Summary
Cell separation, or abscission, is a highly specialized process in plants that facilitates remodeling of their architecture and reproductive success. Because few genes are known to be essential for organ abscission, we conducted a screen for mutations that alter floral organ shedding in Arabidopsis. Nine recessive mutations that block shedding were found to disrupt the function of an ADP-ribosylation factor-GTPase-activating protein (ARF-GAP) we have named NEVERSHED (NEV). As predicted by its homology to the yeast Age2 ARF-GAP and transcriptional profile, NEV influences other aspects of plant development, including fruit growth. Co-localization experiments carried out with NEV-specific antiserum and a set of plant endomembrane markers revealed that NEV localizes to the trans-Golgi network and endosomes in Arabidopsis root epidermal cells. Interestingly, transmission electron micrographs of abscission zone regions from wild-type and nev flowers reveal defects in the structure of the Golgi apparatus and extensive accumulation of vesicles adjacent to the cell walls. Our results suggest that NEV ARF-GAP activity at the trans-Golgi network and distinct endosomal compartments is required for the proper trafficking of cargo molecules required for cell separation.
Keywords: Membrane trafficking, ARF-GAP, Abscission, Floral organ shedding, Flower development, Arabidopsis, Golgi, Paramural, Exosome
INTRODUCTION
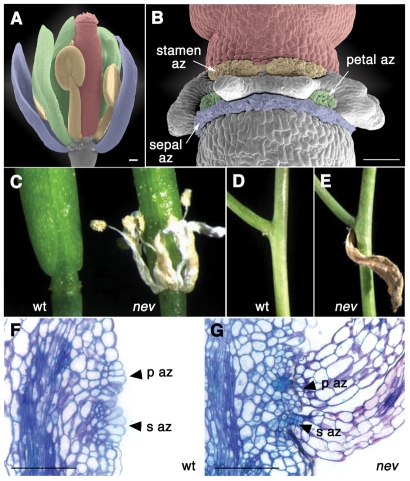
A distinctive feature of plants is their remarkable ability to release entire organs such as leaves, flowers, fruit and seeds by modifying cellular adhesion. Organ separation occurs through the trafficking and secretion of enzymes that locally alter cell walls and dissolve the pectin-rich middle lamella between cells in an abscission zone. The Arabidopsis flower has become a model system to investigate several independently regulated separation events, including stamen dehiscence, floral organ separation, fruit opening and seed abscission (Aalen et al., 2006; Lewis et al., 2006; Roberts et al., 2002). Through organ abscission, the three outer whorls of floral organs - the sepals, petals and stamens - are shed after the stamens release their pollen and fertilization occurs (see Fig. 1A,B). The abscission zones consist of a few cell layers at the base of each of these organs adjacent to the floral stem.
Fig. 1.
Mutations in NEV prevent organ separation. (A) Scanning electron micrograph (SEM) of a wild-type Arabidopsis flower before organ separation (stage 13). The sepals, petals, stamens and gynoecium are colored purple, green, yellow and red, respectively. A few organs have been removed for clarity. (B) SEM of a wild-type flower after organ separation (stage 17). The remaining abscission zone (az) cells of each organ are colored as in A. (C) Floral organs remain attached in nev flowers (stage 17) compared with wild type (wt). (D,E) Cauline leaves in nev plants fail to detach after senescence (E), as compared with wild type (D). (F,G) Longitudinal sections of wild-type (F) and nev (G) flowers at the time of shedding (stage 16) stained with Toluidine Blue. Adjacent petal (p az) and sepal (s az) abscission zones at the base of each flower are indicated. In wild-type flowers, the remaining abscission zone cells have expanded, which does not occur in nev flowers in the absence of organ separation. Scale bars: 100 μm.
Genetic analysis in Arabidopsis is revealing an expanding set of factors that are required for, or affect the timing of, organ separation. Integral components include a pair of closely related, redundant leucine-rich repeat receptor-like kinases, HAESA (HAE) and HAESA-LIKE2 (HSL2), which are probably activated by the IDA and IDA-LIKE secreted peptides, and which in turn switch on a conserved mitogen-activated protein kinase signal transduction pathway essential for activating the separation process (Butenko et al., 2003; Cho et al., 2008; Jinn et al., 2000; Stenvik et al., 2008). Two of the polygalacturonase enzymes that contribute to the cell wall remodeling required for floral organ shedding have been identified (González-Carranza et al., 2007; Ogawa et al., 2009). Although factors that directly influence abscission zone differentiation have remained elusive, a pair of redundant transcription factors, BLADE-ON-PETIOLE1 (BOP1) and BOP2, is known to affect the patterning of leaf and floral organ proximal zones, a prerequisite for abscission zone specification (Hepworth et al., 2005; McKim et al., 2008; Norberg et al., 2005). The timing of organ abscission is influenced by signaling via the plant hormones ethylene and auxin (Ellis et al., 2005; Okushima et al., 2005; Patterson and Bleecker, 2004; Patterson et al., 2007) and by transcriptional regulation and chromatin remodeling, which may globally affect both senescence and abscission (Fernandez et al., 2000; Kandasamy et al., 2005a, Kandasamy et al., 2005b).
In contrast to animal cells that undergo frequent changes in adhesion to facilitate cell migration and the development of a multilayered body plan, plant cells rarely separate from one another. When separation does occur, unique changes in membrane trafficking can be expected to play an essential role in modifying the highly cross-linked cell walls. Although numerous Arabidopsis mutants that alter membrane trafficking have been found to affect cell expansion and polarized growth (Lycett, 2008; Yang, 2008), none have yet been reported to affect cell separation.
Key regulators of membrane trafficking in yeast, animal and plant cells include members of two families that influence the activity and localization of the small ADP-ribosylation factor (ARF) G-proteins during vesicle formation and cargo recruitment (D'Souza-Schorey and Chavrier, 2007; Inoue and Randazzo, 2007; Nielsen et al., 2008). ARF-guanine exchange factors (ARF-GEFs) increase levels of the active GTP-bound ARF protein, which is tethered to the membrane by a myristoylated tail. By stimulating GTP hydrolysis, ARF-GTPase-activating proteins (ARF-GAPs) regenerate the inactive, soluble GDP-bound ARF protein. In addition, interactions of ARF-GAPs with lipids, coat proteins and cargo receptors promote efficient loading of cargo into vesicles and remodeling of the actin cytoskeleton (Inoue and Randazzo, 2007).
Functional characterization of Arabidopsis ARF-GAP and ARF-GEF families is revealing roles for these trafficking regulators during specific phases of plant development. The ARF-GAP VASCULAR NETWORK DEFECTIVE3/SCARFACE (VAN3/SCF) and three closely related homologs - ARF-GAP DOMAIN1 (AGD1), AGD2 and AGD4 - together control vein patterning in developing leaves, potentially by affecting auxin signaling and transport (Koizumi et al., 2005; Sieburth et al., 2006). AGD1 also plays a key role in the orientation of root hair growth by influencing cytoskeletal organization (Yoo et al., 2008). ROOT AND POLLEN ARF-GAP (RPA) affects polarized tip growth of root hairs and of pollen tubes (Song et al., 2006). The ARF-GEF GNOM plays a crucial role during embryo development by regulating endosomal cycling of the PIN1 auxin efflux carrier and thereby controlling auxin transport (Geldner et al., 2003; Steinmann et al., 1999). Together with its homolog GNOM-LIKE1 (GNL1), GNOM also functions during gametophytic development and root growth (Richter et al., 2007). With the Arabidopsis genome predicted to encode 15 ARF-GAPs and eight ARF-GEFs, potentially regulating 21 ARF-GTPases (Vernoud et al., 2003), much work remains to uncover the unique and overlapping functions of each member of these families in membrane trafficking and their impact on plant development.
Here, we report that the ARF-GAP protein NEVERSHED (NEV) is required for floral organ abscission. Our results suggest that mutations in NEV alter the organization of the Golgi apparatus, and block the movement of molecules required for cell separation, providing a link between membrane trafficking and cell separation in plants.
