Summary
The nucleus is one of the most prominent cellular organelles, yet surprisingly little is known about how it is formed, what determines its shape and what defines its size. As the nuclear envelope (NE) disassembles in each and every cell cycle in metazoans, the process of rebuilding the nucleus is crucial for proper development and cell proliferation. In this Commentary, we summarize what is known about the regulation of nuclear shape and size, and highlight recent findings that shed light on the process of building a nucleus, including new discoveries related to NE assembly and the relationship between the NE and the endoplasmic reticulum (ER). Throughout our discussion, we note interesting aspects of nuclear structure that have yet to be resolved. Finally, we present an idea – which we refer to as `the limited flat membrane hypothesis' – to explain the formation of a single nucleus that encompasses of all of the cell's chromosomes following mitosis.
Keywords: ER membrane, Reticulons, ER tubules, ER sheets, Lamina, Nuclear envelope assembly, Nuclear pore complex, Lipin
Introduction
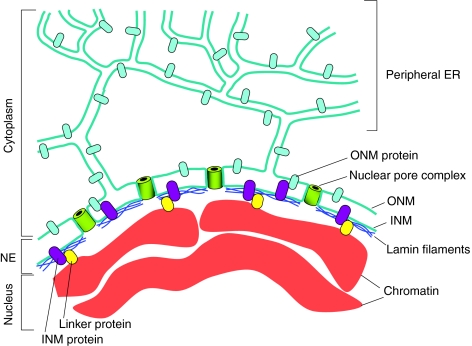
Most eukaryotic cells have a single nucleus in which a nuclear envelope (NE) separates the chromosomes from the cytoplasm. The NE (Fig. 1) is made of a double membrane that is perforated by nuclear-pore complexes (NPCs). The outer nuclear membrane, which is continuous with the ER, connects with the inner nuclear membrane at the curved membrane regions that surround each NPC. In metazoans, the NE also contains a meshwork of proteins, which are collectively called the nuclear lamina, that underlies the inner nuclear membrane and interacts with portions of the chromatin. The nuclear lamina is made predominantly of intermediate filaments called lamins, of which there are two main types: type A and type B (for a review, see Dechat et al., 2008). In mammals, there are two major A-type lamins, lamin A and lamin C, which are generated by alternative splicing of the LMNA gene. There are also two major B-type lamins, lamin B1 and lamin B2, each encoded by its own gene. Lamin A and the two lamin Bs have a C-terminal domain that is lipid modified (farnesylated), thereby promoting their attachment to the inner nuclear membrane. This domain is missing in lamin C and is normally removed by proteolytic cleavage in lamin A. In addition to their location at the nuclear periphery, lamins are also present in the nucleoplasm (Dechat et al., 2008).
Fig. 1.
The nuclear envelope. The NE is an integral part of the ER-membrane network (in blue-green). The inner nuclear membrane (INM) and outer nuclear membrane (ONM) connect at sites of NPCs (green barrels) where the membrane curves as it surrounds the NPC. The ONM is continuous with the peripheral ER. The NE contains a variety of proteins that are embedded in the INM (purple) or the ONM (light blue). Most ONM proteins are also found in the peripheral ER. INM proteins can interact with the underlying nuclear lamina (dark blue), with ONM proteins or with chromatin (red), often through linker proteins (yellow). For a detailed description of the various proteins associated with the NE see recent reviews (Crisp and Burke, 2008; Guttinger et al., 2009).
The nuclear lamina also contains a variety of lamin-associated proteins and other proteins that are embedded in the inner nuclear membrane (reviewed by Wilhelmsen et al., 2006). The nuclear lamina might also play a role in the internal organization of the nucleus. For example, within the nucleus, chromosomes are organized in chromosome territories: rather than being distributed throughout the nucleus, each chromosome appears to occupy a discrete region, or territory (for a review, see Cremer et al., 2006). In addition, heterochromatic chromosomal regions, which are chromosome domains that are transcriptionally inactive, tend to localize at the nuclear periphery (Akhtar and Gasser, 2007). It is likely that the nuclear lamina contributes to these types of intranuclear chromosome organization. Unicellular eukaryotes and plant cells do not have lamins, although they might have proteins that function as a nuclear lamina.
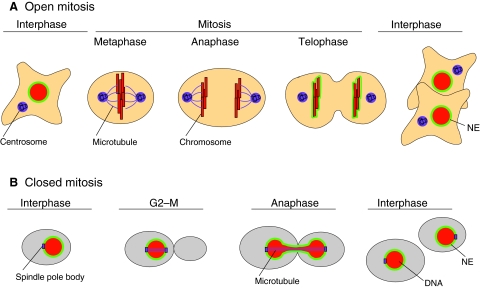
During mitosis, a parent cell gives rise to two daughter cells, each with its own nucleus. Two main strategies have evolved to successfully carry out this task: open mitosis and closed mitosis (Fig. 2). Open mitosis occurs in most eukaryotic cells, whereas closed mitosis occurs in certain species of fungi. In open mitosis, the NE disassembles early in mitosis, allowing microtubules that emanate from cytoplasmic centrosomes to contact the chromosomes and promote their segregation (reviewed by Prunuske and Ullman, 2006). At the end of open mitosis, the NE reassembles around the two segregated DNA masses to form the two daughter nuclei. In closed mitosis, the NE does not disassemble and chromosome segregation takes place entirely within the confines of the nucleus. This strategy works in cell types where the centrosome equivalents, known as spindle-pole bodies, are embedded in the NE, allowing microtubules to associate with chromosomes without the need for NE disassembly. Some organisms, such as Aspergillus nidulans, undergo semi-open mitosis, in which the partial disassembly of the NPCs creates large holes in the NE, but the envelope itself does not completely disassemble (for a review, see De Souza and Osmani, 2007).
Fig. 2.
Open and closed mitosis. (A) Open mitosis is so named because of the disassembly of the NE (green) during mitosis, which opens up the nucleus and exposes the chromosomes (red) to the cytoplasm. The NE breaks down early in mitosis, as the chromosomes condense, allowing microtubules (purple filaments) that emanate from centrosomes (purple structures) to associate with the chromosomes. During mitosis, the chromosomes congress to the metaphase plate, followed by separation of sister chromatids in anaphase. The NE begins to reassemble shortly thereafter, in telophase. Once the NE is completely assembled, the nucleus expands and the chromosomes return to their decondensed state in interphase. (B) Closed mitosis is so named because of the persistence of the NE throughout the cell cycle, such that the nucleus never `opens' to the cytoplasm. This type of mitosis occurs in certain fungi (such as budding yeast, shown here), in which the centrosome equivalents, called the spindle-pole bodies (purple), are embedded in the NE. During closed mitosis, the spindle-pole bodies nucleate microtubules within the nucleus, but as the DNA (red) begins to segregate, the nucleus has to elongate. Once segregation is completed, the nucleus divides and re-establishes a spherical shape. Note that, in budding yeast, chromosome condensation and a metaphase plate are not visible by microscopy.
In all forms of mitosis – open, closed or semi-open – the NE undergoes dramatic structural changes. During open mitosis, the NE has to reform around all of the chromosomes, rather than around a subset of chromosomes, and it then must expand to attain its final size and shape. During closed mitosis, the NE expands to accommodate the movement of the segregating chromosomes, and it must then get cleaved, resealed and restructured to form two round daughter nuclei. In the past few years, numerous studies have shed light on the molecular mechanisms that underlie these processes. To understand how these nuclear gymnastics take place, we first discuss the factors that contribute to nuclear shape and size. We then examine how the NE reassembles to form a nucleus with the proper attributes following mitosis.
