1996; Crumpton et al. 2000a, 2000b; Das and Barone 1999; Flaskos et al. 1994; Jameson et al. 2006, 2007; Li and Casida 1998; Nagata et al. 1997; Qiao et al. 2001, 2005; Slotkin et al. 2007a, 2007b, 2008d, 2008e; Song et al. 1998; Tuler et al. 1989), thus providing validation of the in vitro approach. The second factor in these studies is our use of planned comparisons, a distinctly different approach from the global examination of the tens of thousands of genes present on the microarrays. Planned comparisons are based on testing a specific hypothesis that centers around a defined set of gene families and rests on known, validated outcomes from prior work, in this case involving both in vivo and in vitro demonstrations of oxidative stress from the organophosphates. With examination of the entire genome, verification via reverse transcriptase polymerase chain reaction and other techniques is required because the enormous number of comparisons generates numerous false-positive findings (e.g., the > 2,000 genes that would be false positives if we had considered all 42,000 probes on the array). For our study, we compared only a handful of genes that would generate only three false positives, and we found alterations in most of these genes; for interpretation, we relied on multiple gene changes in a given pathway, as well as effects that repeated across different treatments and/or different time points, rather than changes in any one gene. The odds of all those genes being false positives are astronomically small. However, even for individual genes, a given array used multiple probes and multiple spots (see “Materials and Methods”), so the changes cannot be “chance.” Unlike many array studies, where a single mRNA set derived from multiple samples might be evaluated, we evaluated up to eight separate samples for each treatment condition, so again, it is inconceivable statistically that we could produce these outcomes by accident. Indeed, one of the key points of this study is to demonstrate that a planned comparison approach may provide a superior strategy for the use of microarray data, provided that the relevant target pathways are known in advance.
In conclusion, our findings reinforce the growing body of evidence that the various organophosphates differ in their underlying mechanisms of developmental neurotoxicity, over and above their shared property as cholinesterase inhibitors, culminating in distinct outcomes at the levels of synaptic function (Jameson et al. 2007; Qiao et al. 2001; Roegge et al. 2008; Slotkin et al. 2006a, 2006b, 2007b, 2007c, 2008a, 2008b, 2008c, 2008d; Slotkin and Seidler 2007; Timofeeva et al. 2008a, 2008b). As shown here, these mechanisms are likely to include selective effects on oxidative stress and excitotoxicity, as well as enhanced vulnerability to oxidative stress during a critical period centered around differentiation into neurotransmitter phenotypes and the development of neuritic projections. At the same time, the concordance of overall effects between diazinon and dieldrin, and for effects of dieldrin and Ni2+ on glutamate receptors, indicate that agents from apparently unrelated classes of toxicants can nonetheless converge on common final outcomes, despite differences in underlying targets or originating mechanisms. Finally, the results obtained here illustrate how a combined use of cell culture systems and microarrays can guide future studies toward specific end points that can distinguish similarities and disparities in the effects of diverse developmental neurotoxicants.
Footnotes
T.A.S. has provided expert witness testimony in the past 3 years at the behest of the following law firms: The Calwell Practice (Charleston, WV), Frost Brown Todd (Charleston, WV), Weiner & Weltchek (Lutherville, MD), Frommer Lawrence Haug (Washington, DC), Carter Law (Peoria, IL), Corneille Law (Madison, WI), Angelos Law (Baltimore, MD), and Kopff, Nardelli & Dopf (New York, NY). F.J.S. has provided expert witness testimony on behalf of government agencies, corporations, and/or individuals.
This research was supported by grant ES10356 from the National Institutes of Health.