MATERIALS AND METHODS
Plants
Mutant alleles of NEV were obtained through ethyl methanesulfonate (EMS) screens as described (Liljegren et al., 2000). The nev-1, nev-3 and nev-9 alleles contain nucleotide substitutions within codons 51, 59 and 34, which change a cysteine to a tyrosine, an arginine to a lysine, and a cysteine to a tyrosine, respectively. The nev-2, nev-4 and nev-5 alleles contain nucleotide substitutions within codons 198, 260 and 122, which change a glutamine, a tryptophan and a tryptophan to stop codons, causing production of truncated proteins of 197, 259 and 121 amino acids, respectively. The nev-8 allele contains a nucleotide substitution at the splice acceptor site of the seventh intron. The nev-6 (SALK_075680) and nev-7 (SALK_079928) alleles contain T-DNA insertions in the first intron. With the exception of nev-6 and nev-7, all alleles identified are of the Ler ecotype. The nev-3 mutant was backcrossed to Ler three times before phenotypic analyses; homozygous mutants can be detected by a BspCNI site introduced by the dCAPs oligo 5′-CTGCATGCAATGTTCTGGGATTCTCA-3′.
Mapping
To map NEV, nev-1 and nev-2 mutants were crossed to wild type (Col). Using DNA isolated from 515 nev F2 plants and PCR-based markers, including several designed from Monsanto Ler polymorphisms (http://www.arabidopsis.org/Cereon/), both mutations were mapped to chromosome 5 between CER436970 and CER465330. The coding regions of 8 of 13 predicted genes in this region were sequenced from nev genomic DNA.
Generation of NEV-specific antiserum and immunodetection
NEV cDNA (GenBank Accession number FJ794601) was PCR amplified with 5′-AGATATTGAAGAGCGCTGAAGGCG-3′ and 5′-TCCGTTTAAGTCTTTTTCAGAGAGAGAG-3′ using Col RNA as template. After cloning the cDNA into pCR2.1 (Life Technologies, Carlsbad, CA), a fragment encoding the unique C-terminus corresponding to amino acids 266-483 was amplified with 5′-CACCGCTGGAAGTGGTCAAACG-3′ and 5′-TCAATGTTTTGTGAACATTCCATCC-3′ and inserted into pENTR/D-TOPO (Life Technologies, Carlsbad, CA). Recombination of this construct with pDEST17 (Life Technologies, Carlsbad, CA) was used to express truncated, 6×His-tagged NEV protein in Escherichia coli. Recombinant protein purified by Ni2+ affinity chromatography (HisTrap: GE Healthcare, Chalfont St Giles, UK) was used to immunize rabbits (Covance Research Products, Denver, PA).
Protein extracts were prepared by grinding floral tissue, incubating samples with extraction buffer (50 mM TrisHCl pH 8.5, 2% SDS) at 60°C for 20 minutes, pelleting by centrifugation at 16,000 g for 5 minutes, and resuspending the supernatant in loading buffer [5 mM EDTA (pH 8.5), 10% glycerol, 50 mM mercaptoethanol, 0.05% Bromophenol Blue]. Proteins were separated by SDS-PAGE and analyzed by western blotting. Anti-NEV antibody was used at a 1:20,000 dilution.
Microscopy
Wild-type flowers were analyzed by scanning electron microscopy as described (Liljegren et al., 2000). For transmission electron microscopy, flowers were fixed in 2% paraformaldehyde/1% glutaraldehyde in 0.05 M sodium phosphate buffer overnight at 4°C. After fixation, samples were stained with 1% osmium tetroxide, dehydrated through an ethanol series and embedded in Spurr's resin. Sections (60-70 nm) were cut with a diamond knife, mounted on copper mesh grids, stained with 4% uranyl acetate followed by Reynolds lead citrate, and examined with an LEO EM-910 transmission electron microscope (LEO Electron Microscopy, Thornwood, NY) at 80 kV with images recorded using a Gatan Orius SC1000 CCD camera (Gatan, Pleasanton, CA). Sections (1 μm) from the above samples were stained with 1% Toluidine Blue for light microscopy. To determine the extent of paramural body accumulation, individual cells were examined for the presence of plasma membrane-associated clusters of ten or more vesicles. Plasma membrane perimeters were determined with NIH ImageJ software.
For immunofluorescence assays, seedlings were grown vertically on MS media plates for 5-10 days then prepared as described (Lauber et al., 1997) with minor modifications. Primary antibodies were used at the following concentrations: anti-NEV, 1:2500; chicken anti-GFP, 1:500 (Abcam, Cambridge, MA). Fluorochrome-conjugated secondary antibodies were used at a 1:500 concentration: Alexa Fluor 488 anti-rabbit F(ab′)2 and Alexa Fluor 647 anti-rabbit (Molecular Probes, Eugene, OR); anti-chicken Cy2 (Abcam, Cambridge, MA). Confocal laser scanning microscopy was performed with a Zeiss LSM-410 or 510 (Carl Zeiss, Thornwood, NY). Image brightness and contrast were adjusted with Photoshop 7.0 (Adobe, Mountain View, CA). For Brefeldin A (BFA) treatment, 5- to 10-day-old seedlings were incubated in MS liquid media with or without 100 μM BFA for 50-60 minutes, fixed in 4% formaldehyde and prepared for immunofluorescence detection as described above.
ARF-GTPase-activating assay
The NEV coding region was amplified with 5′-CACCATGAACGAGAAAGCCAACGTC-3′ and 5′-TCAATGTTTTGTGAACATTCCATCC-3′ and inserted into pENTR/D-TOPO (Life Technologies, Carlsbad, CA). Recombination of this construct with pDEST17 was used to express full-length 6×His-tagged NEV protein in E. coli. Recombinant NEV protein was purified as described above. Non-myristoylated Arf1 was prepared as described (Randazzo et al., 1992) for use in the GTPase-activating assay performed as described using recombinant AGAP1 as a positive control (Che et al., 2006). Briefly, α32PGTP was exchanged for GDP bound to Arf1. α32PGTP-Arf1 was incubated with the indicated concentration of NEV for 3 minutes at 30°C. The reaction was terminated by dilution and shifting to 4°C. Arf1 was separated from free nucleotide by filtering on nitrocellulose. Nucleotide was released from Arf1 with formic acid and the relative level of GDP and GTP was determined by quantifying P32 following thin layer chromatographic separation of the nucleotides. The data are presented as the percentage of GTP converted to GDP. No conversion was observed in the absence of a GAP.
Generation of transgenic lines
A 1.8 kb genomic region of NEV, from 908 nucleotides upstream of the predicted translational start site into exon two, was amplified with 5′-GAGCCAGCGAGAGACCACTTCTC-3′ and 5′-CTCTAGAAGAATCTGCAGATAAGTAGCAC-3′ using Col DNA as template. This fragment was cloned into pCR2.1, excised as XbaI fragment, and cloned into pDW137 (Blázquez et al., 1997) to create a translational fusion of NEV to the β-glucuronidase (GUS) reporter. GUS expression was analyzed as described (Blázquez et al., 1997) with minor modifications.
The transgenic marker lines YFP-VTI12, YFP-RabA1e, YFP-RabA1g, YFP-RabF2a, YFP-RabD2a, YFP-NIP1;1 and YFP-NPSN12 used in co-localization studies are part of the `Wave' marker collection (Geldner et al., 2009). The VHA-a1-RFP and NAG-GFP markers were described previously (von der Fecht-Bartenbach et al., 2007; Grebe et al., 2003).
RESULTS
Mutations in NEV prevent floral organ shedding
To identify novel loci required for organ abscission, we carried out mutant screens of Arabidopsis plants as flowers matured. Six mutants that retain their floral organs indefinitely represent a unique complementation group that we have named nevershed (nev) (Fig. 1C). A seventh nev mutant allele was kindly provided by Pataradawn Pinyopich (UCSD).
As each of the identified mutant alleles blocks floral organ abscission, we characterized nev-3 as a representative allele. In examining sections of nev flowers at a range of developmental stages compared to wild type (Fig. 1F,G; data not shown), differences in abscission zone morphology were first detected when organ separation normally occurs in wild-type flowers (stage 16). At this point, abscission zone cells expand and become vacuolated in wild type (Fig. 1F), whereas in nev flowers the organs stay attached and cells within the sepal and petal abscission zones remain small and densely cytoplasmic (Fig. 1G). As abscission zones are thought to be specified in the proximal regions of the floral organs as they differentiate, enhancer trap lines with early expression profiles at the bases of wild-type floral organs (Campisi et al., 1999) were examined in nev flowers (see Fig. S1 in the supplementary material). No temporal or spatial differences were detected for four different markers in nev flowers compared to wild type. Taken together, these results suggest that NEV acts during the separation phase of organ abscission, rather than during the initial patterning and differentiation of abscission zone cells.
