Summary
MRE11-RAD50-NBS1 (MRN) is a conserved nuclease complex that exhibits properties of a DNA damage sensor and is critical in regulating cellular responses to DNA double-strand breaks. NBS1, which is mutated in the human genetic disease Nijmegen breakage syndrome, serves as the regulatory subunit of MRN. of NBS1 by the ATM kinase is necessary for both activation of the S-phase checkpoint and for efficient DNA damage repair response. Here, we report that NBS1 is an acetylated protein and that the acetylation level is tightly regulated by the SIRT1 deacetylase. SIRT1 associates with the MRN complex and, importantly, maintains NBS1 in a hypoacetylated state, which is required for ionizing radiation-induced NBS1 Ser343 phosphorylation. Our results demonstrate the presence of crosstalk between two different posttranslational modifications in NBS1 and strongly suggest that deacetylation of NBS1 by SIRT1 plays a key role in the dynamic regulation of the DNA damage response and in the maintenance of genomic stability.
Introduction
Nijmegen breakage syndrome (NBS) is a rare autosomal recessive condition of chromosomal instability that is clinically manifested by symptoms including microcephaly, a distinct facial appearance, growth retardation, immunodeficiency, radiation sensitivity, and a strong predisposition to lymphoid malignancy (van der Burgt et al., 1996; Shiloh, 1997; Digweed and Sperling, 2004). Cells from NBS patients exhibit radiation hypersensitivity, radioresistant DNA synthesis (RDS), chromosomal instability, and cell cycle checkpoint defect (Tauchi et al., 2002). Mutations in NBS1 (also known as nibrin or p95), the product of the Nijmegen breakage syndrome gene, are responsible for NBS (Varon et al., 1998). The N-terminus of NBS1 protein contains a forkhead-associated (FHA) domain adjacent to a breast cancer carboxy-terminal (BRCT) domain, both of which are commonly found in cell cycle checkpoint proteins. The C-terminus of NBS1 is required for induction of MRN complex-mediated apoptosis in response to irradiation (Stracker et al., 2007).
The ATM protein kinase, a multi-tasking DNA damage sensor, is mutated in individuals with the radiosensitivity disorder ataxia-telangiectasia. Following cellular exposure to ionizing radiation (IR), ATM undergoes rapid autophosphorylation at Ser1981, resulting in the conversion of the inactive dimer form to active monomers (Bakkenist and Kastan, 2003). Activated ATM phosphorylates a number of cellular substrates including NBS1 (Lim et al., 2000; Wu et al., 2000; Zhao et al., 2000). NBS1 associates with MRE11 and RAD50 to form a protein complex (MRN complex) involved in detection, signaling, and repair of DNA damage. Using an Nbs1 knockout cell line, NBS1 was shown to be essential for homologous recombination DNA repair in vertebrate cells (Tauchi et al., 2002). Although phosphorylation of NBS1 does not affect MRN association, this modification is functionally important since mutant NBS1 (S343A) cannot completely complement radiosensitivity in cell lines lacking functional NBS1 (NBS cells) (Gatei et al., 2000; Lim et al., 2000; Zhao et al., 2000). In addition to serving as a downstream effector of ATM, NBS1 may function in activating ATM (Cerosaletti et al., 2006; Lee and Paull, 2005; You et al., 2005). In fact, NBS1 phosphorylation may be required for activation of the S-phase checkpoint by stimulating ATM-mediated phosphorylation of Chk2 (Lee and Paull, 2004).
Besides phosphorylation, the functions and activities of an increasing number of proteins have been found to be regulated by posttranslational acetylation on the ε-amino group of lysines (Glozak et al., 2005; Kouzarides, 2000; Yang, 2004). This modification prevents positive charges from forming on the amino group of lysines and, as a result, has a significant impact on the electrostatic properties of the protein. Over thirty proteins have been reported to possess lysine acetyltransferase activity, and many of these enzymes were first thought to specifically acetylate histones, but later were found to have a wide range of protein substrates in addition to histones (Sterner and Berger, 2000; Roth et al., 2001; Yang, 2004). Also, many acetyltransferases, including p300, CBP (CREB-binding protein), and PCAF (p300/CBP-associated factor) are transcriptional co-activators.
Like many covalent protein modifications, posttranslational lysine acetylation is highly reversible, and increasing evidences suggest that acetylation/deacetylation, like phosphorylation, is important in the regulation of a number of biological processes (Kouzarides, 2000). Thus, in order to fully understand the pathways that modulate the functions of NBS1, it is important to determine whether NBS1 undergoes acetylation and if so, whether this modification is reversibly regulated by deacetylation.
Histone deacetylases (HDACs) are enzymes that catalyze the removal of acetyl moieties from the ε-amino groups of conserved lysine residues in the amino terminal tail of histones. The removal of this modification strengthens histone-DNA interactions and may generate specific docking surfaces for proteins that regulate chromatin folding and/or transcription. Results from numerous studies overwhelmingly support the prediction that HDACs play crucial roles in gene transcription and most likely affect all eukaryotic biological processes that involve chromatin. Recent studies have shown that many non-histone proteins can serve as HDAC substrates and many HDACs, like histone acetyltransferases (HATs), regulate important biological processes that extend beyond histones and gene transcription (Glozak and Seto, 2005).
In humans, HDACs are divided into three categories: the class I RPD3-like proteins (HDAC1, HDAC2, HDAC3, HDAC8); the class II HDA1-like proteins (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10); and the class III Sir2-like proteins (SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6, and SIRT7). The class III proteins do not exhibit any sequence similarity to the other HDAC family members and differ from the other HDACs in that they require the cofactor NAD+ for activity. Whereas the NAD+-dependent class III deacetylases are specifically inhibited by nicotinamide, class I and II HDACs are specifically sensitive to the inhibitor tricohostatin A (TSA).
Of the seven human Sir2-like proteins (sirtuins), SIRT1 is most similar to the yeast Sir2 protein, which is the prototypic class III HDAC (Guarente, 2006; Blander and Guarente, 2004). In vitro, SIRT1 preferentially deacetylates histones H4K16 and H3K9, interacts with and deacetylates histone H1 at K26, and may mediate heterochromatin formation (Vaquero et al., 2004). Besides histones, more than a dozen nonhistone proteins have been found to serve as substrates for SIRT1 (Blander and Guarente, 2004; Haigis and Guarente, 2006). SIRT1 regulates the tumor suppressor protein p53 and FOXO3 to suppress apoptosis and promote cell survival (Brunet et al., 2004; Giannakou and Partridge, 2004; Luo et al., 2001; Vaziri et al., 2001). SIRT1 plays a role in several biological processes including stress resistance, metabolism, differentiation, and aging (Haigis and Guarente, 2006). In addition, thymocytes derived from mice lacking SIRT1 exhibit increased sensitivity to γ-irradiation (Cheng et al., 2003).
In the present study, we show that the NBS1 protein is acetylated. Furthermore, we found that SIRT1 binds to and deacetylates NBS1 in vitro and in vivo and, importantly, maintains NBS1 in a hypoacetylated state that is required for IR-induced NBS1 phosphorylation. Induction of NBS1 hyperacetylation greatly reduces NBS1 phosphorylation. Consistent with the importance of hypoacetylated NBS1 in DNA damage repair, cell survival decreases and RDS increases in NBS1-deficient cells complemented with hyperacetylated NBS1. Together, our findings strongly implicate crosstalk between two different posttranslational modifications in the function of NBS1. Additionally, our results uncovered NBS1 as a substrate for SIRT1 and provide convincing evidence that SIRT1 acts upstream of NBS1.
