Abstract
Williams–Beuren syndrome (WBS) is a neurodevelopmental and multisystemic disease that results from hemizygosity of approximately 25 genes mapping to chromosomal region 7q11.23. We report here the preliminary description of eight novel genes mapping within the WBS critical region and/or its syntenic mouse region. Three of these genes, TRIM50, TRIM73 and TRIM74, belong to the TRIpartite motif gene family, members of which were shown to be associated to several human genetic diseases. We describe the preliminary functional characterization of these genes and show that Trim50 encodes an E3 ubiquitin ligase, opening the interesting hypothesis that the ubiquitin-mediated proteasome pathway might be involved in the WBS phenotype.
Keywords: Williams-Beuren syndrome, tripartite motif protein, ubiquitin ligase, contiguous gene syndrome
Introduction
The Williams–Beuren syndrome (OMIM#194050) is characterized by mental retardation with unique cognitive and personality profile and multiple dysmorphic and metabolic features.1-5 WBS is caused by a microdeletion of a 1.55 or 1.84 Mb region mapping to chromosome band 7q11.23,6 that contains about two dozen of genes.7-11 The hemizygosity of this region is almost exclusively due to sporadic de novo deletions.1,3,12 Mouse models of LIMK1, CYLN2 and GTF2IRD1 suggest that hemizygosity of these genes might play a role in some aspects of this phenotype and/or in craniofacial anomalies13-15 although no absolute association was established. On the contrary, the hemizygosity of the ELN gene was unequivocally linked to the supravalvular aortic stenosis phenotype.16 Further genotype–phenotype correlations have been proposed following diagnosis of atypical patients presenting smaller or ‘shifted’ deletions.13,17-26 Nevertheless further understanding of the molecular pathogenesis of WBS requires the identification and functional characterization of all the genes mapping within the 7q11.23 WBS region.
The TRIpartite motif (TRIM) proteins (also known as RBCC proteins) harbor from their N- to their C-terminal end, a RING (R), one or two B-boxes (B) and a predicted Coiled coil (CC).27-30 The TRIM motif is usually followed by either one or two C-terminal domains, which are specific for each member of the family. To date, more than 68 genes encoding TRIM proteins have been identified in the human genome.31 Alteration of these proteins causes pathological conditions that range from Mendelian diseases to cancer development and viral infection.32-40 In this study, we report the preliminary description of eight mammalian genes mapping in the WBS critical region (WBSCR) and/or the mouse syntenic 5G1 band and experimental evidence showing that one of those genes, TRIM50 encodes an E3 ubiquitin ligase.
Materials and methods
Gene identification and cDNAs
A fully detailed description of the procedure followed for gene identification and sequencing can be viewed in the Supplementary online section. Briefly, we screened the human and mouse dbEST databases to create clusters of ESTs mapping to the genomic interval of interest. These contigs were repeatedly extended by rescreening of dbEST until no new matching ESTs were found.41,42 To identify the 5′-end of the TRIM50-like genes 5′-rapid amplification of cDNA ends was performed on polyA+ RNAs from 12 human tissues as described.43
Northern blot
Mouse northern RNA blot-12 major tissues (Origene) were hybridized following the manufacturers' recommendations with the inserts of IMAGE clones 3484221 (Wbscr24), 602660 (Wbscr25), 336334 (Wbscr26), 4950109 (Wbscr27), with the 1452 bp Trim50 ORF fragment, or with the 464 bp Wbscr28 partial ORF fragment. Sample loading was assessed using an actin probe (not shown).
Fusion plasmids
The full-length ORF of mouse Trim50 was cloned into pCDNA3 vectors modified to include either an myc-EGFP or an HA-tag.30 Trim50 deletion mutants were created using appropriate oligonucleotides and amplification followed by in-frame insertion into the above vectors. The Trim50 A251X that correspond to a murine Trim73-like cDNA was engineered with the Quick-change directed mutagenesis kit (Stratagene).
Cell culture and transfection
293T, COS-7, HeLa, U2OS and HepG2 cells (ATCC) were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum and 1% antibiotics. Fugene 6 (Roche) was used for transfection according to the manufacturers' instructions.
Immunoprecipitation and western blot
Co-immunoprecipitation experiments were performed as described previously.44,45 Complexes were analyzed by western blotting using appropriate antibodies (anti-GFP and anti-myc (Santa Cruz Biotechnology), anti-HA (Covance) and anti-FLAG (Sigma). Horseradish peroxidase-conjugated anti-mouse and anti-rabbit antibodies (GE Healthcare) and the ECL chemiluminescence system (GE Healthcare) were used for detection. Where indicated, the MG132 proteasome inhibitor (Santa Cruz Biotechnology) was added at 10 μm for 4 h.
Interaction mating
Interaction mating is a variation of the classic two-hybrid screening. The bait plasmids express the cDNA directionally fused to the first 202 residues of LexA under the control of the constitutive ADH promoter. Prey plasmids express cDNAs fused to the B42 activation domain, the SV40 T nuclear localization signal (NLS) and an HA tag under the control of the inducible GAL1 promoter. The two plasmids are introduced separately in the two yeast haploid strains EGY48 and EGY42. The pSH18-34 vector with 6 LexA-operators lacZ and a genome integrated 4 LexA-operators LEU2 are used as reporters. EGY48/EGY42 diploids for every pairwise combination were generated by mating.46
Cross-linking and turnover rate assay
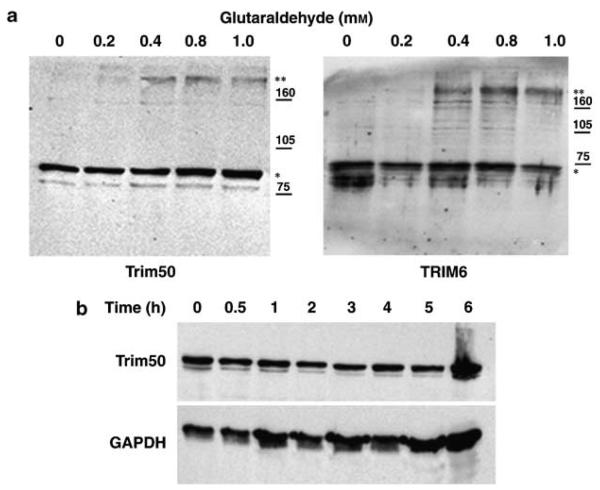
Lysates of U20S cells transfected with myc-EGFP-Trim50 were incubated with varying concentrations (0, 0.2, 0.4, 0.8, 2.0 mm) of glutaraldehyde (Sigma) at room temperature for 5 min, followed by the addition of an excess of glycine to quench the reaction. The cross-linked lysates were then subjected to western blot using anti-GFP antibody. Transfected U20S with myc-EGFP-Trim50 were treated with 100 μg/ml of cycloheximide at different time points and cell lysates containing equal amount of total proteins were subjected to western blot using anti-GFP and GAPDH antibodies.
