Abstract
Evidence has accumulated that p53, a prototypical tumor suppressor, may also influence aspects of organismal aging. We have previously described a p53 mutant mouse model, the p53+/m mouse, which is cancer resistant yet exhibits reduced longevity and premature aging phenotypes. p53+/m mice express one full length p53 allele and one truncated p53 allele that is translated into a C-terminal fragment of p53 termed the M protein. The augmented cancer resistance and premature aging phenotypes in the p53+/m mice are consistent with a hyperactive p53 state. To determine how the M protein could increase p53 activity, we examined the M protein in various cellular contexts. Here, we show that embryo fibroblasts from p53+/m mice exhibit reduced proliferation and cell cycle progression compared to embryo fibroblasts from p53+/− mice (with equivalent wild-type p53 dosage). The M protein interacts with wild-type p53, increases its stability, and facilitates its nuclear localization in the absence of stress. Despite increasing p53 stability, the M protein does not disrupt p53-Mdm2 interactions and does not prevent p53 ubiquitination. These results suggest molecular mechanisms by which the M protein could influence the aging and cancer resistance phenotypes in the p53+/m mouse.
Keywords: p53, Mdm2, premature aging, mouse aging model
1. Introduction
The p53 tumor suppressor responds to a wide range of stresses, including DNA damage and deregulated oncogenes (Giaccia and Kastan, 1998; Horn and Vousden, 2007). The outcome of p53 activation depends on both the type and severity of the stress encountered, and may result in cell cycle arrest, senescence, or apoptosis (Vousden and Lu, 2002). Due to its potent anti-proliferative effects, p53 must be tightly regulated. In unstressed cells p53 is unstable and inactive due to rapid proteosomal degradation mediated in part by the E3 ubiquitin ligase MDM2 (Haupt et al., 1997; Fuchs et al., 1998; Giaccia and Kastan, 1998; Moll and Petrenko, 2003). Phosphorylation of p53 and MDM2 in response to stress leads to their dissociation and subsequent p53 stabilization and activation (Craig et al., 1999; Alarcon-Vargas and Ronai, 2002). p53 can also be regulated at the level of cellular localization. p53 contains both a nuclear localization signal (NLS) and a nuclear export signal (NES) and shuttles between the nucleus and cytoplasm (Ferrigno and Silver, 1999). The translocation between the nucleus and cytoplasm can be regulated in several ways, including post-translational modifications and tetramerization (Fabbro and Henderson, 2003). Nuclear localization is critical for p53 transactivation of target genes to elicit the p53 response. However, p53 may also have some transcription-independent functions in the cytoplasm by inducing apoptosis at the mitochondrial membrane (Erster et al., 2004).
Mutations in p53 are found in nearly 50% of human cancers (Vousden and Lu, 2002). p53 deficient mice are considerably more tumor prone than their wild-type counterparts, and almost all p53 null mice succumb to tumors by 6 months of age (Donehower et al., 1992). The anti-proliferative functions of p53 make it a potent anti-cancer molecule, but could these same functions also promote some aspects of the aging process? Global overexpression of wild-type or truncated variants of p53 in the mouse has consistently resulted in augmented cancer resistance, but has generated mixed results with respect to aging/longevity phenotypes. Addition of one or two normally regulated wild-type copies of p53 to the mouse germline resulted in normal longevity (Garcia-Cao et al., 2002). A similar result was achieved with mice containing MDM2 hypomorphic alleles (Mendrysa et al., 2006). However, two genetically engineered mice expressing truncated C-terminal variants of p53 both displayed reduced longevity and a constellation of premature aging phenotypes. Scrable and colleagues described a transgenic mouse overexpressing a modestly truncated, naturally occurring isoform of p53 called p44 that exhibited reduced longevity, early bone loss, reduced body mass, premature loss of fertility and testicular degeneration, and altered IGF-1 signaling (Maier et al., 2004). The p53 mutant mice generated in our lab resulted in expression of a truncated C-terminal p53 variant as a result of an aberrant gene targeting event in embryonic stem cells. These “p53+/m mice” have one wild-type p53 germ line allele and one truncated “m” allele missing p53 exons 1−6, but retaining exons 7−11 (Tyner et al., 2002). Despite having only one intact p53 allele, the p53+/m mice are highly resistant to tumors yet have a 23% reduction in median longevity compared to p53+/+ counterparts of the same genetic background. Additionally, these mice display early onset of numerous traits associated with aging such as reduced body weight, muscle atrophy, lordokyphosis, osteoporosis, and a decreased tolerance to stress (Tyner et al., 2002). Early studies to examine the effect of the M protein on wild-type p53 showed that the M protein could increase p53-dependent transactivation of the p21 promoter. Moreover, treatment of p53+/m mice with ionizing radiation resulted in prolonged, elevated levels of tissue p53 protein levels that were more pronounced than in p53+/− and even p53+/+ tissues (Tyner et al., 2002). These observations led us and others to view the p53+/m mouse as a model of chronic hyperactive p53 (de Stanchina and Lowe, 2002; Klatt and Serrano, 2003; Dumble et al., 2004).
Despite the fact that the p53+/m mice display cancer resistance and premature aging phenotypes consistent with a hyperactive p53 response, a caveat should be mentioned. In these mice, the m allele message is initiated not from the p53 promoter, but from an upstream region of the Rho guanine exchange factor 15 (Arhgef15) and contains a short leader sequence derived from that gene prior to splicing into exon 7 of the p53 gene (which contains a functional translational initiation codon) (Gentry and Venkatachalam, 2005). The chimeric transcript that results is translated into a truncated 24 kDa C-terminal p53 variant and is the result of a 600 kb deletion event between the Arhgef15 gene and the p53 gene. The p53+/m mice are haploid for a region of mouse chromosome 11 containing 24 genes. Thus, the possibility exists that some of the cancer resistance and accelerated aging phenotypes could result from haploinsufficiency of genes within the m-associated deletion rather than from expression of a truncated M protein.
In this paper, we present further evidence for our hypothesis that the cancer resistance and early aging phenotypes in the p53+/m mice are driven primarily by the truncated m allele gene product resulting in chronic p53 hyperactivity. We show that fibroblasts derived from p53+/m embryos show decreased proliferation and cell cycle progression compared to p53+/− embryo fibroblasts that also contain a single wild-type p53 allele. We also demonstrate that the M protein has modest growth inhibiting activity even in the absence of wild-type p53. Finally, we show that the M protein interacts with wild-type p53 protein, induces nuclear localization of p53 and increases p53 stability, attributes that normally take place only after exposure to stress. The aberrant nuclear localization and increased stability of p53 in the presence of the M protein may drive alterations in p53 function leading to the phenotypes displayed by the p53+/m mice.
