Abstract
Background
The semicircular canals, a subdivision of the vestibular system of the vertebrate inner ear, function as sensors of angular acceleration. Little is currently known, however, regarding the underlying molecular mechanisms that govern the development of this intricate structure. Zebrafish represent a particularly tractable model system for the study of inner ear development. This is because the ear can be easily visualized during early embryogenesis, and both forward and reverse genetic techniques are available that can be applied to the discovery of novel genes that contribute to proper ear development. We have previously shown that in zebrafish, the calcium sensing molecule neuronal calcium sensor-1 (NCS-1) is required for semicircular canal formation. The function of NCS-1 in regulating semicircular canal formation has not yet been elucidated.
Results
We initiated a multistep functional proteomic strategy to identify neuronal calcium sensor-1 (NCS-1) binding partners (NBPs) that contribute to inner ear development in zebrafish. By performing a Y2H screen in combination with literature and database searches, we identified 10 human NBPs. BLAST searches of the zebrafish EST and genomic databases allowed us to clone zebrafish orthologs of each of the human NBPs. By investigating the expression profiles of zebrafish NBP mRNAs, we identified seven that were expressed in the developing inner ear and overlapped with the ncs-1a expression profile. GST pulldown experiments confirmed that selected NBPs interacted with NCS-1, while morpholino-mediated knockdown experiments demonstrated an essential role for arf1, pi4kβ, dan, and pink1 in semicircular canal formation.
Conclusion
Based on their functional profiles, the hypothesis is presented that Ncs-1a/Pi4kβ/Arf1 form a signaling pathway that regulates secretion of molecular components, including Dan and Bmp4, that are required for development of the vestibular apparatus. A second set of NBPs, consisting of Pink1, Hint2, and Slc25a25, are destined for localization in mitochondria. Our findings reveal a novel signalling pathway involved in development of the semicircular canal system, and suggest a previously unrecognized role for NCS-1 in mitochondrial function via its association with several mitochondrial proteins.
Background
The vestibular system of the vertebrate inner ear is a highly complex set of structures that relays information about motion and spatial orientation to the brain. The structural and functional properties of the vestibular apparatus are highly conserved among all vertebrate species. The semicircular canals, one subdivision of the vestibular system, function as sensors of angular acceleration. These structures enhance survival by implementing postural and visual stabilization during motion in a three-dimensional environment [1-3].
Zebrafish has emerged as an important model system for the study of ear development. Zebrafish represent an especially attractive system for analyzing otogenesis, owing to the fact that the ear can be easily visualized during early embryogenesis, and that many mutations in ear development have been described in zebrafish that appear to resemble those that cause human hearing and balance disorders (reviewed in [4]). In zebrafish, as well as other vertebrates, the semicircular canal system consists of three non-sensory components, the anterior, posterior, and lateral canal ducts (reviewed in [5]) which are positioned in three different planes. Sensory regions lie at the end of each canal and detect differential flow of fluid through the non-sensory structures during angular rotation of the head.
The formation of the semicircular canals in zebrafish begins at 45 hpf with the development of three epithelial outpocketings: one anterior protrusion, one posterior protrusion, and one lateral protrusion [6]. At 60 hpf, the lateral protrusion bifurcates into an anterior branch and a posterior branch that fuse with the anterior and posterior protrusions, respectively [7]. Shortly after the formation of the horizontal hub, a ventral protrusion develops at the bottom of the vesicle and fuses with the lateral protrusion. Finally, a thin septum arises from the dorsal wall forming a dorsolateral partition. At 72 hpf, the four hubs are completely formed and appear as a cross shaped structure that spans the lumen of the otic vesicle. Little is currently known, however, regarding the underlying molecular mechanisms that govern the development of this intricate structure.
Mutagenesis screens in zebrafish have uncovered several types of mutants with semicircular canal defects [8-10]. For example, a mutation in the jekyll gene leads to interrupted ear columns (epithelial pillars which form the hubs of the semicircular canals) and malformations of pharyngeal cartilage [11]. The jekyll gene encodes Ugdh, an enzyme required for the production of several components of the extracellular matrix including hyaluronic acid [12]. Localized secretion of hyaluronic acid has been proposed to drive propulsion of the non-sensory epithelium during canal morphogenesis, a process shown to be necessary for proper semicircular canal formation in Xenopus embryos [13]. The zebrafish ortholog of the human deafness gene dfna5, was subsequently shown to be necessary for ugdh expression and proper semicircular canal and jaw development [14]. Another zebrafish mutant, gallery, displays only a lateral semicircular canal protrusion and an immature anterior protrusion [15]. Although the gallery gene has not yet been identified, the defects in this mutant are believed to be caused by overexpression of Bmp4 (bone morphogenetic protein 4) in the presumptive cristae.
Several other genes required for proper semicircular canal formation in zebrafish have recently been identified. These include sparc, a gene encoding a matricellular glycoprotein (osteonectin) that functions in the genetic network regulating pharyngeal and inner ear development [16], tmie (transmembrane inner ear), a gene coding for a protein of unknown function that is important for normal hearing [17], and the Na, K-ATPase α subunit gene α1a.2 [18].
We have recently found that in zebrafish, the gene encoding neuronal calcium sensor-1a (ncs-1a) is required for semicircular canal formation [19]. NCS-1 is the mammalian ortholog of Drosophila frequenin, a protein critical for synaptic transmission within the Drosophila nervous system [20]. In mammalian cells, NCS-1 has been shown to promote exocytosis from dense core vesicles in both neurons and neuroendocrine cells [21], and plays a key role in G-protein coupled receptor desensitization via a direct interaction with the D2 dopamine receptor [22]. In zebrafish, morpholino knockdown of ncs-1a blocks normal development of the non-sensory portion of the semicircular canals [19]. However, the biochemical function of NCS-1 in regulating semicircular canal formation has not been elucidated.
To gain a better understanding of the role of NCS-1 in regulating inner ear development, we initiated a proteomic screen to identify NCS-1 binding partners (NBPs). Using yeast two-hybrid and bioinformatic approaches, we identified a cohort of human NBPs and their zebrafish orthologs. We examined expression of ten NBPs and identified seven that are expressed in the developing zebrafish inner ear. Based on their functional profiles, the hypothesis is presented that Ncs-1a/Pi4kβ/Arf1 form a signaling pathway that is required for semicircular canal formation. A second set of NBPs, consisting of pink1, hint2, and slc25a25, are destined for localization in mitochondria. Using antisense morpholinos, we show that pink1 is also required for proper development of the semicircular canals. Our findings reveal a cohort of novel intracellular NBPs. NCS-1/NBP interactions are likely to contribute to the molecular mechanisms that regulate the development of the vestibular apparatus.
Results
Identification of NCS-1 Binding Proteins (NBPs)
To identify NCS-1 binding proteins (NBPs), we used full-length human NCS-1 as bait in a Y2H screen of an adult human brain cDNA library. Candidate NCS-1 binding partners are listed in Table 1. Based on their functional properties we chose to pursue DAN (differential screening-selected gene aberrant in neuroblastoma), a secreted Bmp antagonist that has previously been shown to play a role in semicircular canal formation [23]; VAMP2, a synaptosomal protein involved in exocytosis [24-26]; and the mitochondrial proteins HINT2, SLC25A25 (SCAMC2), and PINK1, a mitochondrial kinase which has been found to be mutated in inherited forms of Parkinson's disease [27]. Additional interacting proteins identified in the Y2H screen were not examined further in this study. By conducting literature and protein database searches, we also identified several previously characterized NBPs (Table 1) whose potential role in ear development was analyzed. These include the ADP-ribosylation protein ARF1 [28], PI4Kβ [29], the inositol 1,4,5 triphosphate receptor IP3R [30], and TRPC (transient receptor potential channel) subtypes 1 [31] and 5 [32].