To determine whether NEV function is specific to abscission, we examined whether other aspects of plant development are affected by mutations in NEV. While floral organ separation is tightly linked to the initiation of organ senescence (Fang and Fernandez, 2002), the aerial (cauline) leaves of wild-type Arabidopsis plants can be shed well after senescence has occurred (Fig. 1D). Detachment of these leaves is blocked in nev mutants (Fig. 1E). Fruit (silique) growth is also significantly affected by mutations in NEV (see Fig. S2A,B in the supplementary material). These results indicate that NEV plays a role in multiple developmental processes.
NEV encodes an ADP-ribosylation factor GTPase-activating protein
Using standard mapping procedures, the nev-1 and nev-2 mutations were located within a 39 kb interval on the lower arm of chromosome 5, including the BACs MDK4 and GA469. Within one of 13 predicted genes in this interval, At5g54310, a single nucleotide change was identified in the nev-2 mutant that would introduce a stop codon into the corresponding transcript (Fig. 2A). This gene includes 11 exons and encodes a predicted 483-amino acid protein with a zinc finger (CX2CX16CX2C) characteristic of ARF-GAP proteins located near its N-terminus (Fig. 2A). Three of the nine nev alleles introduce missense mutations within the highly conserved 115-amino acid ARF-GAP domain, with two, nev-1 and nev-9, altering crucial cysteine residues in the zinc finger motif to tyrosines, and a third, nev-3, changing an invariant arginine in close proximity to the zinc finger to a lysine (Fig. 2B). This particular arginine has been demonstrated to be essential for ARF-GAP activity for all ARF-GAPs examined (Luo et al., 2007), and structural analysis of the PAPβ/ARF1 complex suggests that it may act as an `arginine finger' directly contacting the active site of the ARF substrate during GTP hydrolysis (Mandiyan et al., 1999). Four additional alleles are predicted to affect the C-terminal region of the protein by introducing premature stop codons (nev-2, nev-4, nev-5) or affecting splicing (nev-8) (Fig. 2A). Two alleles of NEV identified in the Salk T-DNA mutant collection (Alonso et al., 2003), nev-6 and nev-7, contain insertions in the first intron and are predicted to encode a truncated protein without an ARF-GAP domain (Fig. 2A). As plants carrying either of these null alleles are phenotypically indistinguishable from the other nev mutants, all of the identified mutations appear to completely disrupt the function of NEV and are considered to be strong loss-of-function alleles.
Fig. 2.
NEV encodes an ARF GAP. (A) The At5g54310 locus, showing the sites and sequence alterations of characterized nev mutations. Exons are shown as boxes, and the translated regions corresponding to the ARF-GAP domain and the rest of the corresponding protein are indicated in black and gray, respectively. Point mutations are marked by arrows, and T-DNA insertions by arrowheads. (B) Sequence alignment of the ARF-GAP domain from NEV and related proteins from plants (OsI_025582, AGD15), yeast (Age2), worm (W09D10.1) and mouse (Smap1). Amino acids conserved between NEV and other proteins are shaded, and the four cysteine residues that constitute the zinc finger are indicated with asterisks. The sites of nev missense mutations that affect two of these cysteines and a crucial arginine residue are marked by arrows above the alignment. Characterized ARF-GAP proteins in the alignment include yeast Age2 (Zhang et al., 1998) and mouse Smap1 (Sato et al., 1998). Uncharacterized proteins with closely related ARF-GAP regions include OsI_025582, AGD15 (Vernoud et al., 2003) and W09D10.1, predicted from Oryza sativa, Arabidopsis and Caenorhabditis elegans sequences, respectively. (C) NEV promotes GTP hydrolysis of mammalian ARF1. The ARF-GAP activity of recombinant full-length NEV protein was measured using AGAP1, a mammalian ARF-GAP, as a positive control (inset graph). The percentage of GTP bound to Arf1 that was converted to GDP is presented. The data are the summary of two experiments. (D,E) NEV regulatory regions direct broad expression of β-glucuronidase in flowers and shoot inflorescence stems (D), and in developing leaves and vascular strands (E).
Comparison of NEV with other eukaryotic ARF-GAPs has revealed that NEV shares 49% amino acid identity within its ARF-GAP domain with yeast ArfGAP effector2 (Age2), 50% identity with Age2-like ARF-GAPs in worms and mice, and 73% identity with another Age2-like ARF-GAP in Arabidopsis, AGD15 (Fig. 2B). In contrast to NEV, Age2 activity has been found to be entirely redundant with that of another yeast ARF-GAP, Growth cold sensitive1 (Gcs1) (Poon et al., 2001). The yeast age2 gcs1 double mutant disrupts the morphology of the Golgi, and membrane trafficking from the trans-Golgi network (TGN) to the endosome, vacuole and plasma membrane, and is lethal. In mammals, two Age2-related ARF-GAPs, SMAP1 and SMAP2, regulate clathrin-mediated endocytosis from the plasma membrane and retrograde traffic from the early endosome to the TGN, respectively (Natsume et al., 2006; Tanabe et al., 2005). Outside of the ARF-GAP domain, the NEV protein is poorly conserved with Age2 and Age2-like ARF-GAPs, and the conserved clathrin-binding domains of SMAP1 and SMAP2 are not present in the C-terminal regions of either NEV or Age2.
Homology searches also revealed predicted proteins with strong sequence similarity to NEV in diverse species of flowering plants, such as Oryza sativa (rice). A candidate rice ortholog shares 90% amino acid identity within its ARF-GAP domain and 48% amino acid identity overall with NEV; several conserved regions outside the ARF-GAP domain are apparent (Fig. 2B; see Fig. S3 in the supplementary material).
To determine whether NEV is able to function as an ARF-GAP, we tested its ability to promote GTP hydrolysis of the mammalian ARF1 G-protein substrate in vitro (Fig. 2C). Recombinant full-length NEV protein was expressed in E. coli, purified and used in an ARF-GAP assay, along with a mammalian ARF-GAP, AGAP1, as a positive control. NEV promoted GTP hydrolysis of ARF1 in a concentration-dependent manner with enzymatic activity levels similar to those of other Arabidopsis ARF-GAPs acting on heterologous ARF substrates (Koizumi et al., 2005).
NEV is expressed broadly during development
Global analysis of Arabidopsis gene expression profiles revealed that NEV is expressed ubiquitously during development in floral, leaf, stem and root tissue (see Fig. S4 in the supplementary material) (Schmid et al., 2005). To determine whether expression of NEV might be enriched in certain cell types within these tissues, transgenic plants were generated that express a translational fusion of NEV, including 908 nucleotides upstream of the predicted translational start site through part of the second exon, to the complete coding region of β-Glucuronidase (GUS). Of 19 T1 plants analyzed, nine showed broad expression patterns of GUS in the inflorescence stem, abscission zones, stigma, floral and/or leaf vasculature (Fig. 2D,E), eight showed expression profiles in a subset of these tissues, and two showed no apparent expression. These results together with our phenotypic analysis suggest that NEV probably functions broadly during plant development.
NEV localizes to the TGN and endosomes
ARF-GAPs modulate membrane trafficking from multiple sites, including the Golgi, TGN and recycling endosome (Inoue et al., 2008; Min et al., 2008; Poon et al., 1999; Poon et al., 2001). Analysis of NEV using WoLF PSORT (Horton et al., 2007) showed a possible monopartite nuclear localization signal between amino acids 14 and 17 (RHRK) and no other known sorting motifs. To determine the subcellular site of NEV action, we examined its localization using NEV polyclonal antiserum raised against its unique C-terminal region. This antiserum recognized a ∼55 kDa protein in wild-type flower extracts, consistent with the predicted size of NEV (53 kDa) (Fig. 3A). Because this protein was not recognized in extracts from nev-2 flowers containing truncated nev protein, the antiserum appears to be specific for NEV (Fig. 2A; Fig. 3A). Immunolocalization and confocal laser-scanning microscopy revealed that the NEV antiserum labels punctate spots in wild-type root epidermal cells (Fig. 3B). As expected, the signal profile observed in wild-type roots is not consistent with nuclear localization and a substantial reduction of signal is observed in nev-2 roots (Fig. 3B,C).