Nuclear shape
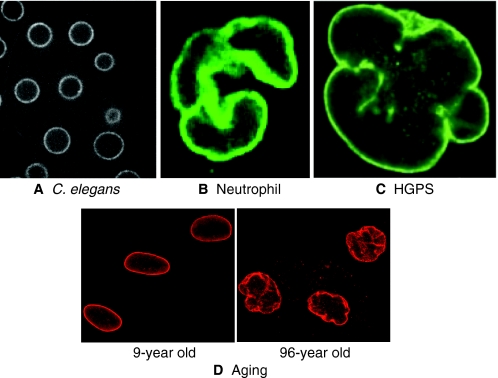
The nuclei of most cells are either round or oval. This, in itself, is hardly remarkable except for the fact that various diseases, as well as aging, are associated with alterations in nuclear shape (Fig. 3). Moreover, in certain specialized cell types, altered nuclear shape is important for cell function. But what determines nuclear shape, and how does shape affect function? In many cell types, altered nuclear shape is due to changes in the nuclear lamina. In some cases, however, the shape of the nucleus is altered by forces that act from the cytoplasm. In either case, it is still not entirely clear how nuclear shape affects function, although two main hypotheses exist. The first hypothesis posits that changes in nuclear shape alter the rigidity of the nucleus; this could be beneficial for cells that need to squeeze through tight spaces, but deleterious to cells that are under mechanical duress. The second hypothesis proposes that changes in nuclear shape result in chromatin reorganization and thereby affect gene expression. It is important to note that these two hypotheses are not mutually exclusive. In addition, because nuclear shape changes are often accompanied by an altered nuclear lamina, it is possible that the dramatic effect on cell function is due to aberrant properties of the lamina rather than nuclear shape changes per se. In this section, we examine some of the cell types and conditions that are associated with irregular nuclear shape, and we discuss, when known, the causes of these shape changes and how they affect cell function.
Fig. 3.
Variation in nuclear shape. The nuclei of most cells, such as those of the C. elegans embryo (A), are either oval or round. However, various cell types or conditions display non-round nuclei. Shown are the nuclei of neutrophils (B), of cells from a patient with HGPS (C) and of cells from a 96-year-old individual (D, right panel) compared with nuclei of cells from a 9-year-old individual (D, left panel). Visualization of nuclei was performed with a GFP-tagged NPC component, NPP-1 (A), an antibody specific for lamin B (B), an antibody specific for emerin (a lamina-associated protein, C) and an antibody specific for lamin A and lamin C (D). The image in B was reprinted with permission from Ada Olins and Donald Olins (Olins and Olins, 2005). The image in C was reprinted with permission from Goldman (Goldman et al., 2004). The images in D were provided by Tom Misteli and Paola Scaffdi (NCI, Bethesda MD) (see also Scaffidi and Misteli, 2006). Nuclei are not shown to scale.
Normal cells with abnormal nuclei
Of all the cell types that normally exhibit unusual nuclear shape, neutrophils have been studied the most thoroughly. Neutrophils are cells of the immune system that migrate through tissue towards sites of infection. They are characterized by their multi-lobed nuclei, typically exhibiting three or four lobes that are connected by thin DNA-containing filaments (Fig. 3B) (reviewed by Hoffmann et al., 2007). Neutrophils with hypolobulated nuclei are associated with Pelger-Huet anomaly, caused by a mutation in the lamin B receptor (Hoffmann et al., 2002). Hypolobulated neutrophils have deficiencies in various cellular functions, including the ability to migrate through small openings. This suggests that nuclear lobulation is an adaptation to fit cell function (Hoffmann et al., 2007). Nuclear lobulation is dependent not only on lamin B receptor, but it is also associated with a paucity of various nuclear lamina proteins (Olins et al., 2008), and it depends on microtubules: treatment of differentiating neutrophils with microtubule-depolymerizing drugs reduced the extent of lobulatons (Olins and Olins, 2004). Thus, in neutrophils, both intranuclear and cytoplasmic proteins affect nuclear shape.
The involvement of microtubules in altering nuclear shape can also be seen during cellularization in the Drosophila embryo when nuclei change shape from spherical to ellipsoid. This change is dependent on both cytoplasmic microtubules and an inner nuclear membrane protein called kugelkern or charleston (Brandt et al., 2006; Pilot et al., 2006). Presumably, kugelkern/charleston is necessary to generate chromatin-NE associations that transduce the forces exerted by cytoplasmic microtubules into nuclear shape changes. Whereas some cell types use microtubule-generated forces to actively alter the shape of their nuclei, others must counteract microtubule-generated forces to maintain their normal nuclear shape. For example, during interphase in fission yeast, microtubule-generated forces in the cytoplasm can alter nuclear shape if any one of several inner nuclear membrane proteins is inactive (King et al., 2008). In this case, NE proteins that normally make the nucleus more rigid must resist nuclear distortions that are caused by cytoplasmic microtubule-associated forces.
Pathologies and conditions associated with altered nuclear shape
It has long been known that reducing the levels of lamina proteins, either by mutation or RNA interference (RNAi), leads to alteration in nuclear shape (Furukawa et al., 2003; Lammerding et al., 2005; Liu et al., 2000). Indeed, cells with abnormally shaped nuclei are often seen in diseases in which lamina proteins are mutated (collectively called laminopathies) (Capell and Collins, 2006) (see Fig. 3C). Thus, the lamina has an active role in maintaining the spherical shape of the nucleus. Mutations in lamina proteins cause various types of lipodystrophies, resulting in abnormal fat tissue, and myopathies, which affect muscle function. For the most part, the link between the lamina defect and disease manifestation is not known, although both an inability to withstand mechanical stress and altered gene expression can be envisioned. A defect in nuclear shape can result not only from reduced levels of lamina proteins, but also from aberrant processing. One of the best-characterized examples of such a case is the premature aging syndrome Hutchison-Gilford progeria syndrome (HGPS). The mutation causing HGPS was mapped to the gene encoding lamin A (De Sandre-Giovannoli et al., 2003; Eriksson et al., 2003); in the mutated protein, the activation of a cryptic splice site generates an aberrant form of lamin A, called progerin, which is constitutively lipid-modified (Rusinol and Sinensky, 2006). This presumably causes lamin A to remain associated with the inner nuclear membrane. It is likely that this membrane retention is the cause of the abnormal nuclear morphology, because treating cells from HGPS patients with compounds that inhibit this lipid modification reverses the abnormalities in nuclear shape (Capell et al., 2005; Glynn and Glover, 2005; Mallampalli et al., 2005; Toth et al., 2005). Other disease-causing mutations that lead to the retention of the lipid modification of lamin A, such as inactivation of the lamin A protease Zmpste24, also result in premature aging and aberrant nuclear morphology (reviewed by Rusinol and Sinensky, 2006).
Similarly to premature aging, normal aging is also associated with abnormal nuclear shape, both in humans (Fig. 3D) and in model organisms (Brandt et al., 2008; Haithcock et al., 2005; Scaffidi and Misteli, 2006). In humans, the aging-dependent change in nuclear shape has been linked to the nuclear lamina, and in particular to progerin, the altered form of lamin A seen in patients with HGPS (Scaffidi and Misteli, 2006). Although the link between the membrane retention of lamin A and aging remains to be discovered, recent studies suggests that an altered nuclear lamina might lead to aging by affecting the transcription profile of stem cells, thereby interfering with their ability to retain an undifferentiated state and reducing the overall stem cell pool and proliferation capacity (Espada et al., 2008; Scaffidi and Misteli, 2008).
An abnormal nuclear shape is also associated with cancer (Zink et al., 2004). In fact, altered nuclear shape is one of the key diagnostic tools used in identifying cancerous cells, and it is the basis for the Pap smear, which is widely used for the early detection of cervical cancer. The functional relationship between altered nuclear shape and cellular transformation – or even the underlying cause of altered nuclear morphology – is often not known, although it has been speculated that changes in nuclear shape lead to changes in chromosome organization, which in turn can affect gene expression (He et al., 2008). Others have proposed that the altered nuclear shape in cancer cells facilitates the formation of metastases because of reduced nuclear stiffness, which could increase the ability of transformed cells to penetrate tissue (Dahl et al., 2008).
In addition to the studies described above, depletion studies and the characterization of numerous mutations have linked other proteins to abnormal nuclear morphology, but the mechanisms involved are mostly unknown. It is plausible, at least in some cases, that the relationship between protein inactivation and altered nuclear shape is indirect. For example, studies in the past decade have shown that the inactivation of proteins that are associated with the ER affects nuclear shape (Higashio et al., 2000; Matynia et al., 2002). These findings suggest that there is an intimate relationship between the ER and the shape of the NE; this relationship will be further explored when we discuss NE assembly (see below).
Finally, nuclear shape can be affected by lipid synthesis. This has been shown in both yeast and C. elegans, where the inactivation of a lipid phosphatase that is homologous to the mammalian lipin (Reue and Zhang, 2008) was shown to cause expansion of the ER membrane and alteration in NE shape (Campbell et al., 2006; Golden et al., 2009; Gorjanacz and Mattaj, 2009; Siniossoglou et al., 1998; Tange et al., 2002). Interestingly, at least in budding yeast, this expansion was confined to the region of the NE adjacent to the nucleolus, whereas the NE associated with the bulk of the DNA remained unchanged (Campbell et al., 2006). This observation suggests that some NE domains are more sensitive than others to shape disruption caused by changes in lipid biosynthesis or membrane composition. We will revisit the relationship between lipid synthesis and the NE when we discuss how a single nucleus is formed at the end of mitosis.