Table 1.
Identification, cloning, and function of NBPs
| Human Gene | Gene Name | Function | Interaction | Zebrafish Ortholog | mRNA Accession Number (riboprobe NTs) |
| Arf1 | ADP-ribosylation factor 1 | Trafficking/Exocytosis | GST-pulldown [28,42] | NM_201452 |
NM_201452 (99–643) |
| Pi4kβ (Pik4cb) | Phosphatidylinositol 4-kinase beta | Trafficking/Exocytosis | Immuno-precipitation [29] | BM095643 |
FJ032032 (1–2083) |
| Vamp2 | Vesicle associated membrane protein 2/Synaptobrevin 2 | Exocytosis | Y2H | BC059626 |
NM_200005 (1–894) |
| Hint2 | Histidine triad protein | Mitochondrial AMP-lysine hydrolase | Y2H | CN504819 |
FJ032031 (1–514) |
| Slc25a25 (SCaMC2) | Ca2+-dependent mitochondrial carrier | Mitochondrial ATP-Mg/Pi carrier | Y2H | NW_001513403 |
NM_213257 (65–1249) |
| Pink1 | PTEN-induced kinase | Mitochondrial Kinase | Y2H | NW_001513662 |
FJ032033 (1–1725) |
| Trpc1 | Classical transient receptor potential-1 | Cation channel | GST-pulldown [31] | EE302043 |
XM_694363.3 (1097–2334) |
| Trpc5 | Classical transient receptor potential-5 | Cation channel | Immuno-precipitation and GST-pulldown [32] | AW343190 |
NM_001044827 (1–2395) |
| Ip3r (Itpr) | Inositol 1,4,5 trisphosphate receptor | Ca2+ release channel | Immuno-precipitation [30] | CK017406 |
XM_691322 (6591–8187) |
| Dan (Nbl-1) | Differential screening-selected gene aberrant in neuroblastoma | Extracellular BMP antagonist | Y2H | BC066387 |
FJ032030 (1–702) |
By using BLAST searches of the zebrafish EST database and genomic sequences available from GenBank and the Zebrafish Sequencing Group at the Sanger Institute http://www.ensembl.org/ we identified zebrafish orthologs of each of the ten human NBPs. The sources and accession numbers for each of the zebrafish orthologs of the human NBPs are listed in Table 1. Zebrafish ESTs were obtained from ATCC (Manassas, VA). In cases where ESTs contained partial sequences, full-length ORFs (open reading frames) were generated by RT-PCR. Alignments of the predicted amino acid sequences of human NBPs and their zebrafish orthologs are presented in Additional File 1.
Expression of NBPs in Zebrafish Inner Ear
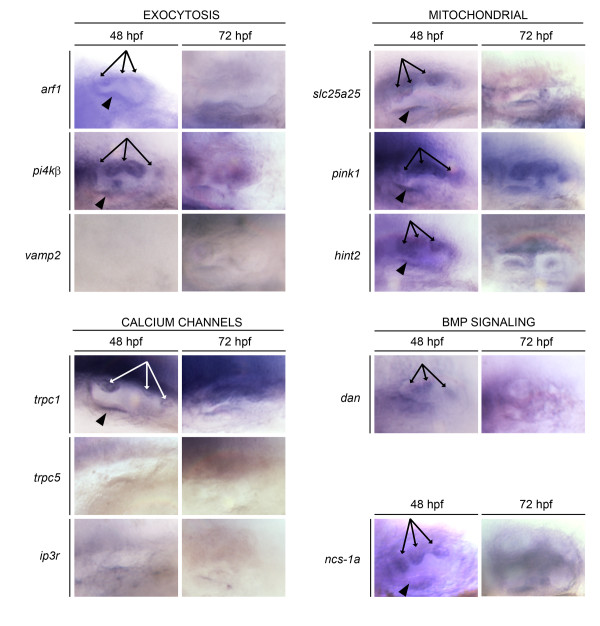
We used whole-mount in situ hybridization to analyze mRNA expression for each of the 10 NBPs (listed in Table 1) during zebrafish otogenesis. The otic expression profile of each candidate NBP gene at 48 hpf and 72 hpf is shown in Fig. 1, while additional file 2 shows expression of these genes at 24, 48 and 72 hpf in the head region. Genes were grouped according to their known functional properties (i.e. exocytosis, calcium channels, Bmp signaling) or intracellular localization (i.e. mitochondria), and their otic expression profiles compared with that of the ncs-1a gene (Fig. 1: lower right panel). All of the genes investigated were expressed within the brain at each time point examined, while some were detected at high levels in additional structures including branchial arches (arf1, pi4kβ, dan, ip3r, hint2, pink1, and slc25a25), somites (hint2, pink1, and slc25a25), and heart (ip3r). Similarly, ncs-1a expression was detected throughout the brain and in the posterior somites and pharyngeal pouches [19].
Figure 1.
Expression of NCS-1 binding partners (NBPs) during zebrafish inner ear development. Whole mount in situ hybridization analysis was performed at 48 and 72 hpf. Genes are grouped according to presumed functional properties. All images are lateral views of the otic vesicle, anterior to the left. Arrows indicate mRNA expression in semicircular canal protrusions at 48 hpf. Arrowheads indicate mRNA expression in the anterior macula at 48 hpf.
At 48 hpf, ncs-1a transcripts can be visualized in the semicircular canal protrusions, and in the fully formed semicircular canal hubs at 72 hpf. We first analyzed expression of arf1, pi4kβ, and vamp2, NBP genes encoding proteins involved in exocytosis and intracellular trafficking. arf1 transcripts were expressed at 48 hpf in the epithelial protrusions of the semicircular canals but were not present in the semicircular canal hubs at 72 hpf, suggesting that arf1 contributes to early events in semicircular canal formation. Expression of pi4kβ mRNA, on the other hand, was very similar to the expression profile seen for ncs-1a mRNA. pi4kβ transcripts were detected at 48 hpf in the semicircular canal protrusions and in the fully formed semicircular canal hubs at 72 hpf. vamp2 mRNA was undetectable in the inner ear at either time point, indicating that vamp2 is unlikely to play a role in otogenesis. Three NBP genes, pink1, hint2, and slc25a25, each encoding a mitochondrial protein, showed very similar expression patterns. Transcripts of each gene were detected at 48 hpf in the semicircular canal protrusions and the anterior macula, and at 72 hpf in the semicircular canal hubs.
Amongst the genes encoding calcium channels, trpc1 transcripts were detectable at 48 hpf and 72 hpf in the semicircular canal protrusions and hubs, respectively. Transcripts of the other two calcium channel genes, trpc5 and ip3r, were undetectable in the inner ear at either 48 hpf or 72 hpf. Expression of dan, a gene encoding a secreted protein involved in Bmp signaling, was similar to that of the pi4kβ gene and was present in the semicircular canals at both 48 hpf and 72 hpf.
Taken together, our data indicate that seven NBP genes including arf1, pi4kβ, trpc1, dan, pink1, hint2, and slc25a25, are expressed in the semicircular canals during zebrafish embryogenesis and are likely to contribute to the formation of the vestibular apparatus.