Results
NBS1 is an Acetylated Protein
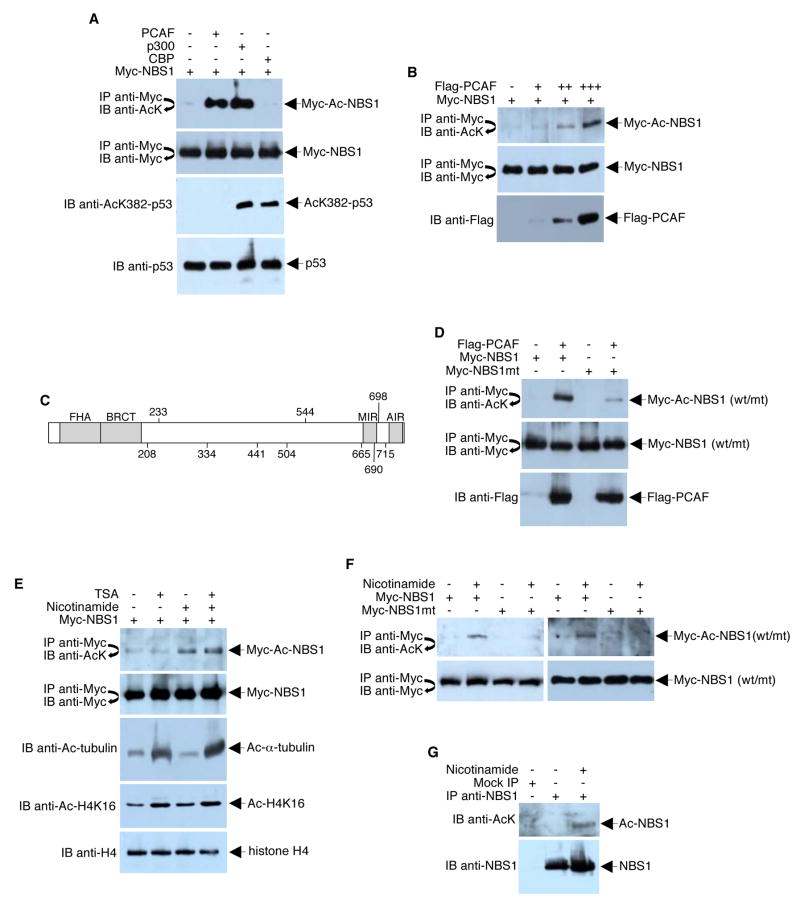
The NBS1 protein (accession BAA28616) contains three lysine acetylation consensus motifs, Kx1–2x/KK (Yang, 2004), at residues 233–237 (KGHKK), 683–687 (KNFKK), and 686–690 (KKFKK). Thus, in addition to phosphorylation, NBS1 is potentially modified by posttranslational lysine acetylation. To determine if NBS1 is indeed a substrate for acetyltransferases, 293T cells were co-transfected with plasmids expressing a Myc-tagged NBS1 protein and three different acetyltransferases (PCAF, p300, or CBP). Acetylated NBS1 was readily detected in cells that overexpressed PCAF or p300 but not in cells that overexpressed CBP (Figure 1A). The level of NBS1 acetylation by PCAF was dose-dependent (Figure 1B). of purified Myc-NBS1 by LC tandem mass spectrometry (LC-MS/MS) revealed that 10 of the 70 lysine residues were acetylated. These included K208, K233, K334, K441, K504, K544, K665, K690, K698, and K715 (Figure 1C). of all ten sites from lysine to arginine resulted in a dramatic decrease in the overall NBS1 acetylation levels as determined by Western blot analysis with anti-acetyl-lysine antibodies (Figure 1D). small amount of residual acetylation can still be detected by Western blotting with anti-acetyl-lysine antibodies, suggesting that there could be minor acetylation sites that escaped mass spectrometry analysis.
Figure 1. NBS1 is Acetylated.
(A) 293T cells were co-transfected with equal amounts (4 μg) of Myc-NBS1 ± one of the following expression plasmids: Flag-PCAF, HA-p300, or HA-CBP. Cell lysates were immunoprecipitated (IP) under high stringency conditions using anti-Myc antibodies. Immunoprecipitates were subjected to Western blot (IB) analysis using anti-acetyl-lysine (AcK) antibodies. The blot was stripped and re-probed with anti-Myc antibodies to confirm equal immunoprecipitation efficiency and loading. As CBP (but not PCAF) is known to acetylate p53-K382, a Western blot was performed with anti-AcK382-p53 to confirm that the expressed CBP was functional. (B) 293T cells were co-transfected with Myc-NBS1 (4 μg) and Flag-PCAF (0, 0.5, 1, or 4 μg). Cell lysates were subjected to immunoprecipitation/Western blot analysis using the indicated antibodies. Western blot analysis of whole extracts was performed to assess PCAF expression (bottom panel). (C) Myc-NBS1, which was expressed and purified from 293T cells, was digested with trypsin and subjected to ITMS. Positions of the unambiguously-identified acetylated lysine residues are shown. FHA, forkhead-associated domain; BRCT, BRCA1 C-terminal domain; MIR, MRE11-interacting domain; AIR, ATM-interacting domain. (D) 293T cells were co-transfected with Myc-NBS1 (wild-type or lysine to arginine mutant [mt]) and Flag-PCAF (4 μg each), as indicated. Total and acetylated NBS1 levels were analyzed by immunoprecipitation/Western blotting. (E) 293T cells were transfected with plasmids that expressed Myc-NBS1 (4 μg). Twenty-four hours post-transfection, cells were left untreated or treated overnight with TSA (1.3 μM), nicotinamide (20 mM), or TSA plus nicotinamide. Cell lysates were subjected to immunoprecipitation/Western blotting. Whole cell extracts were probed with anti-Ac-tubulin and anti-Ac-H4K16 to confirm that TSA is functional under the treatment condition (lower panels). (F) 293T cells were transfected with plasmids (4 μg each) expressing wild-type or mutant Myc-NBS1 as indicated and then treated with 20 mM nicotinamide overnight. Myc-NBS1 acetylation and protein expression were examined by immunoprecipitation/Western blotting. (G) HeLa cells were treated with nicotinamide (20 mM) or left untreated overnight. Cell lysates were then immunoprecipitated under high stringency conditions with monoclonal anti-NBS1 antibodies. Endogenous acetylated NBS1 was analyzed by Western blotting with a mixture of polyclonal anti-acetyl-lysine antibodies.
Class III HDACs Regulate NBS1 Acetylation Level
To examine whether NBS1 might be regulated by deacetylation, we immunoprecipitated Myc-NBS1 from 293T cells treated with HDAC inhibitors. shown in Figure 1E, treatment of cells with TSA, a class I and II HDAC inhibitor, did not affect the acetylation status of NBS1. contrast, cells treated with nicotinamide, a class III (Sir2 family) HDAC inhibitor, resulted in a notable increase in acetylation of NBS1. Treatment with a combination of TSA and nicotinamide was not associated with any additional increase in NBS1 acetylation when compared to treatment with nicotinamide alone. Thus, NBS1 might be deacetylated by class III HDACs but not by class I or II HDACs. Hyperacetylation of NBS1 is specific to nicotinamide treatment since the NBS1-acetylation mutant did not respond to this HDAC inhibitor (Figure 1F). Anti-acetyl-lysine Western blot analysis of anti-NBS1 immunoprecipitates obtained under high stringency conditions in which neither MRE11 nor RAD50 co-precipitated with NBS1, revealed that nicotinamide enhanced the acetylation state of endogenous NBS1 (Figure 1G), providing proof that endogenous NBS1 is regulated by class III HDACs.
NBS1 Interacts with SIRT1
An interesting feature of the class III HDACs is their subcellular location (Michishita et al., 2005). SIRT1, SIRT6, and SIRT7 are present in the nucleus while SIRT2 is located in the cytosol despite the possibility of deacetylating H4K16 during mitosis (Vaquero et al., 2006). SIRT3, SIRT4, and SIRT5 are localized in the mitochondria. Of the three nuclear SIRTs, SIRT1 has been shown to possess robust deacetylase enzymatic activity, while SIRT6 and SIRT7 have been reported to have very weak or no deacetylase activity (Liszt et al., 2005; North et al., 2003). Therefore, SIRT1 is likely responsible for the deacetylation of NBS1.