Nuclear size
The nucleus increases in size from the time of its formation, immediately following NE assembly, to when it reaches its final size in interphase. This raises the question of what controls nuclear size: is it a mere consequence of increase in volume over time, or are there factors that regulate or limit nuclear expansion? Decades of observations suggest that the latter is true, and that nuclear and cytoplasmic volumes are somehow related to each other; this phenomenon is referred to as the karyoplasmic ratio (Gregory, 2005). Moreover, in both budding and fission yeasts, the ratio of nuclear to cellular volume remains constant throughout the cell cycle, even as cell volume increases (Jorgensen et al., 2007; Neumann and Nurse, 2007). This suggests the existence of a mechanism that links nuclear and cellular volumes. If indeed such a mechanism exists, does cellular volume dictate nuclear volume, or does nuclear volume determine cell volume? What cellular factors determine nuclear volume? And finally, why is nuclear volume important?
Factors that affect nuclear volume
There are conflicting reports regarding the dominant cellular factors that determine nuclear volume. One idea, known as the nucleoskeletal theory, is that DNA content influences the volume of the nucleus, which in turn influences the size of the cell (Cavalier-Smith, 2005; Gregory, 2005). Intuitively, DNA may affect nuclear volume, because the size of the nucleus could be directly proportional to amount of DNA it contains and the extent to which that DNA is compacted. Simply comparing genome size to nuclear and cell volume among species supports this theory, because species with larger genomes generally have larger nuclear and cellular volumes (Cavalier-Smith, 2005; Jovtchev et al., 2006). Experiments in mice also give credence to the nucleoskeletal theory: it has been shown that tetraploid mouse embryos have nuclei that are twice as large as those in a diploid control (Henery et al., 1992; Henery and Kaufman, 1992).
However, other data suggest that genome size per se is not the determining factor of nuclear size. Rather, it is likely that there is a nuclear-scaling mechanism whereby nuclear volume is proportional to, and determined by, the levels of one or more cellular factors. Indeed, nuclear transplant experiments support this claim: implanting a small hen erythrocyte nucleus into a HeLa cell results in expansion of the nucleus to the appropriate size for its new environment, without affecting DNA content (Harris, 1967). Moreover, the nucleoskeletal theory does not explain why cells from different tissues in a given organism have the same amount of DNA but varied nuclear sizes (Altman and Katz, 1976). Studies in yeast also contradict the notion that DNA content dictates nuclear and cellular volumes (Jorgensen et al., 2007; Neumann and Nurse, 2007). In neither fission yeast nor budding yeast does nuclear volume increase sharply during S phase, as would be expected if DNA content had a direct affect on nuclear size (Jorgensen et al., 2007; Neumann and Nurse, 2007). Furthermore, even a 16-fold increase in ploidy does not affect nuclear size in fission yeast (Neumann and Nurse, 2007). Instead, the displacement of nuclei by centrifugation in multi-nucleated fission yeast showed that nuclear size adjusted in proportion to the amount of surrounding cytoplasm (Neumann and Nurse, 2007). These studies support a mechanism whereby nuclear size is determined by cytoplasmic volume rather than DNA content.
Assuming that cytoplasmic factors determine nuclear size, what might these be? In cell-free extracts of Xenopus oocytes, an increase in nuclear volume after NE reassembly requires an intact ER (Anderson and Hetzer, 2007). This suggests that the membrane for the newly formed NE is supplied by the ER, and therefore membrane availability could be a limiting factor in determining nuclear size. The ER exists as a continuous meshwork of membrane sheets and membrane tubules. Proteins known as reticulons cause tubule formation in the ER (Voeltz et al., 2006), and high levels of reticulons are inhibitory to nuclear growth, which suggests that the availability of membrane in the form of sheets can put an upper limit on nuclear size (Anderson and Hetzer, 2008; Kiseleva et al., 2007). Work in the Xenopus system has demonstrated a requirement for NPCs and nuclear import in nuclear growth after NE assembly (D'Angelo et al., 2006; Newport et al., 1990), which suggests that the import of one or more nuclear proteins contributes to sizing the nucleus. Indeed, several nuclear lamina proteins that are transported into the nucleus through the NPCs have been found to affect interphase nuclear growth (e.g. Brandt et al., 2006; Dittmer et al., 2007; Newport et al., 1990). However, many questions remain. For example, how do yeast – which lack lamins and lamin-associated proteins – adjust nuclear volume in response to changes in cytoplasmic volume? Also, what is the mechanism, in any organism, that establishes the upper limit to nuclear growth?
Does size matter for nuclear function?
Although the mechanisms that control nuclear volume remain unclear, the existence of a karyoplasmic ratio suggests that nuclear size is important for cell function. Disturbance of this ratio is associated with certain types of cancers (Slater et al., 2005; Zink et al., 2004), suggesting that the ratio between nuclear and cytoplasmic volumes is crucial for cell integrity. Moreover, it has been proposed that cell-cycle progression depends on nuclear size (Roca-Cusachs et al., 2008; Yen and Pardee, 1979), and that cells monitor the ratio between cytoplasmic and nuclear volume to gauge the proper time to enter the cell cycle (Futcher, 1996). In addition, a strong correlation between nuclear size, RNA transcription levels and cell size has been found (e.g. Sato et al., 1994; Schmidt and Schibler, 1995). It is therefore possible that larger nuclei facilitate the increase in transcription that is required in larger cells. Additionally, the volume of the nucleus might be important for maintaining nuclear compartments, such as the nucleolus, and the activity of enzymes such as DNA polymerase, which are sensitive to macromolecular crowding (Hancock, 2004; Miyoshi and Sugimoto, 2008; Sasaki et al., 2006). An increasingly popular view of molecular dynamics within the nucleus favors self-organization of complex structures – a process that depends on biochemical and physical interactions between numerous proteins (Misteli, 2001). A recent example is the assembly of Cajal bodies, which are nuclear structures involved in the biogenesis of small nuclear ribonucleoproteins (snRNPs). Cajal bodies assemble by self-organization through stochastic interactions between the building blocks of which they are composed (Kaiser et al., 2008; Misteli, 2008). Because self-organization may be acutely sensitive to the concentration of the individual components, the regulation of nuclear volume might have an important role in enabling this process.
Nuclear-envelope assembly
For over two decades, NE assembly has been studied using an invaluable in vitro system based on Xenopus egg extract (Lohka and Masui, 1983; Newport, 1987). This extract contains all of the components that are necessary for the assembly of a functional NE when mixed with demembranated sperm chromatin. In this system, the NE assembles from vesicles that contain NE proteins. It was assumed that these vesicles form during NE breakdown earlier in the cell cycle. Moreover, NE assembly in vitro was shown to require the small GTPase Ran, GTP hydrolysis, and the AAA ATPase p97, which is involved in membrane fusion (Hetzer et al., 2001). These observations led to the idea that the NE is assembled by the fusion of mitotic vesicles that contain NE proteins. However, more recent studies raise the possibility that such vesicles are a consequence of ER fragmentation during extract preparation (Collas and Courvalin, 2000). Indeed, experiments in mammalian cells strongly suggest that transmembrane NE proteins are not sequestered into vesicles, but rather are incorporated into the ER during mitosis (Daigle et al., 2001; Ellenberg and Lippincott-Schwartz, 1999; Ellenberg et al., 1997; Yang et al., 1997). It is currently unclear whether NE components disperse completely within the ER network or whether they cluster into specific ER subdomains. Intriguing models propose that NE proteins remain in ER subcompartments during mitosis, and that these subdomains serve as the precursors for NE reassembly. The requirement for GTP hydrolysis in vitro can be explained by the need to form an intact ER before NE assembly (Anderson and Hetzer, 2007). In addition, the requirement for p97 can be explained by its ability to extract proteins from chromosomes: chromatin-bound aurora kinase inhibits NE formation, and p97 relieves this inhibition by extracting aurora kinase from chromosomes in telophase (Ramadan et al., 2007). Nonetheless, at present, the possibility that some vesicles are involved in NE assembly cannot be ruled out.