Validation of NCS-1/NBP Interactions
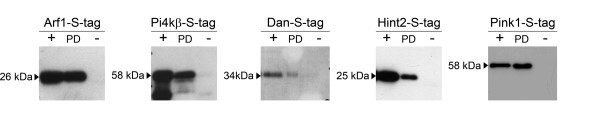
We used pulldown techniques to confirm the interaction between NCS-1 and several of the NBPs expressed in ear. Lysates from bacteria expressing S-tagged zebrafish proteins Arf1 (residues 1–180), Dan (residues 1–183), Pi4kβ (residues 1–423), Pink1 (residues 1–574), or Hint2 (residues 1–161) were tested for the ability to associate with an NCS-1-GST fusion protein containing full-length (residues 1–190) human NCS-1. In this context it is important to note that zebrafish ncs1a and human NCS-1 differ at only 8 of 190 positions [19]. Pulldowns were not performed for Slc25a25 or Trpc1. As shown in Fig. 2, a Western blot containing lysate from bacteria expressing S-tagged Arf1 produced an immunoreactive band of ~26 kDa when probed with an anti-S-protein antibody. This band corresponds to the expected size of the Arf1 fragment encoded by the cDNA construct. The same band was detected by pulldown after the bacterial lysate was incubated with the NCS-1-GST fusion protein, but not when the lysate was incubated with GST-containing beads alone. We next used the GST-pulldown assay to test whether NCS-1 also interacted with Dan, Pi4kβ, Hint2, and Pink1 (Fig. 2). Lysates from bacteria expressing S-tagged fragments of Dan (~34 kDa), Pi4kβ (~58 kDa), Hint2 (~25 kDa), and Pink1 (~58 kDa), each produced an immunoreactive band of the expected size (left lane in each panel). In each case, the same band was detected by pulldown after incubation with the NCS-1-GST fusion protein (middle lane of each panel). Virtually no immunoreactivity was detected when the lysates were incubated with beads containing GST alone (right lane in each panel). These results support the validity of the interaction between NCS-1 and Arf1, Dan, Pi4kβ, Pink1, and Hint2.
Figure 2.
GST-pulldowns. GST pulldowns were performed to validate NCS-1/NBP interactions. A human NCS-1-GST fusion protein was used to pull down S-tagged zebrafish NBPs. In each panel, the left (+) lane contains a bacterial lysate expressing the designated S-tagged NBP, the middle lane (PD), contains NBPs pulled down from bacterial lysates after incubation with the NCS-1-GST fusion protein, and the right lane (-) shows proteins pulled down from lysates after incubation with GST alone. Blots were probed with an HRP-conjugated S-tag antibody to detect each S-tagged NBP. Molecular weights (kDa) of S-tagged NPB fragments are indicated to the left of each panel.
Knockdowns of NBPs Involved in Exocytosis and Secretion
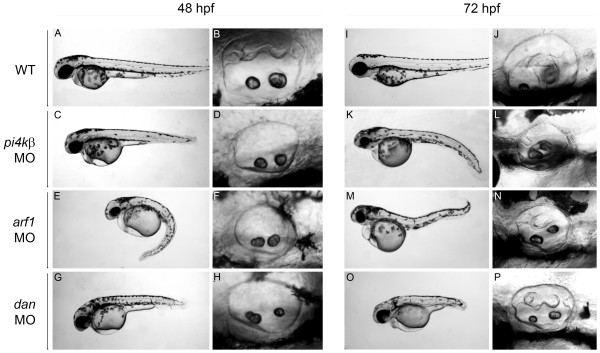
To gain an understanding of the function of NBPs during otogenesis, we used morpholino antisense oligonucleotides to knock down translation of selected NBP mRNAs in developing zebrafish. Our initial experiments involved knocking down translation of arf1 and pi4kβ mRNAs, encoding NBPs involved in exocytosis, and dan mRNA, encoding a secreted NBP known to play a role in semicircular canal formation [23]. We generated a set of non-overlapping morpholinos (MOs) targeted against either the initiating methionine (ATG-MO) or the 5'-UTR (UTR-MO) of pi4kβ, arf1, and dan mRNAs (Table 2), and microinjected the MOs into one cell-stage embryos. Representative examples of the NBP knockdowns are shown in Fig. 3. In wild type embryos, saccular and utricular otoliths are present at 48 hpf, and the semicircular canals protrusions are clearly visible (Fig. 3A). By 72 hpf, the epithelial pillars have fused to form the semicircular canal hubs exhibiting the characteristic cruciform pattern (Fig. 3J). Microinjection of MOs against each of the three NBP genes produced noticeable effects on inner ear development. MOs targeted against either pi4kβ (Fig 3C, K), arf1 (Fig. 3E, M), or dan (Fig. 3G, O), produced morphants with a shortened body axis, reduced eye size, and curved tails. These defects were most pronounced in dan morphants (Fig. 3G, O), even at low doses (1 ng) of the dan MO. At 48 hpf, semicircular canal protrusions were missing in > 90% of pi4kβ (Fig. 3D), arf1 (Fig. 3F), and dan (Fig. 3H) morphants (n = 50–60/group). Analysis of the developing ear through 48 hpf revealed that each of the NBP morphants exhibited normal otic placode induction and otolith formation compared to control embryos.
Table 2.
Morpholino sequences
| Morpholino | Sequence |
| ncs1a-MO | 5'-TAGTTTGCTGTTGGATTTGCCCATC-3' |
| arf1-ATG MO | 5'-TAAAGAGGTTTGCGAATATGTTTCC-3' |
| arf1-UTR MO | 5'-CGCCTTGTTCACACCAAGTTCCAAG-3' |
| pi4kβ-ATG MO | 5'-AAGCTCCAGCTCTGTATCACCCATG-3' |
| pi4kβ-UTR MO | 5'-CACTTCAGGCCCCTCAAAATAAACC-3' |
| dan-ATG MO | 5'-CGCGCACACACATCACCATACCTTC-3' |
| dan-UTR MO | 5'-TCAGTGGATTAGCAGCTCGCGGTGT-3' |
| pink1-ATG MO | 5'-GGCTGAGAACATGCTTTACTGACAT-3' |
Figure 3.
Knockdown of arf-1, pi4kβ, and dan mRNA expression. Antisense MOs targeted against arf-1, pi4kβ, or dan mRNA were injected into single cell embryos, and morphants analyzed at 48 (C-H) and 72 hpf (K-P). For comparison, control (buffer injected) embryos are shown at 48 (A-B) and 72 (I-J) hpf. Embryos were injected with either 4 ng pi4kβ-ATG MO (C-D, K-L), 2 ng arf1-UTR MO (E-F, M-N), or 1ng of dan-ATG MO (G-H, O-P). Images of otic vesicle (B, D, F, H, J, L, N, P) are all lateral views with anterior to the left. Lateral views of wild-type embryos are shown at 48 (A) and 72 (I) hpf. Lateral views of morphant embryos are shown at 48 (C, E, G) and 72 (K, M, O) hpf.
At 72 hpf, we observed that many of the morphants had begun to develop semicircular canals. However, the canals exhibited a range of aberrant morphologies including missing protrusions (Fig. 3L, N), failure of the pillars to fuse (Fig. 3P), as well as the presence of disorganized cell masses in the otic vesicle (Fig. 3L, N). For all genes tested, injection of a second, independent MO (either the ATG-MO or UTR-MO) phenocopied the ear defects generated by the primary MO (data not shown). In general, higher doses of the secondary MO were required to achieve effects comparable to those produced by the primary MO on semicircular canal formation. Together, these MO knockdown experiments provide compelling evidence that in addition to ncs-1a, expression of arf1, pi4kβ, and dan genes is required for semicircular canal formation in zebrafish.