Fig. 3.
NEV localizes to the trans-Golgi network (TGN) and endosomes. (A) NEV antiserum recognizes a ∼55 kDa protein in wild-type flower extracts. As the nev-2 allele encodes a truncated protein lacking the C-terminal recognition sequence of the NEV antiserum, no protein is detected in nev-2 flower extracts. Ponceau Red staining was used as a protein-loading control. (B-O) Immunofluorescent localization of NEV and endomembrane markers in primary root epidermal cells of wild-type (B), nev (C) and transgenic marker (D-O) plants. In M-O, the primary roots of YFP-RabA1e plants were incubated for 1 hour with 100 μM BFA before fixation and immunofluorescent staining. (B,C) NEV antiserum detects NEV protein in punctate structures in wild-type cells (B). Background fluorescence is shown for nev-2 cells (C). (D-I) NEV localization is distinct from the Golgi (D-F), and shows more precise localization with YFP-VTI12, a marker of the TGN (arrows, G-I). Occasional spots labeled by NEV (arrowheads, G-I) do not co-localize with VTI12. (J-O) NEV also co-localizes with YFP-RabA1e, a novel endosomal marker proposed to localize to the recycling endosome (arrows, J-L). BFA treatment of the primary root causes both NEV and YFP-RabA1e to localize to BFA bodies (M-O). Whereas the localization of YFP-RabA1e is confined to the core of the BFA bodies, NEV is also found at the periphery (arrows, O). Individual channels: YFP markers, green; NEV, red. Scale bars: 5 μm.
We next used a set of markers for specific plant endomembrane compartments (see Table S1 in the supplementary material) to explore the precise localization of NEV (Fig. 3; see Fig. S5 in the supplementary material). Because Age2 localizes to and regulates traffic from the TGN in yeast (Poon et al., 2001), we first examined the co-localization of NEV with markers of the TGN and Golgi (Fig. 3). Whereas NEV localization is distinct from the Golgi marker NAG-GFP (Fig. 3D-F) (Grebe et al., 2003), the punctate spots labeled by NEV antiserum co-localize with spots of a similar size and structure labeled by the TGN/early endosome (EE) marker YFP-VTI12 (Fig. 3G-I, arrows) (Sanderfoot et al., 2001). To further confirm that this marker labels the TGN/EE, co-localization of YFP-VTI12 with the TGN/EE marker VHA-a1-RFP was shown (see Fig. S6 in the supplementary material) (von der Fecht-Bartenbach et al., 2007). In plants, accumulating evidence exists for a unified TGN/EE compartment (Dettmer et al., 2006; Müller et al., 2007; Robinson et al., 2008). These results suggest that NEV is localized at the TGN/EE.
As we observed occasional sites at which NEV fluorescence does not overlap with the YFP-VTI12 marker (Fig. 3G-I, arrowheads), a wider range of endomembrane markers was tested for co-localization with NEV. We discovered that NEV also co-localizes with two novel endosomal markers, YFP-RabA1e and YFP-RabA1g (Fig. 3J-L, arrows; data not shown) proposed to label recycling endosomes (Geldner et al., 2009). The endosomal compartment labeled by these markers is distinct from the TGN/EE and shows a high sensitivity to BFA (Geldner et al., 2009). Rab11, the closest mammalian homolog of the expanded RabA class in Arabidopsis, regulates trafficking through the recycling endosome; other characterized RabA family members in Arabidopsis and tomato are thought to localize at the TGN/EE (Chow et al., 2008; Nielsen et al., 2008; Rehman et al., 2008). NEV did not co-localize with the YFP-RabF2A marker of the late endosome/pre-vacuolar complex (see Fig. S5A in the supplementary material) (Lee et al., 2004; Ueda et al., 2004) or the YFP-NIP1;1 marker of the ER network (see Fig. S5B in the supplementary material) (Geldner et al., 2009). Vesicles labeled by NEV occasionally overlapped with YFP-NPSN12, a marker of the plasma membrane (see Fig. S5C in the supplementary material) (Zheng et al., 2002). NEV also partially localized with YFP-RabD2a, a marker associated with the Golgi network and endosome (see Fig. S5D in the supplementary material) (Zheng et al., 2005). Taken together, these results are consistent with roles for NEV in regulating trafficking at the TGN/EE and a distinct endosomal compartment, which may correspond to recycling endosomes.
To further explore NEV localization, we used BFA, a fungal toxin that interferes with the activity of a subset of BFA-sensitive ARF-GEFs, inhibiting ARF-mediated vesicular transport from the target membrane (Peyroche et al., 1999). In Arabidopsis roots, BFA inhibits recycling of vesicles back to the plasma membrane without blocking endocytosis or ER-Golgi traffic, resulting in the accumulation of the TGN/EE and rapidly cycling proteins such as PIN1 into BFA bodies (Geldner et al., 2003; Grebe et al., 2003; Richter et al., 2007; Robinson et al., 2008; Teh and Moore, 2007). Golgi, by contrast, tend to cluster around the BFA bodies while remaining functionally intact. When we treated primary roots with 100 μM BFA before immunodetection of NEV, NEV localized to structures resembling BFA bodies rather than the punctate spots characteristic of untreated cells (Fig. 3N; see Fig. S7 in the supplementary material). In the presence of BFA, NEV localization within the BFA compartment overlaps with but is more disperse than that of the endosomal marker YFP-RabA1e found at the core of the BFA compartment (Fig. 3N-O, arrows). These results suggest that NEV is not only localized to the TGN/EE and distinct endosomes, but also to additional BFA-resistant membranes.
Mutations in NEV disrupt Golgi structure, alter location of the TGN, and cause accumulation of paramural vesicles
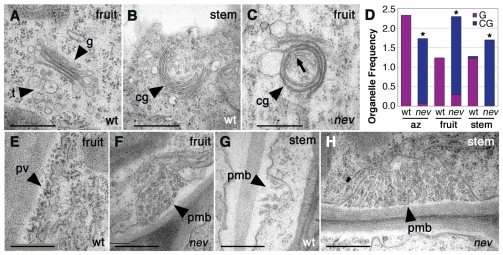
As nev mutations affect a member of a protein family known to regulate membrane trafficking, we examined whether subcellular defects could be detected in nev mutant cells. Transmission electron microscopy was performed on nev flowers compared to wild type at the time of organ shedding (stage 16). We discovered that cells in the region of nev sepal abscission zones predominantly exhibited circularized or cup-shaped multilamellar structures (Fig. 4B; Fig. 5D; see Fig. S8D-F in the supplementary material) rather than the flat stacks of Golgi cisternae typical of wild type (Fig. 4A; Fig. 5D; see Fig. S8A-C in the supplementary material). We also often observed the TGN closely associated with the trans-face of the Golgi in wild-type cells (Fig. 4A; see Fig. S8A-C in the supplementary material), but did not readily observe these tubular-vesicular networks near the circularized structures in nev abscission zone cells (Fig. 4B; see Fig. S8D-F in the supplementary material). In older nev flowers (stage 17), a mixture of Golgi with a wild-type appearance and circularized structures were observed in sepal abscission zones (data not shown). These unusual compartments were also observed in the two other regions of nev flowers (stage 16) examined - fruit walls and floral stems (pedicels) - although vesicular structures resembling the TGN were more apparent in the fruit cells examined (Fig. 5C,D). Taken together, these results suggest that defects in NEV-regulated trafficking can severely affect the structure of the Golgi apparatus and alter the location of the TGN.
Fig. 4.
Mutations in NEV alter Golgi structure and location of the TGN and cause an accumulation of paramural vesicles. Transmission electron micrographs of cells at the base of wild-type and nev sepals at the time of shedding (stage 16). (A,B) Cup-shaped and circularized multilamellar structures are predominantly found in nev cells (B) instead of the typically flat Golgi cisternae of wild-type cells (A). Whereas the TGN is frequently observed (84%, n=19) near the Golgi (0.041±0.018 μm, n=16) in wild-type cells (A), these tubular-vesicular compartments are not clearly identifiable near the circularized structures of nev cells (B). (C,D) Whereas paramural vesicles are infrequently observed between the plasma membrane and cell wall of wild-type cells (C), numerous vesicles are frequently observed in large paramural bodies in nev cells (D). (E,F) Distribution of paramural vesicles in cells at the base of wild-type (E) and nev (F) sepals. Abscission zone regions are indicated by arrowheads, analyzed cells are colored light blue, and paramural bodies with more than 30 vesicles or 10-30 vesicles are indicated by pink and blue circles, respectively. cg, circularized multilamellar structures; cw, cell wall; g, Golgi cisternae; pm, plasma membrane; pmb, paramural body; t, TGN. Scale bars: 0.5 μm in A-D; 10 μm in E,F.