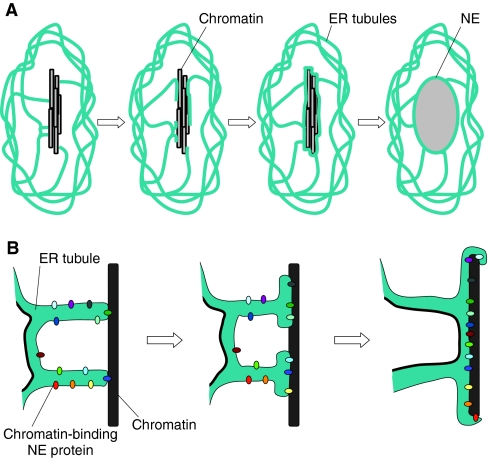
Nuclear-envelope formation: from ER tubules to membrane sheets
During mitosis in mammalian cells, a large portion of the ER is converted into tubules (Puhka et al., 2007). Given that the membrane for the NE comes from the ER, how are ER tubules converted into an intact NE, which is essentially a membrane sheet? Data from both in vitro and mammalian systems argue that early in telophase, the tips of ER tubules contact chromatin, initiating the process of NE assembly (Anderson and Hetzer, 2007; Anderson and Hetzer, 2008) (Fig. 4). This interaction requires neither energy nor the cytosol, and is made possible by the fact that the multiple ER-associated inner nuclear membrane proteins have a high affinity for DNA (Anderson and Hetzer, 2007; Antonin et al., 2005; Mansfeld et al., 2006; Ulbert et al., 2006). These proteins bind DNA early in NE assembly, thereby recruiting ER tubules to chromatin. Following this initial binding, the membrane tubules flatten into sheets, which spread across the chromatin and re-organize into a sealed NE (Anderson and Hetzer, 2007; Anderson and Hetzer, 2008) (Fig. 4). Inner nuclear membrane proteins are also required for the sealing steps (Anderson and Hetzer, 2007; Antonin et al., 2008; Chi et al., 2007; Gorjanacz et al., 2007; Mansfeld et al., 2006), although the exact mechanism by which they are involved remains to be unveiled. In mammalian cells, membrane flattening is inhibited by the overexpression of reticulons and made more efficient by reticulon depletion, suggesting that the equilibrium between flat and curved membrane is vital for this step (Antonin et al., 2008). Reticulon depletion has also been shown to interfere with NE disassembly in Caenorhabditis elegans (Audhya et al., 2007), underscoring the importance of ER-membrane structure in NE dynamics. The final step in membrane sealing requires closing the holes that remain after the membrane expands across the chromatin surface. The mechanistic details of this step are still a mystery. The completion of NE assembly also requires the insertion of NPCs. Although it is tempting to speculate that NPC insertion serves to `plug' the holes left over from membrane flattening (Anderson and Hetzer, 2007), there is currently no evidence that this is the case; in addition, it is clear that NPCs can be inserted into intact NE, for example, in cells that undergo closed mitosis (D'Angelo and Hetzer, 2008). So how are NPCs introduced into the NE?
Fig. 4.
Nuclear-envelope assembly. (A) A general view of NE assembly. Initial contacts with the chromatin (black and gray bars) are thought to be made by tips of ER tubules (blue-green). These tubules then flatten to form an intact NE, which then expands and the chromosomes decondense. (B) A closer view of NE assembly. The ER tubules are decorated with chromatin-binding NE proteins (shown in multiple colors), which are thought to mediate the interaction between the membrane and the chromatin (black). These proteins are eventually located on the inner nuclear membrane. As NE assembly progresses, the membrane flattens onto the chromatin (note the progressive accumulation of chromatin-binding proteins at the interface between the membrane and chromatin). Because the ER membrane is one continuous membrane, the gap between two adjacent tubules will be filled by this membrane-flattening process.
NPC insertion into the NE
Post-mitotic NPC assembly occurs in a step-wise process that begins early in anaphase, with soluble NPC proteins positioning on the chromatin even before membrane reformation, followed by the later recruitment of transmembrane nucleoporins (e.g. Bodoor et al., 1999; Dultz et al., 2008; Rasala et al., 2008); for a detailed discussion, the reader is directed to two excellent recent reviews (Antonin et al., 2008; D'Angelo and Hetzer, 2008). But NPC addition is not restricted to cells undergoing open mitosis. The number of NPCs increases during interphase of dividing cells, well after NE assembly, and during yeast closed mitosis, indicating that another mechanism must exist for insertion of NPCs into a fully formed NE (Maul et al., 1971; Maul et al., 1973; Winey et al., 1997). Studies in the cell-free Xenopus system revealed that interphase addition of NPCs requires the addition of nucleoporins from both the cytoplasmic and nuclear side of the NE (D'Angelo et al., 2006). More recently, reticulon proteins have been implicated in NPC assembly in yeast and Xenopus systems, presumably by inducing curved membranes around the inserted NPC (Dawson et al., 2009). This role is strikingly separate from their function in tubular ER formation, and might reflect a novel use of their membrane-shaping properties to form or stabilize the pore membrane (Dawson et al., 2009). Biochemical analysis of Xenopus extract capable of post-mitotic NPC addition unexpectedly uncovered a role for the major vault protein, suggesting that the vault ribonucleoprotein also facilitates membrane distortions required for NPC assembly (Vollmar et al., 2009). Finally, D'Angelo and co-workers reported that the transmembrane nucleoporins, which serve as the NPC scaffold, are inserted into the NE only in dividing cells, although other NPC subunits are exchanged with newly synthesized ones in nondividing cells (D'Angelo et al., 2009). This implies that the NPC scaffold in nondividing cells, such as neurons, must remain functional for years. Indeed, NPCs of cells from old individuals are more `leaky' than NPCs of young individuals, suggesting that the permeability barrier of NPCs deteriorates over time (D'Angelo et al., 2009).
Organizing the nucleus after NE assembly
Once a sealed NE complete with functional NPCs is formed, the NE expands to its final size and shape. An interesting but still unanswered question is how soon after NE assembly do chromosomes organize into their characteristic nuclear positioning, including chromosome territories and the peripheral localization of heterochromatin? At least two scenarios are possible: in the first, the chromatin is organized nonrandomly in telophase (for example, with regions of heterochromatin facing the exterior of the telophase chromatin mass), such that the reassembling NE makes contacts preferentially with chromatin that will be targeted to the nuclear periphery in the newly formed nucleus. In the second scenario, the chromatin in telophase is not in a particular arrangement, and NE assembly begins with the indiscriminate binding of membranes and lamina proteins to whatever chromatin they encounter. If the latter scenario is correct, then as the NE expands, the nuclear lamina must eventually detach from the chromatin to which it is initially bound and reattach to chromatin that is destined to be at the nuclear periphery. Consistent with this idea, Thomson and colleagues (Thomson et al., 2004) found that peripheral chromatin localization is established during early G1 phase, after NE formation, suggesting that remodeling of NE-chromatin binding takes place early after nuclear expansion. However, this observation does not exclude the possibility that some specific NE-chromatin interaction occurs at the time of NE assembly. Whether the NE assembly process has an inherent specificity to it – either in chromosome configuration or the regions of initial NE-chromatin contact – awaits further research.
The formation of a single nucleus at the end of mitosis
The goal of NE reassembly is the construction of a single nucleus of the appropriate size and shape. Factors that influence nuclear shape and size are discussed above, but what determines the formation of a single NE around the entire chromatin mass? In fact, some organisms undergo stages of development that are characterized by the initial formation of multiple smaller nuclei. For instance, during early embryonic divisions of Xenopus, sea urchins and polychaetes, separate NEs form around the individual chromosomes, creating structures called karyomeres (Montag et al., 1988). Each karyomere is capable of DNA replication and has a complete NE with a nuclear lamina and functional NPCs (Lemaitre et al., 1998). Once the blastula stage is reached, the karyomeres fuse into a single nucleus. Why karyomeres form remains a mystery; perhaps they facilitate rapid entry into S phase during the extremely short cell cycles of early embryonic development (Lemaitre et al., 1998; Lenart and Ellenberg, 2003). Equally fascinating is the question of what dictates separate compartmentalization of the chromosomes in the early embryo. Is there a mechanism that holds chromosomes apart in order to prevent their capture inside a single envelope? Relevant factors might include proteins that are involved in chromosome compaction, the length of the mitotic spindle in these extremely large cells and the copious lipids and membrane proteins that are synthesized during early embryogenesis (Lemaitre et al., 1998). It is tempting to speculate that changes in ER membrane structure (i.e. the ratio of tubules to sheets) during development could also contribute, as discussed below.