ncs-1a, pi4kβ, and arf1 Functionally Interact to Regulate Formation of Semicircular Canals
NCS-1, Arf1, and Pi4kβ have been shown to functionally interact in PC12 cells to regulate neuronal secretion [28]. We wished to determine whether these three genes functionally interact within the zebrafish inner ear to regulate semicircular canal development. To test this idea, we co-injected subeffective doses of ncs-1a-ATG (0.5 ng) plus arf1-UTR (0.5 ng) MOs, ncs-1a-ATG plus pi4kβ-ATG (2 ng) MOs, and arf1-UTR plus pi4kβ-ATG MOs, and examined the effect on semicircular canal formation. The results are shown in Fig. 4. At a concentration of 0.5 ng, neither the ncs-1a-ATG MO nor the arf1-UTR MO produced severe semicircular canal development defects, although a small percentage of fish (< 5%) displayed no epithelial protrusions at 48 hpf. 100% of embryos injected with 2 ng of the pi4kβ-ATG MO exhibited normal semicircular canal development at 48 hpf. However, co-injection of subeffective doses of ncs-1a-ATG plus arf1-UTR MOs caused an absence of epithelial protrusions in 53% of embryos at 48 hpf. Co-injection of ncs-1a-ATG plus pi4kβ-ATG MOs caused an absence of epithelial protrusions in 40% of embryos, while co-injection of subeffective doses of arf1-UTR plus pi4kβ-ATG MOs blocked outgrowth of epithelial protrusions in 44% of embryos. At 72 hpf, < 7% of embryos injected with subeffective doses of either ncs-1a-ATG, arf1-UTR, or pi4kβ-ATG MOs alone exhibited abnormal semicircular canal hub formation. Abnormal semicircular canal hub formation was observed in 58% of embryos co-injected with subeffective doses of ncs-1a plus arf1 MOs, in 38% of embryos co-injected with ncs-1a-ATG plus pi4kβ-ATG MOs, and in 49% of embryos co-injected with subeffective doses of arf1-UTR plus pi4kβ-ATG MOs. Co-injection of each pair of MOs thus appears to amplify the semicircular canal defects observed when subeffective doses of either ncs-1a-ATG, arf1-UTR, or pi4kβ-ATG MOs are injected alone. The functional interaction observed in this assay, in addition to the demonstrated physical interaction of their encoded proteins, suggests that ncs1a, arf1, and pi4kβ may act in a common pathway leading to formation of the semicircular canal system.
Figure 4.
Phenotypes of embryos co-injected with subeffective doses of NBP MOs. Bar graph depicts percent of fish at 48 hpf or 72 hpf that displayed normal epithelial protrusions or semicircular canal hub formation, abnormal epithelial protrusions or semicircular canal hub formation, or absence of epithelial protrusions. Embryos were injected with 0.5 ng of ncs-1a-ATG MO alone, 0.5 ng of arf1-UTR MO alone, or 2 ng of pi4kβ-ATG MO alone. Alternatively, embryos were co-injected with 0.5 ng of ncs-1a-ATG MO plus 0.5 ng of arf1-UTR MO, 0.5 ng of ncs-1a-ATG MO plus 2 ng of pi4kβ-ATG MO, or 0.5 ng arf1-UTR plus 2 ng pi4kβ-ATG MO. The number of fish assayed for each treatment is displayed above the bars.
pink1 is Required for Semicircular Canal Formation
PINK1 was initially implicated as an NBP in a Y2H screen (Table 1), while the interaction of zebrafish pink1 with NCS-1 was verified via pulldown with an NCS-1-GST fusion protein (Fig. 2). Whole-mount in situ hybridization analysis revealed expression of pink1 mRNA sequences in the epithelial pillars and hubs of the semicircular canals during zebrafish otogenesis (Fig. 1). To directly determine whether pink1 expression was necessary for ear development, we knocked down pink1 mRNA translation with a pink1-ATG MO and examined the effect on semicircular canal formation.
A representative set of pink1 morphants is shown in Fig. 5. In control embryos at 48 hpf (Fig. 5A, B), otoliths are present and the epithelial protrusions of the semicircular canals are clearly visible. In 48 hpf MO-injected embryos, otoliths appear to have developed normally, although the overall size of the otocyst is somewhat smaller compared to control embryos. Microinjection of either 2 ng (Fig. 5C, D) or 4 ng (Fig. 5E, F) of a pink1-ATG MO produced morphants that were missing semicircular canal protrusions at 48 hpf in > 95% of embryos examined (n = 30). These results strongly support the idea that expression of the pink1 gene is required for semicircular canal formation in zebrafish.
Figure 5.
Pink1 expression is required for semicircular canal development. An antisense pink1 MO was used to knock down pink1 mRNA translation. Images are all lateral views of 48 hpf embryos with anterior to the right. (A) head of wild type embryo; (B) otic vesicle of wild type embryo. Head of pink1 morphant injected with 2 ng (C) or 4 ng (E) of pink1-ATG MO. Otic vesicle of pink1 morphant injected with either 2 ng (D) or 4 ng (F) of pink1-ATG MO.
Discussion
Ncs-1 is a calcium sensing molecule that is required for semicircular canal formation in zebrafish [19]. To better understand of the role of NCS-1 in ear development, we employed a multistep functional proteomic strategy to identify molecular partners of NCS-1 that contribute to the development of the vestibular apparatus. First, by performing a Y2H screen in combination with literature and database searches, we identified 10 human NBPs. Second, zebrafish orthologs of each of these NBPs were identified using BLAST searches of the zebrafish EST and genomic databases. Third, by investigating the expression profiles of NBPs, we were able to deduce which candidate NBPs were likely to interact with ncs-1a in the ear and contribute to semicircular canal formation. We found that mRNA sequences encoding seven of the putative zebrafish NBPs were detected in the developing ear and exhibited an overlap in expression with that of the ncs-1a gene. On the other hand, three NBP genes (vamp2, trpc5, and ip3r) were not expressed in the ear, and thus were eliminated from further analysis. Fourth, GST pulldown experiments were carried out to confirm that candidate zebrafish NBPs also interacted with NCS-1. Finally, MO-mediated knockdowns confirmed that four selected NBPs were essential for semicircular canal formation in zebrafish.
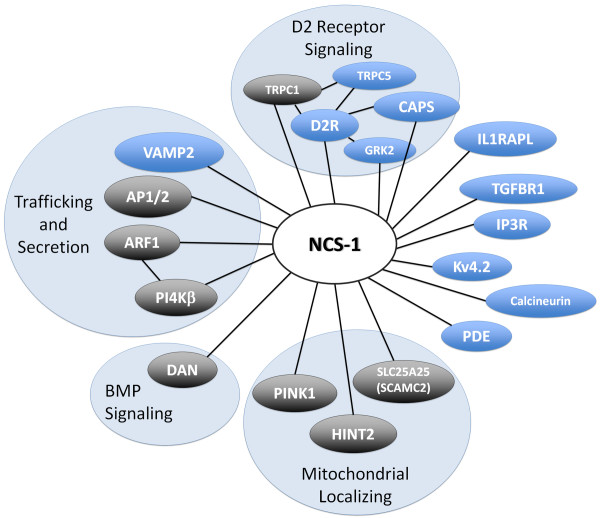
The strategy outlined above has proven to be a fruitful method for identifying novel genes that contribute to inner ear development. Using this approach, we show that two previously identified (arf1, pi4kβ) and two novel (dan, pink1) NBP genes are required for semicircular canal formation (see interaction map, Fig. 6[33]). In the present study, we did not examine the role of hint2, slc25a25, or trpc1 using a morpholino knockdown approach. Further studies will be required to analyze the role of these and other NBPs in ear development. Mutagenesis screens in zebrafish combined with positional cloning approaches have traditionally been used to identify genes that contribute to ear development [8-10]. For example, the zebrafish dog-eared mutation was identified in a large-scale ENU screen by abnormal morphology of the inner ear [10]. The identity of this gene was then determined to be zebrafish eya1 via positional cloning [34]. While mutagenesis screens provide an unbiased strategy for gene discovery, this approach is generally extremely labor intensive and time consuming. Our strategy, on the other hand, circumvents some of the limitations inherent in an unbiased screen, by rapidly identifying interacting partners of genes already known to regulate inner ear development. This approach, in combination with the reverse genetic techniques available in zebrafish, should make it feasible to identify in a systematic fashion additional genes whose roles in ear development were previously unknown or overlooked.