References
- Bagchi D, Bagchi M, Hassoun EA, Stohs SJ. In vitro and in vivo generation of reactive oxygen species, DNA damage and lactate dehydrogenase leakage by selected pesticides. Toxicology. 1995;104:129–140. doi: 10.1016/0300-483x(95)03156-a. [DOI] [PubMed] [Google Scholar]
- Bagchi D, Bhattacharya G, Stohs SJ. In vitro and in vivo induction of heat shock (stress) protein (Hsp) gene expression by selected pesticides. Toxicology. 1996;112:57–68. doi: 10.1016/0300-483x(96)03350-1. [DOI] [PubMed] [Google Scholar]
- Barr DB, Allen R, Olsson AO, Bravo R, Caltabiano LM, Montesano A, et al. Concentrations of selective metabolites of organophosphorus pesticides in the United States population. Environ Res. 2005;99:314–326. doi: 10.1016/j.envres.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Benters J, Schafer T, Beyersmann D, Hechtenberg S. Agonist-stimulated calcium transients in PC12 cells are affected differentially by cadmium and nickel. Cell Calcium. 1996;20:441–446. doi: 10.1016/s0143-4160(96)90007-x. [DOI] [PubMed] [Google Scholar]
- Bouvier G, Seta N, Vigouroux-Villard A, Blanchard O, Momas I. Insecticide urinary metabolites in nonoccupationally exposed populations. J Toxicol Environ Health. 2005;8:485–512. doi: 10.1080/10937400591007284. [DOI] [PubMed] [Google Scholar]
- Brannen KC, Devaud LL, Liu JP, Lauder JM. Prenatal exposure to neurotoxicants dieldrin or lindane alters tert-butyl-bicyclophosphorothionate binding to GABA(A) receptors in fetal rat brainstem. Dev Neurosci. 1998;20:34–41. doi: 10.1159/000017296. [DOI] [PubMed] [Google Scholar]
- Casey CE, Robinson MF. Copper, manganese, zinc, nickel, cadmium and lead in human foetal tissues. Br J Nutr. 1978;39:639–646. doi: 10.1079/bjn19780079. [DOI] [PubMed] [Google Scholar]
- Casida JE, Quistad GB. Organophosphate toxicology: safety aspects of nonacetylcholinesterase secondary targets. Chem Res Toxicol. 2004;17:983–998. doi: 10.1021/tx0499259. [DOI] [PubMed] [Google Scholar]
- Chebabo SR, Santos MD, Albuquerque EX. The organophosphate sarin, at low concentrations, inhibits the evoked release of GABA in rat hippocampal slices. Neurotoxicology. 1999;20:871–882. [PubMed] [Google Scholar]
- Choi DW, Rothman SM. The role of glutamate neurotoxicity in hypoxic-ischemic neuronal death. Annu Rev Neurosci. 1990;13:171–182. doi: 10.1146/annurev.ne.13.030190.001131. [DOI] [PubMed] [Google Scholar]
- Crumpton TL, Seidler FJ, Slotkin TA. Developmental neurotoxicity of chlorpyrifos in vivo and in vitro: effects on nuclear transcription factor involved in cell replication and differentiation. Brain Res. 2000a;857:87–98. doi: 10.1016/s0006-8993(99)02357-4. [DOI] [PubMed] [Google Scholar]
- Crumpton TL, Seidler FJ, Slotkin TA. Is oxidative stress involved in the developmental neurotoxicity of chlorpyrifos? Dev Brain Res. 2000b;121:189–195. doi: 10.1016/s0165-3806(00)00045-6. [DOI] [PubMed] [Google Scholar]
- Damodaran TV, Patel AG, Greenfield ST, Dressman HK, Lin SM, Abou-Donia MB. Gene expression profiles of the rat brain both immediately and 3 months following acute sarin exposure. Biochem Pharmacol. 2006;71:497–520. doi: 10.1016/j.bcp.2005.10.051. [DOI] [PubMed] [Google Scholar]
- Das KP, Barone S. Neuronal differentiation in PC12 cells is inhibited by chlorpyrifos and its metabolites: is acetyl-cholinesterase inhibition the site of action? Toxicol Appl Pharmacol. 1999;160:217–230. doi: 10.1006/taap.1999.8767. [DOI] [PubMed] [Google Scholar]
- Dekundy A, Kaminski RM, Zielinska E, Turski WA. NMDA antagonists exert distinct effects in experimental organo-phosphate or carbamate poisoning in mice. Toxicol Appl Pharmacol. 2007;219:114–121. doi: 10.1016/j.taap.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Faber JJ, Anderson DF, Morton MJ, Parks CM, Pinson CW, Thornburg KL, et al. Birth, its physiology, and the problems it creates. In: Jones CT, Nathanielsz PW, editors. The Physiological Development of the Fetus and Newborn. London: Academic Press; 1985. pp. 371–380. [Google Scholar]