Subcellular localization
Transfected cells were fixed in 4% formaldehyde for 15 min at room temperature, washed, permeabilized with 0.1% Triton X-100 and blocked 1 h with 1% bovine serum albumin. The anti-HA antibody (Covance) and the secondary anti-mouse AlexaFluor 568 (Molecular Probes) were used at 1:1000 and 1:500 dilutions, respectively. Cells were incubated for 1 h with the primary antibody at room temperature. After extensive washing, cells were incubated with the secondary antibody for 1 h at room temperature. Nuclei were stained with DAPI (0.5 μg/ml). MitoTracker, ER-Tracker, LysoTracker and SelectFX 488 Peroxisome Labeling Kit (all Molecular Probes) and ADLP (Chemicon) were used for staining of mitochondria, endoplasmic reticulum, lysosomes, PMP70 and ADLP peroxisomal proteins, respectively.
Results
Genes from the WBSCR
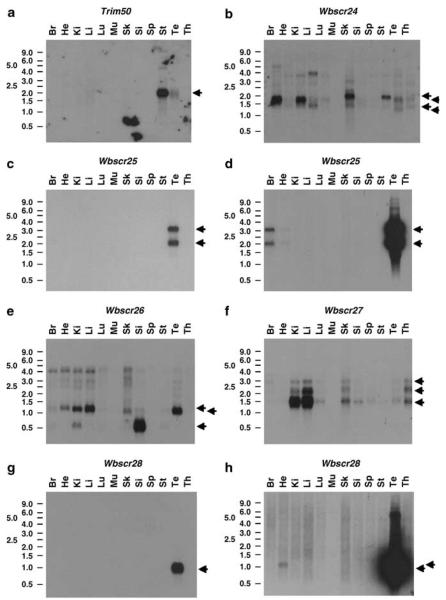
To refine the WBSCR annotation, we identified, cloned and sequenced the human WBSCR24, WBSCR26, WBSCR27, WBSCR28, TRIM50, TRIM73 and TRIM74 genes and the murine Wbscr24, Wbscr25, Wbscr26, Wbscr27, Wbscr28 and Trim50 genes (nomenclature committee approved symbols, ie, WBSCR transcript 24–28, TRIM gene 50, 73 and 74). A schematic representation of the region is depicted in Figure 1. All sequences were deposited in GenBank (AF498998–999, AK008014, AY081947–954, AY352519, AY354925–928, AY369079-080, AY372053–054 and BM008899). Mouse tissue northern blots were performed to confirm these as bona fide genes (Figure 2). The size of the Wbscr24-, Wbscr25-, Wbscr26-, Wbscr27- and Wbscr28-deposited sequences is analogous to the estimated sizes determined by northern blotting for these transcripts, suggesting that we have identified and sequenced the entire mouse cDNAs (Supplementary Table S1), while we confirmed the entire Trim50 ORF by RT–PCR followed by sequencing (see ‘Materials and methods’).
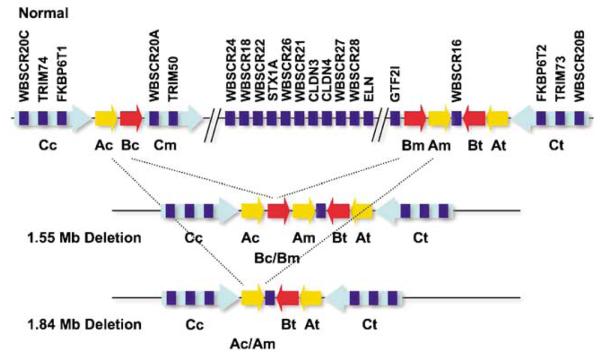
Figure 1.
Update of the schematic partial transcript map of the 7q11.23 region published by Bayes et al6 with the results discussed in this report in normal chromosome (top) and in chromosomes bearing the classical Williams–Beuren syndrome (WBS) deletions (1.55 and 1.84 Mb deletions, center and bottom, respectively). The different centromeric (c), middle (m) and telomeric (t) duplicons within the low-copy repeats are represented by specific arrows that specify their relative orientation and type (BLOCK A: yellow arrow (A); BLOCK B: red arrow (B); BLOCK C: light blue arrow (C)). Transcript units are depicted by dark blue rectangles and named above.
Figure 2.
Northern blot analyses of mouse adult tissues probed with Trim50 (a), Wbscr24 (b), Wbscr25 (c, d), Wbscr26 (e), Wbscr27 (f) and Wbscr28 (g, h). The 18 h exposures are shown for Trim50, Wbscr24, Wbscr27 and Wbscr28, while both short (left column, 30 min) and long (right column, 18 h) exposures are presented for Wbscr25 (c, d) and Wbscr28 (g, h). Arrowheads on the right mark the position of the detected mRNAs. Bars on the left indicate the position of the RNA size markers 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 4.0, 5.0, 6.0 and 9.0 kb. Br, brain; He, heart; Ki, kidney; Li, liver; Lu, lung; Mu, muscle; Sk, skin; Si, small intestine; Sp, spleen; St, stomach; Te, testis; Th, thymus.