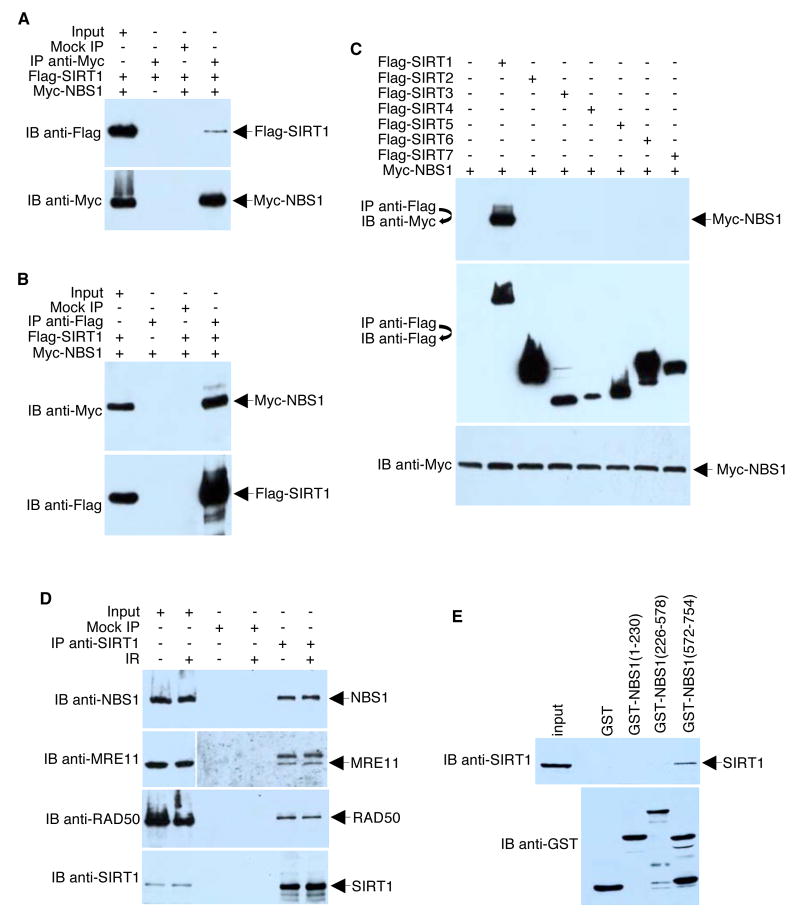
Since most, if not all, deacetylase substrates interact with their respective enzymes, we first tested the ability of NBS1 to interact with SIRT1. As shown in Figure 2A, Flag-SIRT1 co-precipitated with Myc-NBS1. Similarly, Myc-NBS1 co-precipitated with Flag-SIRT1 (Figure 2B). The SIRT1-NBS1 interaction is highly specific since none of the other Sir2 family members (SIRT2, SIRT3, SIRT4, SIRT5, SIRT6, or SIRT7) co-precipitated with NBS1 under identical conditions (Figure 2C). This association of SIRT1 and NBS1 also occurs in the absence of overexpression, epitope tagging of either protein, or IR treatment (Figure 2D). Because the subunits of the MRN complex most likely do not act individually, MRE11 and RAD50 also co-precipitated with SIRT1 as expected, suggesting that SIRT1 associates with the whole MRN complex. Analyses of three different GST-NBS1 deletions indicated that SIRT1 interacts with the C-terminus of NBS1 (residues 572–754), a segment that contains the MRE11 and ATM interaction region (Figure 2E).
Figure 2. SIRT1 Interacts With NBS1.
(A, B, C) 293T cells were co-transfected with plasmids (4 μg each) encoding the indicated Myc and Flag fusion proteins. Anti-Myc and anti-Flag immunoprecipitates obtained under low stringency conditions were analyzed by Western blotting with the indicated antibodies. (D) HeLa cells were irradiated (10 Gy) or left untreated. One hour after irradiation, endogenous SIRT1 was immunoprecipitated under low stringency conditions with anti-SIRT1 polyclonal antibodies. Immune complexes were analyzed by Western blotting with anti-NBS1, anti-MRE11, anti-RAD50, or anti-SIRT1 antibodies. For maximum clarity, the image gathered for the left upper middle panel (IB anti-MRE11) was under-exposed compared to the right upper middle panel. (E) GST-NBS1 deletion mutants coupled to Sepharose beads were incubated with whole HeLa cell extracts. After the beads were washed, bound proteins were eluted and analyzed by Western blotting with an anti-SIRT1 antibody. The blot was stripped and re-probed with anti-GST antibodies to confirm equal quantities of GST proteins in each reaction.
SIRT1 Deacetylates NBS1
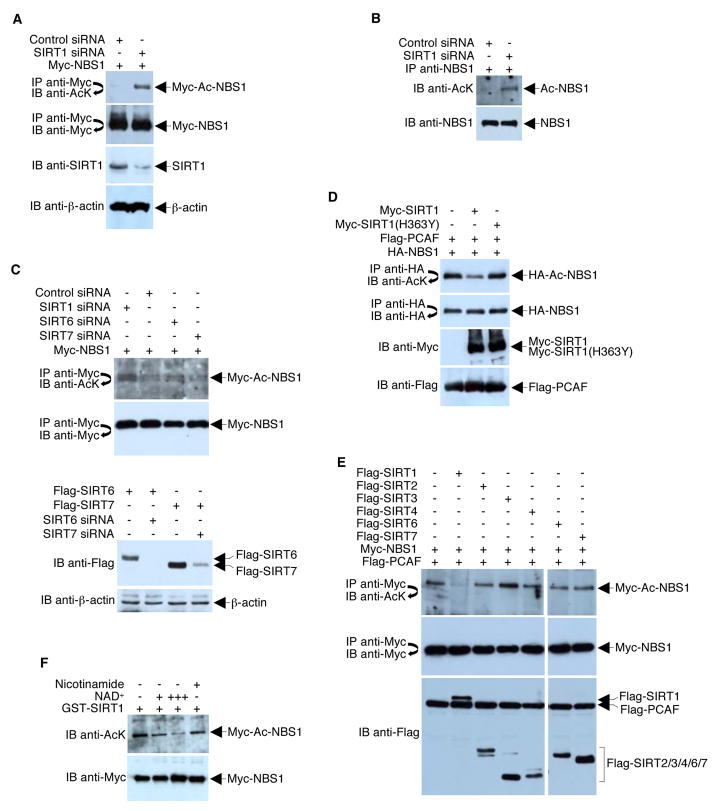
To test whether SIRT1 deacetylates NBS1, we examined the effect of RNAi-mediated SIRT1 knockdown on NBS1 acetylation in 293T cells. BS/U6-templated siRNA efficiently silenced expression of SIRT1 but not the control protein β-actin, as monitored by Western blot analysis (Figure 3A). In agreement with the finding that SIRT1 binds NBS1, acetylation of both Myc-NBS1 (Figure 3A) and endogenous NBS1 (Figure 3B) was significantly increased as a result of SIRT1 knockdown. Unlike knock-down of SIRT1, depletion of SIRT6 or SIRT7 did not affect NBS1 acetylation levels (Figure 3C). Furthermore, PCAF-acetylated NBS1 was deacetylated by wild-type SIRT1 (Figure 3D), but not by a catalytic-defective SIRT1 mutant (H363Y) (Figure 3D) or by SIRT2, SIRT3, SIRT4, SIRT6, SIRT7 (Figure 3E). In vitro deacetylation assays using purified SIRT1 in the presence of NAD+ further confirmed that NBS1 is a substrate of SIRT1 (Figure 3F). Collectively, our data strongly suggest that SIRT1 modulates the acetylation status of NBS1 via its deacetylase activity.