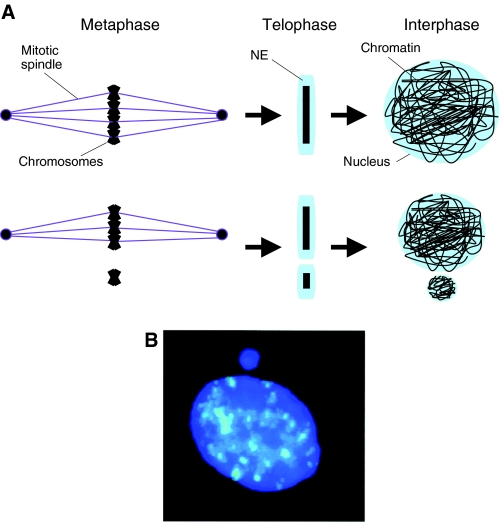
Multiple nuclei within single cells are also observed in disease states and commonly occur as micronuclei in cancer cells. Micronuclei result from either chromosome breakage or imperfect mitosis, when a chromosome fragment or an entire chromosome gets separated from the bulk of the DNA (Fig. 5) (Ford et al., 1988; Norppa and Falck, 2003). Upon NE reassembly, the lagging DNA is excluded from the nucleus and becomes encapsulated in its own NE, complete with a nuclear lamina and NPCs (Walker et al., 1996). Micronuclei spontaneously accumulate in lymphocytes in an age-dependent manner, but they can also be triggered by environmental factors, exposure to genotoxic chemicals (Norppa and Falck, 2003) or depletion of factors required for chromosome segregation and congression to the metaphase plate (e.g. Goshima et al., 2003; Salina et al., 2003). Thus, the physical distance between chromosomes in telophase is clearly important for the encapsulation of all of the chromatin into a single NE.
Fig. 5.
The formation of micronuclei. (A) When all chromosomes (black) congress properly to the metaphase plate via the mitotic spindle (purple), a single NE (blue) is able to form around all of the DNA in telophase (top), resulting in a single nucleus in the ensuing interphase. However, when some DNA remains separate from the metaphase plate, such as lagging chromosomes that are not associated with the spindle, then that DNA fails to become encapsulated into the same NE and a micronucleus forms. (B) Micronucleus formed adjacent to the nucleus of a human buccal cell. DNA is shown in dark blue, and alpha-satellite DNA, which marks centromeres, the chromosomal sites of attachment to spindle microtubules, is shown in light blue. Note that the micronucleus does not contain alpha-satellite sequences, suggesting that the micronucleus was formed because of a failure in attaching to the spindle. Reprinted from Norppa and Falck (Norppa and Falck, 2003) with permission from Oxford University Press.
Recent studies provide insight into how chromosomes might achieve the tight packing that ensures formation of a single nucleus. The highest degree of chromosome axial compaction occurs in late anaphase and requires dynamic microtubules and Aurora kinase. Interfering with this compaction by inhibiting either of these factors causes nuclear morphology defects, including the formation of multi-lobed nuclei (Mora-Bermudez et al., 2007). Interestingly, this defect in compaction did not result in the formation of multiple nuclei, suggesting that other factors are involved in either maintaining chromosome proximity or otherwise limiting the formation of multiple nuclei. More recently, the chromokinesin Kid was reported to be required for maximum compaction of anaphase chromosomes. When Kid was depleted from HeLa cells, chromosome compaction was altered, leading to the formation multi-lobed, wrinkled nuclei. The phenotype was even more severe in Kid–/– mouse zygotes, including the formation of multiple micronuclei (Ohsugi et al., 2008). Whereas Kid affects chromosome compaction in each cell division, it is required to prevent the formation of micronuclei only in oocyte meiosis and in the first few mitotic divisions. This is curious, and might suggest that extreme chromosome compaction is uniquely required to prevent the formation of multiple nuclei during the first few cell cycles. This further suggests that there are other, yet unidentified, factors involved in ensuring that only a single nucleus is formed at the end of mitosis.
The formation of multiple nuclei is also seen in C. elegans after depletion of the nucleoporin gp210, cyclin B, the GTPase Rab-5 or reticulon proteins (Audhya et al., 2007; Galy et al., 2008; Sonnichsen et al., 2005). Interestingly, a subset of these conditions also disrupts ER structure. Multiple nuclei are also formed after depletion of LPIN-1, the C. elegans homolog of lipin, an enzyme that is involved in fat metabolism and lipid synthesis (Golden et al., 2009). Similarly to the effect of Rab-5 or reticulon depletion, downregulation of LPIN-1 expression disrupts ER structure (Golden et al., 2009; Gorjanacz and Mattaj, 2009). Thus, although the distance between chromosomes clearly constitutes an important consideration in the formation of a single nucleus during telophase, the amount of membrane available, and its potential to adopt proper sheet and tubule structures, might also have an important role.
The limited flat membrane hypothesis
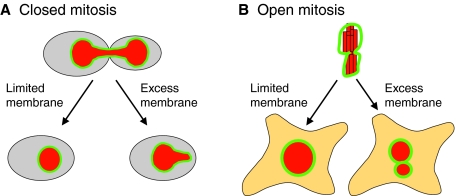
The above observations regarding NE assembly led us to propose the `limited flat membrane hypothesis' (Fig. 6). According to this hypothesis, the limiting factor for the surface area of the NE is the amount of ER membrane that can be converted into membrane sheets, and it is this requirement that contributes to the formation of a single nucleus at the end of open mitosis, or the formation of a round nucleus after closed mitosis. This hypothesis is based on the following suppositions: first, that there is a constant ratio between nuclear size and cell size; second, that the membrane used for NE formation originates in the ER; and third, that only a fraction of the ER membrane – that which is not captured in specialized structures such as tubules – is available for NE formation. There is good evidence in support of the first two suppositions, whereas the third is more speculative and is based on the observed effects of altered reticulon levels and altered lipid synthesis on NE formation (described above).
Fig. 6.
The limited flat membrane hypothesis. During closed mitosis (A), excess membrane in the form of sheets results in a failure to reform a spherical nucleus, suggesting that limited membrane availability drives nuclear shape change at the end of mitosis. During open mitosis (B), excess flat membrane might facilitate the formation of multiple nuclei that collectively have the same volume as a single nucleus that would form under conditions of limited flat membrane availability. The NE is shown in green and the DNA in red. See text for more details.
In the case of closed mitosis, altered lipid synthesis due to the inactivation of the lipin pathway results in extensive ER membrane sheets and the inability to form a spherical nucleus after nuclear division (Campbell et al., 2006; Siniossoglou et al., 1998; Tange et al., 2002). Because lipid synthesis is decreased as cells exit mitosis (Santos-Rosa et al., 2005), we propose that, in closed mitosis, limited lipid synthesis, and specifically limiting amounts of membrane in the form of sheets (i.e. `flat' membrane), drives the nuclear shape change from elongated to round. When the amount of flat membrane is not limiting, such as when lipin is inactive, this transition does not occur (Fig. 6). To see how this hypothesis applies to open mitosis, let us assume that, at the time of NE reassembly, a membrane can form around all of the chromosomes, leading to the formation of a single nucleus, or it can form around a subset of chromosomes, leading to the generation of multiple nuclei that together can expand to the same volume as a single nucleus. Importantly, in the latter case, the combined surface area of the multiple nuclei would be greater than the surface area of a single nucleus with the same volume. If the amount of flat membrane is limited and much of the ER is captured in the form of tubules, the NE assembly reaction will be pushed towards the formation of a single nucleus. If this were the case, it would explain the observation that multiple nuclei are formed under a variety of conditions in which more flat membrane is available. Additionally, it is tempting to speculate that in cancer cells the ratio of ER sheets to tubules is altered, or that the rules that link nuclear volume to cell size are relaxed, thereby facilitating the formation of multiple nuclei. Further research into the relationship between ER structure, lipid synthesis and NE dynamics will be useful for testing the validity of this hypothesis.