2. Materials and Methods
2.1 Plasmids
The human MDM2 expression vector, pCMV6-XL4-MDM2, was obtained from Origene. Human GFP-M expression vector (C-24−7) was modified from pp53-EGFP. The human GFP-p53 expression vector, pp53-EGFP (Clontech), and human GFP-M expression vectors were a generous gift from David Sassoon at Mt. Sinai Medical School. For osteosarcoma colony suppression assays, the pTracer-CMV2 vector (Invitrogen) was used to clone the murine wild-type p53 cDNA, point mutant murine p53 cDNA (R172H), and a truncated cDNA corresponding to the m allele containing p53 exons 7−11. Exon 7 of p53 contains an ATG codon with a strong Kozak sequence allowing efficient translation of a truncated C-terminal p53 fragment of 24 kDa. Plasmid vectors for generation of the retroviral recombination assay vectors have been described (Lu et al., 2003).
2.2 Site-Directed Mutagenesis
Mutations in wild-type p53 (pp53-EGFP) and M (C-24−7) were introduced by site-directed mutagenesis using the Quick-Change II Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer's protocol. The following primers were used:
- 305KR to AA:
- Sense: 5'-CTGCCCCCAGGGAGCACTGCGGCAGCACTGCCCAACAACACCAGC-3'
- Antisense: 5'-GCTGGTGTTGTTGGGCAGTGCTGCCGCAGTGCTCCCTGGGGGCAG-3'
- 319KKK to AAA:
- Sense: 5'-CCAGCTCCTCTCCCCAGCCAGCGGCGGCACCACTGGATGGAGAATATTTCAC-3'
- Antisense: 5'-GTGAAATATTCTCCATCCAGTGGTGCCGCCGCTGGCTGGGGAGAGGAGCTGG-3'
- L348A:
- Sense: 5'-GCTGAATGAGGCCGCGGAACTCAAGGATG-3'
- Antisense: 5'-CATCCTTGAGTTCCGCGGCCTCATTCAGC-3'
- L350A:
- Sense: 5'-GAGGCCGCGGAAGCCAAGGATGCCCAGG-3'
- Antisense: 5'-CCTGGGCATCCTTGGCTTCCGCGGCCTC-3'
To reconstitute the NLS in the M NLS mutant (KR-AA, KKK-AAA), the NLS from the SV40 Large T Antigen was fused to the C-terminal using the following oligonucleotides:
Sense: 5'Phos/-GATCCCCAAAGAAAAAGCGCAAGGTGG-3'
Antisense: 5'Phos/-GATCCCACCTTGCGCTTTTTCTTTGGG-3'
The oligos were boiled for 5 minutes an allowed to anneal at room temperature for 2 hours before ligating into the BamHI site between the last amino acid of p53 and the start of GFP. All mutations were verified by DNA sequencing (BCM Sequencing Core Facility).
2.3 Cell Lines, Culture Conditions and Cell Transfections
Human U2OS and Saos-2 cells are human osteosarcoma cell lines obtained from the American Type Tissue Collection. U2OS cells retain wild-type p53 and Saos-2 cells are deleted for both p53 alleles (Chen et al., 1990; Florenes et al., 1994). TE85 osteosarcoma cells were obtained from Ronald Javier and contain a mutant R156P p53 allele (and no wild-type allele) (Romano et al., 1989). p53+/+ and p53−/− HCT116 cells were a kind gift from Bert Vogelstein (Bunz et al., 1998). All cells were cultured in a 37°C incubator with 5% CO2 in Dulbecco's modified Eagle Medium (DMEM) with 10% fetal bovine serum. Mouse embryonic fibroblasts (MEFs) were maintained in a 37°C incubator with 5% C02 in DMEM with 15% fetal bovine serum. Transient transfection was accomplished using Lipofectamine Plus (Invitrogen) according to the manufacturer's protocol.
2.4 Cycloheximide Assays
Cycloheximide (Sigma) was added to the cell culture medium to a final concentration of 40 ug/ml 24 hours following transfection. At the indicated time points, cells were washed in PBS and scraped into lysis buffer buffer (50mM Tris pH 7.5, 150mM NaCl, 1% NP-40, supplemented with a protease inhibitor tablet). Cell lysates were resolved by SDS-PAGE and transferred to PVDF membranes. Membranes were probed for p53 (DO-1, Santa Cruz or IC12, Cell Signaling) and actin (Ab5, Neomarkers). Western blots were quantitated using a Storm 860 Phosphorimager (U2OS) or a Densitometer (MEFs) (GE Healthcare) and ImageQuant software (Molecular Dynamics).
2.5 Immunoprecipitation
Cell lysates were collected 24 hours following transfection as described above. For most experiments, 500 ug of total protein was used for each sample. For co-immunoprecipitation of p53 with M, p53 was immunoprecipitated using a mouse monoclonal p53 antibody to the N-terminus (DO-1, Santa Cruz). For co-immunoprecipitation of p53 with MDM2, p53 was immunoprecipitated using a rabbit polyclonal p53 antibody (FL393, Santa Cruz). All immunoprecipitations were incubated overnight at 4°C followed by the addition of Protein GAgarose beads for 1 hour. Immunoprecipitated complexes were resolved by SDS-PAGE and transfered to PVDF membranes. Membranes were probed with p53 antibodies (FL393 or DO-1, Santa Cruz) or GFP (B-2, Santa Cruz) to detect GFP tagged p53 and M. To detect MDM2, membranes were probed with a mixture of Ab-4 and Ab-5 (Calbiochem).
2.6 Analysis of p53 Ubiquitination
Ubiquitination assays were performed as described previously (Grossman et al., 2003). Briefly, 24 hours following transfection, cells were treated with 25 uM each of the proteasome inhibitors MG101 (Sigma) and MG132 (Sigma) for 6 hours prior to harvest. Cells were washed with PBS and lysed with IPB-240 (20mM HEPES pH 7.4, 240mM NaCl, 0.1 mM EDTA, 0.5% TritonX-100, 1mM PMSF, 1mM DTT and supplemented with a protease inhibitor tablet). p53 was immunoprecipitated with p53 antibody (FL393, Santa Cruz) as described above. Beads were washed 5 times with IPB-240. After resolving the samples by SDS-PAGE, membranes were probed with mouse monoclonal antibodies to p53 (DO-1, Santa Cruz), Ubiquitin (P4D1, Santa Cruz), and MDM2 (Ab4 and Ab5, Calbiochem).