Figure 6.
NCS-1 interaction map. Web of newly identified and previously known NCS-1 binding partners. Interaction map was adapted from Haynes et al. 2006 [33]. NBPs were grouped based on previously characterized functional properties or cellular localization. NBPs depicted in gray shaded ovals are those whose genes are expressed within the developing zebrafish inner ear.
The specificity of the phenotypes obtained by MO knockdown is supported by a number of considerations. First, morpholinos targeted against pi4kβ, arf1, and dan all caused similar phenotypes including vestibular malformation, shortened body axis, reduced head and eye size, and curved tails. Since each of these genes is expressed throughout the brain and jaw during embryonic development, defects in head formation were not unexpected. These defects are not likely due to off-target effects, since for each of the genes tested, two independent non-overlapping MOs produced identical phenotypes. Second, injection of buffer alone did not produce morphant phenotypes. Third, a MO targeted against otopetrin, a gene previously shown to be required for otolith formation [35], blocked development of otoliths but did not affect semicircular canal development. Fourth, injection of subeffective doses of pi4kβ, arf1, and ncs-1a MOs alone did not produce inner ear, jaw, or brain defects. Co-injection of subeffective doses of MOs against a pair of genes was synergistic and phenocopied the ear, jaw, and brain defects obtained using an effective dose of each of MO alone. Finally, brain and ear development appear normal in pi4kβ, arf1, and dan morphants until 48 hpf, indicating that disruption of early patterning events is not likely to have caused vestibular phenotypes or developmental delay. Together, these results support the idea that the phenotypes produced by NBP knockdowns are specific, and do not result from non-specific cytotoxic or off-target gene effects.
Our double MO knockdown studies, together with the pulldown experiments, indicate that NCS-1, Arf1 and Pi4kβ form a small interaction network (Fig. 6 adapted from [33]) that is likely to have functional significance for development of the zebrafish semicircular canal system. We now propose a model in which these three proteins regulate the targeted secretion of molecules, such as Dan, that are required for formation of the semicircular canals (Fig. 7). This model is based on the following lines of evidence. Pi4kβ is the enzyme responsible for the synthesis of phosphatidylinositol 4-phosphate, a lipid species that regulates trafficking of cargo from the Golgi apparatus to the plasma membrane in yeast and mammalian cells [36-38]. Pi4kβ has also been shown to regulate secretion in both PC12 cells and pancreatic β cells [39,40]. NCS-1 [41] and Arf1 [41] are known activators of Pi4kβ, however a direct interaction between NCS-1 and Arf1 has been shown to antagonize the enzymatic activity of Pi4kβ [28,42]. Bidirectional control of Pi4kβ activation/inhibition may thus serve to regulate the trafficking of molecules involved in inner ear development from the trans-golgi network to the plasma membrane during otogenesis.
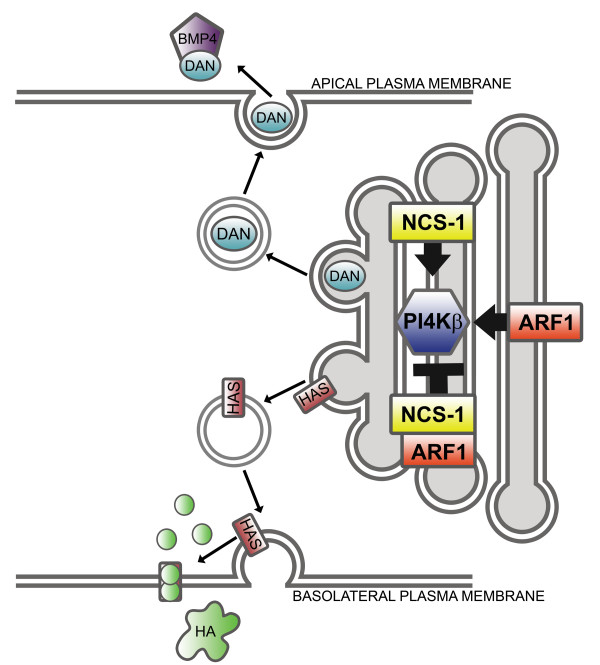
Figure 7.
Proposed model for functional interaction between Ncs-1a, Pi4kβ, and Arf1 in trafficking and secretion. Dan, Bmp4, and HA are secreted molecules that contribute to proper semicircular canal development and are predicted to require the action of an Ncs-1/Pi4kb/Arf1 signaling pathway for their secretion. Bold arrows indicate activation, whereas the bold T indicates inhibition, of this signaling pathway. Thin arrows indicate directional flow of vesicles or proteins.
Dan, Bmp4, and Hyaluronic acid represent secreted molecules that play critical roles in semicircular canal formation [13-15,23,43] and may require an ncs-1a/arf1/pi4kβ pathway for secretion. BMPs are expressed in the developing vertebrate inner ear and participate in axial patterning of the otic vesicle and differentiation of the sensory epithelium [23,44-48]. Bmp4 is secreted from the apical membrane of cells located in the cristae of the otic vesicle and its regulated expression is essential for proper development of the non-sensory portion of the semicircular canals [15,43]. Dan is a Bmp antagonist that plays a key role in the patterning of the chick inner ear, and is also secreted from the apical membrane [23]. In chick, loss of Dan leads to structural deformities in the semicircular canals and the endolymphatic duct and sac. The MO knockdown experiments we performed show that Dan also contributes to semicircular canal formation in zebrafish. We have also demonstrated a direct interaction between Dan and NCS-1. Although the significance of this interaction is currently unknown, it is possible that the Dan/NCS-1 interaction may be required to deliver Dan to the Golgi apparatus. This idea is supported by previous studies which showed that the yeast ortholog of NCS-1 (Frq1p) can recruit Pik1p (yeast ortholog of PI4kβ) from the endoplasmic reticulum to the trans-Golgi network [49].
Hyaluronic acid (HA) is an extracellular matrix molecule that is secreted from the basolateral membrane of otic epithelium during semicircular canal formation and is thought to drive the outgrowth of the developing protrusions [13]. Hyaluronan synthases (HAS) are the enzymes responsible for HA synthesis and secretion at the plasma membrane [50]. It is possible that trafficking of HAS from the Golgi apparatus to the plasma membrane may also be regulated by Pi4kβ activity. Since HA secretion from epithelial pillars in zebrafish inner ear can be monitored histochemically [14], it should be possible to test this hypothesis by determining whether knockdown of Ncs-1a, Arf1, or Pi4kβ inhibits HA secretion.
Our yeast two-hybrid screen revealed interaction of NCS-1 with three proteins, HINT2, SLC25a25, and PINK1, that are destined to localize to mitochondria. The interaction of NCS-1 with Pink1 (phosphatase/tensin homolog on chromosome 10 (PTEN)- induced putative kinase 1) is of particular interest. Mutations in the human PINK1 gene are the second most common cause of autosomal recessive Parkinson's disease [51], and are also implicated in sporadic cases of the disease [52]. MO knockdown studies we performed indicate that pink1 morphants exhibit severe brain abnormalities as well as defects in development of the semicircular canal system. Interestingly, a recent study in zebrafish also showed that pink1 morphants display severe defects in brain morphology, as well an impaired escape response [53]. In this context, it is important to consider that no direct link has been established between mutations in Pink1 and defects in inner ear development in mammalian systems. It is therefore possible that Pink1 has evolved to perform a critical role in brain and ear development in zebrafish, while mammals may have developed compensatory mechanisms that regulate development of these organs.
The interaction of NCS-1 with zebrafish Pink1 raises another interesting issue. NCS-1 does not possess a defined mitochondrial targeting sequence, and its expression in mitochondria has not been documented. In mammalian cells, NCS-1 is constitutively associated with the plasma membrane and Golgi complex [54]. It will be important to determine whether NCS-1 serves to regulate Pink1 function, either within mitochondria or other cellular compartments, and whether the NCS-1/Pink1 interaction contributes to the etiology of Parkinson's disease.