- Flaskos J, McLean WG, Hargreaves AJ. The toxicity of organophosphate compounds towards cultured PC12 cells. Toxicol Lett. 1994;70:71–76. doi: 10.1016/0378-4274(94)90146-5. [DOI] [PubMed] [Google Scholar]
- Fujita K, Lazarovici P, Guroff G. Regulation of the differentiation of PC12 pheochromocytoma cells. Environ Health Perspect. 1989;80:127–142. doi: 10.1289/ehp.8980127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia SJ, Seidler FJ, Slotkin TA. Developmental neurotoxicity of chlorpyrifos: targeting glial cells. Environ Toxicol Pharmacol. 2005;19:455–461. doi: 10.1016/j.etap.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Giordano G, Afsharinejad Z, Guizzetti M, Vitalone A, Kavanagh TJ, Costa LG. Organophosphorus insecticides chlorpyrifos and diazinon and oxidative stress in neuronal cells in a genetic model of glutathione deficiency. Toxicol Appl Pharmacol. 2007;219:181–189. doi: 10.1016/j.taap.2006.09.016. [DOI] [PubMed] [Google Scholar]
- Gitto E, Reiter RJ, Karbownik M, Tan DX, Gitto P, Barberi S, et al. Causes of oxidative stress in the pre- and perinatal period. Biol Neonate. 2002;81:146–157. doi: 10.1159/000051527. [DOI] [PubMed] [Google Scholar]
- Gupta RC. Brain regional heterogeneity and toxicological mechanisms of organophosphates and carbamates. Toxicol Mech Methods. 2004;14:103–143. doi: 10.1080/15376520490429175. [DOI] [PubMed] [Google Scholar]
- Henderson G, Chen J, Schenker S. Ethanol, oxidative stress, reactive aldehydes and the fetus. Front Biosci. 1999;4:541–550. doi: 10.2741/henderson. [DOI] [PubMed] [Google Scholar]
- Jameson RR, Seidler FJ, Qiao D, Slotkin TA. Chlorpyrifos affects phenotypic outcomes in a model of mammalian neurodevelopment: critical stages targeting differentiation in PC12 cells. Environ Health Perspect. 2006;114:667–672. doi: 10.1289/ehp.8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson RR, Seidler FJ, Slotkin TA. Nonenzymatic functions of acetylcholinesterase splice variants in the developmental neurotoxicity of organophosphates: chlorpyrifos, chlorpyrifos oxon and diazinon. Environ Health Perspect. 2007;115:65–70. doi: 10.1289/ehp.9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jett DA, Navoa RV. In vitro and in vivo effects of chlorpyrifos on glutathione peroxidase and catalase in developing rat brain. Neurotoxicology. 2000;21:141–145. [PubMed] [Google Scholar]
- Katz EJ, Cortes VI, Eldefrawi ME, Eldefrawi AT. Chlorpyrifos, parathion, and their oxons bind to and desensitize a nicotinic acetylcholine receptor: relevance to their toxicities. Toxicol Appl Pharmacol. 1997;146:227–236. doi: 10.1006/taap.1997.8201. [DOI] [PubMed] [Google Scholar]
- Kern JK, Jones AM. Evidence of toxicity, oxidative stress, and neuronal insult in autism. J Toxicol Environ Health B. 2006;9:485–499. doi: 10.1080/10937400600882079. [DOI] [PubMed] [Google Scholar]
- Kitazawa M, Anantharam V, Kanthasamy AG. Dieldrin-induced oxidative stress and neurochemical changes contribute to apoptopic cell death in dopaminergic cells. Free Radical Biol Med. 2001;31:1473–1485. doi: 10.1016/s0891-5849(01)00726-2. [DOI] [PubMed] [Google Scholar]
- Kitazawa M, Anantharam V, Kanthasamy AG. Dieldrin induces apoptosis by promoting caspase-3-dependent proteolytic cleavage of protein kinase Cδ in dopaminergic cells: relevance to oxidative stress and dopaminergic degeneration. Neuroscience. 2003;119:945–964. doi: 10.1016/s0306-4522(03)00226-4. [DOI] [PubMed] [Google Scholar]
- Lagercrantz H, Slotkin TA. The “stress” of being born. Sci Am. 1986;254:100–107. doi: 10.1038/scientificamerican0486-100. [DOI] [PubMed] [Google Scholar]
- Li WW, Casida JE. Organophosphorus neuropathy target esterase inhibitors selectively block outgrowth of neurite-like and cell processes in cultured cells. Toxicol Lett. 1998;98:139–146. doi: 10.1016/s0378-4274(98)00116-7. [DOI] [PubMed] [Google Scholar]