Conclusions and Perspectives
The existence of diseases associated with altered nuclear shape and size underscores the importance of uncovering the mechanisms that control NE dynamics. In the past few years, the field of nuclear architecture has witnessed an explosion of knowledge, from the understanding of how nuclear lamina proteins function to the basic principles of NE assembly. We still need to determine the link between nuclear shape and nuclear function, and to distinguish between cases where an altered cellular function is a direct consequence of altered nuclear shape and those where both altered function and shape are independent consequences of defects in a particular structure (e.g. the nuclear lamina). Equally interesting is how nuclear size is determined; although a link between nuclear size and cytoplasmic volume has been suspected for many years, recent studies in yeast have introduced a tractable genetic system in which this question could be answered. Likewise, mutant and RNAi screens in other organisms could uncover proteins that are involved in nuclear size determination. Finally, the finding that NE assembly begins with ER tubules, rather than vesicles, has opened a new avenue of investigation that is focused on understanding the properties of the ER and of the proteins that contribute to ER-membrane dynamics. In the coming years, it is likely that we will learn more about the relationship between the ER and NE dynamics; specifically, how the balance between membrane tubules and membrane sheets is maintained and regulated, and how lipid synthesis contributes to both ER and NE structure.
We thank Will Prinz, Paula Fearon, Daphna Joseph-Strauss (National Institute of Diabetes and Digestive and Kidney Diseases, NIH, Bethesda, MD), and Donald Olins and Ada Olins (Bowdoin College, Brunswick ME) for comments on the manuscript. We also thank Tom Misteli and Paola Scaffidi (National Cancer Institute, Bethesda, MD) for generously providing the images in Fig. 3D, and Ada Olins and Donald Olins for allowing us to use the image in Fig. 3B. M.W., K.L.W. and O.C.F. are funded by an intramural grant from the National Institute of Diabetes and Digestive and Kidney diseases.
References
- Akhtar, A. and Gasser, S. M. (2007). The nuclear envelope and transcriptional control. Nat. Rev. Genet. 8, 507-517. [DOI] [PubMed] [Google Scholar]
- Altman, P. L. and Katz, D. D. (1976). Cell Biology. Bethesda, MD: Federation of American Societies for Experimental Biology.
- Anderson, D. J. and Hetzer, M. W. (2007). Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat. Cell Biol. 9, 1160-1166. [DOI] [PubMed] [Google Scholar]
- Anderson, D. J. and Hetzer, M. W. (2008). Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J. Cell Biol. 182, 911-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin, W., Franz, C., Haselmann, U., Antony, C. and Mattaj, I. W. (2005). The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol. Cell 17, 83-92. [DOI] [PubMed] [Google Scholar]
- Antonin, W., Ellenberg, J. and Dultz, E. (2008). Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 582, 2004-2016. [DOI] [PubMed] [Google Scholar]
- Audhya, A., Desai, A. and Oegema, K. (2007). A role for Rab5 in structuring the endoplasmic reticulum. J. Cell Biol. 178, 43-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodoor, K., Shaikh, S., Salina, D., Raharjo, W. H., Bastos, R., Lohka, M. and Burke, B. (1999). Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 112, 2253-2264. [DOI] [PubMed] [Google Scholar]
- Brandt, A., Papagiannouli, F., Wagner, N., Wilsch-Brauninger, M., Braun, M., Furlong, E. E., Loserth, S., Wenzl, C., Pilot, F., Vogt, N. et al. (2006). Developmental control of nuclear size and shape by Kugelkern and Kurzkern. Curr. Biol. 16, 543-552. [DOI] [PubMed] [Google Scholar]
- Brandt, A., Krohne, G. and Grosshans, J. (2008). The farnesylated nuclear proteins KUGELKERN and LAMIN B promote aging-like phenotypes in Drosophila flies. Aging Cell 7, 541-551. [DOI] [PubMed] [Google Scholar]
- Campbell, J. L., Lorenz, A., Witkin, K. L., Hays, T., Loidl, J. and Cohen-Fix, O. (2006). Yeast nuclear envelope subdomains with distinct abilities to resist membrane expansion. Mol. Biol. Cell 17, 1768-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell, B. C. and Collins, F. S. (2006). Human laminopathies: nuclei gone genetically awry. Nat. Rev. Genet. 7, 940-952. [DOI] [PubMed] [Google Scholar]
- Capell, B. C., Erdos, M. R., Madigan, J. P., Fiordalisi, J. J., Varga, R., Conneely, K. N., Gordon, L. B., Der, C. J., Cox, A. D. and Collins, F. S. (2005). Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA 102, 12879-12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith, T. (2005). Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Ann. Bot. (Lond.) 95, 147-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, Y. H., Haller, K., Peloponese, J. M., Jr and Jeang, K. T. (2007). Histone acetyltransferase hALP and nuclear membrane protein hsSUN1 function in de-condensation of mitotic chromosomes. J. Biol. Chem. 282, 27447-27458. [DOI] [PubMed] [Google Scholar]
- Collas, P. and Courvalin, J. C. (2000). Sorting nuclear membrane proteins at mitosis. Trends Cell Biol. 10, 5-8. [DOI] [PubMed] [Google Scholar]
- Cremer, T., Cremer, M., Dietzel, S., Muller, S., Solovei, I. and Fakan, S. (2006). Chromosome territories-a functional nuclear landscape. Curr. Opin. Cell Biol. 18, 307-316. [DOI] [PubMed] [Google Scholar]
- Crisp, M. and Burke, B. (2008). The nuclear envelope as an integrator of nuclear and cytoplasmic architecture. FEBS Lett. 582, 2023-2032. [DOI] [PubMed] [Google Scholar]
- D'Angelo, M. A. and Hetzer, M. W. (2008). Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 18, 456-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo, M. A., Anderson, D. J., Richard, E. and Hetzer, M. W. (2006). Nuclear pores form de novo from both sides of the nuclear envelope. Science 312, 440-443. [DOI] [PubMed] [Google Scholar]
- D'Angelo, M. A., Raices, M., Panowski, S. H. and Hetzer, M. W. (2009). Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136, 284-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl, K. N., Ribeiro, A. J. and Lammerding, J. (2008). Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 102, 1307-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle, N., Beaudouin, J., Hartnell, L., Imreh, G., Hallberg, E., Lippincott-Schwartz, J. and Ellenberg, J. (2001). Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154, 71-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, T. R., Lazarus, M. D., Hetzer, M. W. and Wente, S. R. (2009). ER membrane-bending proteins are necessary for de novo nuclear pore formation. J. Cell Biol. 184, 659-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sandre-Giovannoli, A., Bernard, R., Cau, P., Navarro, C., Amiel, J., Boccaccio, I., Lyonnet, S., Stewart, C. L., Munnich, A., Le Merrer, M. et al. (2003). Lamin a truncation in Hutchinson-Gilford progeria. Science 300, 2055. [DOI] [PubMed] [Google Scholar]
- De Souza, C. P. and Osmani, S. A. (2007). Mitosis, not just open or closed. Eukaryot. Cell 6, 1521-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat, T., Pfleghaar, K., Sengupta, K., Shimi, T., Shumaker, D. K., Solimando, L. and Goldman, R. D. (2008). Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 22, 832-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer, T. A., Stacey, N. J., Sugimoto-Shirasu, K. and Richards, E. J. (2007). LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell 19, 2793-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz, E., Zanin, E., Wurzenberger, C., Braun, M., Rabut, G., Sironi, L. and Ellenberg, J. (2008). Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 180, 857-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg, J. and Lippincott-Schwartz, J. (1999). Dynamics and mobility of nuclear envelope proteins in interphase and mitotic cells revealed by green fluorescent protein chimeras. Methods 19, 362-372. [DOI] [PubMed] [Google Scholar]