Conclusion
In this study, we show that several new and previously identified NBPs appear to play an important role in the development of the zebrafish inner ear. We suggest a model in which Ncs-1a/Arf1/Pi4kβ interactions may form a signaling pathway that mediates trafficking and export of secreted molecules that are required for semicircular canal formation. We also made an unexpected observation that NCS-1 interacts with several mitochondrial proteins, including Pink1, a gene that contributes to familial Parkinson's disease. These findings suggest a previously unrecognized role for NCS-1 in mitochondrial function via its association with mitochondrial proteins, and reveal a new and potentially important signaling pathway involved in development of the vestibular apparatus.
Methods
Yeast two-hybrid Screen
A yeast two-hybrid (Y2H) screen was performed as previously described [55] using human NCS-1 as bait to screen an adult human brain cDNA library. Full length human NCS-1 was subcloned into the yeast GAL4 DNA-binding domain expression vector pAS2-1 (Clontech, Palo Alto, CA), while the human brain cDNA library was subcloned into the GAL4 activation domain vector pACT2 (Clontech). Bait and prey plasmids were simultaneously co-transformed into yeast strain MaV103 [55]. Positive clones were identified by growth on Leu-/Trp-/His-/Ura- selection plates. Protein interaction was assayed for by β-galactosidase activity via the nitrocellulose filter lift method [55]. cDNAs were extracted from yeast colonies, sequenced, and subjected to a BLAST search against the human genome to determine the identity of the NCS-1 interacting cDNA clones.
Identification and Cloning of Zebrafish NBP Orthologs
Zebrafish orthologs of human NBPs were identified by using BLAST searches of the zebrafish EST database and genomic sequences available from GenBank http://www.ncbi.nlm.nih.gov/Genbank/index.html and the Zebrafish Sequencing Group at the Sanger Institute http://www.ensembl.org/. The sources and accession numbers for each of the zebrafish orthologs of the human NBPs are listed in Table 1. Zebrafish ESTs were obtained from ATCC (Manassas, VA). For RT-PCR (reverse transcriptase-polymerase chain reaction), 48 hpf (hours post fertilization) zebrafish embryos were collected and homogenized in TRIzol Reagent (Invitrogen; Carlsbad, CA). Total RNA was extracted as previously described [56], and 0.5 μg of RNA was used as template to generate single stranded cDNA using the SuperScript First Strand Synthesis kit (Invitrogen), and RT-PCR performed using REDTaq DNA polymerase (Sigma; St. Louis, MO) and a RoboCycler Gradient Temperature Cycler (Stratagene; La Jolla, CA). Primers used for RT-PCR are listed in Table 3.
Table 3.
Primers for cloning NBPs
| Zebrafish Gene | RT-PCR Forward Primer | RT-PCR Reverse Primer |
| arf1 | 5'-GATGGGAAACATATTCGCAAACCT-3' | 5'-TCATTTCTGGTTTTTCAGCTGATTGGACA-ACC-3' |
| pi4kβ | 5'-GTGGCCATGGGTGATACAGAGCTGGAG--CTTTCTC-3' | 5'-TCACATGATACCGTTGGTCAGATACTGG-AAGCCGT-3' |
| hint2 | 5'-ACGGTCACGAACGTAGTTTGTCC-3' | 5'-CTATCCTGGAGGCCATTTCATTTGCC-3' |
| pink1 | 5'-CAGATGACAGTGGAGGAGCTTCTGC-3' | 5'-CTATGGCTGAGAGTTAGACATCAGCAGGT-ATC-3' |
| trpc1 | 5'-CTCTCTCCGTTATAATGGCTGCTC-3' | 5'-GCTCAGCTGGTTCCGTGACTCTTC-3' |
| dan | 5'-TCAGCATCTGAGATCCGTCAGCGGGAC-GTC-3' | 5'-GTTTCACCCAGCCTCGGGCGCGTGCAT-GTC-3' |
Whole-mount in situ Hybridization
48 and 72 hpf zebrafish embryos were preserved in 4% paraformaldehyde in PBS. Whole-mount in situ hybridization analysis was performed as previously described [57]. Riboprobes were synthesized for the following genes (Table 1): arf1, dan, hint2, ip3r, pink1, pi4kβ, slc25a25, trpc1, trpc5, and vamp2.
Morpholino Knockdowns
Antisense morpholino oligonucleotides (MOs) were obtained from Gene Tools LLC (Philomath, OR). Two non-overlapping MOs were targeted against either the initiating methionine or the 5' untranslated region of pi4kβ, arf1, ncs-1a, dan, and pink1 mRNAs. The sequence for each morpholino is listed in Table 2. MOs were resuspended in 1× Danieau buffer (58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, 5 mM HEPES, pH 7.6), and microinjected directly into the yolk sac of single cell embryos.
Glutathione S-transferase Pulldown
An NCS-1-GST (glutathione S-transferase) fusion protein was constructed by fusing amino acids 1–190 of human NCS-1 to GST in the expression vector pGEX-4T-1 (Amersham Biosciences, Piscataway, NJ). The NCS-1-GST-fusion protein was induced in Escherichia coli strain BL21 (DE3) using the ZYP-5052 auto-induction media described previously [58,59], then purified on glutathione sepharose beads (Amersham) according to manufacturer's instructions. cDNAs encoding zebrafish arf1 (amino acids 1–180), pi4kβ (residues 1–423), dan (residues 1 – 183), pink1 (residues 1–574), and hint2 (residues 1–161) were subcloned into the pET30C expression vector containing an S-tag. Constructs were induced using auto-induction media as described above. NCS-1-GST fusion protein was used to pull down NBPs from bacterial lysates as previously described [60]. Eluted proteins were separated by SDS-PAGE and transferred to a PVDF (polyvinylidene fluoride) filter. The filter was probed with a horseradish peroxidase conjugated S-tag antibody (1:5000 dilution, Santa Cruz Biotechnology). Immunoreactivity was detected by enhanced chemiluminescence with an ECL Plus kit (Amersham).
Abbreviations
NBP: NCS-1 binding protein; GST: glutathione S-transferase; HPF: hours post fertilization; HA: hyaluronic acid; Y2H: yeast 2 hybrid; EST: expressed sequence tag; RT-PCR: reverse transcriptase polymerase chain reaction; ORF: open reading frame; cDNA: copy DNA; MO: morpholino; ENU: N-ethyl-N-nitrosourea; PVDF: polyvinylidene fluoride; SDS-PAGE: sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Authors' contributions
JAP identified and generated sequence alignments of the zebrafish orthologs of human NBP's, performed all molecular cloning procedures, in situ hybridizations, GST-pulldowns, and morpholino knockdowns, and drafted the manuscript. NK performed the yeast two-hybrid screen and assisted in the preparation of the manuscript. CF, MW, KH, and MC assisted with GST-pulldowns. VAC assisted with bioinformatics, identification of zebrafish NBP orthologs, and preparation of the manuscript. RL conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Comparison of human and zebrafish NBPs. Amino acid alignments of the human (h) and zebrafish (zf) NBP orthologs listed in Table 1. Identical amino acids are highlighted in black, and conserved amino acids are highlighted in gray. Amino acids are numbered to the left. File is a pdf and can be viewed with adobe acrobat reader. Genbank accession numbers for sequences used in the alignments are as follows: h_Arf1 (CAI23120), zf_Arf1 (NP_958860), h_Dan (BAA92265), zf_Dan (ACH92116), h_Hint2 (CAI10991), zf_Hint2 (ACH92117), h_Ip3r (NP_002215), zf_Ip3r (XP_696414), h_Pi4kβ (AAH00029), zf_Pi4kβ (ACH92118), h_Pink1 (AAQ89316), zf_Pink1 (ACH92119), h_Slc25a25 (CAI13838), zf_Slc25a25 (NP_998422), h_Trpc1 (CAA61447), zf_Trpc1 (XP_699455), h_Trpc5 (NP_036603), zf_Trpc5 (NP_001038292), h_Vamp2 (NP_055047), zf_Vamp2 (NP_956299).