TRIM50, TRIM73 and TRIM74
We identified three TRIM50-like copies on the human genome, TRIM50 (also known as TRIM50A), TRIM73 (TRIM50B) and TRIM74 (TRIM50C). All three are expressed, as demonstrated by the identification of spliced ESTs specific to each transcript. TRIM50 maps between WBSCR20A and FKBP6 within repeat BLOCK C-mid, TRIM73 between WBSCR20B and FKBP6T2 in BLOCK C-tel, while TRIM74 maps to the BLOCK C-cen interval, between WBSCR20C and FKBP6T1 (Figure 1).8,10,47 WBS patients are therefore hemizygotes for TRIM50, but not for TRIM73 and TRIM74 (Figure 1).6 TRIM50, TRIM73 and TRIM74 share five orthologous exons. Stronger sequence conservation is observed between the telomeric and centromeric TRIM50-like copies (TRIM73 and TRIM74, 947/949 bp identity (99.8%)), than between these copies and the central copy (TRIM50, 913/949 (96.2%) and 914/949 bp (96.3%) identity, respectively) (Figure 1). The sequences of the three TRIM50-like copies diverge after exon 5, as exons 6 and 7 are absent from BLOCK C-cen and BLOCK C-tel.
The TRIM50 transcript encodes a putative protein of 487 residues 88% identical to its murine orthologue (Trim50). The presence of a single copy of TRIM50-like genes in rodents is consistent with the absence of duplications in the WBS-corresponding region of mouse chromosome 5.11,48 TRIM50 and Trim50 harbor, from their N- to their C-terminal end, a RING, a B-box type 2, a CC and an RFP-like/B30.2 domain. TRIM73 and TRIM74 encode almost identical putative proteins of 250 residues (99.6% identical, 249/250) containing only the R, B2 and CC domains. These peptides are 93 (233/250) and 94% (234/250) identical to the first 250 amino-acid residues encoded by TRIM50. They show 80% identity to Trim50 (201/250), in a region where TRIM50 and Trim50 are 85% identical (212/250), an observation consistent with the notion that speciation preceded duplication.
Trim50 encodes an E3 ubiquitin ligase
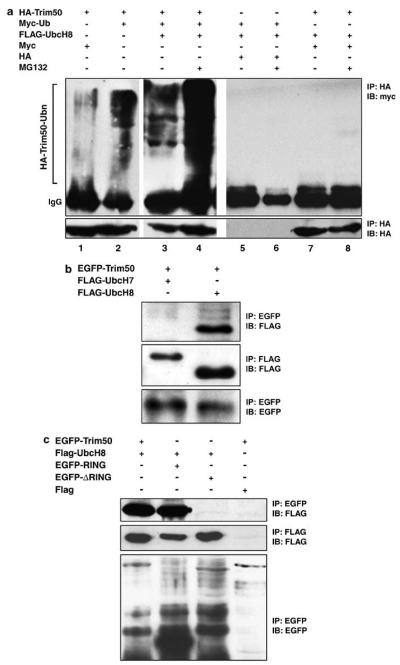
Because recent data suggest that some TRIM proteins act as E3 ubiquitin ligase31,49-55 we investigated whether Trim50 retains this ability. First, we co-transfected HEK293T cells with HA-tagged Trim50 (HA-Trim50) and myc-tagged ubiquitin-expressing plasmids or corresponding empty vectors. Total cell lysates were immunoprecipitated with anti-HA antibody, separated by gel electrophoresis and subjected to western blot analysis with anti-myc antibody (Figure 3a). The precipitated HA-Trim50 is polyubiquitinated at varying degree. These polypeptides are detected as a high-molecular weight smear. Specificity of this signal is assessed by transfection with the myc-tag vector only (compare lane 2 with lane 1 and lane 4 with lane 8). The formation of the Trim50-Ub polypeptides is increased in presence of UbcH8 (also known as UBE2L6), an E2 ubiquitin-conjugating enzyme (compare lanes 3 and 2). Further intensification of the signal is achieved through inactivation of the proteasome with the MG132 inhibitor (compare lanes 4 and 3). Similar results were obtained using transfected COS-7 cells (data not shown).
Figure 3.
Trim50 is a RING-finger-dependent E3 ubiquitin ligase. (a) The interaction between tagged Trim50 and ubiquitin was assayed in transiently transfected 293T cells expressing the indicated fusion proteins. Total cell lysates immunoprecipitated with anti-HA and immunoblotted with anti-myc revealed several Trim50-ubiquitylated proteins (lanes 2–4). (b) 293T cells co-transfected with EGFP-Trim50 and FLAG-tagged UbcH7 or UbcH8 were immunoprecipitated with anti-GFP and detected with anti-FLAG. (c) 293T cells expressing the indicated fusion proteins were immunoprecipitated with anti-GFP and revealed with anti-FLAG.