- Liu JP, Brannen KC, Grayson DR, Morrow AL, Devaud LL, Lauder JM. Prenatal exposure to the pesticide dieldrin or the GABA(A) receptor antagonist bicuculline differentially alters expression of GABA(A) receptor subunit mRNAs in fetal rat brainstem. Dev Neurosci. 1998;20:83–92. doi: 10.1159/000017302. [DOI] [PubMed] [Google Scholar]
- Mulholland PJ, Stepanyan TD, Self RL, Hensley AK, Harris BR, Kowalski A, et al. Corticosterone and dexamethasone potentiate cytotoxicity associated with oxygen- glucose deprivation in organotypic cerebellar slice cultures. Neuroscience. 2005;136:259–267. doi: 10.1016/j.neuroscience.2005.07.043. [DOI] [PubMed] [Google Scholar]
- Nagata K, Huang CS, Song JH, Narahashi T. Direct actions of anticholinesterases on the neuronal nicotinic acetylcholine receptor channels. Brain Res. 1997;769:211–218. doi: 10.1016/s0006-8993(97)00707-5. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information. GenBank Overview. 2008. [[accessed 27 February 2009]]. Available: http://www.ncbi.nlm.nih.gov/Genbank/
- National Library of Medicine. Superfund Chemicals in TOXMAP. 2006. [[accessed 9 August 2006]]. Available: http://toxmap.nlm.nih.gov/toxmap/main/sfChemicals.jsp.
- Nikodijevic B, Guroff G. Nerve growth factor-stimulated calcium uptake into PC12 cells: uniqueness of the channel and evidence for phosphorylation. J Neurosci Res. 1992;31:591–599. doi: 10.1002/jnr.490310402. [DOI] [PubMed] [Google Scholar]
- Ohtsuka K, Suzuki T. Roles of molecular chaperones in the nervous system. Brain Res Bull. 2000;53:141–146. doi: 10.1016/s0361-9230(00)00325-7. [DOI] [PubMed] [Google Scholar]
- Olanow CW, Arendash GW. Metals and free radicals in neurodegeneration. Curr Opin Neurol. 1994;7:548–558. doi: 10.1097/00019052-199412000-00013. [DOI] [PubMed] [Google Scholar]
- Ostrea EM, Morales V, Ngoumgna E, Prescilla R, Tan E, Hernandez E, et al. Prevalence of fetal exposure to environmental toxins as determined by meconium analysis. Neurotoxicology. 2002;23(3):329–339. doi: 10.1016/s0161-813x(02)00077-3. [DOI] [PubMed] [Google Scholar]
- Pardo CA, Eberhart CG. The neurobiology of autism. Brain Pathol. 2007;17:434–447. doi: 10.1111/j.1750-3639.2007.00102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope CN. Organophosphorus pesticides: do they all have the same mechanism of toxicity? J Toxicol Environ Health. 1999;2:161–181. doi: 10.1080/109374099281205. [DOI] [PubMed] [Google Scholar]
- Qiao D, Seidler FJ, Slotkin TA. Developmental neurotoxicity of chlorpyrifos modeled in vitro: comparative effects of metabolites and other cholinesterase inhibitors on DNA synthesis in PC12 and C6 cells. Environ Health Perspect. 2001;109:909–913. doi: 10.1289/ehp.01109909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao D, Seidler FJ, Slotkin TA. Oxidative mechanisms contributing to the developmental neurotoxicity of nicotine and chlorpyrifos. Toxicol Appl Pharmacol. 2005;206:17–26. doi: 10.1016/j.taap.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Qiao D, Seidler FJ, Violin JD, Slotkin TA. Nicotine is a developmental neurotoxicant and neuroprotectant: stage-selective inhibition of DNA synthesis coincident with shielding from effects of chlorpyrifos. Dev Brain Res. 2003;147:183–190. doi: 10.1016/s0165-3806(03)00222-0. [DOI] [PubMed] [Google Scholar]
- Roegge CS, Timofeeva OA, Seidler FJ, Slotkin TA, Levin ED. Developmental diazinon neurotoxicity in rats: later effects on emotional response. Brain Res Bull. 2008;75:166–172. doi: 10.1016/j.brainresbull.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha C, Seth K, Islam F, Chaturvedi RK, Shukla S, Mathur N, et al. Behavioral and neurochemical effects induced by pyrethroid-based mosquito repellent exposure in rat offsprings during prenatal and early postnatal period. Neurotoxicol Teratol. 2006;28:472–481. doi: 10.1016/j.ntt.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Slotkin TA. Developmental cholinotoxicants: nicotine and chlorpyrifos. Environ Health Perspect. 1999;107(suppl 1):71–80. doi: 10.1289/ehp.99107s171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA. Cholinergic systems in brain development and disruption by neurotoxicants: nicotine, environmental tobacco smoke, organophosphates. Toxicol Appl Pharmacol. 2004;198:132–151. doi: 10.1016/j.taap.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Slotkin TA. Developmental neurotoxicity of organophosphates: a case study of chlorpyrifos. In: Gupta RC, editor. Toxicity of Organophosphate and Carbamate Pesticides. San Diego: Elsevier; Academic Press; 2005. pp. 293–314. [Google Scholar]