Expression of zebrafish NBPs in head region. Whole mount in situ hybridization analysis was performed at 24, 48, and 72 hpf. Expression profiles in head region are shown for all of the NBPs represented in Figure 1. Genes are grouped according to presumed functional properties. All images are lateral views of the head, anterior to the left.
Acknowledgments
Acknowledgements
We thank Joe Bednarczyk (Molecular Genetics Core Facility, Penn State College of Medicine) for DNA sequencing. This project was funded by an NIH Conte Center Grant (MH068789) and a grant from the Pennsylvania Department of Health using Tobacco Settlement Funds.
Contributor Information
Jessica A Petko, Email: jac554@psu.edu.
Nadine Kabbani, Email: nkabbani@gmu.edu.
Colleen Frey, Email: chenry2@hmc.psu.edu.
Matthew Woll, Email: mpw149@psu.edu.
Katharine Hickey, Email: hickka02@gettysburg.edu.
Michael Craig, Email: mtc1@cec.wustl.edu.
Victor A Canfield, Email: vac3@psu.edu.
Robert Levenson, Email: rlevenson@hmc.psu.edu.
References
- Spoor F, et al. Vestibular evidence for the evolution of aquatic behaviour in early cetaceans. Nature. 2002;417:163–6. doi: 10.1038/417163a. [DOI] [PubMed] [Google Scholar]
- Land MF. Motion and vision: why animals move their eyes. J Comp Physiol [A] 1999;185:341–52. doi: 10.1007/s003590050393. [DOI] [PubMed] [Google Scholar]
- Walls GL. The evolutionary history of eye movements. Vision Research. 1962;2:69–80. doi: 10.1016/0042-6989(62)90064-0. [DOI] [Google Scholar]
- Whitfield TT, et al. Development of the zebrafish inner ear. Dev Dyn. 2002;223:427–58. doi: 10.1002/dvdy.10073. [DOI] [PubMed] [Google Scholar]
- Nicolson T. The genetics of hearing and balance in zebrafish. Annu Rev Genet. 2005;39:9–22. doi: 10.1146/annurev.genet.39.073003.105049. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, et al. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Haddon C, Lewis J. Early ear development in the embryo of the zebrafish, Danio rerio. J Comp Neurol. 1996;365:113–28. doi: 10.1002/(SICI)1096-9861(19960129)365:1<113::AID-CNE9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Malicki J, et al. Mutations affecting development of the zebrafish ear. Development. 1996;123:275–83. doi: 10.1242/dev.123.1.275. [DOI] [PubMed] [Google Scholar]
- Schibler A, Malicki J. A screen for genetic defects of the zebrafish ear. Mech Dev. 2007;124:592–604. doi: 10.1016/j.mod.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Whitfield TT, et al. Mutations affecting development of the zebrafish inner ear and lateral line. Development. 1996;123:241–54. doi: 10.1242/dev.123.1.241. [DOI] [PubMed] [Google Scholar]
- Neuhauss SC, et al. Mutations affecting craniofacial development in zebrafish. Development. 1996;123:357–67. doi: 10.1242/dev.123.1.357. [DOI] [PubMed] [Google Scholar]
- Walsh EC, Stainier DY. UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science. 2001;293:1670–3. doi: 10.1126/science.293.5535.1670. [DOI] [PubMed] [Google Scholar]
- Haddon CM, Lewis JH. Hyaluronan as a propellant for epithelial movement: the development of semicircular canals in the inner ear of Xenopus. Development. 1991;112:541–50. doi: 10.1242/dev.112.2.541. [DOI] [PubMed] [Google Scholar]
- Busch-Nentwich E, et al. The deafness gene dfna5 is crucial for ugdh expression and HA production in the developing ear in zebrafish. Development. 2004;131:943–51. doi: 10.1242/dev.00961. [DOI] [PubMed] [Google Scholar]
- Omata Y, et al. Role of Bone morphogenetic protein 4 in zebrafish semicircular canal development. Dev Growth Differ. 2007;49:711–9. doi: 10.1111/j.1440-169X.2007.00964.x. [DOI] [PubMed] [Google Scholar]
- Rotllant J, et al. Sparc (Osteonectin) functions in morphogenesis of the pharyngeal skeleton and inner ear. Matrix Biol. 2008;27:561–72. doi: 10.1016/j.matbio.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YC, et al. The transmembrane inner ear (tmie) gene contributes to vestibular and lateral line development and function in the zebrafish (Danio rerio) Dev Dyn. 2008;237:941–52. doi: 10.1002/dvdy.21486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasiole B, et al. Separate Na, K-ATPase genes are required for otolith formation and semicircular canal development in zebrafish. Dev Biol. 2006;294:148–60. doi: 10.1016/j.ydbio.2006.02.034. [DOI] [PubMed] [Google Scholar]
- Blasiole B, et al. Neuronal calcium sensor-1 gene ncs-1a is essential for semicircular canal formation in zebrafish inner ear. J Neurobiol. 2005;64:285–97. doi: 10.1002/neu.20138. [DOI] [PubMed] [Google Scholar]
- Pongs O, et al. Frequenin – a novel calcium-binding protein that modulates synaptic efficacy in the Drosophila nervous system. Neuron. 1993;11:15–28. doi: 10.1016/0896-6273(93)90267-U. [DOI] [PubMed] [Google Scholar]
- McFerran BW, Graham ME, Burgoyne RD. Neuronal Ca2+ sensor 1, the mammalian homologue of frequenin, is expressed in chromaffin and PC12 cells and regulates neurosecretion from dense-core granules. J Biol Chem. 1998;273:22768–72. doi: 10.1074/jbc.273.35.22768. [DOI] [PubMed] [Google Scholar]
- Kabbani N, et al. Interaction with neuronal calcium sensor NCS-1 mediates desensitization of the D2 dopamine receptor. J Neurosci. 2002;22:8476–86. doi: 10.1523/JNEUROSCI.22-19-08476.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanishi T, et al. Dan is required for normal morphogenesis and patterning in the developing chick inner ear. Dev Growth Differ. 2007;49:13–26. doi: 10.1111/j.1440-169X.2007.00900.x. [DOI] [PubMed] [Google Scholar]
- Schiavo G, et al. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–5. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- Schiavo G, et al. Identification of the nerve terminal targets of botulinum neurotoxin serotypes A, D, and E. J Biol Chem. 1993;268:23784–7. [PubMed] [Google Scholar]
- Schiavo G, et al. Botulinum neurotoxin serotype F is a zinc endopeptidase specific for VAMP/synaptobrevin. J Biol Chem. 1993;268:11516–9. [PubMed] [Google Scholar]
- Valente EM, et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–60. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Haynes LP, Thomas GM, Burgoyne RD. Interaction of neuronal calcium sensor-1 and ADP-ribosylation factor 1 allows bidirectional control of phosphatidylinositol 4-kinase beta and trans-Golgi network-plasma membrane traffic. J Biol Chem. 2005;280:6047–54. doi: 10.1074/jbc.M413090200. [DOI] [PubMed] [Google Scholar]
- Zhao X, et al. Interaction of neuronal calcium sensor-1 (NCS-1) with phosphatidylinositol 4-kinase beta stimulates lipid kinase activity and affects membrane trafficking in COS-7 cells. J Biol Chem. 2001;276:40183–9. doi: 10.1074/jbc.M010281200. [DOI] [PubMed] [Google Scholar]