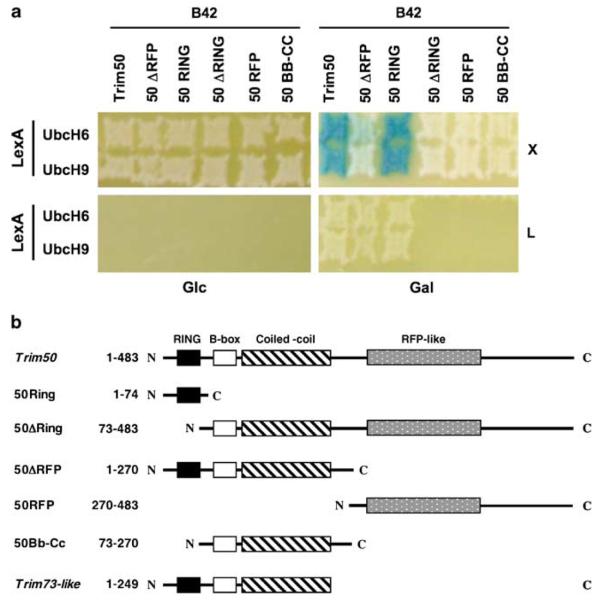
E3 ubiquitin ligase enzymes directly interact with E2 ubiquitin-conjugating enzymes and their substrate.56 To further assess the E3 ubiquitin ligase activity of Trim50, we therefore investigated if this protein was able to interact with some E2 ubiquitin-conjugating enzymes. We found that Trim50 preferentially binds to ubiquitin-conjugating enzyme UbcH8 (UBE2L6) rather than UbcH7 (UBE2L3; Figure 3b) in immunoprecipitation experiments. Moreover, we took advantage of the yeast interaction-mating technology46 to determine if Trim50 was able to bind to other E2 enzymes. The cDNAs encoding Trim50 and several E2 enzymes were expressed as fusion proteins with the LexA-DNA-binding domain (baits) and tested for interaction in a binary assay with the same proteins fused to the B42-acidic moiety (preys). Strong binding was observed with UbcH6 (UBE2E1) and UbcH9 (UBE2E3; Figure 4a). Weak binding was detected with an additional E2 enzyme, UbcH5B (UBE2D2), while no interaction was detected with UbcH2 (UBE2H) (data not shown). We then sought to investigate whether, as reported for other RING-containing E3 ubiquitin ligases, the RING domain of Trim50 was responsible of the specific interaction with the E2-conjugating enzyme.57 To assess the contribution of each TRIM domain we created a set of Trim50 deletion mutants (Figure 4b) and tested their ability to retain the binding with the E2 ubiquitin-conjugating enzymes in interaction-mating and co-immunoprecipitation experiments. We confirmed that the RING domain is necessary and sufficient for binding to E2 enzymes (Figures 3c and 4a).
Figure 4.
Interaction-mating assays show that Trim50 interacts with E2 ubiquitin-conjugating enzymes. (a) Bait strains containing plasmids that expressed LexA-fusions to E2 ubiquitin-conjugating enzymes were mated to EGY48 derivatives that contained B42 fusions to Trim50 and Trim50 deletion mutants. Plates contain either glucose (Glc) or galactose/raffinose (Gal) and are devoid of leucine (L) or supplemented with X-Gal (X). The Trim50 deletion mutants used in (a) are schematically depicted and compared to full-length Trim50 in (b).
Together these results indicate that Trim50 is a novel E3 ubiquitin ligase that through the RING domain catalyzes ubiquitination in presence of UbcH8 and possibly other E2s and that Trim50 is itself ubiquitinated.
Trim50 is a stable protein that trimerizes
TRIM proteins were shown to define specific subcellular compartments as a consequence of their propensity to form higher-order molecular weight structures and oligomerization seems to be a common feature of cytoplasmic TRIM family members.30,58,59 To investigate whether also Trim50 has this propensity, we first performed yeast interaction-mating assays and showed that Trim50 is able to homodimerize (data not shown). In addition lysates from U2OS cells transfected with myc-EGFP-Trim50 were incubated with increasing concentration of glutaraldehyde and analyzed by western blotting. As reported in Figure 5a, Trim50 is cross-linked into gel-stable complexes with high molecular weight of approximately 250 kDa consistent with the formation of trimers.
Figure 5.
(a) Trim50 forms oligomers. Lysates of U2OS cells expressing myc-EGFP-tagged Trim50 and treated with the indicated amount of glutaraldehyde were separated on SDS–PAGE and resolved by immunoblotting. TRIM6 was used as positive control. The position of the molecular weight markers is indicated in kD on the right, while the single and double asterisks mark the TRIM monomer and trimers, respectively. (b) Trim50 is a stable protein. Lysates of U20S cells transfected with myc-EGFP-Trim50 were treated with cycloheximide for a period of 6 h. Equal amounts of cell lysates were analyzed by immunoblotting with anti-GFP (top) and anti-GAPDH as control (bottom).
Next, we examined the half-life of Trim50 in U20S-transfected cell lines treated with cycloheximide and assayed for protein expression level at different points. These experiments demonstrate that Trim50 is a stable protein also 6 h after treatment (Figure 5b).
Trim50 subcellular localization
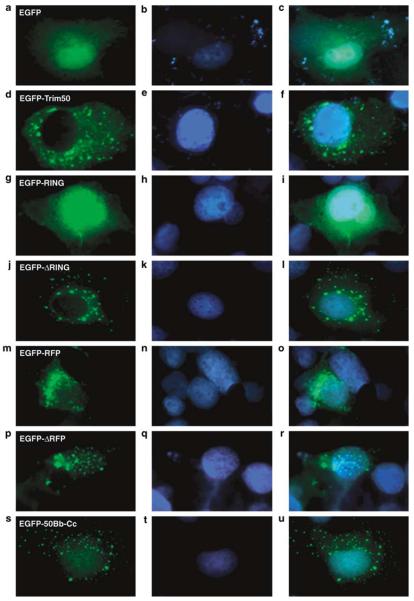
We investigated the subcellular localization of Trim50 in mammalian cell lines. Ectopically expressed Trim50 localizes into discrete cytoplasmic structures in COS-7, HeLa, HepG2 and 293T cells (Figure 6; data not shown). Next, we investigated the contribution of the TRIM domains to this localization (Figures 4b and 6). Independent expression of the RING and the RFP domain shows a diffuse nuclear and a vast perinuclear subcellular distribution, respectively (Figures 6g and m), suggesting that these domains include an NLS and a nuclear export signal, respectively. Consistently, deletion of the RFP domain induces relocalization of the mutant protein to discrete nuclear structures, while the RING domain deletion mutant localizes like the full-length protein (Figures 6j and p). The expression of a construct carrying only the B-box and the CC domains defines the same discrete cytoplasmic structures as the full-length and the RING domain-deleted protein (Figure 6s). This construct also shows a weak diffuse signal in the nucleus, suggestive of a decreased capacity of this protein to be exported outside of the nucleus. These results confirm that the central region of the tripartite proteins (B-box and CC domains) is indispensable for the proper localization to subcellular compartments,30 while the RING and RFP domains cooperate to this localization.