2.7 Indirect Immunofluorescence
Cells were cultured directly on glass coverslips. 24 hours following transfection, cells were fixed on ice in 4% paraformaldehyde for 20 minutes. After washing, cells were permeablized in 0.1% Triton-X 100 on ice for 10 minutes, followed by blocking with 1% BSA. Cells were incubated with mouse anti-p53 (DO-1, Santa Cruz), followed by incubation with Alexa-594 conjugated goat anti-mouse secondary antibody (Molecular Probes) and stained with DAPI. Cells were visualized on a Zeiss Axioplan II Fluorescent Microscope (Zeiss, BCM Microscopy Core Facility).
2.8 Cell Cycle Analysis
MEFs of each p53 genotype were cultured in DMEM supplemented with 15% FBS. 2.5 hours prior to harvest, cells were labeled with 10uM/mL of bromodeoxyuridine (BrdU). Cells were then fixed in 70% ethanol and stained with an anti-BrdU FITC-conjugated antibody (Becton Dickenson) and 50ug/mL of propidium iodide (PI). The stained cells were analyzed by flow cytometry (Coulter Epics XL-MCL).
2.9 Recombination Assays
Homologous recombination assays were performed as described previously (Lu et al., 2003). Briefly, MEFs of each p53 genotype were infected with 1−2 × 106 G418 colony forming units of the retroviral vector pLNCX-GZ per 106 cells. After infection, at each passage, each p53 genotype of infected MEFs was split into two populations of identical numbers of cells and selected in G418 alone (0.75−1.0 mg/ml) or G418 plus zeocin (100 μg/ml). Two weeks after initiation of selection, cells were rinsed with phosphate buffered saline and fixed and stained in 0.5% crystal violet, 10% acetic acid, and 25% methanol. G418 and G418/zeocin resistant colonies were counted and recombination frequencies determined. Recombination frequency was estimated by dividing the number of G418 plus zeo resistant colonies by the number of G418 resistant colonies in each genotype group.
2.10 Osteosarcoma Cell Suppression Assays
1 × 106 to 5 ×106 cells were transfected with 25 μg plasmid. For stable transfections, cells were transfected with zeocin resistance gene containing plasmids (and which contained either wild-type p53, truncated m allele p53, or intact point mutant p53 driven by a CMV promoter) into TE85 (point mutant p53), Saos2 (p53 null), or U2OS (wild-type p53) osteosarcoma cells, then after 48 hours, cells were split 1:4 or 1:5 into medium containing 50mg/ml zeocin (Invitrogen). After 4−6 weeks of selection, plates were stained with methylene blue and colonies counted.
3. Results
3.1 p53+/m MEFs exhibit slower growth kinetics, enhanced cell cycle arrest and increased recombination suppression compared to p53+/− MEFs
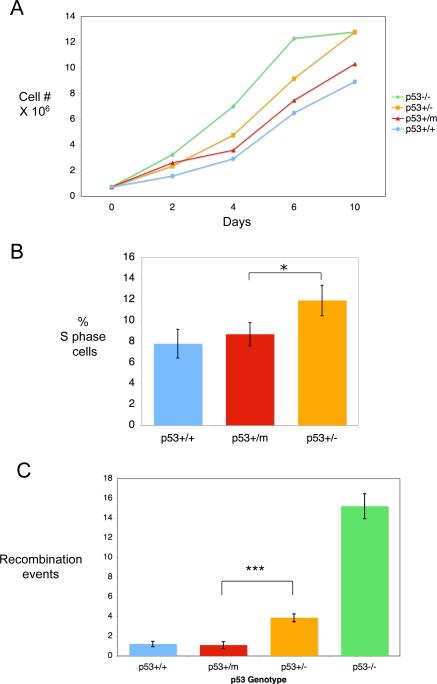
The growth kinetics of p53+/+, p53+/−, p53+/m, and p53−/− MEFs in culture were analyzed over a period of 10 days (Figure 1A). p53−/− MEFs displayed a clear growth advantage as has been previously reported (Harvey et al., 1993). MEFs with only one copy of p53 (p53+/−) also have a growth advantage over p53+/+ MEFs, but less than that of p53−/− MEFs. p53+/m MEFs displayed growth kinetics similar to or slightly more robust than p53+/+ MEFs (Figure 1A). The reduction of growth rate of p53+/m MEFs relative to p53+/− MEFs was determined to be significant by t-test (P = 0.003). This was further confirmed by labeling actively dividing, sub-confluent p53+/+, p53+/− and p53+/m MEFs with BrdU and analyzing the number of cells in S phase by flow cytometry (Figure 1B). The percentage of cells in S phase in the p53+/m MEFs was significantly reduced compared to p53+/− MEFs. Finally, we examined the ability of p53+/+, p53+/−, p53+/m, and p53−/− MEFs to suppress recombination (Figure 1C). MEFs from each of the p53 genotypes were infected with a retroviral vector containing a neomycin resistance marker and two tandem repeats of mutant versions of a GFP-Zeocin (GFP-Zeo) resistance fusion gene (Lu et al., 2003). The GFP-Zeo is non-functional until an intrachromosomal homologous recombination event takes place that can regenerate a single intact copy of the GFP-Zeo fusion gene. Such events can be scored by cells showing GFP fluorescence or zeocin plus G418 drug resistance. This vector, when transduced as a single copy into target cells, is an indicator of genomic instability and is particularly relevant in this context since wild-type p53 has been shown to suppress homologous recombination events (Sturzbecher et al., 1996; Mekeel et al., 1997; Bertrand et al., 1997; Saintigny et al., 1999; Lu et al., 2003). After infection with the Neo-GFP-Zeo retroviral vector, MEFs were selected in G418 or G418 plus zeocin and the number of zeocin resistant colonies per 106 G418 colonies was calculated. It has been previously shown that p53−/− MEFs have significantly higher levels of homologous recombination than p53+/+ MEFs, and that p53+/− MEFs have an intermediate level of recombination, suggesting that p53 deficiency may promote recombination (Lu et al., 2003). Here we found that the p53+/m MEFs were significantly more efficient in suppressing recombination than the p53−/− and p53+/− MEFs. Despite the fact that the p53+/− and p53+/m MEFs both contain only one intact copy of p53, the p53+/m MEFs have slower growth kinetics, an enhanced cell cycle arrest response, and are better able to suppress recombination, suggesting that the M protein may provide the p53+/m MEFs with increased p53 activity compared to p53+/− MEFs.
Figure 1.