Fig. 5.
The fruit and stems of nev flowers also show membrane trafficking defects. (A-C,E-H) Transmission electron micrographs of cells in the fruit wall and pedicel (stem) of wild-type and nev flowers (stage 16). (A-C) Circularized multilamellar structures are enriched in nev fruit and stem cells (C,D) by comparison with the linear Golgi cisternae of wild-type cells (A,D). Rarely, Golgi cisternae with a curved appearance were observed in wild-type cells (B,D). Vesicular structures resembling the TGN of wild-type cells (A) are often observed in the vicinity or inside the multilamellar structures of nev cells (C, arrow) (73%, n=15). (D) Frequency of Golgi with a wild-type appearance (G, purple) and circularized multilamellar structures (CG, blue) per cell in sections of nev and wild-type sepal AZ regions, fruit walls and pedicels. For each type of tissue analyzed, n (cells) ≥15. Statistical differences between nev and wild-type tissues are indicated by asterisks (Fisher's exact test, P<0.0001). (E-H) Large paramural bodies are found in nev fruit (F) and stem (H) cells. Paramural vesicles and paramural bodies were also observed in wild-type fruit (E) and stem (G) cells. cg, circularized multilamellar structures; g, Golgi cisternae; pmb, paramural body; pv, paramural vesicles; t, TGN. Scale bars: 0.5 μm.
Extensive accumulation of vesicles between the plasma membrane and cell wall was also observed in the sepal abscission zone regions of nev flowers (Fig. 4D). Vesicle clusters associated with pockets of the plasma membrane have been termed paramural bodies (PMBs) by researchers who have also observed them in the embryos of the Arabidopsis vps9a Rab-GEF mutant and in barley leaf cells upon fungal infection (An et al., 2007; Goh et al., 2007). Although PMBs are present in wild-type cells (Fig. 4C,E), far more PMBs containing numerous vesicles were found in nev flowers (Fig. 4D,F) [0.002±0.01 PMB/μm plasma membrane in wild-type cells (n=20) compared with 0.082±0.066 PMB/μm plasma membrane in nev cells (n=28)]. Some PMBs in nev flowers have the appearance of a recent fusion of a multivesicular body to the plasma membrane (Fig. 4D). Additional PMBs were also observed in the fruit walls and pedicels of nev flowers (Fig. 5F,H; see Fig. S9B,D in the supplementary material) compared with wild type (Fig. 5E,G; see Fig. S9A,C in the supplementary material). These results suggest that NEV may play a role in PMB turnover or biogenesis.
DISCUSSION
We report here the identification and characterization of NEV, an Arabidopsis Age2-like ARF-GAP that is required for floral organ abscission. Our studies suggest that NEV plays an essential role in membrane trafficking during the separation stage of organ abscission by facilitating the movement of key cargo molecules through the TGN/EE and other distinct endosomes. Although mutations in NEV do not appear to affect the early differentiation of abscission zone cells, they do alter their fate. Instead of undergoing cell separation and expansion, cells in the abscission zone regions of nev flowers remain small and cytoplasmic (Figs 1 and 4). Disruption of membrane trafficking in nev flowers could affect the localization of signaling molecules required to initiate the separation process, the subsequent secretion of cell wall modifying enzymes, or the endocytosis of cell wall materials. Key molecules in the signaling network thought to activate the separation phase of organ abscission include the IDA and IDA-LIKE peptides, and their proposed receptors, the transmembrane HAE and HSL2 leucine-rich repeat receptor-like kinases (Butenko et al., 2003; Cho et al., 2008; Jinn et al., 2000; Stenvik et al., 2008). Cell wall modifying and hydrolytic enzymes - polygalacturonases, cellulases, expansins and others - are secreted by abscission zone cells to loosen their cell walls and and dissolve the pectin-rich middle lamellae. Identifying which cargo molecules are incorrectly trafficked in nev flowers will be central to understanding further the role of NEV in abscission.
We discovered two unusual subcellular defects in the cells of nev mutant flowers: circularized multilamellar structures and extensive PMBs. Either or both of these defects may be responsible for blocking organ abscission. Because of their resemblance to the Golgi cisternae, and the corresponding lack of Golgi with a wild-type morphology and closely associated TGN (Figs 4 and 5; see Fig. S8 in the supplementary material), we believe the multilamellar structures are Golgi-derived and may represent TGN-Golgi fusions. It is noteworthy that localization of NEV at the TGN and its role in maintaining normal Golgi morphology further support a conserved function with its closest yeast ArfGAP relative, Age2. Although the age2 single mutant is unaffected, the age2 gcs1 double mutant accumulates atypical membrane-bound structures due to a fragmentation of the TGN (Poon et al., 2001). The appearance of these structures is clearly distinct from the circularized structures we see in nev flowers, which is not surprising as wild-type Saccharomyces cerevisiae Golgi are composed of a single disk, in contrast to the multilamellar organization of the plant Golgi system (Preuss et al., 1992). In the future, we plan to test the potential chimeric nature of the aberrant multilamellar structures we have observed.
Interestingly, the multilamellar structures that we see in nev flower cells (Fig. 4; see Fig. S8 in the supplementary material) are similar in appearance to Golgi-derived structures that form when the activity of Arabidopsis vacuolar H+-ATPase (V-ATPase) complexes are reduced (Brüx et al., 2008; Dettmer et al., 2005; Dettmer et al., 2006; Neubert et al., 2008). The eukaryotic V-ATPase complex is a multi-subunit proton transporter that is crucial for acidification of intracellular organelles and vesicles, thereby affecting secretory and endocytic trafficking (Dettmer et al., 2006; Marshansky and Futai, 2008). The characteristic changes in Golgi organization that are seen in cases of reduced V-ATPase activity (bending of the stacks and swelling at the ends of the cisternae) are thought to be due to defects in vesicle trafficking (Dettmer et al., 2006). Disruption of V-ATPase activity has severe effects on development, as illustrated by the block in pollen development associated with hypomorphic mutations in the TGN/early endosome-localized VHA-a1 subunit (Dettmer et al., 2005; Dettmer et al., 2006). V-ATPases are known to be required for the recruitment of proteins involved in vesicle budding, including ARF G-proteins, ARF-GEFs, and coat components (Aniento et al., 1996; Hurtado-Lorenzo et al., 2006; Maranda et al., 2001; Zeuzem et al., 1992). As ARF-GAPs modulate vesicle formation by promoting GTP hydrolysis of ARF G-proteins and interacting with coat components, the similarity between the multilamellar structures found in nev and V-ATPase mutants suggests the possibility that TGN-localized NEV and V-ATPase complexes may jointly regulate the recruitment and activity of the same ARF G-protein(s).
The hyperaccumulation of paramural vesicles in nev flowers, fruit and pedicels (Fig. 4F; see Fig. S8B,D in the supplementary material) is a surprising discovery, and would appear to represent a role for NEV in membrane trafficking that is not conserved with yeast Age2. NEV and the Rab-GEF VPS9a are the first reported Arabidopsis proteins with potential roles in PMB biogenesis or turnover (Goh et al., 2007). The functions and origin of PMBs and paramural vesicles in plants are unknown, although their appearance has been associated with cell-plate formation, secondary cell wall thickening and exposure to pathogens (An et al., 2007). One possibility, previously offered by Goh et al. (Goh et al., 2007), is that PMBs could represent a novel form of endocytosis, by which cell wall materials are taken up into vesicles and transported into the cell for recycling or degradation. Whereas this model is congruent with the need for extensive cell wall modifications during plant cell separation events, the currently favored model is that paramural vesicles in plants may be analogous to the exosomes produced by other higher eukaryotes (An et al., 2007). In animals, exosomes are formed in multivesicular bodies by the reverse budding of membrane into the lumen of late endosomal compartments, and secreted by fusion of the multivesicular body with the plasma membrane rather than with a lytic compartment (van Niel et al., 2006). Exosomes can carry transmembrane signaling molecules and are known to play roles in immunity, development and diseases such as cancer; the multivesicular body pathway of a host cell can also be hijacked by viruses to release viral particles within exosomes (Gould et al., 2003; van Niel et al., 2006; Lakkaraju and Rodriguez-Boulan, 2008). Based on the current understanding of multivesicular body biogenesis, interlumenal vesicles destined for exocytosis versus degradation are indistinguishable in size yet differ in membrane composition (Trajkovic et al., 2008). Analysis of the orientation, content and membrane composition of nev and wild-type paramural vesicles will provide further insights into the origin and intended destination of these structures, as well as their potential role in cell separation.