Figure 6.
Representative examples of subcellular localizations of exogenously expressed EGFP (a–c), EGFP-TRIM50 full-length (d–f) and deletion mutants (g–u) in fixed DAPI-counterstained COS-7 cultured cells, as visualized by indirect immunofluorescence analysis. Merged images of the first two columns are presented in the third column (c–u). A schematic representation of the Trim50 deletion mutants can be viewed in Figure 4a.
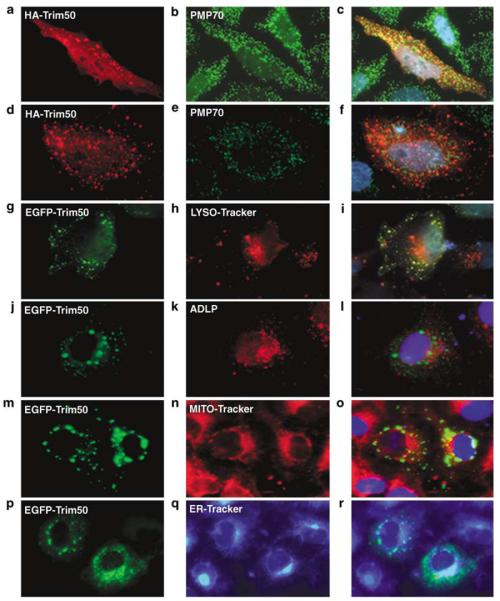
Next, we compared the localization of myc-EGFP-Trim50 with compartment-specific cellular markers. No co-localization of Trim50 staining was observed with endoplasmic reticulum (Figures 7p–r), mitochondria (Figures 7m–o) and lysosomes (Figures 7g–i). Interestingly, we observed a partial colocalization with peroxisomes (Figures 7a–f and j–l). These results are in agreement with the notion that TRIM proteins mainly define novel subcellular compartments.30
Figure 7.
Distribution of exogenously expressed full-length tagged Trim50 and endogenous cell compartments. HeLa cells transfected with HA-Trim50 (a, d) and counterstained with the PMP70 peroxisomal marker (b, e) were fixed and analyzed 24 h (a–c) and 16 h (d–f) post transfection. HepG2 cells transfected with EGFP-Trim50 (g, j, m, p) were counterstained with the LYSO-tracker lysosomial marker (h), the ALDP peroxisomal marker (k), the MITO-tracker mitochondrial marker (n) or the ER-tracker endoplasmic reticulum marker (q) 16 h post transfection. Merged images of the first two columns are presented in the third column (c–r).
Discussion
To identify candidate genes for the multiple WBS phenotypes and contribute to the update of the WBSCR annotation, we analyzed ESTs and cDNAs corresponding to previously identified.11 and novel transcription units. Our analysis allowed the mapping and sequencing of eight mammalian genes mapping to 7q11.23 (WBSCR24, WBSCR26, WBSCR27, WBSCR28, TRIM50, TRIM73, TRIM74) or the mouse syntenic 5G1 band (Wbscr24, Wbscr25, Wbscr26, Wbscr27, Wbscr28, Trim50). They encode TRIM-containing proteins (TRIM50, TRIM73, TRIM74 and Trim50), proteins containing a ‘modifier of rudimentary’ (WBSCR24 and Wbscr24) or a methyltransferase domain (WBSCR27 and Wbscr27). The role of these novel genes in the pathology of WBS remains to be determined; however we present here the preliminary genomic and functional characterization of Trim50.
Trim50 specifically interacts with E2 ubiquitin-conjugating enzymes and autoubiquitinates showing that it can act as an E3 ubiquitin ligase. This enzymatic activity was already reported for other TRIM protein members, namely TRIM5δ, TRIM18/MID1, TRIM23/ARD1, TRIM25/Efp and TRIM32/HT2A, TRIM37 targeting the degradation of proteins such as phosphatase 2A and 14-3-3σ,49-51,53-55 The interaction of Trim50 with the E2 enzymes is mediated by the RING domain as already reported for other RING-containing proteins.54,55,57,60 This E3 enzyme localizes to cytoplasmic aggregates that do not correspond to organelles such as the endoplasmic reticulum, mitochondria, lysosomes, although a very partial localization was shown with peroxisomes (Figure 7). This localization is dependent on the integrity of a central region that includes the B-box type 2 and the CC domains (Figure 6). Many other TRIM family proteins showed co-localization with cytoplasmic aggregates similar to those observed for Trim50.30 For example, TRIM20/pyrin, defective in familial Mediterranean fever, forms perinuclear aggregates when transiently expressed in cell culture.61 Similarly aggregates were reported for TRIM37, defective in mulibrey nanism,50 supporting the notion that their formation is a common propensity of TRIM proteins.31 These structures, named aggresomes, are defined as perinuclear regions where misfolded and aggregated proteins are sequestered for proteasomal degradation.62 Hence, we can speculate that the TRIM E3 ubiquitin ligases localize to these sites to control the ubiquitination and eventually the degradation of specific targets.
WBS patients are hemizygous for TRIM50, but not for the paralogous TRIM73 and TRIM74 copies (Figure 1)6 and the expression of this gene appears to be reduced accordingly in lymphoblastoid cell lines established with patient blood.63
Thus hemizygosity of the TRIM50 E3 ubiquitin ligase possibly plays a role in the WBS phenotype as the result of accumulation of specific TRIM50 target substrates.