- Ellenberg, J., Siggia, E. D., Moreira, J. E., Smith, C. L., Presley, J. F., Worman, H. J. and Lippincott-Schwartz, J. (1997). Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138, 1193-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson, M., Brown, W. T., Gordon, L. B., Glynn, M. W., Singer, J., Scott, L., Erdos, M. R., Robbins, C. M., Moses, T. Y., Berglund, P. et al. (2003). Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 423, 293-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada, J., Varela, I., Flores, I., Ugalde, A. P., Cadinanos, J., Pendas, A. M., Stewart, C. L., Tryggvason, K., Blasco, M. A., Freije, J. M. et al. (2008). Nuclear envelope defects cause stem cell dysfunction in premature-aging mice. J. Cell Biol. 181, 27-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, J. H., Schultz, C. J. and Correll, A. T. (1988). Chromosome elimination in micronuclei: a common cause of hypoploidy. Am. J. Hum. Genet. 43, 733-740. [PMC free article] [PubMed] [Google Scholar]
- Furukawa, K., Sugiyama, S., Osouda, S., Goto, H., Inagaki, M., Horigome, T., Omata, S., McConnell, M., Fisher, P. A. and Nishida, Y. (2003). Barrier-to-autointegration factor plays crucial roles in cell cycle progression and nuclear organization in Drosophila. J. Cell Sci. 116, 3811-3823. [DOI] [PubMed] [Google Scholar]
- Futcher, B. (1996). Cyclins and the wiring of the yeast cell cycle. Yeast 12, 1635-1646. [DOI] [PubMed] [Google Scholar]
- Galy, V., Antonin, W., Jaedicke, A., Sachse, M., Santarella, R., Haselmann, U. and Mattaj, I. (2008). A role for gp210 in mitotic nuclear-envelope breakdown. J. Cell Sci. 121, 317-328. [DOI] [PubMed] [Google Scholar]
- Glynn, M. W. and Glover, T. W. (2005). Incomplete processing of mutant lamin A in Hutchinson-Gilford progeria leads to nuclear abnormalities, which are reversed by farnesyltransferase inhibition. Hum. Mol. Genet. 14, 2959-2969. [DOI] [PubMed] [Google Scholar]
- Golden, A., Liu, J. and Cohen-Fix, O. (2009). Inactivation of the C. elegans homolog of lipin leads to endoplasmic reticulum disorganization and defects in nuclear envelope breakdown and reassembly. J. Cell Sci. (in press) doi: 10.1242/jcs.044743. [DOI] [PMC free article] [PubMed]
- Goldman, R. D., Shumaker, D. K., Erdos, M. R., Eriksson, M., Goldman, A. E., Gordon, L. B., Gruenbaum, Y., Khuon, S., Mendez, M., Varga, R. et al. (2004). Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA 101, 8963-8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjanacz, M. and Mattaj, I. W. (2009). Lpin-1 is required for efficient nuclear envelope breakdown in Caenorhabditis elegans. J. Cell Sci. (in press) doi: 10.1242/jcs.044750. [DOI] [PubMed]
- Gorjanacz, M., Klerkx, E. P., Galy, V., Santarella, R., Lopez-Iglesias, C., Askjaer, P. and Mattaj, I. W. (2007). Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly. EMBO J. 26, 132-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., Kiyomitsu, T., Yoda, K. and Yanagida, M. (2003). Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160, 25-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, T. (2005). Genome Size Evolution in Animals, vol. 1, pp. 4-87. London: Elsevier Academic Press. [Google Scholar]
- Guttinger, S., Laurell, E. and Kutay, U. (2009). Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell. Biol. 10, 178-191. [DOI] [PubMed] [Google Scholar]
- Haithcock, E., Dayani, Y., Neufeld, E., Zahand, A. J., Feinstein, N., Mattout, A., Gruenbaum, Y. and Liu, J. (2005). Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 102, 16690-16695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, R. (2004). A role for macromolecular crowding effects in the assembly and function of compartments in the nucleus. J. Struct. Biol. 146, 281-290. [DOI] [PubMed] [Google Scholar]
- Harris, H. (1967). The reactivation of the red cell nucleus. J. Cell Sci. 2, 23-32. [DOI] [PubMed] [Google Scholar]
- He, S., Dunn, K. L., Espino, P. S., Drobic, B., Li, L., Yu, J., Sun, J. M., Chen, H. Y., Pritchard, S. and Davie, J. R. (2008). Chromatin organization and nuclear microenvironments in cancer cells. J. Cell Biochem. 104, 2004-2015. [DOI] [PubMed] [Google Scholar]
- Henery, C. C. and Kaufman, M. H. (1992). Relationship between cell size and nuclear volume in nucleated red blood cells of developmentally matched diploid and tetraploid mouse embryos. J. Exp. Zool. 261, 472-478. [DOI] [PubMed] [Google Scholar]
- Henery, C. C., Bard, J. B. and Kaufman, M. H. (1992). Tetraploidy in mice, embryonic cell number, and the grain of the developmental map. Dev. Biol. 152, 233-241. [DOI] [PubMed] [Google Scholar]
- Hetzer, M., Meyer, H. H., Walther, T. C., Bilbao-Cortes, D., Warren, G. and Mattaj, I. W. (2001). Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat. Cell Biol. 3, 1086-1091. [DOI] [PubMed] [Google Scholar]
- Higashio, H., Kimata, Y., Kiriyama, T., Hirata, A. and Kohno, K. (2000). Sfb2p, a yeast protein related to Sec24p, can function as a constituent of COPII coats required for vesicle budding from the endoplasmic reticulum. J. Biol. Chem. 275, 17900-17908. [DOI] [PubMed] [Google Scholar]
- Hoffmann, K., Dreger, C. K., Olins, A. L., Olins, D. E., Shultz, L. D., Lucke, B., Karl, H., Kaps, R., Muller, D., Vaya, A. et al. (2002). Mutations in the gene encoding the lamin B receptor produce an altered nuclear morphology in granulocytes (Pelger-Huet anomaly). Nat. Genet. 31, 410-414. [DOI] [PubMed] [Google Scholar]
- Hoffmann, K., Sperling, K., Olins, A. L. and Olins, D. E. (2007). The granulocyte nucleus and lamin B receptor: avoiding the ovoid. Chromosoma 116, 227-235. [DOI] [PubMed] [Google Scholar]
- Jorgensen, P., Edgington, N. P., Schneider, B. L., Rupes, I., Tyers, M. and Futcher, B. (2007). The size of the nucleus increases as yeast cells grow. Mol. Biol. Cell 18, 3523-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovtchev, G., Schubert, V., Meister, A., Barow, M. and Schubert, I. (2006). Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Cytogenet. Genome Res. 114, 77-82. [DOI] [PubMed] [Google Scholar]
- Kaiser, T. E., Intine, R. V. and Dundr, M. (2008). De novo formation of a subnuclear body. Science 322, 1713-1717. [DOI] [PubMed] [Google Scholar]
- King, M. C., Drivas, T. G. and Blobel, G. (2008). A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell 134, 427-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva, E., Morozova, K. N., Voeltz, G. K., Allen, T. D. and Goldberg, M. W. (2007). Reticulon 4a/NogoA locates to regions of high membrane curvature and may have a role in nuclear envelope growth. J. Struct. Biol. 160, 224-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding, J., Hsiao, J., Schulze, P. C., Kozlov, S., Stewart, C. L. and Lee, R. T. (2005). Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J. Cell Biol. 170, 781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre, J. M., Geraud, G. and Mechali, M. (1998). Dynamics of the genome during early Xenopus laevis development: karyomeres as independent units of replication. J. Cell Biol. 142, 1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart, P. and Ellenberg, J. (2003). Nuclear envelope dynamics in oocytes: from germinal vesicle breakdown to mitosis. Curr. Opin. Cell Biol. 15, 88-95. [DOI] [PubMed] [Google Scholar]
- Liu, J., Rolef Ben-Shahar, T., Riemer, D., Treinin, M., Spann, P., Weber, K., Fire, A. and Gruenbaum, Y. (2000). Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol. Biol. Cell 11, 3937-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohka, M. J. and Masui, Y. (1983). Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science 220, 719-721. [DOI] [PubMed] [Google Scholar]
- Mallampalli, M. P., Huyer, G., Bendale, P., Gelb, M. H. and Michaelis, S. (2005). Inhibiting farnesylation reverses the nuclear morphology defect in a HeLa cell model for Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA 102, 14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld, J., Guttinger, S., Hawryluk-Gara, L. A., Pante, N., Mall, M., Galy, V., Haselmann, U., Muhlhausser, P., Wozniak, R. W., Mattaj, I. W. et al. (2006). The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol. Cell 22, 93-103. [DOI] [PubMed] [Google Scholar]
- Matynia, A., Salus, S. S. and Sazer, S. (2002). Three proteins required for early steps in the protein secretory pathway also affect nuclear envelope structure and cell cycle progression in fission yeast. J. Cell Sci. 115, 421-431. [DOI] [PubMed] [Google Scholar]
- Maul, G. G., Price, J. W. and Lieberman, M. W. (1971). Formation and distribution of nuclear pore complexes in interphase. J. Cell Biol. 51, 405-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul, H. M., Hsu, B. Y., Borun, T. M. and Maul, G. G. (1973). Effect of metabolic inhibitors on nuclear pore formation during the HeLa S3 cell cycle. J. Cell Biol. 59, 669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T. (2001). The concept of self-organization in cellular architecture. J. Cell Biol. 155, 181-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T. (2008). Cell biology: nuclear order out of chaos. Nature 456, 333-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi, D. and Sugimoto, N. (2008). Molecular crowding effects on structure and stability of DNA. Biochimie 90, 1040-1051. [DOI] [PubMed] [Google Scholar]