Figure 3. SIRT1 Deacetylates NBS1.
(A) 293T cells were co-transfected with equal amounts (4 μg) of Myc-NBS1 plasmid and either a plasmid encoding pBS/U6-SIRT1 or control siRNA. Western blots were performed with the indicated antibodies to assess the levels of SIRT1, β-actin, total Myc-NBS1, and acetylated Myc-NBS1. (B) 293T cells were infected with either adenovirus that expresses control siRNA or adenovirus that expresses SIRT1 siRNA. Western blots were performed with the indicated antibodies to assess the acetylation of endogenous NBS1 and NBS1 immunoprecipitation efficiency. (C) Upper panels, 293T cells were co-transfected with equal amounts (4 μg) of Myc-NBS1 plasmid and either a plasmid encoding SIRT1 siRNA, SIRT6 siRNA, SIRT7 siRNA, or control siRNA. Western blots were performed with the indicated antibodies to assess the levels of total Myc-NBS1 and acetylated Myc-NBS1. Bottom panels, similar experiments were performed with over-expression of Flag-SIRT6 and Flag-SIRT7 to show that SIRT6 and SIRT7 siRNAs are functional. D) 293T cells were co-transfected with plasmids that express HA-NBS1, Flag-PCAF, and either a wild-type or a catalytically-defective Myc-SIRT1. Acetylation of HA-NBS1 and all protein levels were determined with direct Western blotting or immunoprecipitations followed by Western blotting using the indicated antibodies. (E) 293T cells were co-transfected with plasmids that express Myc-NBS1, Flag-PCAF, and different Flag-tagged SIRTs. Acetylation of Myc-NBS1 and all protein levels were determined with direct Western blotting or immunoprecipitations followed by Western blotting using the indicated antibodies. (F) 293T cells were transfected with Myc-NBS1 and Flag-PCAF. Anti-Myc immunoprecipitates were incubated with recombinant GST-SIRT1 in the presence or absence of NAD+ (+, 1mM; +++, 10 mM) and nicotinamide (10 mM) at 30°C for 1 h. Western blot analysis of acetylated Myc-NBS1 was then performed using anti-acetyl-lysine (AcK) antibodies.
Acetylation Inhibits NBS1 Phosphorylation
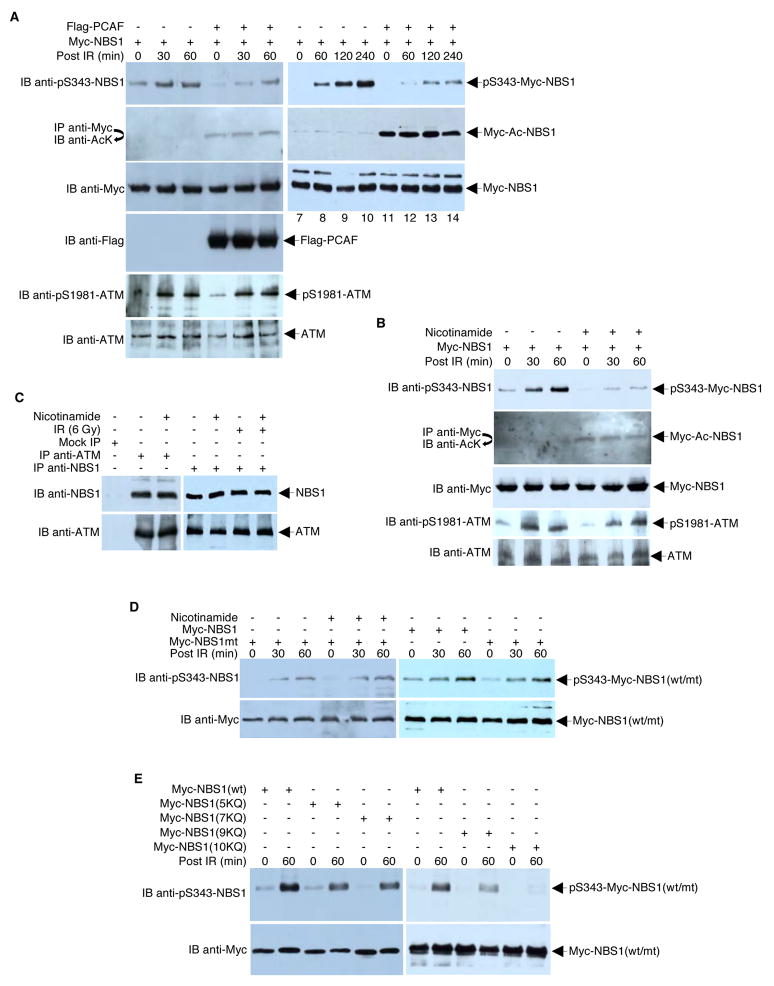
The balance between acetylation and deacetylation may have functional consequences for NBS1. Previous work has shown that ATM-mediated phosphorylation of NBS1 at Ser343 in response to DNA damage is critical for the DNA damage responses and for activation of the cell cycle S-phase checkpoint (Lim et al., 2000; Wu et al., 2000; Zhao et al., 2000). We hypothesized that different posttranslational modifications of NBS1 affect the phosphorylation of NBS1 at Ser343. NBS cells (GM07166) expressing Myc-NBS1 alone or in combination with Flag-PCAF were irradiated, and the time course of NBS1 Ser343 phosphorylation was examined via Western blot analysis. As shown in Figure 4A, overexpression of PCAF, which acetylates NBS1, reduced the levels of NBS1 phosphorylation at all times examined. Similar results were obtained when NBS cells, complemented with Myc-NBS1, were treated with nicotinamide to induce NBS1 hyperacetylation (Figure 4B). The decrease in NBS1 Ser343 phosphorylation was not due to a decrease in ATM protein levels (Figures 4A and 4B), a decrease in activated ATM (as measured by phospho-S1981 of ATM; Figures 4A and 4B), or a decrease in ATM-NBS1 interaction (Figure 4C). Treatment with nicotinamide had no effect on NBS1 Ser343 phosphorylation in an NBS1 mutant that was refractory to acetylation/deacetylation (Figure 4D).
Figure 4. NBS1 Acetylation Status Affects IR-induced NBS1 Phosphorylation.
(A) The NBS cell line GM07166 was transfected with equal amounts (4 μg) of Myc-NBS1 expression plasmid and Flag-PCAF expression plasmid or empty vector. Twenty-four hours post-transfection, cells were irradiated (6 Gy) or left untreated. Cell lysates were subjected to Western blot analysis or to high stringency immunoprecipitation/Western blot analysis, as indicated. (B) NBS cells transfected with Myc-NBS1 expression plasmid (4 μg) were treated overnight with 20 mM of nicotinamide or left untreated. The cells were then irradiated (6 Gy) or left untreated. Cell lysates were prepared after the indicated time and subjected to direct Western blot analysis or high stringency immunoprecipitation/Western blot analysis using the indicated antibodies. (C) To examine the effect of nicotinamide on ATM-NBS1 interactions, ATM and NBS1 were immunoprecipitated with anti-ATM antibodies and anti-NBS1 antibodies, respectively, from extracts prepared from HeLa cells treated ± nicotinamide. NBS1 and ATM were identified in the immunoprecipitates by Western blotting with anti-NBS1 and anti-ATM, respectively. (D) NBS cells were transfected with plasmids encoding either wild-type or acetylation-defective Myc-NBS1 (4 μg). Parallel cultures were then irradiated or left unirradiated in the presence or absence of nicotinamide. Lysates were analyzed by Western blots with the indicated antibodies. (E) NBS cells were transfected with plasmids encoding either wild-type Myc-NBS1 or acetylation-mimic Myc-NBS1 mutants (4 μg). Parallel cultures were then irradiated or left unirradiated and lysates were analyzed by Western blots with the indicated antibodies.