The expression profile of NEV and its homology to Age2 suggest that it probably plays a more fundamental role in development than is indicated by the nev single mutant phenotype (Fig. 1; see Fig. S2 in the supplementary material). As Age2 activity is redundant with that of the Gcs1 ArfGAP in controlling traffic in yeast cells from the TGN to the endosome, plasma membrane and vacuole (Poon et al., 2001), further functional analysis of the Arabidopsis Age2-like and Gcs1-like family members NEV, AGD15, AGD6 and AGD7 may reveal broad, overlapping requirements for these proteins in plant growth and development.
If NEV is indeed regulating membrane trafficking in multiple plant cell types, why do mutations in NEV have such a profound effect in abscission zone cells? One possibility is that because these cells rely on a tremendous surge of vesicle trafficking to carry out the extensive cell wall remodeling necessary for cell separation within a short time frame, even a minor delay in traffic could affect this process. More than 30 years ago, ultrastructural studies of leaf abscission zone cells revealed evidence of the importance of vesicle trafficking and protein synthesis in cell separation, with dramatic increases in the area of cytoplasm occupied by the ER and Golgi bodies observed upon induction of abscission (Sexton and Hall, 1974; Sexton et al., 1977). Furthermore, the response of abscission zone cells to nev-mediated trafficking defects could be exacerbated by a feedback loop pushing increased vesicle traffic through the system in response to the lack of cell separation. The potential of increased traffic in nev mutant flowers suggest that it may represent a sensitized background to screen for mutations in the signaling components NEV may regulate.
To date, analysis of distinct cell separation events during plant development has not uncovered any common regulatory factors (Lewis et al., 2006). Leaf and floral organ abscission may prove to be an exception. Floral organs and leaves have long been considered to be derived from the same basal structure, and many of the same regulatory networks act to establish their early pattern and polarity (Bowman et al., 2002; Smyth, 2005). It is intriguing that neither floral organ shedding nor aerial leaf detachment occurs in Arabidopsis plants carrying mutations in NEV. Because Arabidopsis leaf detachment is not considered a classical form of abscission, future studies to test whether candidate orthologs of NEV control the shedding of both types of lateral organs in other angiosperms will be informative.
Throughout the history of crop domestication, breeders have selected varieties of grains with reduced shattering characteristics and adopted plants such as the `jointless' tomato mutant that lack fruit abscission (Butler, 1936; Doebley, 2004; Li et al., 2006). Chemicals that control abscission are currently sprayed on fruit trees to prevent preharvest fruit drop, and are used to extend the lifetime of cut flowers and potted plants. Alternative strategies that target specific cell separation events and allow fine-tuning of the timing of separation are expected to arise from increased knowledge of the molecular basis of domestication-related traits and the mechanisms that model plants use to orchestrate this remarkable process.
Supplementary material
Supplementary material available online at http://dev.biologists.org/cgi/content/full/136/11/1909/DC1
Supplementary Material
We thank M. Peifer, J. Reed, J. Kieber, M. Duncan, V. Bautch, S. Patterson, V. Bankaitis and C. Jones for helpful discussions; V. Madden (UNC Microscopy Services) and T. Perdue (UNC Biology Microscopy) for assistance with TEM, SEM and confocal microscopy; S. Guimil, C. Habbersett, A. Imler, D. Reyes and R. Weaver for assistance with mutant screens and mapping; P. Pinyopich for the nev-3 allele; T. Jack, Y. Eshed and J. Bowman for enhancer trap lines; and Monsanto for access to Ler polymorphisms. This research was primarily supported by an NSF grant and UNC startup funds to S.J.L.; the early stages of the project were funded by grants from USDA to S.J.L., NSF and DOE to J.R.E., and NSF to M.F.Y.; generation of the Wave markers was supported by funding from NSF, USDA and HHMI to J.C. M.E.L. was supported by a William R. Kenan Fellowship and the UNC Curriculum in Genetics and Molecular Biology, and M.W.L. by the NIH-funded UNC Developmental Biology training program. P.A.R. and R.L. are supported by the Intramural Program of the National Cancer Institute, NIH, Department of Health and Human Services. Deposited in PMC for release after 6 months.
References
- Aalen, R. B., Butenko, M. A., Stenvik, G. E., Tandstad, N. M. and Patterson, S. E. (2006). Genetic control of floral abscission. In Floriculture, Ornamental and Plant Biotechnology: Advances and Topical Issues, vol. I (ed. J. A. Teixeira da Silva), pp. 101-108. Isleworth, UK: Global Science Books. [Google Scholar]
- Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., Stevenson, D. K., Zimmerman, J., Barajas, P., Cheuk, R. et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653-657. [DOI] [PubMed] [Google Scholar]
- An, Q., van Bel, A. J. E. and Hückelhoven, R. (2007). Do plant cells secrete exosomes derived from multivesicular bodies? Plant Signal. Behav. 2, 4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aniento, F., Gu, F., Parton, R. G. and Gruenberg, J. (1996). An endosomal beta COP is involved in the pH-dependent formation of transport vesicles destined for late endosomes. J. Cell Biol. 133, 29-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez, M. A., Soowal, L. N., Lee, I. and Weigel, D. (1997). LEAFY expression and flower initiation in Arabidopsis. Development 124, 3835-3844. [DOI] [PubMed] [Google Scholar]
- Bowman, J. L., Eshed, Y. and Baum, S. F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18, 134-141. [DOI] [PubMed] [Google Scholar]
- Brüx, A., Liu, T. Y., Krebs, M., Stierhof, Y. H., Lohmann, J. U., Miersch, O., Wasternack, C. and Schumacher, K. (2008). Reduced V-ATPase activity in the trans-Golgi network causes oxylipin-dependent hypocotyl growth inhibition in Arabidopsis. Plant Cell 20, 1088-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko, M. A., Patterson, S. E., Grini, P. E., Stenvik, G. E., Amundsen, S. S., Mandal, A. and Aalen, R. B. (2003). INFLORESCENCE DEFICIENT IN ABSCISSION controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15, 2296-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, L. (1936). Inherited characters of tomato. II. Jointless pedicel. J. Hered. 37, 25-26. [Google Scholar]
- Campisi, L., Yang, Y., Yi, Y., Helig, E., Herman, B., Cassista, A. J., Allen, D. W., Xiang, H. and Jack, T. (1999). Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 17, 699-707. [DOI] [PubMed] [Google Scholar]
- Che, M. M., Nie, Z. and Randazzo, P. A. (2006). Assays and properties of the Arf GAPs AGAP1, ASAP1 and Arf GAP1. Methods Enzymol. 404, 147-163. [DOI] [PubMed] [Google Scholar]
- Cho, S. K., Larue, C. T., Chevalier, D., Wang, H., Jinn, T. L., Zhang, S. and Walker, J. C. (2008). Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105, 15629-15634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow, C. M., Neto, H., Foucart, C. and Moore, I. (2008). Rab-A2 and Rab-A3 GTPases define a trans-golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20, 101-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer, J., Schubert, D., Calvo-Weimar, O., York-Dieter, S., Schmidt, R. and Schumacher, K. (2005). Essential role of the V-ATPase in male gametophyte development. Plant J. 41, 117-124. [DOI] [PubMed] [Google Scholar]
- Dettmer, J., Hong-Hermesdorf, A., Stierhof, Y. H. and Schumacher, K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18, 715-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J. (2004). The genetics of maize evolution. Annu. Rev. Genet. 38, 37-59. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey, C. and Chavrier, P. (2006). ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347-358. [DOI] [PubMed] [Google Scholar]
- Ellis, C. M., Nagpal, P., Young, J. C., Hagen, G., Guilfoyle, T. J. and Reed, J. W. (2005). AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132, 4563-4574. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S. F. and Bowman, J. L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99, 199-209. [DOI] [PubMed] [Google Scholar]