Considering the myriad of substrates that can be targeted by an E3 ligase, it is not surprising that mutations in these genes result in several pathological conditions. Consistently, several syndromes associated with mental impairment such as Opitz syndrome (OMIM #300000), mulibrey nanism (OMIM #253250), Bardet–Biedl syndrome (OMIM #209900) and limb-girdle muscular dystrophy type 2H (OMIM #254110) are caused by mutations of TRIM family E3 ubiquitin ligases.34-36,40 For example, TRIM18/MID1, the gene mutated in Opitz syndrome, targets the catalytic subunit (PP2Ac) of the microtubule-associated phosphatase PP2A for degradation. Mutations in its B30.2 domain abolish microtubule binding leading to accumulation of PP2Ac and hypophosphorylation of microtubule-associated proteins.34,49
It is unclear how a defective TRIM50 E3 ligase activity could influence some of the clinical manifestations of WBS. The specific expression of TRIM50 and Trim50 in stomach, intestine, liver and brain (Figure 2; L Micale et al, unpublished data) suggest a possible involvement of TRIM50 haploinsufficiency in the gastrointestinal pathologies and/or the cognitive profile of WBS patients. The identification of TRIM50-interacting proteins, as well as its substrates should provide a better understanding of the biological function of this E3 ligase, as potentially an insight into the molecular pathogenesis of WBS.
It is likely, although not experimentally verified in this study, that both TRIM73 and TRIM74 proteins retain their ability to act as E3 ligase because they have the CC and RING domains. However, these proteins might not be fully redundant with TRIM50, because the RFP-like domain (also known as B30.2), that is absent in TRIM73 and TRIM74, was suggested to be important for interaction with proteasome subunits.64 Furthermore, our Trim73-like construct, which mimics TRIM73 and TRIM74, also shows a nuclear staining absent in Trim50 (Figure 4b; Supplementary Figure S1). Consistently, analysis of MID1 (TRIM18) mutations detected in Opitz syndrome patients demonstrated the importance of the RFP-like domain in the proper localization of the protein.58,65
In conclusion, the present report increases the number of identified genes mapping within the region commonly deleted in WBS patients and thus putatively involved in phenotype determination. Having shown that one of these novel genes, TRIM50, acts as an E3 ubiquitin ligase opens the interesting hypothesis of a direct involvement of ubiquitination in the WBS pathology.
Supplementary Material
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Acknowledgements
We thank Satoshi Inoue for reagents and Stylianos E Antonarakis and Samuel Deutsch for discussion. This work was supported by grants from the Jérôme Lejeune Foundation, the Désirée and Niels Yde Foundation, the Novartis Foundation, the Swiss National Science Foundation and European commission (grant 037627) to AR, grants from the Italian Ministry of Health (RC 2006–2007), the Fondazione Banca del Monte di Foggia ‘Domenico Siniscalco Ceci’, Italian Telethon Foundation (Grant N. GGP06122) and the Jérôme Lejeune Foundation to GiM, and the Italian Telethon Foundation to GeM.
Footnotes
References
- 1.Osborne LR. Williams–Beuren syndrome: unraveling the mysteries of a microdeletion disorder. Mol Genet Metab. 1999;67:1–10. doi: 10.1006/mgme.1999.2844. [DOI] [PubMed] [Google Scholar]
- 2.Morris CA, Demsey SA, Leonard CO, Dilts C, Blackburn BL. Natural history of Williams syndrome: physical characteristics. J Pediatr. 1988;113:318–326. doi: 10.1016/s0022-3476(88)80272-5. [DOI] [PubMed] [Google Scholar]
- 3.Francke U. Williams–Beuren syndrome: genes and mechanisms. Hum Mol Genet. 1999;8:1947–1954. doi: 10.1093/hmg/8.10.1947. [DOI] [PubMed] [Google Scholar]
- 4.Cherniske EM, Carpenter TO, Klaiman C, et al. Multisystem study of 20 older adults with Williams syndrome. Am J Med Genet. 2004;131A:255–264. doi: 10.1002/ajmg.a.30400. [DOI] [PubMed] [Google Scholar]
- 5.Stagi S, Bindi G, Neri AS, et al. Thyroid function and morphology in patients affected by Williams syndrome. Clin Endocrinol (Oxf) 2005;63:456–460. doi: 10.1111/j.1365-2265.2005.02365.x. [DOI] [PubMed] [Google Scholar]
- 6.Bayes M, Magano LF, Rivera N, Flores R, Perez Jurado LA. Mutational mechanisms of Williams–Beuren syndrome deletions. Am J Hum Genet. 2003;73:131–151. doi: 10.1086/376565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairo S, Merla G, Urbinati F, Ballabio A, Reymond A. WBSCR14, a gene mapping to the Williams–Beuren syndrome deleted region, is a new member of the Mlx transcription factor network. Hum Mol Genet. 2001;10:617–627. doi: 10.1093/hmg/10.6.617. [DOI] [PubMed] [Google Scholar]
- 8.Merla G, Ucla C, Guipponi M, Reymond A. Identification of additional transcripts in the Williams–Beuren syndrome critical region. Hum Genet. 2002;110:429–438. doi: 10.1007/s00439-002-0710-x. [DOI] [PubMed] [Google Scholar]
- 9.Makeyev AV, Erdenechimeg L, Mungunsukh O, et al. GTF2IRD2 is located in the Williams–Beuren syndrome critical region 7q11.23 and encodes a protein with two TFII-I-like helix-loop-helix repeats. Proc Natl Acad Sci USA. 2004;101:11052–11057. doi: 10.1073/pnas.0404150101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doll A, Grzeschik KH. Characterization of two novel genes, WBSCR20 and WBSCR22, deleted in Williams–Beuren syndrome. Cytogenet Cell Genet. 2001;95:20–27. doi: 10.1159/000057012. [DOI] [PubMed] [Google Scholar]