To further examine the effect of acetylation/deacetylation on NBS1 Ser343 phosphorylation, we constructed four plasmids that express NBS1 acetylation-mimic mutants, with lysine to glutamine changes, and introduced them into NBS cells. As shown in Figure 4E, consistent with our observation that acetylation of NBS1 inhibits phosphorylation of NBS1, mutation of lysine to glutamine in NBS1 resulted in a significant decrease in NBS1 Ser343 phosphorylation. Importantly, the decrease in NBS1 phosphorylation is strictly proportional to the number of lysine mutations (i.e., the more lysine to glutamine change, the more decrease in NBS1 phosphorylation).
Presence of SIRT1 Affects NBS1 phosphorylation
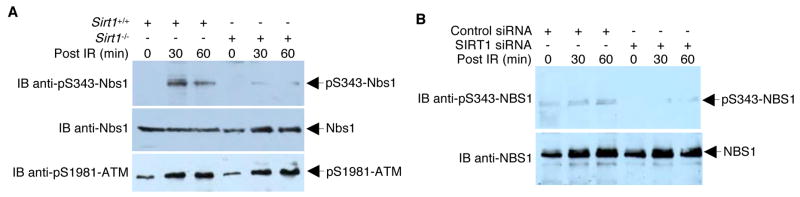
Because SIRT1 is the chief regulator of the acetylation status of NBS1 (Figure 3), we determined whether NBS1 Ser343 phosphorylation was directly affected by the presence of SIRT1. As shown in Figure 5A, Nbs1 Ser343 was hypophosphorylated in Sirt1−/− MEFs compared to wild-type MEFs. Likewise, depletion of SIRT1 by siRNA resulted in a significant decrease in NBS1 Ser343 phosphorylation (Figure 5B).
Figure 5. Presence of SIRT1 affects NBS1 Ser343 phosphorylation.
(A) Murine Sirt1+/+ and Sirt1−/− fibroblasts were irradiated (6 Gy) or left unirradiated. At different time points after IR, cell lysates were collected and analyzed by Western blotting with anti-pS343-mNbs1 antibodies. The blot was sequentially stripped and re-probed with anti-mNbs1 and pS1981-ATM antibodies to confirm equal mNbs1 protein levels and intact ATM activation. (B) 293T cells infected with adenoviruses expressing SIRT1 siRNA or control viruses were irradiated (6 Gy) or left unirradiated. At different time points after IR, cell lysates were collected and analyzed by Western blotting with anti-pS343-NBS1 antibodies. The blot was stripped and re-probed with anti-NBS1.
Acetylation of NBS1 Affects RDS
Although results from one study suggest that ATM phosphorylation of NBS1 contributes to the formation of IR-induced foci in cells (Zhao et al., 2000), other studies convincingly demonstrated that NBS1 phosphorylation does not affect formation of IR-induced foci and that the MRN complex is recruited to foci independently of ATM (Lim et al., 2000). We examined the ability of the NBS1 acetylation-defective mutant to associate with MRE11 and RAD50 and found that no significant differences exist between mutant and wild-type NBS1 (Figure S1). Furthermore, no significant differences in NBS1 foci formation could be detected between Sirt1+/+ and Sirt1−/− MEFs following IR treatment (data not shown). Therefore, our data suggest that, similar to phosphorylation, acetylation of NBS1 does not play a role in the recruitment of MRN to γH2AX domains surrounding DNA DSBs.
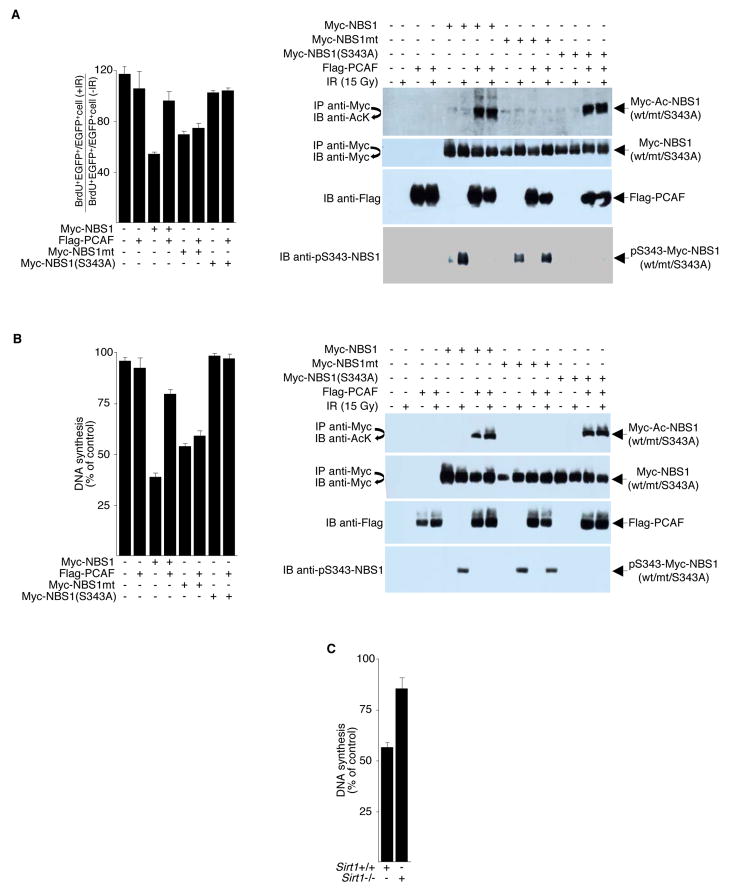
Unlike IR-induced foci formation, phosphorylation of NBS1 is clearly involved in regulating the intra-S-phase checkpoint (Lim et al., 2000; Zhao et al., 2000). A common phenotype of NBS cells is the loss of the IR-induced S-phase checkpoint, leading to RDS. Using an RDS assay, we examined the effects of NBS1 acetylation on cell cycle checkpoints activated by DNA damage. NBS cells expressing Myc-NBS1 and enhanced green fluorescent protein (EGFP) were irradiated or mock-treated immediately after 25-bromodeoxyuridine (BrdU) treatment, and the ratios of BrdU-positive/EGFP-positive cells to EGFP-positive cells were determined. Compared to wild-type NBS1, PCAF-hyperacetylated NBS1 was ineffective at inhibiting RDS, suggesting that hypoacetylation of NBS1 is crucial for intra-S-phase checkpoint activation (Figure 6A). In contrast, PCAF did not alter the ability of the acetylation-refractory mutant NBS1 to inhibit RDS. Similarly, in control experiments, PCAF had no effect on the NBS1 phosphorylation mutant (S343A) in inhibiting RDS. Western blot analysis confirmed that Ser343 phosphorylation of wildtype (Myc-NBS1), but not the acetylation mutant (Myc-NBS1mt), decreased with increased acetylation in NBS cells.
Figure 6. NBS1 Acetylation Affects Radioresistant DNA synthesis (RDS).
(A) NBS cells were transfected with plasmids encoding the indicated proteins. Twenty-four hours after transfection, cells were irradiated (15 Gy) or left unirradiated. Left panel, RDS was determined by comparing the ratio of BrdU+/EGFP+ cells to EGFP+ cells. The bar represents the ratio in the IR-treated cells divided by the ratio in the untreated cells. Right panels, to assess immunoprecipitation efficiency, protein expression, and NBS1 acetylation/phosphorylation, cell lysates were subjected to Western blot analysis or to high stringency immunoprecipitation/Western blot analysis, as indicated. Representative blots are shown. (B) Left panel, stably transfected NBS cells (GM 07166) were treated with 15 Gy of IR or left untreated, cells were radiolabeled with thymidine for RDS analysis. Right panels, representative blots to monitor immunoprecipitation efficiency, protein expression, and NBS1 acetylation/phosphorylation. (C) 2 × 104 Sirt1+/+ or Sirt1−/− MEFs were plated on 35 mm dishes for 48 h. RDS assays were performed using the radio-labeling method to determine inhibition of DNA synthesis after 15 Gy of IR. Error bars denote the standard deviation (SD). The data are expressed as mean ± SD from three separate experiments.