Growth kinetics, cell cycle checkpoint control, and suppression of recombination of p53+/+, p53+/−, p53+/m, and p53−/− MEFs. A. Representative growth kinetics of p53+/+, p53+/−, p53+/m, and p53−/− MEFs. 750,000 cells were plated onto 10cm dishes and collected and counted on the days indicated. T-tests on multiple growth experiments indicated no significant difference between p53+/+ and p53+/m growth rates (P = 0.61), while growth rate differences between p53+/m and p53+/− MEFs were significantly different (P = 0.003). B. S phase fractions in rapidly dividing MEFs of different p53 genotypes. MEFs were analyzed for DNA content and BrdU incorporation by flow cytometry and the percentage of cells in S phase was calculated. T-tests indicated the S phase fractions of p53+/m and p53+/+ MEFs were not significantly different (P = 0.47), while S phase fractions of p53+/m and p53+/− MEFs were significantly different and are indicated by an asterisk (P = 0.05). C. Suppression of recombination in p53+/+, p53+/−, p53+/m, and p53−/− passage 1−3 MEFs. p53+/+, p53+/−, p53+/m, and p53−/− MEFs were infected with a retrovirus expressing two tandem copies of mutant forms of a GFP-Zeocin gene as well as a neomycin resistance marker. MEFs were selected with G418 or G418 plus Zeocin to determine the recombination frequencies. The p53+/m MEFs have a recombination frequency roughly equal to that of p53+/+ MEFs. The difference in recombination frequency between p53+/m and p53+/− MEFs is highly significant (P < .0001) and is indicated by three asterisks. Differences between p53+/+ and p53+/− MEFs are also statistically significant (P < .0001).
3.2 The M protein displays modest p53-independent growth suppression
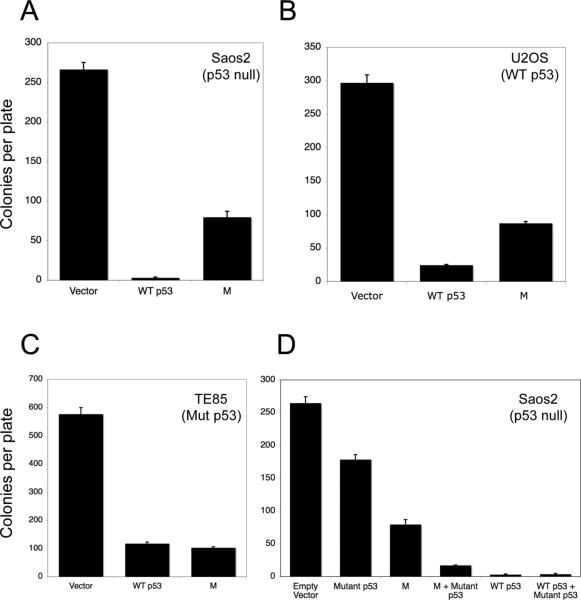
To further assess the anti-proliferative functions of the M protein in the presence and absence of p53, colony suppression assays were performed in three human osteosarcoma cell lines that differ in their p53 status. In p53 null Saos-2 cells, introduction of wild-type p53 suppresses colony formation almost completely. When the M protein is introduced into these cells, there was a significant 70% suppression of colony formation, suggesting that the M protein may have some growth suppressive functions independent of p53 (Figure 2A). In U2OS cells which contain wild-type p53, introducing additional p53 results in a 90% suppression of colony formation (Figure 2B). Introduction of the M protein into U2OS cells results in a significant 75% reduction in colony formation, though whether this M-induced suppression is largely wild-type p53-dependent or independent is not clear from this assay (Figure 2B). In TE85 cells that contain a dominant negative point mutant form of p53 (R156P) and no wild-type p53 (Romano et al., 1989), the introduction of either additional wild-type p53 or the M protein results in significant growth suppression, over 80% reduction in colony formation (Figure 2C). The capacity of the M protein to suppress TE85 colony formation to the same extent as wild-type p53 is surprising, but is consistent with a model in which M protein interacts with endogenous mutant p53 and converts it into a wild-type configuration as has previously been shown for other C-terminal p53 fragments and peptides (Selivanova et al., 1997; Selivanova et al., 1999). Finally, to determine in another context whether the M protein could cooperate with mutant p53 protein to enhance growth suppression, we examined the effect of mutant p53 in combination with either wild-type p53 or the M protein in p53 null Saos-2 cells (Figure 2D). Mutant p53 (R172H) had only a marginal suppressive effect on Saos-2 cell colony formation. However, introduction of both mutant p53 and wild-type p53 (or wild-type p53 by itself) led to almost complete suppression of colony formation. While the M protein by itself caused a 70% reduction in colony formation, the M protein in combination with mutant p53 resulted in a 94% reduction in colony formation. These results suggest that the M protein can enhance the growth suppressive functions of mutant p53.
Figure 2.
Effects of wild-type p53, mutant p53, and the p53 m allele on growth suppression of human osteosarcoma cells. A-C. Colony formation in osteosarcoma cells. Saos-2 cells (null for p53) (A), U2OS cells (wild-type for p53) (B), and TE85 (containing mutant p53) (C), were transfected with empty vector (zeocin resistance) constructs, wild-type p53 (zeocin resistance) expression constructs, and m allele expression (zeocin resistance) constructs. Forty-eight hours after transfection, osteosarcoma cells were selected in zeocin for two weeks and zeocin resistant colonies fixed, stained and counted. D. Saos-2 cells were transfected with a zeocin resistance gene construct expressing either no insert (empty vector), a mutant version of p53 (codon 172 arg→his), wild-type p53, or the truncated p53 m allele, or the indicated combination of vectors. Zeocin resistant colonies were identified and counted as described for panels A-C.