- Fang, S.-C. and Fernandez, D. E. (2002). Effect of regulated overexpression of the MADS domain factor AGL15 on flower senescence and fruit maturation. Plant Physiol. 130, 78-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, D. E., Heck, G. R., Perry, S. E., Patterson, S. E., Bleecker, A. B. and Fang, S.-C. (2000). The embryo MADS domain factor AGL15 acts postembryonically. Inhibition of perianth senescence and abscission via constitutive expression. Plant Cell 12, 183-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A. and Jurgens, G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-mediated plant growth. Cell 112, 219-230. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Dénervaud-Tendon, V., Hyman, D. L., Mayer, U., Stierhof, Y. D. and Chory, J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. doi: 10.1111/j.1365-313X.2009.03851.x. [DOI] [PMC free article] [PubMed]
- Goh, T., Uchida, W., Arakawa, S., Ito, E., Dainobu, T., Ebine, K., Takeuchi, M., Sato, K., Ueda, T. and Nakano, A. (2007). VPS9a, the common activator for two distinct types of Rab5 GTPases, is essential for the development of Arabidopsis thaliana. Plant Cell 19, 3504-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Carranza, Z. H., Elliott, K. A. and Roberts, J. A. (2007). Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J. Exp. Bot. 58, 3719-3730. [DOI] [PubMed] [Google Scholar]
- Gould, S. J., Booth, A. M. and Hildreth, J. E. K. (2003). The Trojan exosome hypothesis. Proc. Natl. Acad. Sci. USA 100, 10592-10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe, M., Xu, J., Mobius, W., Ueda, T., Nakano, A., Geuze, H. J., Rook, M. B. and Scheres, B. (2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13, 1378-1387. [DOI] [PubMed] [Google Scholar]
- Hepworth, S. R., Zhang, Y., McKim, S., Li, X. and Haughn, G. W. (2005). BLADE-ON-PETIOLE1 dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17, 1434-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J. and Nakai, K. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585-W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado-Lorenzo, A., Skinner, M., El Annan, J., Futai, M., Sun-Wada, G. H., Bourgoin, S., Casanova, J., Wildeman, A., Bechoua, S., Ausiello, D. A. et al. (2006). V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat. Cell Biol. 8, 124-136. [DOI] [PubMed] [Google Scholar]
- Inoue, H. and Randazzo, P. A. (2007). Arf GAPs and their interacting proteins. Traffic 8, 1465-1475. [DOI] [PubMed] [Google Scholar]
- Inoue, H., Ha, V. L., Prekeris, R. and Randazzo, P. A. (2008). Arf GTPase-activating protein ASAP1 interacts with Rab11 effector FIP3 and regulates pericentrosomal localization of transferrin receptor-positive recycling endosome. Mol. Biol. Cell 19, 4224-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn, T. L., Stone, J. M. and Walker, J. C. (2000). HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14, 108-117. [PMC free article] [PubMed] [Google Scholar]
- Kandasamy, M. K., Deal, R. B., McKinney, E. C. and Meagher, R. B. (2005a). Silencing the nuclear actin-related protein AtARP4 in Arabidopsis has multiple effects on plant development, including early flowering and delayed floral senescence. Plant J. 41, 845-858. [DOI] [PubMed] [Google Scholar]
- Kandasamy, M. K., McKinney, E. C., Deal, R. B. and Meagher, R. B. (2005b). Arabidopsis ARP7 is an essential actin-related protein required for normal embryogenesis, plant architecture and floral organ abscission. Plant Physiol. 138, 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi, K., Naramoto, S., Sawa, S., Yahara, N., Ueda, T., Nakano, A., Sugiyama, M. and Fukuda, H. (2005). VAN3 ARF-GAP-mediated vesicle transport is involved in leaf vascular network formation. Development 132, 1699-1711. [DOI] [PubMed] [Google Scholar]
- Lakkaraju, A. and Rodriguez-Boulan, E. (2008). Itinerant exosomes: emerging roles in cell and tissue polarity. Trends Cell Biol. 18, 199-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber, M. H., Waizenegger, I., Steinmann, T., Schwarz, H., Mayer, U., Hwang, I., Lukowitz, W. and Jurgens, G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139, 1485-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, G. J., Sohn, E. J., Lee, M. H. and Hwang, I. (2004). The Arabidopsis rab5 homologs rha1 and ara7 localize to the prevacuolar compartment. Plant Cell Physiol. 45, 1211-1220. [DOI] [PubMed] [Google Scholar]
- Lewis, M. W., Leslie, M. E. and Liljegren, S. J. (2006). Plant cell separation: 50 ways to leave your mother. Curr. Opin. Plant Biol. 9, 59-65. [DOI] [PubMed] [Google Scholar]
- Li, C., Zhou, A. and Sang, T. (2006). Rice domestication by reduced shattering. Science 311, 1936-1939. [DOI] [PubMed] [Google Scholar]
- Liljegren, S. J., Ditta, G. S., Eshed, Y., Savidge, B., Bowman, J. L. and Yanofsky, M. F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766-770. [DOI] [PubMed] [Google Scholar]
- Luo, R., Ahvazi, B., Amariei, D., Shroder, D., Burrola, B., Losert, W. and Randazzo, P. A. (2007). Kinetic analysis of GTP hydrolysis catalysed by the Arf1-GTP-ASAP1 complex. Biochem. J. 402, 439-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycett, G. (2008). The role of Rab GTPases in cell wall metabolism. J. Exp. Bot. 59, 4061-4074. [DOI] [PubMed] [Google Scholar]
- Mandiyan, V., Andreev, J., Schlessinger, J. and Hubbard, S. R. (1999). Crystal structure of the ARF-GAP domain and ankyrin repeats of PYK2-associated protein β. EMBO J. 18, 6890-6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maranda, B., Brown, D., Bourgoin, S., Casanova, J. E., Vinay, P., Ausiello, D. A. and Marshansky, V. (2001). Intra-endosomal pH-sensitive recruitment of the Arf-nucleotide exchange factor ARNO and Arf6 from cytoplasm to proximal tubule endosomes. J. Biol. Chem. 276, 18540-18550. [DOI] [PubMed] [Google Scholar]
- Marshansky, V. and Futai, M. (2008). The V-type H+-ATPase in vesicular trafficking: targeting, regulation and function. Curr. Opin. Cell Biol. 20, 415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim, S. M., Stenvik, G. E., Butenko, M. A., Kristiansen, W., Cho, S. K., Hepworth, S. R., Aalen, R. B. and Haughn, G. W. (2008). The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 135, 1537-1546. [DOI] [PubMed] [Google Scholar]
- Min, M. K., Kim, S. J., Miao, Y., Shin, J., Jiang, L. and Hwang, I. (2008). Overexpression of Arabidopsis AGD7 causes relocation of Golgi-localized proteins to the endoplasmic reticulum and inhibits protein trafficking to plant cells. Plant Physiol. 143, 1601-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, J., Mettbach, U., Menzel, D. and Sǎmaj, J. (2007). Molecular dissection of endosomal compartments in plants. Plant Physiol. 145, 293-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume, W., Tanabe, K., Kon, S., Yoshida, N., Watanabe, T., Torii, T. and Satake, M. (2006). SMAP2, a novel ARF GTPase-activating protein, interacts with clathrin and clathrin assembly protein and functions on the AP1-positive early endosome/trans-Golgi network. Mol. Biol. Cell 17, 2592-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubert, C., Graham, L. A., Black-Maier, E. W., Coonrod, E. M., Liu, T. Y., Stierhof, Y. D., Seidel, T., Stevens, T. H. and Schumacher, K. (2008). Arabidopsis has two functional orthologs of the yeast V-ATPase assembly factor Vma21p. Traffic 9, 1618-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, E., Cheung, A. Y. and Ueda, T. (2008). The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiol. 147, 1516-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norberg, M., Holmlund, M. and Nilsson, O. (2005). The BLADE ON PETIOLE genes act redundantly to control growth and development of lateral organs. Development 132, 2203-2213. [DOI] [PubMed] [Google Scholar]
- Ogawa, M., Kay, P., Wilson, S. and Swain, S. M. (2009). ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21, 216-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima, Y., Mitina, I., Quach, H. L. and Theologis, A. (2005). AUXIN RESPONSE FACTOR2 (ARF2): a pleiotropic developmental regulator. Plant J. 43, 29-46. [DOI] [PubMed] [Google Scholar]
- Patterson, S. E. and Bleecker, A. B. (2004). Ethylene-dependent and independent processes associated with floral organ abscission in Arabidopsis. Plant Physiol. 134, 194-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, S., Butenko, M. and Kim, J. (2007). Ethylene responses in abscission and other processes of cell separation in Arabidopsis. In Advances in Plant Ethylene Research (ed. A. Ramina, C. Chang, J. Giovannoni, H. Klee, P. Perata and E. Woltering), pp. 271-278. Netherlands: Springer.