- Schlecker C, et al. Neuronal calcium sensor-1 enhancement of InsP3 receptor activity is inhibited by therapeutic levels of lithium. J Clin Invest. 2006;116:1668–74. doi: 10.1172/JCI22466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan MA, et al. Interaction with dopamine D2 receptor enhances expression of transient receptor potential channel 1 at the cell surface. Biochim Biophys Acta. 2008;1778:974–82. doi: 10.1016/j.bbamem.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui H, et al. Calcium-sensing mechanism in TRPC5 channels contributing to retardation of neurite outgrowth. J Physiol. 2006;572:165–72. doi: 10.1113/jphysiol.2005.102889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes LP, et al. Analysis of the interacting partners of the neuronal calcium-binding proteins L-CaBP1, hippocalcin, NCS-1 and neurocalcin delta. Proteomics. 2006;6:1822–32. doi: 10.1002/pmic.200500489. [DOI] [PubMed] [Google Scholar]
- Kozlowski DJ, et al. The zebrafish dog-eared mutation disrupts eya1, a gene required for cell survival and differentiation in the inner ear and lateral line. Dev Biol. 2005;277:27–41. doi: 10.1016/j.ydbio.2004.08.033. [DOI] [PubMed] [Google Scholar]
- Hughes I, et al. Otopetrin 1 is required for otolith formation in the zebrafish Danio rerio. Dev Biol. 2004;276:391–402. doi: 10.1016/j.ydbio.2004.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns JR, et al. Multiple roles for phosphatidylinositol 4-kinase in biosynthetic transport in polarized Madin-Darby canine kidney cells. J Biol Chem. 2002;277:2012–8. doi: 10.1074/jbc.M108571200. [DOI] [PubMed] [Google Scholar]
- Gromada J, et al. Neuronal calcium sensor-1 potentiates glucose-dependent exocytosis in pancreatic beta cells through activation of phosphatidylinositol 4-kinase beta. Proc Natl Acad Sci USA. 2005;102:10303–8. doi: 10.1073/pnas.0504487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat Cell Biol. 1999;1:523–5. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- Weisz OA, et al. Overexpression of frequenin, a modulator of phosphatidylinositol 4-kinase, inhibits biosynthetic delivery of an apical protein in polarized madin-darby canine kidney cells. J Biol Chem. 2000;275:24341–7. doi: 10.1074/jbc.M000671200. [DOI] [PubMed] [Google Scholar]
- Hendricks KB, et al. Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase. Nat Cell Biol. 1999;1:234–41. doi: 10.1038/12058. [DOI] [PubMed] [Google Scholar]
- Godi A, et al. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat Cell Biol. 1999;1:280–7. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- Haynes LP, et al. Specificity, promiscuity and localization of ARF protein interactions with NCS-1 and phosphatidylinositol-4 kinase-III beta. Traffic. 2007;8:1080–92. doi: 10.1111/j.1600-0854.2007.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, et al. Bmp4 is essential for the formation of the vestibular apparatus that detects angular head movements. PLoS Genet. 2008;4:e1000050. doi: 10.1371/journal.pgen.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. BMP4 signaling is involved in the generation of inner ear sensory epithelia. BMC Dev Biol. 2005;5:16. doi: 10.1186/1471-213X-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, ten Dijke P, Wu DK. BMP pathways are involved in otic capsule formation and epithelial-mesenchymal signaling in the developing chicken inner ear. Dev Biol. 2002;251:380–94. doi: 10.1006/dbio.2002.0822. [DOI] [PubMed] [Google Scholar]
- Mowbray C, Hammerschmidt M, Whitfield TT. Expression of BMP signalling pathway members in the developing zebrafish inner ear and lateral line. Mech Dev. 2001;108:179–84. doi: 10.1016/S0925-4773(01)00479-8. [DOI] [PubMed] [Google Scholar]
- Oh SH, Johnson R, Wu DK. Differential expression of bone morphogenetic proteins in the developing vestibular and auditory sensory organs. J Neurosci. 1996;16:6463–75. doi: 10.1523/JNEUROSCI.16-20-06463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach LM, et al. Addition of the BMP4 antagonist, noggin, disrupts avian inner ear development. Development. 2000;127:45–54. doi: 10.1242/dev.127.1.45. [DOI] [PubMed] [Google Scholar]
- Strahl T, et al. Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. J Cell Biol. 2005;171:967–79. doi: 10.1083/jcb.200504104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Yamaguchi Y. Molecular identification of a putative human hyaluronan synthase. J Biol Chem. 1996;271:22945–8. doi: 10.1074/jbc.271.38.22945. [DOI] [PubMed] [Google Scholar]
- Li Y, et al. Clinicogenetic study of PINK1 mutations in autosomal recessive early-onset parkinsonism. Neurology. 2005;64:1955–7. doi: 10.1212/01.WNL.0000164009.36740.4E. [DOI] [PubMed] [Google Scholar]
- Valente EM, et al. PINK1 mutations are associated with sporadic early-onset parkinsonism. Ann Neurol. 2004;56:336–41. doi: 10.1002/ana.20256. [DOI] [PubMed] [Google Scholar]
- Anichtchik O, et al. Loss of PINK1 function affects development and results in neurodegeneration in zebrafish. J Neurosci. 2008;28:8199–207. doi: 10.1523/JNEUROSCI.0979-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan DW, et al. Differential use of myristoyl groups on neuronal calcium sensor proteins as a determinant of spatio-temporal aspects of Ca2+ signal transduction. J Biol Chem. 2002;277:14227–37. doi: 10.1074/jbc.M111750200. [DOI] [PubMed] [Google Scholar]
- Lin R, et al. Use of the Yeast Two-Hybrid System to Detect Receptor-Protein Interactions. In: George SR, O'Dowd BF, editor. G Protein-Coupled Receptor-Protein Interactions. John Wiley & Sons, Inc.: Hoboken; 2005. pp. 201–220. [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- De Cotiis DA, et al. Optimized expression and purification of myristoylated human neuronal calcium sensor 1 in E. coli. Protein Expr Purif. 2008;61:103–12. doi: 10.1016/j.pep.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–34. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Lin R, et al. Dopamine D2 and D3 receptors are linked to the actin cytoskeleton via interaction with filamin A. Proc Natl Acad Sci USA. 2001;98:5258–63. doi: 10.1073/pnas.011538198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of human and zebrafish NBPs. Amino acid alignments of the human (h) and zebrafish (zf) NBP orthologs listed in Table 1. Identical amino acids are highlighted in black, and conserved amino acids are highlighted in gray. Amino acids are numbered to the left. File is a pdf and can be viewed with adobe acrobat reader. Genbank accession numbers for sequences used in the alignments are as follows: h_Arf1 (CAI23120), zf_Arf1 (NP_958860), h_Dan (BAA92265), zf_Dan (ACH92116), h_Hint2 (CAI10991), zf_Hint2 (ACH92117), h_Ip3r (NP_002215), zf_Ip3r (XP_696414), h_Pi4kβ (AAH00029), zf_Pi4kβ (ACH92118), h_Pink1 (AAQ89316), zf_Pink1 (ACH92119), h_Slc25a25 (CAI13838), zf_Slc25a25 (NP_998422), h_Trpc1 (CAA61447), zf_Trpc1 (XP_699455), h_Trpc5 (NP_036603), zf_Trpc5 (NP_001038292), h_Vamp2 (NP_055047), zf_Vamp2 (NP_956299).
Expression of zebrafish NBPs in head region. Whole mount in situ hybridization analysis was performed at 24, 48, and 72 hpf. Expression profiles in head region are shown for all of the NBPs represented in Figure 1. Genes are grouped according to presumed functional properties. All images are lateral views of the head, anterior to the left.