3.3 The M protein localizes to the nucleus and interacts with full-length p53
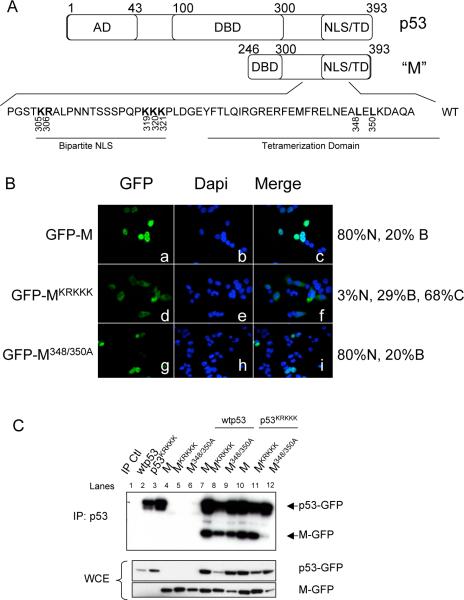
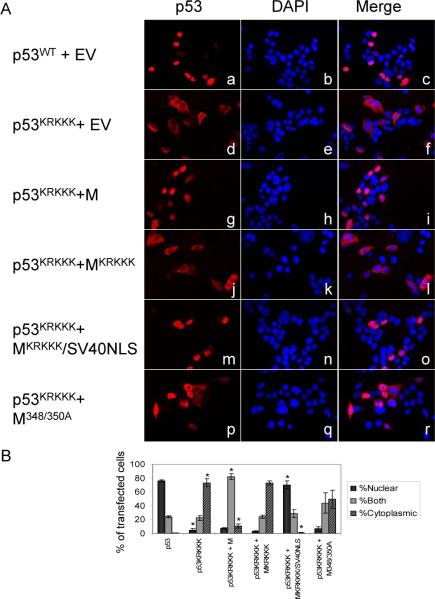
The M protein is a C-terminal fragment of p53 that is missing the transactivation domain and most of the DNA binding domain (Tyner et al., 2002). The M protein therefore consists of a C-terminal p53 fragment that includes the oligomerization domain and nuclear localization and export signals (Figure 3A). To examine the cellular localization of the M protein, a construct encoding the M protein fused at the C-terminus to GFP (GFP-M) was transfected into p53−/− HCT116 cells (Figure 3B, panels a-c). 80% of transfected cells expressed the M protein predominantly in the nucleus and the remaining 20% of transfected cells expressed the M protein in both the nucleus and cytoplasm. The nuclear localization of the M protein is due to a bipartite NLS in the C-terminal region of the protein (see Figure 3A). Key lysine and arginine residues in the NLS were mutated to alanine to generate an M NLS mutant, MKRKKK (O'Keefe et al., 2003). When this mutant is transfected into p53−/− HCT116 cells, 68% of transfected cells express the mutant protein predominantly in the cytoplasm (Figure 3B, panels d-f). Point mutations were also introduced to change the leucines at positions 348 and 350 in the oligomerization domain of the M protein to alanines to generate M348/350A (Mateu and Fersht, 1998; Stommel et al., 1999). The M348/350A mutant is also predominantly nuclear in 80% of transfected cells, similar to the localization of M (Figure 3B, panels g-i).
Figure 3.
The M protein localizes to the nucleus and interacts with wild-type p53. A. Diagram of human p53 and the M protein. Residues that were mutated to alanine in the NLS and tetramerization domain are shown in bold. AD: Activation Domain. DBD: DNA Binding Domain. NLS/TD: Nuclear Localization Signal/Tetramerization Domain. B. Localization of M and M mutants. Cells were transfected with GFP tagged M (panels a-c), GFP tagged MKRKKK (panels d-f), or GFP tagged M348/350A (panels g-i) as indicated. The localization of M was determined by immunofluorescence and nuclei visualized by DAPI staining. 300 cells were counted and scored as having GFP-M mostly nuclear (N), mostly cytoplasmic (C), or both nuclear and cytoplasmic (B). Percentages listed are an average of three independent experiments. C. Interaction between p53 and M mutants. p53−/− HCT116 cells were transfected with GFP tagged p53 and M expression constructs as indicated. p53 was immunoprecipitated with an antibody specific to the N-terminus of p53. Membranes were probed with an antibody to GFP to detect both full-length p53 and the M protein.
As the M protein contains the oligomerization domain of p53, we next examined the ability of the M protein to interact with full-length p53 (Figure 3C). p53−/− HCT116 cells were transfected with p53 and M as indicated and p53 immunoprecipitated with an antibody specific to the N-terminal domain of p53 (DO-1, Santa Cruz) to examine the interaction between p53 and M. The M protein retains the ability to bind to full-length, wild-type p53 (Figure 3C, lane 7). Mutation of the NLS in the M protein (MKRKKK) does not interfere with the interaction between p53 and the M protein (lanes 8 and 11). Despite the mutations in the oligomerization domain, the M348/350A mutant retained the ability to interact with wild-type p53 (lane 9). This result suggests that the M348/350 mutations were insufficient to fully disrupt M-p53 oligomerization through the tetramerization domain. We also examined the ability of the M protein and M mutants to interact with an NLS mutant of the full-length p53 (p53KRKKK). The M protein and MKRKKK were able to interact with p53KRKKK (lanes 10 and 11). However, the M348/350A mutant was immunoprecipitated less efficiently with p53KRKKK (lane 12).
3.4 The M protein promotes nuclear accumulation of full-length p53
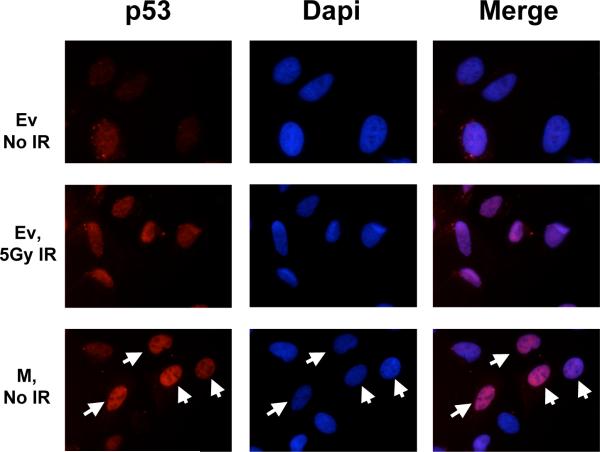
Prior to activation, p53 exists at low levels in an inactive state. In response to stress, p53 rapidly translocates to the nucleus where it is able to function as a transcription factor (Giaccia and Kastan, 1998). Previous studies have indicated that short, C-terminal fragments of p53 are capable of inducing the nuclear localization of p53 in the absence of stress (Ostermeyer et al., 1996). In order to determine the effect of the M protein on p53 localization, U2OS cells expressing wild-type p53 were transfected with M and the localization of the full-length p53 determined by indirect immunofluorescence (Figure 4). p53 levels are low in cells transfected with empty vector (top panels), but p53 accumulates in the nucleus of these cells after irradiation (middle panels). Cells transfected with M show increased nuclear p53 in the absence of irradiation compared to untransfected, unirradiated cells (bottom panels, arrows indicate cells transfected with M).
Figure 4.
The M protein induces nuclear accumulation of p53. U2OS cells were transfected with empty vector (Ev) or a plasmid encoding M. Cells were irradiated with 5 Grays of ionizing radiation or mock irradiated four hours prior to fixing and immunostaining for full-length p53. Top: Empty vector transfected cells. Middle: Empty vector transfected cells 4 hours following irradiation. Bottom: M transfected cells. Cells that are transfected with M (indicated by white arrows) have increased nuclear p53 compared to untransfected cells.