To be certain that NBS1 acetylation/deacetylation affects RDS, we transfected NBS cells (GM07166) with plasmids expressing wild-type or mutated NBS1, in the presence or absence of plasmids that express PCAF. Stable clones were selected by growth in G418 plus or minus puromycin, and cells stably expressing NBS1 and/or PCAF were treated with IR and examined for DNA synthesis using radiolabeled thymidine. Consistent with transient transfection results, acetylation of wild-type, but not acetylation-defective, NBS1 by PCAF significantly reduced its ability to inhibit RDS (Figure 6B).
Finally, consistent with observations that NBS1 hyperacetylation prevents its ability to inhibit RDS in transfected NBS cells, we found that Sirt1−/− MEFs, which contain hypophosphorylated and hyperacetylated Nbs1, display a higher level of RDS compared to Sirt1+/+ cells (Figure 6C).
Acetylation of NBS1 Affects Cell Survival
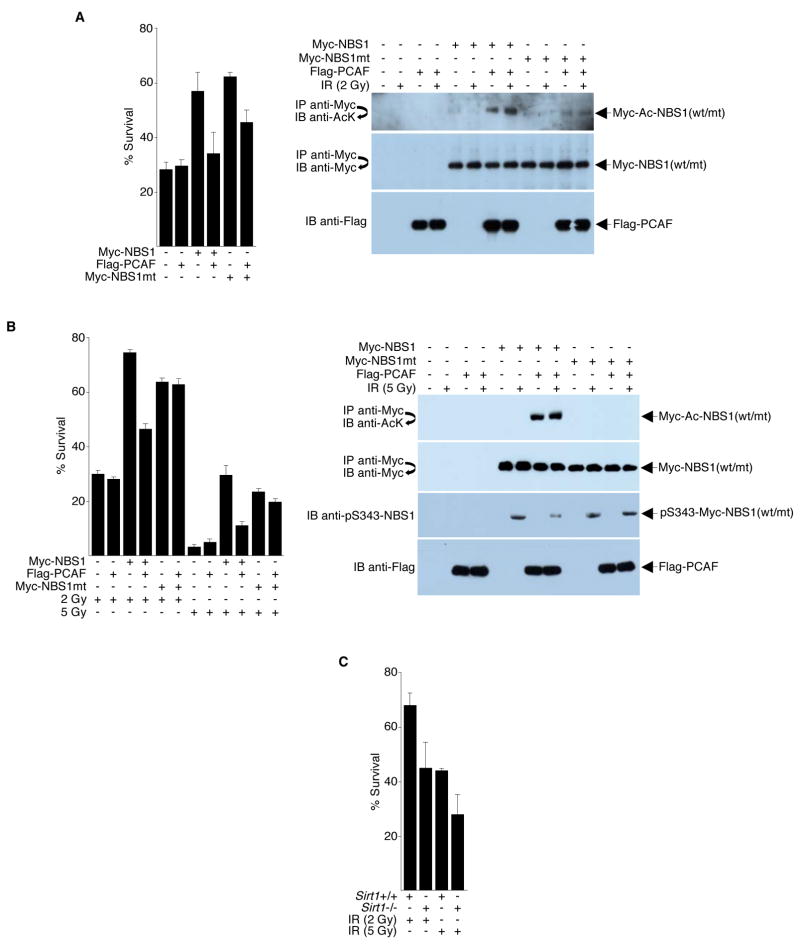
Unsuccessful DNA damage responses are reflected in an overall decrease in cell survival. Therefore, clonogenic cell survival assays were employed to examine the importance of NBS1 acetylation in DNA damage responses. NBS cells were exposed to a sublethal dose of γ-IR, and colonies were counted two weeks later. Cell transfection and immunoprecipitation efficiency, protein expression levels, and NBS1/NBS1mt acetylation levels were monitored by immunoprecipitation followed with Western blots or by direct Western blots (Figure 7A). Consistent with the finding that hyperacetylated NBS1 negatively affects cell cycle checkpoint activation, PCAF-hyperacetylated NBS1 was much less effective in reducing the IR sensitivity of NBS cells (increasing cell survival) than non-hyperacetylated NBS1. As expected, in the presence of PCAF, survival among cells expressing mutant NBS1, which is less efficiently acetylated by PCAF, was higher compared to cells expressing wild-type NBS1.
Figure 7. NBS1 Acetylation Affects Cell Survival.
(A) Left panel, NBS cells transfected with plasmids (4 μg each) encoding the indicated proteins were irradiated (2 Gy) or left unirradiated. The surviving colonies were counted two weeks later. Right panels, direct Western blots and high stringency immunoprecipitation/Western blot analyses were performed to determine NBS1 acetylation, immunoprecipitation efficiency, and protein expression levels. Representative blots are shown. (B) Left panel, stably transfected NBS cells (GM 07166) were treated with 2 or 5 Gy of IR or left untreated and surviving colonies were counted two weeks later. Right panels, direct Western blots and high stringency immunoprecipitation/Western blot analyses were performed to determine NBS1 acetylation/phosphorylation, immunoprecipitation efficiency, and protein expression levels. Although similar results were obtained from 2 Gy and 5 Gy IR treatments, representative blots from 5 Gy treatments only are shown for simplicity. (C) Sirt1+/+ and Sirt1−/− MEFs were irradiated with 2 or 5 Gy of IR or left unirradiated. The surviving colonies were counted two weeks later. Fractional cell survival is the fraction of colonies surviving irradiation divided by the total number of colonies in the unirradiated parallel culture. Error bars denote the standard deviation (SD). The data are expressed as mean ± SD from three separate experiments.
To confirm that hyperacetylation of NBS1 negatively affects cell survival, we repeated the colony survival assays in cells stably expressing wild-type or mutant NBS1 and PCAF. As shown in Figure 7B, the presence of PCAF reduced the ability of wild-type NBS1 to increase cell survival after IR treatment in stably transfected NBS cells. Likewise, in complementary experiments, the absence of Sirt1 decreased survival rate following γ-IR compared to wild-type MEFs (Figure 7C).
Discussion
HATs and HDACs play critical roles in diverse biological functions. In humans, two well-characterized HATs, hGCN5 and TIP60, play important roles in DNA repair (Brand et al., 2001; Ikura et al., 2000). In the current study, we show that NBS1 can be acetylated by PCAF and p300 and that acetylation of NBS1 inhibits Ser343 phosphorylation. At this time, the exact mechanism by which acetylation/deacetylation of NBS1 modulates phosphorylation is unclear. One plausible model is that hypoacetylation of lysines on NBS1 induces an intramolecular structural change in NBS1 that favors phosphorylation. Our data clearly indicate that NBS1 acetylation results in a decrease in cell survival and an increase in RDS most likely due to decreased NBS1 phosphorylation.
Mass spectrometry analysis of NBS1 indicated that ten of the 70 lysine residues in NBS1 were acetylated. The NBS1 protein possesses three lysine acetylation consensus motifs, Kx1–2x/KK, (previously described by Yang, 2004), at residues 233–237 (KGHKK), 683–687 (KNFKK), and 686–690 (KKFKK). However, we found that only two out of the ten potential acetylation sites identified by sequence analysis were actually acetylated (residues 233 and 690). Moreover, eight other acetylation sites detected on NBS1 by mass spectrometry analysis do not conform to any consensuses. Therefore, not only does substrate recognition by SIRT1 not dependent on the amino acid sequence proximate to the acetylated lysine (Blander et al., 2005), it appears that the NBS1 acetyltransferase(s) similarly does not have strict sequence requirements.