- Montag, M., Spring, H. and Trendelenburg, M. F. (1988). Structural analysis of the mitotic cycle in pre-gastrula Xenopus embryos. Chromosoma 96, 187-196. [DOI] [PubMed] [Google Scholar]
- Mora-Bermudez, F., Gerlich, D. and Ellenberg, J. (2007). Maximal chromosome compaction occurs by axial shortening in anaphase and depends on Aurora kinase. Nat. Cell Biol. 9, 822-831. [DOI] [PubMed] [Google Scholar]
- Neumann, F. R. and Nurse, P. (2007). Nuclear size control in fission yeast. J. Cell Biol. 179, 593-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport, J. (1987). Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell 48, 205-217. [DOI] [PubMed] [Google Scholar]
- Newport, J. W., Wilson, K. L. and Dunphy, W. G. (1990). A lamin-independent pathway for nuclear envelope assembly. J. Cell Biol. 111, 2247-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norppa, H. and Falck, G. C. (2003). What do human micronuclei contain? Mutagenesis 18, 221-233. [DOI] [PubMed] [Google Scholar]
- Ohsugi, M., Adachi, K., Horai, R., Kakuta, S., Sudo, K., Kotaki, H., Tokai-Nishizumi, N., Sagara, H., Iwakura, Y. and Yamamoto, T. (2008). Kid-mediated chromosome compaction ensures proper nuclear envelope formation. Cell 132, 771-782. [DOI] [PubMed] [Google Scholar]
- Olins, A. L. and Olins, D. E. (2004). Cytoskeletal influences on nuclear shape in granulocytic HL-60 cells. BMC Cell Biol. 5, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins, D. E. and Olins, A. L. (2005). Granulocyte heterochromatin: defining the epigenome. BMC Cell Biol. 6, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins, A. L., Zwerger, M., Herrmann, H., Zentgraf, H., Simon, A. J., Monestier, M. and Olins, D. E. (2008). The human granulocyte nucleus: unusual nuclear envelope and heterochromatin composition. Eur. J. Cell Biol. 87, 279-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot, F., Philippe, J. M., Lemmers, C., Chauvin, J. P. and Lecuit, T. (2006). Developmental control of nuclear morphogenesis and anchoring by charleston, identified in a functional genomic screen of Drosophila cellularisation. Development 133, 711-723. [DOI] [PubMed] [Google Scholar]
- Prunuske, A. J. and Ullman, K. S. (2006). The nuclear envelope: form and reformation. Curr. Opin. Cell Biol. 18, 108-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhka, M., Vihinen, H., Joensuu, M. and Jokitalo, E. (2007). Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J. Cell Biol. 179, 895-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan, K., Bruderer, R., Spiga, F. M., Popp, O., Baur, T., Gotta, M. and Meyer, H. H. (2007). Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature 450, 1258-1262. [DOI] [PubMed] [Google Scholar]
- Rasala, B. A., Ramos, C., Harel, A. and Forbes, D. J. (2008). Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol. Biol. Cell 19, 3982-3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reue, K. and Zhang, P. (2008). The lipin protein family: dual roles in lipid biosynthesis and gene expression. FEBS Lett. 582, 90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs, P., Alcaraz, J., Sunyer, R., Samitier, J., Farre, R. and Navajas, D. (2008). Micropatterning of single endothelial cell shape reveals a tight coupling between nuclear volume in G1 and proliferation. Biophys. J. 94, 4984-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinol, A. E. and Sinensky, M. S. (2006). Farnesylated lamins, progeroid syndromes and farnesyl transferase inhibitors. J. Cell Sci. 119, 3265-3272. [DOI] [PubMed] [Google Scholar]
- Salina, D., Enarson, P., Rattner, J. B. and Burke, B. (2003). Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 162, 991-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa, H., Leung, J., Grimsey, N., Peak-Chew, S. and Siniossoglou, S. (2005). The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24, 1931-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, Y., Miyoshi, D. and Sugimoto, N. (2006). Effect of molecular crowding on DNA polymerase activity. Biotechnol. J. 1, 440-446. [DOI] [PubMed] [Google Scholar]
- Sato, S., Burgess, S. B. and McIlwain, D. L. (1994). Transcription and motoneuron size. J. Neurochem. 63, 1609-1615. [DOI] [PubMed] [Google Scholar]
- Scaffidi, P. and Misteli, T. (2006). Lamin A-dependent nuclear defects in human aging. Science 312, 1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi, P. and Misteli, T. (2008). Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat. Cell Biol. 10, 452-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, E. E. and Schibler, U. (1995). Cell size regulation, a mechanism that controls cellular RNA accumulation: consequences on regulation of the ubiquitous transcription factors Oct1 and NF-Y and the liver-enriched transcription factor DBP. J. Cell Biol. 128, 467-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou, S., Santos-Rosa, H., Rappsilber, J., Mann, M. and Hurt, E. (1998). A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17, 6449-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater, D. N., Rice, S., Stewart, R., Melling, S. E., Hewer, E. M. and Smith, J. H. (2005). Proposed Sheffield quantitative criteria in cervical cytology to assist the grading of squamous cell dyskaryosis, as the British Society for Clinical Cytology definitions require amendment. Cytopathology 16, 179-192. [DOI] [PubMed] [Google Scholar]
- Sonnichsen, B., Koski, L. B., Walsh, A., Marschall, P., Neumann, B., Brehm, M., Alleaume, A. M., Artelt, J., Bettencourt, P., Cassin, E. et al. (2005). Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434, 462-469. [DOI] [PubMed] [Google Scholar]
- Tange, Y., Hirata, A. and Niwa, O. (2002). An evolutionarily conserved fission yeast protein, Ned1, implicated in normal nuclear morphology and chromosome stability, interacts with Dis3, Pim1/RCC1 and an essential nucleoporin. J. Cell Sci. 115, 4375-4385. [DOI] [PubMed] [Google Scholar]
- Thomson, I., Gilchrist, S., Bickmore, W. A. and Chubb, J. R. (2004). The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr. Biol. 14, 166-172. [DOI] [PubMed] [Google Scholar]
- Toth, J. I., Yang, S. H., Qiao, X., Beigneux, A. P., Gelb, M. H., Moulson, C. L., Miner, J. H., Young, S. G. and Fong, L. G. (2005). Blocking protein farnesyltransferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc. Natl. Acad. Sci. USA 102, 12873-12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbert, S., Platani, M., Boue, S. and Mattaj, I. W. (2006). Direct membrane protein-DNA interactions required early in nuclear envelope assembly. J. Cell Biol. 173, 469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz, G. K., Prinz, W. A., Shibata, Y., Rist, J. M. and Rapoport, T. A. (2006). A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573-586. [DOI] [PubMed] [Google Scholar]
- Vollmar, F., Hacker, C., Zahedi, R. P., Sickmann, A., Ewald, A. U., S. and Dabauvalle, M. C. (2009). Assembly of nuclear pore complexes mediated by major vault protein. J. Cell Sci. 122, 780-786. [DOI] [PubMed] [Google Scholar]
- Walker, J. A., Boreham, D. R., Unrau, P. and Duncan, A. M. (1996). Chromosome content and ultrastructure of radiation-induced micronuclei. Mutagenesis 11, 419-424. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen, K., Ketema, M., Truong, H. and Sonnenberg, A. (2006). KASH-domain proteins in nuclear migration, anchorage and other processes. J. Cell Sci. 119, 5021-5029. [DOI] [PubMed] [Google Scholar]
- Winey, M., Yarar, D., Giddings, T. H., Jr and Mastronarde, D. N. (1997). Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol. Biol. Cell 8, 2119-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., Guan, T. and Gerace, L. (1997). Lamin-binding fragment of LAP2 inhibits increase in nuclear volume during the cell cycle and progression into S phase. J. Cell Biol. 139, 1077-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, A. and Pardee, A. B. (1979). Role of nuclear size in cell growth initiation. Science 204, 1315-1317. [DOI] [PubMed] [Google Scholar]
- Zink, D., Fischer, A. H. and Nickerson, J. A. (2004). Nuclear structure in cancer cells. Nat. Rev. Cancer 4, 677-687. [DOI] [PubMed] [Google Scholar]