To further confirm the ability of the M protein to induce p53 nuclear localization, we utilized the mutants of the M protein described above. Expression constructs encoding the M protein and the M mutants were co-transfected with wild-type p53 or p53KRKKK expression constructs into p53−/− HCT116 cells and the localization of full-length p53 determined by immunofluorescence using an antibody that recognizes the N-terminus of p53 (Figure 5). When transfected alone into p53−/− HCT116 cells, wild-type p53 is almost exclusively nuclear (Figure 5A, panels a-c and 5B). Co-transfection of wild-type p53 and M has no effect on p53 localization (data not shown). In contrast to wild-type p53, p53KRKKK localizes predominantly to the cytoplasm in more than 70% of the transfected cells (5A, panels d-f and 3B). When p53KRKKK is co-expressed with wild-type M, p53KRKKK is localized to the nucleus or both the cytoplasm and the nucleus in 90% of the transfected cells, indicating the ability of M to induce p53 nuclear accumulation (Figure 5A, panels g-i and Figure 5B). This is dependent on M having an intact NLS of its own, as co-expression of p53KRKKK with MKRKKK results in p53 localizing predominantly to the cytoplasm in more than 70% of the transfected cells, similar to the results obtained by transfecting p53KRKKK alone (Figure 5A, panels j-l and 5B). To further confirm the specific effect of the nuclear localization signal of M, a construct was generated that fused the NLS from the SV40 Large T-antigen to the MKRKKK NLS mutant. Co-transfection of this construct with p53KRKKK again resulted in the nuclear accumulation of p53 (Figure 5A, panels mo and 5B).
Figure 5.
M induces nuclear localization of a p53 NLS mutant (p53KRKKK) and this is dependent on the nuclear localization signal of M and efficient interaction between p53 and the M protein. A. p53−/− HCT116 cells were transfected with the plasmids indicated. Localization of full-length p53 was determined by immunofluorescence. B. Quantification of immunofluorescence staining in A. 300 cells per transfection condition were scored as having primarily nuclear p53, primarily cytoplasmic p53, or both nuclear and cytoplasmic p53. Values are the average of three independent experiments and error bars represent standard error. * p<.001; p53KRKKK is compared to p53wt and all others are compared to p53KRKKK.
We also wanted to determine if the effect of the M protein on p53 localization was dependent on the interaction between the two proteins. As mentioned above, the M348/350A mutant retained the ability to interact with wild-type p53 but was reduced in its ability to interact with p53KRKKK (Figure 3C). Co-transfection of p53KRKKK with M348/350A indicated that M348/350A was less efficient than wild-type M at inducing nuclear accumulation of p53KRKKK, with only about 45% of transfected cells expressing p53KRKKK in the nucleus or both the nucleus and cytoplasm (Figure 5A panels p-r, 5B). These results indicate that the M protein is capable of inducing p53 nuclear accumulation in the absence of stress such as irradiation. This ability is dependent on M having an intact and functional nuclear localization signal and is aided by efficient interaction between the M protein and full-length p53.
3.5 The M protein enhances the stability of p53
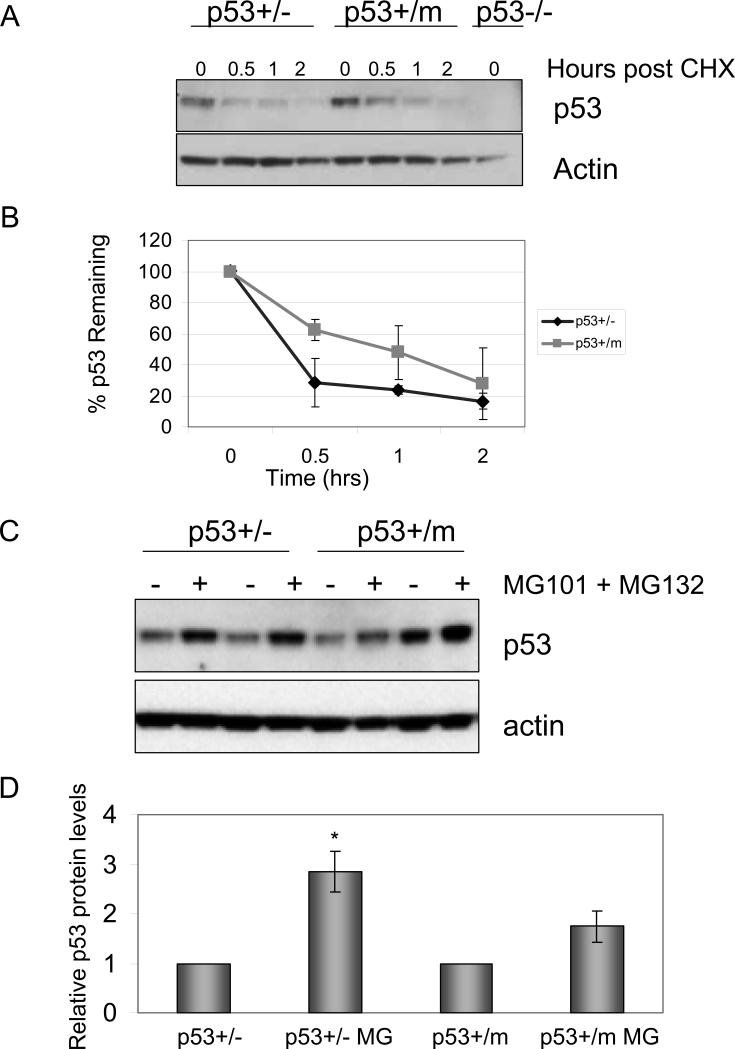
Activation of p53 is also concurrent with an increase in p53 protein stability due to the disruption of the interaction between p53 and its negative regulator, MDM2 (Moll and Petrenko, 2003). In order to examine the stability of p53 in p53+/m MEFs, cycloheximide based translation suppression time course experiments were performed (Figure 6A, 6B). Low passage p53+/− and p53+/m MEFs were treated with cycloheximide for the indicated timepoints to examine p53 protein levels in the absence of de novo protein synthesis. Western blot analysis indicated that p53 protein levels declined more rapidly in the p53+/− MEFs compared to p53+/m MEFs. The Western blots were quantitated by densitometry and graphed as the percent of p53 remaining versus time (Figure 6B). To further confirm the increase in p53 protein stability in p53+/m MEFs, p53 protein levels were examined in the presence and absence of the proteasome inhibitors, MG101 and MG132 (Figure 6C). Low passage p53+/− and p53+/m MEFs were untreated, or treated with proteasome inhibitors for 6 hours prior to collecting cell lysates. p53 protein levels accumulate significantly upon treatment with proteasome inhibitors in p53+/− MEFs. In contrast, p53 protein levels are not significantly increased in response to proteasome inhibitor treatment in p53+/m MEFs, suggesting that p53 is protected from proteasomal degradation in p53+/m MEFs.