Although we cannot rule out the possibility that in addition to p300 and PCAF more acetyltransferases can modify NBS1, our data clearly demonstrate that NBS1 is deacetylated predominantly, if not exclusively, by SIRT1. Our results are consistent with a model in which SIRT1 is required to maintain hypoacetylated NBS1, which promotes efficient IR-induced NBS1 phosphorylation and the ensuing cellular responses to DNA damage. Thus, an additional function of SIRT1 may be to maintain NBS1 in a constant hypoacetylated state. Alternatively, IR may induce SIRT1 activity in order to regulate the acetylation status of NBS1; however, this possibility cannot be tested directly due to the low basal levels of acetylated NBS1. Despite this dilemma, these results are in accordance with the notion that SIRT1, in addition to silencing transcription, plays an important role in suppressing genomic instability and, consequently, in regulating aging in mammals (Lombard et al., 2005).
Many evidences suggest a close link between histone deacetylation and DNA repair. For example, the yeast Sin3/Rpd3 complex has been shown to modulate DNA DSB repair through the deacetylation of histone H4K16 (Jazayeri et al., 2004). The Sir2 and Rpd3 proteins are recruited to HO lesions during homology-directed repair (Tamburini and Tyler, 2005). RPD3 and HOS2 are required for the activation of DNA damage-inducible genes RNR3 and HUG1 (Sharma et al., 2007). Furthermore, HDAC4 is recruited to DSBs in vivo and, therefore, is implicated in DNA repair in mammalian cells (Kao et al., 2003). Knockdown of HDAC4 expression by RNAi increases radiosensitivity and blocks G2/M checkpoint maintenance. Also, it was reported that the SMRT-HDAC3 complex is involved in cellular recovery from DNA DSBs in mammalian cells (Yu et al., 2006). SMRT and HDAC3 are required for transcriptional repression by Ku70, a subunit of the DNA-PK DSB repair complex.
Although p53 is known to be a substrate for SIRT1 (Luo et al., 2001; Vaziri et al., 2001), the contribution of SIRT1 to p53-mediated DNA damage repair is controversial. While one study suggests that SIRT1 modulates p53-dependent DNA-damage responses (Chen et al., 2005), other reports argue that SIRT1 does not affect p53-mediated biological activities (Kamel et al., 2006; Solomon et al., 2006). More recently, SIRT6 was found to be a chromatin-associated protein that promotes normal base excision repair but does not play a role in regulating cell cycle checkpoints (Mostoslavsky et al., 2006). Our finding that SIRT1 deacetylates NBS1 and is intimately involved in the regulation of the intra-S-phase checkpoint, coupled with the recent report that DNA damage induces SIRT1 expression (Wang et al., 2006), raises the intriguing possibility that HDACs ensure genomic stability through a variety of mechanisms involving multiple pathways.
In the MRN complex, besides acetylation/deacetylation and phosphorylation of NBS1, the activities and functions of MRE11 are regulated by posttranslational modifications. MRE11 undergoes cell cycle-dependent phosphorylation in response to several genotoxic agents, and this modification requires NBS1 but not ATM (Dong et al., 1999; Yuan et al., 2002). In addition, MRE11 is methylated at arginine residues by PRMT1, and mutation of the arginines severely impairs the exonuclease activity of MRE11 although the mutated protein still forms the MRN complex (Boisvert et al., 2005). In sharp contrast to the hypoacetylation of NBS1, cells containing hypomethylated MRE11 display intra-S-phase DNA damage checkpoint defects. In preliminary studies, we found that MRE11, but not RAD50, is acetylated in the cell (data not shown). Because SIRT1 associates with the entire MRN complex, future studies are warranted to determine whether MRE11, like NBS1, is similarly regulated by SIRT1 and if so, whether acetylation/deacetylation of MRE11 interacts with other MRE11 modifications to ultimately affect the DNA DSB repair pathway.
Experimental Procedures
Plasmids, Antibodies, and Viruses
Details of all plasmid constructions and sources of recombinant viruses and antibodies are provided in the Supplementary Experimental Procedures.
Cell Culture, Transfection, and Adenovirus Infection
Sirt1+/− mice were time-mated to screen for homozygous Sirt1−/− mutant embryos by PCR. MEFs were generated from 13.5 dpc embryos using standard methods. HeLa, 293T, and murine Sirt1+/+ and Sirt1−/− fibroblasts were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS) and penicillin/streptomycin (pen/strep). The NBS cell line (GM07166) was obtained from Coriell Cell Repository and grown in minimum essential medium (MEM) with 10% FCS and pen/strep. All transfections were normalized with equal amounts of parental vector DNA. Transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. To generate stable cell lines, NBS cells (GM 07166) transfected with vector alone or various expression plasmids were grown in the presence of G418 (200 μg/ml, for cells that received NBS1 expression plasmids) and puromycin (0.5 μg/ml, for cells that received PCAF expression plasmids) for 10 days. Resistant colonies were isolated, pooled together, and grown for further analysis. viral infection, HeLa cells were infected with adenovirus for 24 h in DMEM with 0.5% BSA as previously described (Rodgers et al., 2005).
Immunoprecipitation and Western Blot Analysis
For immunoprecipitations, cells were lysed in buffer (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1% NP-40, and protease inhibitor cocktail) containing either 500 mM NaCl (high stringency) or 150 mM NaCl (low stringency). The lysates were incubated with the primary antibody overnight at 4°C. The resultant immunocomplexes were collected, washed four times in lysis buffer, and resolved by SDS-PAGE. For immunoblotting, samples were transferred onto nitrocellulose membranes. Membranes were probed with the appropriate antibodies. Proteins of interest were visualized using the Chemiluminescent Detection Kit (Pierce).
Ion Trap Mass Spectrometry
293T cells were transfected with the Myc-NBS1 expression plasmid and treated with TSA (1.3 μM) and nicotinamide (20 mM) overnight. The cells were then lysed in high stringency buffer containing 10 mM NaB and 10 mM nicotinamide. Cell extracts were subjected to immunoprecipitation with anti-Myc antibodies. The immune complexes were resolved by SDS-PAGE and stained with colloidal Blue (Invitrogen). The Myc-NBS1 sequence was analyzed with an in-house algorithm (EnzOpt) for a dual enzyme strategy maximizing proteotypic peptide coverage of all lysines. The appropriate Myc-NBS1 gel band was excised and divided into two parts. Each part was subjected to in-gel reduction and carboxyamidomethylation followed by separate tryptic or chymotryptic digestion. Acetylated peptides from each digest were detected and sequenced using microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry (μLC-MS/MS) on a Thermo LTQ linear quadrupole ion trap mass spectrometer. Data analysis was facilitated with SEQUEST and the Proteomics Browser Suite (Thermo).
GST Pull-Down Assay
GST and GST-NBS1 deletion mutants were expressed and purified from bacteria using standard methods. Equimolar quantities of the various purified proteins were conjugated to glutathione-Sepharose beads and incubated with HeLa whole cell lysates for 1 h at 4°C. After extensive washing, bound proteins were eluted and analyzed by Western blotting with anti-SIRT1 antibodies.
In Vitro Deacetylation Assay
In vitro deacetylation of NBS1 by SIRT1 was performed using an approach similar to that used previously for detection of PGC-1α acetylation (Nemoto et al., 2005).
RDS Assay
The RDS assay was performed as described previously (Zhao et al., 2000) with minor modifications. Briefly, NBS cells were transfected with either pEGFP-C3 (Clontech) or pEGFP-C3 and the Myc-NBS1 expression plasmid at a ratio of 1:10. In some experiments, cells were also transfected with an additional plasmid encoding Flag-PCAF. Cells were -irradiated with 15 Gy, incubated for 1 h at 37°C, and then incubated for an additional 2 h in a medium containing 100 μM of BrdU. BrdU incorporation was detected using an anti-BrdU antibody following immunostaining procedures described previously (Zhang et al., 2004).