- Slotkin TA, Bodwell BE, Levin ED, Seidler FJ. Neonatal exposure to low doses of diazinon: long-term effects on neural cell development and acetylcholine systems. Environ Health Perspect. 2008a;116:340–348. doi: 10.1289/ehp.11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Bodwell BE, Ryde IT, Levin ED, Seidler FJ. Exposure of neonatal rats to parathion elicits sex-selective impairment of acetylcholine systems in brain regions during adolescence and adulthood. Environ Health Perspect. 2008b;116:1308–1314. doi: 10.1289/ehp.11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Levin ED, Seidler FJ. Comparative developmental neurotoxicity of organophosphate insecticides: effects on brain development are separable from systemic toxicity. Environ Health Perspect. 2006a;114:746–751. doi: 10.1289/ehp.8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Levin ED, Seidler FJ. Developmental neurotoxicity of parathion: progressive effects on serotonergic systems in adolescence and adulthood. Neurotoxicol Teratol. 2009;31:11–17. doi: 10.1016/j.ntt.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, MacKillop EA, Ryde IT, Seidler FJ. Ameliorating the developmental neurotoxicity of chlorpyrifos: a mechanisms-based approach in PC12 cells. Environ Health Perspect. 2007a;115:1306–1313. doi: 10.1289/ehp.10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, MacKillop EA, Ryde IT, Tate CA, Seidler FJ. Screening for developmental neurotoxicity using PC12 cells: comparisons of organophosphates with a carbamate, an organochlorine and divalent nickel. Environ Health Perspect. 2007b;115:93–101. doi: 10.1289/ehp.9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Oliver CA, Seidler FJ. Critical periods for the role of oxidative stress in the developmental neurotoxicity of chlorpyrifos and terbutaline, alone or in combination. Dev Brain Res. 2005;157:172–180. doi: 10.1016/j.devbrainres.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Slotkin TA, Ryde IT, Levin ED, Seidler FJ. Developmental neurotoxicity of low-dose diazinon exposure of neonatal rats: effects on serotonin systems in adolescence and adulthood. Brain Res Bull. 2008c;75:640–647. doi: 10.1016/j.brainresbull.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ. Comparative developmental neurotoxicity of organophosphates in vivo: transcriptional responses of pathways for brain cell development, cell signaling, cytotoxicity and neurotransmitter systems. Brain Res Bull. 2007;72:232–274. doi: 10.1016/j.brainresbull.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ. Developmental neuro toxicants target neurodifferentiation into the serotonin phenotype: chlorpyrifos, diazinon, dieldrin and divalent nickel. Toxicol Appl Pharmacol. 2008;233(2):211–219. doi: 10.1016/j.taap.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ. Transcriptional profiles reveal similarities and differences in the effects of developmental neurotoxicants on differentiation into neurotransmitter phenotypes in PC12 cells. Brain Res Bull. 2009;78:211–225. doi: 10.1016/j.brainresbull.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ, Fumagalli F. Exposure to organophosphates reduces the expression of neurotrophic factors in neonatal rat brain regions: similarities and differences in the effects of chlorpyrifos and diazinon on the fibroblast growth factor superfamily. Environ Health Perspect. 