Figure 6.
p53 protein levels are more stable in p53+/m MEFs compared to p53+/− MEFs. A. p53 protein levels in the absence of de novo protein synthesis. Low passage MEFs were treated with cycloheximide and cell lysates collected at the indicated time points. B. p53+/m MEFs exhibit increased p53 stability compared to p53+/− MEFs. Western blots were normalized to actin, quantitated by densitometry and graphed as the percent p53 remaining over time relative to the zero time point. The graph represents the average of three independent experiments (and independent MEF lines). Error bars represent standard error of the mean. Statistical analyses (ANOVA) indicated that cycloheximide treated p53+/m cells exhibited significant increases in p53 stability compared to similarly treated p53+/− cells (P = 0.04). C. p53 protein levels in p53+/m and p53+/− MEFs after treatment with proteasome inhibitors. Two different isolates of of low passage p53+/− MEFs (MEF-1: lanes 1,2; MEF-2: lanes 3,4) and p53+/m MEFs (MEF-3: lanes 5,6; MEF-4: lanes 7,8) were untreated or treated with proteasome inhibitors MG101 and MG132 for 6 hours prior to harvest. p53 protein levels accumulate upon proteasome inhibitor treatment in p53+/− MEFs, but are not significantly increased in p53+/m MEFs implying that p53 is protected from proteasomal degradation in p53+/m MEFs. D. p53 protein levels are less sensitive to proteasome inhibitors in p53+/m MEFs. Western blots in C were normalized to actin and quantitated by densitometry. The values shown are the average p53 levels of three independent MEF lines for each genotype and error bars represent standard error. *P < 0.01 compared to p53+/− untreated control. CHX: cycloheximide.
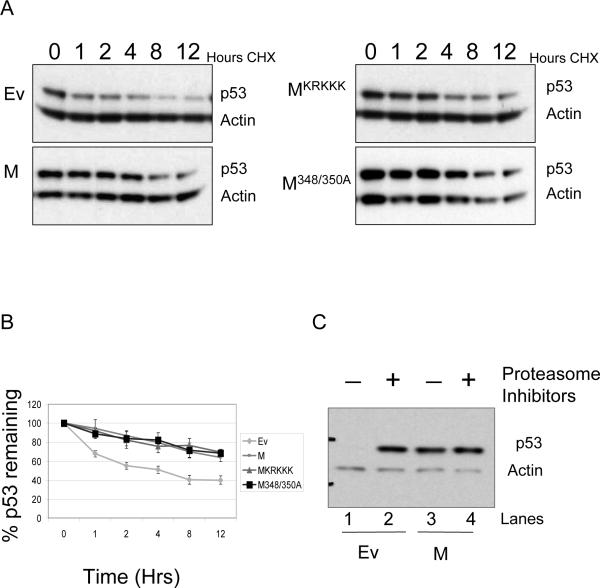
The increased stability of p53 in the p53+/m MEFs compared to p53+/− MEFs was confirmed in similar experiments using U2OS cells (Figure 7). U2OS cells were transfected with empty vector or plasmids encoding M, MKRKKK, or M348/350A and treated with cycloheximide to examine the effect of the M protein on p53 stability in the absence of de novo p53 synthesis. Western blot analysis indicates a more rapid decline in p53 protein levels following addition of cycloheximide in cells transfected with empty vector compared to cells transfected with M (Figure 7A). The MKRKKK and M348/350A mutants stabilized p53 as well as M (Figure 7A). The western blots were quantitated and graphed as the percent p53 remaining versus time (Figure 7B). Increased p53 stability in the presence of M or M mutants was highly significant compared to p53 stability in the absence of M (P < .0001). These results were confirmed by examining p53 protein levels in the presence of proteasome inhibitors (Figure 7C). U2OS cells were transfected with empty vector or M and then treated with proteasome inhibitors for 6 hours. In cells transfected with empty vector, p53 protein levels accumulate upon treatment with proteasome inhibitors (compare lanes 1 and 2). In contrast, p53 protein levels do not change upon treatment of the cells with the proteasome inhibitors in cells transfected with M (compare lanes 3 and 4). It appears that the M protein increases the stability of the full-length p53 protein and hinders p53 proteasomal degradation.
Figure 7.
The M protein enhances the stability of full-length p53. U2OS cells were transfected with plasmids encoding M and M mutants as indicated. A. p53 protein levels in the absence of de novo protein synthesis. Cells were treated with cycloheximide and protein lysates collected at the indicated time points followed by a Western blot for total p53. B. Graphical representation of total p53 levels in A after normalization to actin levels. Blots were scanned on a GE Storm 860 imager and quantitated using ImageQuant software. The graphs indicate the percent p53 remaining relative to the zero time point. Values represent the average of three independent experiments and error bars represent standard error. Statistical analyses (ANOVA) indicated that empty vector transfected U2OS cells exhibited significantly less p53 stability than cells transfected with M or mutant M vectors (P < .0001). C. p53 protein levels after treatment with proteasome inhibitors. Following transfection with empty vector or M, U2OS cells were treated with the proteasome inhibitors MG101 and MG132 for 6 hours. Cells were harvested and p53 protein levels analyzed by Western blot. p53 accumulates in the presence of the proteasome inhibitors in cells transfected with empty vector. However, p53 protein levels are not affected in cells transfected with M, indicating that the M protein protects p53 from proteasomal degradation.
3.6 The M protein does not disrupt the interaction between p53 and MDM2
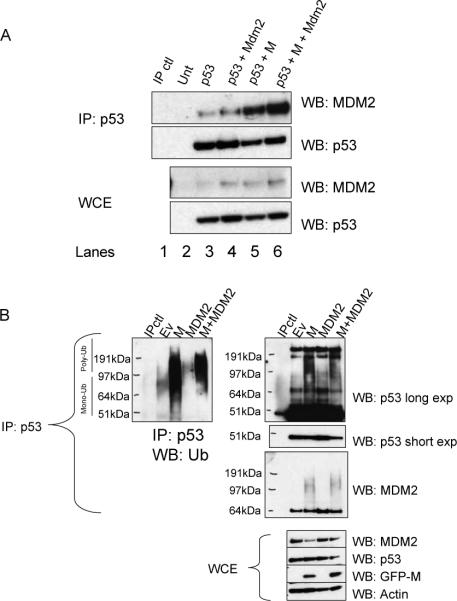
MDM2 acts as one of the key negative regulators of p53. In unstressed cells, MDM2 interacts with p53 and ubiquitinates it, resulting in degradation of p53 (Haupt et al., 1997; Fuchs et al., 1998). To determine whether the M protein interferes with the MDM2-p53 interaction, p53 null Saos-2 cells were transfected with combinations of p53, MDM2, and M and coimmunoprecipitations between p53 and MDM2 were performed (Figure 8A). Surprisingly, the M protein did not disrupt the interaction between p53 and MDM2 (compare lanes 3 to 5 and lanes 4 to 6). In fact, introduction of the M protein seemed to enhance the interaction between p53 and MDM2 (lane 6).