Alternative RDS assays were performed as described by Lim et al. (2000) and by Zhao et al. (2002). Briefly, cells were labeled for 24 h in medium containing 10 nCi/ml of 14C-thymidine. The cells were washed once with PBS and then incubated for 24 h in a nonradioactive medium. After treatment with IR or left untreated, cells were incubated at 37°C for 30 min and pulse-labeled for 30 min in a medium containing 2.5 μCi/ml of 3H-thymidine. Cells were then washed with PBS and lysed with 0.5 ml of 0.2 M NaOH. Radioactivity was quantified in a liquid scintillation counter, and the resulting ratios of 3H/14C were calculated and compared with ratios of non-irradiated cells.
Colony Survival Assay
Colony survival assays were performed as previously described (Zhao et al., 2002) with minor modifications. Briefly, NBS cells were plated in quadruplicate (1,000 cells per 60-mm tissue culture dish). The cells were γ-irradiated with 0, 2, or 5 Gy. After two weeks, dishes were washed with PBS, fixed in ice-cold methanol for 15 min, and then stained with Giemsa stain for 30 min. Colonies on each plate were quantified and expressed as the percentage of the unirradiated control.
Supplementary Material
Acknowledgments
We would like to thank A. Ghosh, R. Goodman, I. Horikawa, M. Kastan, M. Leid, J. Petrini, P. Puigserver, and E. Verdin for providing plasmids. We would also like to thank R. Mostoslavsky and K. Chua for Sirt1+/− mice and J. Neveu and R. Robinson for LC-MS/MS. We also thank J. Koomen, N. Luetteke, A. Monteiro, and H.G. Wang for helpful discussions and for critical reading of the manuscript. We appreciate the assistance from the Moffitt Cancer Center Core Facility. This work was supported by grants from the National Institutes of Health and the Kaul Foundation (E.S.) and by fellowships from the American Heart Association (Z.Y. and N.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Blander G, Olejnik J, Krzymanska-Olejnik E, McDonagh T, Haigis M, Yaffe MB, Guarente L. SIRT1 shows no substrate specificity in vitro. J Biol Chem. 2005;280:9780–9785. doi: 10.1074/jbc.M414080200. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Dery U, Masson JY, Richard S. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 2005;19:671–676. doi: 10.1101/gad.1279805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Moggs JG, Oulad-Abdelghani M, Lejeune F, Dilworth FJ, Stevenin J, Almouzni G, Tora L. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 2001;20:3187–3196. doi: 10.1093/emboj/20.12.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Cerosaletti K, Wright J, Concannon P. Active role for nibrin in the kinetics of atm activation. Mol Cell Biol. 2006;26:1691–1699. doi: 10.1128/MCB.26.5.1691-1699.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digweed M, Sperling K. Nijmegen breakage syndrome: clinical manifestation of defective response to DNA double-strand breaks. DNA Repair. 2004;3:1207–1217. doi: 10.1016/j.dnarep.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Dong Z, Zhong Q, Chen PL. The Nijmegen breakage syndrome protein is essential for Mre11 phosphorylation upon DNA damage. J Biol Chem. 1999;274:19513–19516. doi: 10.1074/jbc.274.28.19513. [DOI] [PubMed] [Google Scholar]
- Gatei M, Young D, Cerosaletti KM, Desai-Mehta A, Spring K, Kozlov S, Lavin MF, Gatti RA, Concannon P, Khanna K. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat Genet. 2000;25:115–119. doi: 10.1038/75508. [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Partridge L. The interaction between FOXO and SIRT1: tipping the balance towards survival. Trends Cell Biol. 2004;14:408–412. doi: 10.1016/j.tcb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444:868–874. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Jazayeri A, McAinsh AD, Jackson SP. Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc Natl Acad Sci USA. 2004;101:1644–1649. doi: 10.1073/pnas.0304797101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel C, Abrol M, Jardine K, He X, McBurney MW. SirT1 fails to affect p53-mediated biological functions. Aging Cell. 2006;5:81–88. doi: 10.1111/j.1474-9726.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- Kao GD, McKenna WG, Guenther MG, Muschel RJ, Lazar MA, Yen TJ. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J Cell Biol. 2003;160:1017–1027. doi: 10.1083/jcb.200209065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayshi J, Tauchi H, Sakamoto S, Nakamura A, Morishima K, Matsuura S, Kobayashi T, Tamai K, Tanimoto K, Komatsu K. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr Biol. 2002;12:1846–1851. doi: 10.1016/s0960-9822(02)01259-9. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, Kastan MB. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzoeva OK, Petrini JH. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol Cell Biol. 2001;21:281–288. doi: 10.1128/MCB.21.1.281-288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Sharma VM, Tomar RS, Dempsey AE, Reese JC. Histone deacetylase RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol Cell Biol. 2007;27:3199–3210. doi: 10.1128/MCB.02311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. Ataxia-telangiectasia and the Nijmegen breakage syndrome: related disorders but genes apart. Annu Rev Genet. 1997;31:635–662. doi: 10.1146/annurev.genet.31.1.635. [DOI] [PubMed] [Google Scholar]
- Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol R. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Morales M, Couto SS, Hussein H, Petrini JHJ. The carboxy terminus of NBS1 is required for induction of apoptosis by the MRE11 complex. Nature. 2007;447:218–221. doi: 10.1038/nature05740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauchi H, Kobayashi J, Morishima K, van Gent DC, Shiraishi T, Verkaik NS, vanHeems D, Ito E, Nakamura A, Sonoda E, et al. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature. 2002;420:93–98. doi: 10.1038/nature01125. [DOI] [PubMed] [Google Scholar]
- Tauchi H, Matsuura S, Kobayashi J, Sakamoto S, Komatsu K. Nijmegen Breakage Syndrome gene, NBS1, and molecular links to factors for genome stability. Oncogene. 2002;21:8967–8980. doi: 10.1038/sj.onc.1206136. [DOI] [PubMed] [Google Scholar]
- van der Burgt I, Chrzanowska KH, Smeets D, Weemaes C. Nijmegen breakage syndrome. J Med Genet. 1996;33:153–156. doi: 10.1136/jmg.33.2.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ, Stumm M, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, Nemoto S, Finkel T, Gu W, Cress WD, Chen J. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- Wu X, Ranganathan V, Weisman DS, Heine WF, Ciccone DN, O’Neill TB, Crick KE, Pierce KA, Lane WS, Rathbun G, et al. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature. 2000;405:477–482. doi: 10.1038/35013089. [DOI] [PubMed] [Google Scholar]
- Yang XJ. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays. 2004;26:1076–1087. doi: 10.1002/bies.20104. [DOI] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Palmer C, Alenghat T, Li Y, Kao G, Lazar MA. The corepressor silencing mediator for retinoid and thyroid hormone receptor facilitates cellular recovery from DNA double-strand breaks. Cancer Res. 2006;66:9316–9322. doi: 10.1158/0008-5472.CAN-06-1902. [DOI] [PubMed] [Google Scholar]
- Yuan SS, Su JH, Hou MF, Yang FW, Zhao S, Lee EY. Arsenic-induced Mre11 phosphorylation is cell cycle-dependent and defective in NBS cells. DNA Repair. 2002;1:137–142. doi: 10.1016/s1568-7864(01)00009-x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wharton W, Yuan Z, Tsai SC, Olashaw N, Seto E. Activation of the growth-differentiation factor 11 gene by the histone deacetylase (HDAC) inhibitor trichostatin A and repression by HDAC3. Mol Cell Biol. 2004;24:5106–5118. doi: 10.1128/MCB.24.12.5106-5118.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Renthal W, Lee EY. Functional analysis of FHA and BRCT domains of NBS1 in chromatin association and DNA damage responses. Nucleic Acids Res. 2002;30:4815–4822. doi: 10.1093/nar/gkf612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Weng YC, Yuan SS, Lin YT, Hsu HC, Lin SC, Gerbino E, Song MH, Zdzienicka MZ, Gatti RA, et al. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature. 2000;405:473–477. doi: 10.1038/35013083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.