2007c;115:909–916. doi: 10.1289/ehp.9901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ, Fumagalli F. Targeting of neurotrophic factors, their receptors, and signaling pathways in the developmental neurotoxicity of organophosphates in vivo and in vitro. Brain Res Bull. 2008d;76:424–438. doi: 10.1016/j.brainresbull.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ, Fumagalli F. Neurotoxicol Teratol. 2008e. Unrelated developmental neurotoxicants elicit similar transcriptional profiles for effects on neurotrophic factors and their receptors in an in vitro model. [Online 13 December 2008] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Tate CA, Ryde IT, Levin ED, Seidler FJ. Organophosphate insecticides target the serotonergic system in developing rat brain regions: disparate effects of diazinon and parathion at doses spanning the threshold for cholinesterase inhibition. Environ Health Perspect. 2006b;114:1542–1546. doi: 10.1289/ehp.9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smulders CJ, Bueters TJ, Vailati S, van Kleef RG, Vijverberg HP. Block of neuronal nicotinic acetylcholine receptors by organophosphate insecticides. Toxicol Sci. 2004;82:545–554. doi: 10.1093/toxsci/kfh269. [DOI] [PubMed] [Google Scholar]
- Song X, Violin JD, Seidler FJ, Slotkin TA. Modeling the developmental neurotoxicity of chlorpyrifos in vitro: macromolecule synthesis in PC12 cells. Toxicol Appl Pharmacol. 1998;151:182–191. doi: 10.1006/taap.1998.8424. [DOI] [PubMed] [Google Scholar]
- Tanaka J, Toku K, Zhang B, Isihara K, Sakanaka M, Maeda N. Astrocytes prevent neuronal death induced by reactive oxygen and nitrogen species. Glia. 1999;28:85–96. doi: 10.1002/(sici)1098-1136(199911)28:2<85::aid-glia1>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Tata DA, Yamamoto BK. Interactions between methamphetamine and environmental stress: role of oxidative stress, glutamate and mitochondrial dysfunction. Addiction. 2007;102:49–60. doi: 10.1111/j.1360-0443.2007.01770.x. [DOI] [PubMed] [Google Scholar]
- Teng KK, Greene LA. Cultured PC12 cells: a model for neuronal function and differentiation. In: Celis JE, editor. Cell Biology: A Laboratory Handbook. San Diego, CA: Academic Press; 1994. pp. 218–224. [Google Scholar]
- Timofeeva OA, Roegge CS, Seidler FJ, Slotkin TA, Levin ED. Persistent cognitive alterations in rats after early postnatal exposure to low doses of the organophosphate pesticide, diazinon. Neurotoxicol Teratol. 2008a;30:38–45. doi: 10.1016/j.ntt.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofeeva OA, Sanders D, Seemann K, Yang L, Hermanson D, Regenbogen S, et al. Persistent behavioral alterations in rats neonatally exposed to low doses of the organophosphate pesticide, parathion. Brain Res Bull. 2008b;77(6):404–111. doi: 10.1016/j.brainresbull.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuler SM, Hazen AA, Bowen JM. Release and metabolism of dopamine in a clonal line of pheochromocytoma (PC12) cells exposed to fenthion. Fundam Appl Toxicol. 1989;13:484–492. doi: 10.1016/0272-0590(89)90284-4. [DOI] [PubMed] [Google Scholar]
- U.S. EPA. Chlorpyrifos: Re-evaluation Report of the FQPA Safety Factor Committee HED Doc No 014077. Washington, DC: U.S. Environmental Protection Agency; 2000. [Google Scholar]
- U.S EPA (U.S. Environmental Protection Agency) Chlorpyrifos: End-Use Products Cancellation Order. 2002. [[accessed 6 December 2004]]. Available: http://www.epa.gov/fedrgstr/EPA-PEST/2002/January/Day-25/p1764.htm.
- U.S EPA (U.S. Environmental Protection Agency) Opportunities to Improve Data Quality and Children’s Health through the Food Quality Protection Act. Report no. 2006-P-00009. 2006. [[accessed 7 July 2006]]. Available: http://www.epa.gov/oig/reports/2006/20060110-2006-P-00009.pdf.
- Uzoukwu M, Sleight SD. Dieldrin toxicosis: fetotoxicosis, tissue concentrations, and microscopic and ultrastructural changes in guinea pigs. Am J Vet Res. 1972;33:579–583. [PubMed] [Google Scholar]