- Peyroche, A., Antonny, B., Robineau, S., Acker, J., Cherfils, J. and Jackson, C. L. (1999). Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3, 275-285. [DOI] [PubMed] [Google Scholar]
- Poon, P. P., Cassel, D., Spang, A., Rotman, M., Pick, E., Singer, R. A. and Johnston, G. C. (1999). Retrograde transport from the yeast Golgi is mediated by two ARF GAP proteins with overlapping function. EMBO J. 18, 555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon, P. P., Nothwehr, S. F., Singer, R. A. and Johnston, G. C. (2001). The Gcs1 and Age2 ArfGAP proteins provide overlapping essential function for transport from the yeast trans-Golgi network. J. Cell Biol. 155, 1239-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss, D., Mulholland, J., Franzusoff, A., Segev, N. and Botstein, D. (1992). Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Mol. Biol. Cell 3, 789-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo, P. A., Weiss, O. and Kahn, R. A. (1992). Preparation of recombinant ADP-ribosylation factor. Methods Enzymol. 219, 362-369. [DOI] [PubMed] [Google Scholar]
- Rehman, R. U., Stigliano, E., Lycett, G. W., Sticher, L., Sbano, F., Faraco, M., Dalessandro, G. and Di Sansebastiano, G. (2008). Tomato Rab11a characterization evidenced a difference between SYP121-dependent and SYP122-dependent exocytosis. Plant Cell Phys. 49, 751-766. [DOI] [PubMed] [Google Scholar]
- Richter, S., Geldner, N., Schrader, J., Wolters, H., York-Dieter, S., Rios, G., Konca, C., Robinson, D. G. and Jürgens, G. (2007). Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 448, 488-492. [DOI] [PubMed] [Google Scholar]
- Roberts, J. A., Elliott, K. A. and González-Carranza, Z. H. (2002). Abscission, dehiscence, and other cell separation processes. Annu. Rev. Plant Biol. 53, 131-158. [DOI] [PubMed] [Google Scholar]
- Robinson, D. G., Jiang, L. and Schumacher, K. (2008). The endosomal system of plants: charting new and familiar territories. Plant Physiol. 147, 1482-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot, A. A., Kovaleva, V., Bassham, D. C. and Raikhel, N. V. (2001). Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol. Biol. Cell 12, 3733-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, Y., Hong, H. N., Yanai, N. and Obinata, M. (1998). Involvement of stromal membrane-associated protein (SMAP-1) in erythropoietic microenvironment. J. Biol. Chem. 124, 209-216. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T. S., Henz, S. R., Pape, U. J., Demar, M., Vingron, M., Schölkopf, B., Weigel, D. and Lohmann, J. U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501-506. [DOI] [PubMed] [Google Scholar]
- Sexton, R. and Hall, J. L. (1974). Fine structure and cytochemistry of the abscission zone cells of Phaseolus leaves. I. Ultrastructural changes occurring during abscission. Ann. Bot. 38, 849-854. [Google Scholar]
- Sexton, R., Jamieson, G. G. and Allan, M. H. (1977). An ultrastructural study of abscission zone cells with special reference to the mechanism of enzyme secretion. Protoplasma 91, 369-387. [Google Scholar]
- Sieburth, L. E., Muday, G. K., King, E. J., Benton, G., Kim, S., Metcalf, K. E., Meyers, L., Seamen, E. and Van Norman, J. M. (2006). SCARFACE encodes an ARF-GAP that is required for normal auxin efflux and vein patterning in Arabidopsis. Plant Cell 18, 1396-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D. R. (2005). Morphogenesis of flowers: our evolving view. Plant Cell 17, 330-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X. F., Yang, C. Y., Liu, J. and Yang, W. C. (2006). RPA, a class II ARFGAP protein, activates ARF1 and U5 and plays a role in root hair development in Arabidopsis. Plant Physiol. 141, 966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C. L., Paris, S., Galweiler, L., Palme, K. and Jurgens, G. (1999). Coordinated polar localization of the auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286, 316-318. [DOI] [PubMed] [Google Scholar]
- Stenvik, G. E., Tandstad, N. M., Guo, Y., Shi, C. L., Kristiansen, W., Holmgren, A., Clark, S. E., Aalen, R. B. and Butenko, M. A. (2008). The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20, 1805-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe, K., Torii, T., Natsume, W., Braesch-Andersen, S., Watanabe, T. and Satake, M. (2005). A novel GTPase-activating protein for ARF6 directly interacts with clathrin and regulates clathrin-dependent endocytosis. Mol. Biol. Cell 16, 1617-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh, O. and Moore, I. (2007). An ARF GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448, 493-496. [DOI] [PubMed] [Google Scholar]
- Trajkovic, K., Hsu, C., Chiantia, S., Rajendran, L., Wenzel, D., Wieland, F., Schwille, P., Brügger, B. and Simons, M. (2008). Ceramide triggers budding of exosome vesicles into multivesciular endosomes. Science 319, 1244-1247. [DOI] [PubMed] [Google Scholar]
- Ueda, T., Uemura, T., Sato, M. H. and Nakano, A. (2004). Functional differentiation of endosomes in Arabidopsis cells. Plant J. 40, 783-789. [DOI] [PubMed] [Google Scholar]
- van Niel, G., Porto-Carreiro, I., Simoes, S. and Raposo, G. (2006). Exosomes: a common pathway for a specialized function. J. Biochem. 140, 13-21. [DOI] [PubMed] [Google Scholar]
- Vernoud, V., Horton, A. C., Yang, Z. and Nielsen, E. (2003). Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131, 1191-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Fecht-Bartenbach, J., Bogner, M., Krebs, M., Stierhof, Y. D., Schumacher, K. and Ludewig, U. (2007). Function of the anion transporter AtCLC-d in the trans-Golgi network. Plant J. 50, 466-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. (2008). Cell polarity signaling in Arabidopsis. Annu. Rev. Cell Dev. Biol. 24, 551-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, C. M., Wen, J., Motes, C. M., Sparks, J. A. and Blancaflor, E. B. (2008). A class one ADP-ribosylation factor GTPase-activating protein is critical for maintaining directional root hair growth in Arabidopsis thaliana. Plant Physiol. 147, 1659-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeuzem, S., Feick, P., Zimmermann, P., Haase, W., Kahn, R. A. and Schulz, I. (1992). Intravesicular acidification correlates with binding of ADP-ribosylation factor to microsomal membranes. Proc. Natl. Acad. Sci. USA 89, 6619-6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C.-J., Cavenagh, M. M. and Kahn, R. A. (1998). A family of Arf effectors defined as suppressors of the loss of Arf function in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273, 19792-19796. [DOI] [PubMed] [Google Scholar]
- Zheng, H., Bednarek, S. Y., Sanderfoot, A. A., Alonso, J., Ecker, J. R. and Raikhel, N. V. (2002). NPSN11 is a cell plate-associated SNARE protein that interacts with the syntaxin KNOLLE. Plant Physiol. 129, 530-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, H., Camacho, L., Wee, E., Batoko, H., Legen, J., Leaver, C. J., Malho, R., Hussey, P. J. and Moore, I. (2005). A rab-E GTPase mutant acts downstream of the rab-D subclass in biosynthetic membrane traffic to the plasma membrane in tobacco leaf epidermis. Plant Cell 17, 2020-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.