- 11.DeSilva U, Elnitski L, Idol JR, et al. Generation and comparative analysis of approximately 3.3 Mb of mouse genomic sequence orthologous to the region of human chromosome 7q11.23 implicated in Williams syndrome. Genome Res. 2002;12:3–15. doi: 10.1101/gr.214802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stromme P, Bjornstad PG, Ramstad K. Prevalence estimation of Williams syndrome. J Child Neurol. 2002;17:269–271. doi: 10.1177/088307380201700406. [DOI] [PubMed] [Google Scholar]
- 13.Morris CA, Mervis CB, Hobart HH, et al. GTF2I hemizygosity implicated in mental retardation in Williams syndrome: genotype-phenotype analysis of five families with deletions in the Williams syndrome region. Am J Med Genet. 2003;123A:45–59. doi: 10.1002/ajmg.a.20496. [DOI] [PubMed] [Google Scholar]
- 14.Hoogenraad CC, Koekkoek B, Akhmanova A, et al. Targeted mutation of Cyln2 in the Williams syndrome critical region links CLIP-115 haploinsufficiency to neurodevelopmental abnormalities in mice. Nat Genet. 2002;32:116–127. doi: 10.1038/ng954. [DOI] [PubMed] [Google Scholar]
- 15.Tassabehji M, Hammond P, Karmiloff-Smith A, et al. GTF2IRD1 in craniofacial development of humans and mice. Science. 2005;310:1184–1187. doi: 10.1126/science.1116142. [DOI] [PubMed] [Google Scholar]
- 16.Ewart AK, Morris CA, Atkinson D, et al. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet. 1993;5:11–16. doi: 10.1038/ng0993-11. [DOI] [PubMed] [Google Scholar]
- 17.Hirota H, Matsuoka R, Chen XN, et al. Williams syndrome deficits in visual spatial processing linked to GTF2IRD1 and GTF2I on chromosome 7q11.23. Genet Med. 2003;5:311–321. doi: 10.1097/01.GIM.0000076975.10224.67. [DOI] [PubMed] [Google Scholar]
- 18.Botta A, Sangiuolo F, Calza L, et al. Expression analysis and protein localization of the human HPC- 1/syntaxin 1A, a gene deleted in Williams syndrome. Genomics. 1999;62:525–528. doi: 10.1006/geno.1999.5987. [DOI] [PubMed] [Google Scholar]
- 19.Howald C, Merla G, Digilio MC, et al. Two high throughput technologies to detect segmental aneuploidies identify new Williams–Beuren syndrome patients with atypical deletions. J Med Genet. 2006;43:266–273. doi: 10.1136/jmg.2005.034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gagliardi C, Bonaglia MC, Selicorni A, Borgatti R, Giorda R. Unusual cognitive and behavioural profile in a Williams syndrome patient with atypical 7q11.23 deletion. J Med Genet. 2003;40:526–530. doi: 10.1136/jmg.40.7.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heller R, Rauch A, Luttgen S, Schroder B, Winterpacht A. Partial deletion of the critical 1.5 Mb interval in Williams–Beuren syndrome. J Med Genet. 2003;40:e99. doi: 10.1136/jmg.40.8.e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karmiloff-Smith A, Grant J, Ewing S, et al. Using case study comparisons to explore genotype-phenotype correlations in Williams–Beuren syndrome. J Med Genet. 2003;40:136–140. doi: 10.1136/jmg.40.2.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korenberg JR, Chen XN, Hirota H, et al. Genome structure and cognitive map of Williams syndrome. J Cogn Neurosci. 2000;12(Suppl 1):89–107. doi: 10.1162/089892900562002. [DOI] [PubMed] [Google Scholar]
- 24.Tassabehji M, Metcalfe K, Karmiloff-Smith A, et al. Williams syndrome: use of chromosomal microdeletions as a tool to dissect cognitive and physical phenotypes. Am J Hum Genet. 1999;64:118–125. doi: 10.1086/302214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edelmann L, Prosnitz A, Pardo S, et al. An atypical deletion of the Williams–Beuren Syndrome interval implicates genes associated with defective visuospatial processing and autism. J Med Genet. 2007;44:136–143. doi: 10.1136/jmg.2006.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schubert C, Laccone F. Williams–Beuren syndrome: determination of deletion size using quantitative real-time PCR. Int J Mol Med. 2006;18:799–806. [PubMed] [Google Scholar]
- 27.Reddy BA, Etkin LD, Freemont PS. A novel zinc finger coiled-coil domain in a family of nuclear proteins. Trends Biochem Sci. 1992;17:344–345. doi: 10.1016/0968-0004(92)90308-v. [DOI] [PubMed] [Google Scholar]
- 28.Cao T, Borden KL, Freemont PS, Etkin LD. Involvement of the rfp tripartite motif in protein-protein interactions and subcellular distribution. J Cell Sci. 1997;110:1563–1571. doi: 10.1242/jcs.110.14.1563. [DOI] [PubMed] [Google Scholar]
- 29.Borden KL. RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem Cell Biol. 1998;76:351–358. doi: 10.1139/bcb-76-2-3-351. [DOI] [PubMed] [Google Scholar]
- 30.Reymond A, Meroni G, Fantozzi A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 32.Consortium A candidate gene for familial Mediterranean fever. The French FMF Consortium. Nat Genet. 1997;17:25–31. doi: 10.1038/ng0997-25. [DOI] [PubMed] [Google Scholar]
- 33.Consortium Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. The International FMF Consortium. Cell. 1997;90:797–807. doi: 10.1016/s0092-8674(00)80539-5. [DOI] [PubMed] [Google Scholar]
- 34.Quaderi NA, Schweiger S, Gaudenz K, et al. Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat Genet. 1997;17:285–291. doi: 10.1038/ng1197-285. [DOI] [PubMed] [Google Scholar]
- 35.Frosk P, Weiler T, Nylen E, et al. Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am J Hum Genet. 2002;70:663–672. doi: 10.1086/339083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Avela K, Lipsanen-Nyman M, Idanheimo N, et al. Gene encoding a new RING-B-box-Coiled-coil protein is mutated in mulibrey nanism. Nat Genet. 2000;25:298–301. doi: 10.1038/77053. [DOI] [PubMed] [Google Scholar]
- 37.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 38.Grignani F, Fagioli M, Alcalay M, et al. Acute promyelocytic leukemia: from genetics to treatment. Blood. 1994;83:10–25. [PubMed] [Google Scholar]
- 39.Le Douarin B, Zechel C, Garnier JM, et al. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang AP, Beck JS, Yen HJ, et al. Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet–Biedl syndrome gene (BBS11) Proc Natl Acad Sci USA. 2006;103:6287–6292. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reymond A, Camargo AA, Deutsch S, et al. Nineteen additional unpredicted transcripts from human chromosome 21. Genomics. 2002;79:824–832. doi: 10.1006/geno.2002.6781. [DOI] [PubMed] [Google Scholar]
- 42.Reymond A, Friedli M, Henrichsen CN, et al. From PREDs and open reading frames to cDNA isolation: revisiting the human chromosome 21 transcription map. Genomics. 2001;78:46–54. doi: 10.1006/geno.2001.6640. [DOI] [PubMed] [Google Scholar]
- 43.Denoeud F, Kapranov P, Ucla C, et al. Prominent use of distal 5′ transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res. 2007;17:746–759. doi: 10.1101/gr.5660607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reymond A, Volorio S, Merla G, et al. Evidence for interaction between human PRUNE and nm23-H1 NDPKinase. Oncogene. 1999;18:7244–7252. doi: 10.1038/sj.onc.1203140. [DOI] [PubMed] [Google Scholar]
- 45.Merla G, Howald C, Antonarakis SE, Reymond A. The subcellular localization of the ChoRE-binding protein, encoded by the Williams–Beuren syndrome critical region gene 14, is regulated by 14-3-3. Hum Mol Genet. 2004;13:1505–1514. doi: 10.1093/hmg/ddh163. [DOI] [PubMed] [Google Scholar]
- 46.Reymond A, Brent R. p16 proteins from melanoma-prone families are deficient in binding to Cdk4. Oncogene. 1995;11:1173–1178. [PubMed] [Google Scholar]
- 47.Valero MC, de Luis O, Cruces J, Perez Jurado LA. Fine-scale comparative mapping of the human 7q11.23 region and the orthologous region on mouse chromosome 5G: the low-copy repeats that flank the Williams–Beuren syndrome deletion arose at breakpoint sites of an evolutionary inversion(s) Genomics. 2000;69:1–13. doi: 10.1006/geno.2000.6312. [DOI] [PubMed] [Google Scholar]
- 48.DeSilva U, Massa H, Trask BJ, Green ED. Comparative mapping of the region of human chromosome 7 deleted in Williams syndrome. Genome Res. 1999;9:428–436. [PMC free article] [PubMed] [Google Scholar]
- 49.Trockenbacher A, Suckow V, Foerster J, et al. MID1, mutated in Opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nat Genet. 2001;29:287–294. doi: 10.1038/ng762. [DOI] [PubMed] [Google Scholar]
- 50.Kallijarvi J, Lahtinen U, Hamalainen R, Lipsanen-Nyman M, Palvimo JJ, Lehesjoki AE. TRIM37 defective in mulibrey nanism is a novel RING finger ubiquitin E3 ligase. Exp Cell Res. 2005;308:146–155. doi: 10.1016/j.yexcr.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 51.Vichi A, Payne DM, Pacheco-Rodriguez G, Moss J, Vaughan M. E3 ubiquitin ligase activity of the trifunctional ARD1 (ADP-ribosylation factor domain protein 1) Proc Natl Acad Sci USA. 2005;102:1945–1950. doi: 10.1073/pnas.0409800102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diaz-Griffero F, Li X, Javanbakht H, et al. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology. 2006;349:300–315. doi: 10.1016/j.virol.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 53.Horn EJ, Albor A, Liu Y, et al. RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis. 2004;25:157–167. doi: 10.1093/carcin/bgh003. [DOI] [PubMed] [Google Scholar]
- 54.Urano T, Saito T, Tsukui T, et al. Efp targets 14-3-3 sigma for proteolysis and promotes breast tumour growth. Nature. 2002;417:871–875. doi: 10.1038/nature00826. [DOI] [PubMed] [Google Scholar]
- 55.Xu L, Yang L, Moitra PK, et al. BTBD1 and BTBD2 colocalize to cytoplasmic bodies with the RBCC/tripartite motif protein, TRIM5delta. Exp Cell Res. 2003;288:84–93. doi: 10.1016/s0014-4827(03)00187-3. [DOI] [PubMed] [Google Scholar]
- 56.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 57.Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 58.Cainarca S, Messali S, Ballabio A, Meroni G. Functional characterization of the Opitz syndrome gene product (midin): evidence for homodimerization and association with microtubules throughout the cell cycle. Hum Mol Genet. 1999;8:1387–1396. doi: 10.1093/hmg/8.8.1387. [DOI] [PubMed] [Google Scholar]
- 59.Li X, Gold B, O'HUigin C, et al. Unique features of TRIM5alpha among closely related human TRIM family members. Virology. 2007;360:419–433. doi: 10.1016/j.virol.2006.10.035. [DOI] [PubMed] [Google Scholar]
- 60.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 61.Tidow N, Chen X, Muller C, et al. Hematopoietic-specific expression of MEFV, the gene mutated in familial Mediterranean fever, and subcellular localization of its corresponding protein, pyrin. Blood. 2000;95:1451–1455. [PubMed] [Google Scholar]
- 62.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Merla G, Howald C, Henrichsen CN, et al. Submicroscopic deletion in patients with Williams–Beuren syndrome influences expression levels of the nonhemizygous flanking genes. Am J Hum Genet. 2006;79:332–341. doi: 10.1086/506371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzumori N, Burns KH, Yan W, Matzuk MM. RFPL4 interacts with oocyte proteins of the ubiquitin-proteasome degradation pathway. Proc Natl Acad Sci USA. 2003;100:550–555. doi: 10.1073/pnas.0234474100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schweiger S, Foerster J, Lehmann T, et al. The Opitz syndrome gene product, MID1, associates with microtubules. Proc Natl Acad Sci USA. 1999;96:2794–2799. doi: 10.1073/pnas.96.6.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)