Figure 8.
The M protein does not enhance p53 stability by disrupting MDM2 function. p53 null Saos-2 cells were transfected with combinations of p53, M, and MDM2 as indicated. A. The M protein increases MDM2-p53 interactions. Lysates from transfected Saos-2 cells were immunoprecipitated with a p53 antibody, and Western blots were probed with antibodies to p53 and MDM2. MDM2 is readily detected in complex with p53 in the presence of M. The Western blot of total lysate prior to immunoprecipitation is also shown. B. Ubiquitination of p53. U2OS cells were transfected with the indicated combinations of M and MDM2. Total p53 was immunoprecipitated and Western blots were probed with antibodies to Ubiquitin, p53, and MDM2. IP: Immunoprecipitate, WB: Western blot, WCE: whole cell extract, Ub: ubiquitin.
We next wanted to determine if ubiquitination of p53 was affected by M. U2OS cells were transfected with combinations of p53, MDM2, and M. p53 was immunoprecipitated and ubiquitination examined by Western blot. The M protein increased the levels of ubiquitinated proteins present in the p53 immunoprecipitate compared to control cells (Figure 8B). The overexpression of MDM2 resulted in a mobility shift of ubiquitinated proteins, suggesting that the increase in MDM2 promotes poly-ubiquitination as has been previously described (Li et al., 2003). In cells co-transfected with M and MDM2, the shift towards poly-ubiquitination was again observed, and it again appears that the M protein promotes increased levels of ubiquitinated proteins. The membranes were then re-probed with an antibody specific to the N-terminus of p53 to specifically examine ubiquitinated, full-length p53 (Figure 8B). In cells transfected with M, both mono- and poly-ubiquitinated p53 appeared to be increased in the presence of M. Overexpression of MDM2 leads to a shift towards poly-ubiquitination that is also enhanced when co-transfected with M. The membranes were then re-probed with an antibody to MDM2, again confirming that the M protein does not prevent the interaction between p53 and MDM2 (Figure 8B). The presence of the higher mobility bands may represent ubiquitinated MDM2 as MDM2 is an E3 ubiquitin ligase for itself as well as p53 (Honda et al., 1997). These higher mobility bands were only visible in samples expressing the M protein, suggesting that the M protein may enhance MDM2 self-ubiquitination in addition to p53 ubiquitination. These results indicate that the M protein in some way increases the ubiquitination of p53. Despite this, p53 protein levels are stabilized suggesting that the M protein may promote the disassociation of p53 ubiquitination from the degradation of p53 by the proteasome.
4. Discussion
p53 is well established as a potent tumor suppressor. However, in recent years a number of papers have appeared, particularly those involving prematurely aging mouse models, that implicate p53 in the regulation of some aging phenotypes (Lim et al., 2000; Tyner et al., 2002; Cao et al., 2003; Wong et al., 2003; Maier et al., 2004; Varela et al., 2005; Baker et al., 2006; Wang et al., 2007). In 2002, our laboratory described a genetically engineered mutant p53 mouse model (p53+/m) that exhibited an enhanced resistance to cancer that was accompanied by reduced longevity and premature onset of a number of aging phenotypes (Tyner et al., 2002). The p53+/m mouse expressed an altered p53 transcript that encoded a C-terminal truncated form of p53 (designated “M”) that we hypothesized could interact with wild-type p53 and promote p53 hyperactivity.
The experiments presented in this paper were designed to provide further mechanistic insights into how the M protein might enhance wild-type p53 activity. The MEF experiments in Figure 1 demonstrate that p53+/m MEFs display proliferation rates and S phase entry rates intermediate between that of p53+/− and p53+/+ MEFs. Given that p53+/m MEFs have only a single wild-type p53 allele, the reduced proliferation and augmented checkpoint responses compared to p53+/− MEFs (also with a single wild-type p53 allele) suggests enhanced p53 activity. However, while growA differences might be ascribed to influences other than p53 activity, it seems less likely that the enhanced recombination suppression observed for the p53+/m MEFs relative to p53+/− MEFs is due to non-p53 influences, since this is a relatively specific activity. We and others have shown that an important function of wild-type p53 is to suppress homologous recombination (Sturzbecher et al., 1996; Mekeel et al., 1997; Bertrand et al., 1997; Saintigny et al., 1999; Lu et al., 2003). The C-terminus of p53 binds directly to recombination intermediates in vitro and both homologous and non-homologous end joining processes are inhibited by p53 (Subramanian and Griffith, 2002; Yang et al., 2002).
We have previously shown that p53+/m MEFs are significantly more resistant to transformation than p53+/+ MEFs, consistent with an underlying mechanism of p53 hyperactivation (Tyner et al., 2002). Because this p53+/m MEF transformation resistance phenotype could potentially be a result of factors other than p53, we wanted to determine whether the M protein could suppress transformation or growth by itself, or in the context of wild-type or mutant p53. In our osteosarcoma colony formation assay, we found that M protein expression by itself resulted in a moderate (70%) suppression of osteosarcoma cell growth in the absence of p53. This was a surprising result and suggested that even though the 24 kDa M protein is missing all of its transcriptional activation domain and most of its DNA binding domain; it retains some functional growth suppression, though it is not nearly as efficient as wild-type p53. This wild-type p53 independent growth suppression by M may also partially explain why p53−/m mice have a slightly delayed tumor incidence compared to p53−/− mice (Tyner et al., 2002). In addition, the M protein appeared to enhance the capacity of endogenous wild-type and even mutant p53 to suppress osteosarcoma cell growth. The potential of the M protein to induce growth suppression by interacting with a point mutant version of p53 is consistent with studies showing that C-terminal p53 peptides and protein fragments can convert point mutant p53 molecules into a wild-type growth suppressive conformation (Selivanova et al., 1997; Selivanova et al., 1999). Thus, such M-associated wild-type p53 dependent and independent growth suppressive activities are likely to be responsible for at least some of the cancer resistance in the p53+/m mice.
We also show here that the M protein localizes predominantly to the nucleus (it retains the major p53 NLS) and interacts efficiently with wild-type p53 (Figure 3). As the M protein retains the tetramerization domain, the interaction likely takes place in part through this domain, as point mutations in the M tetramerization domain reduce M-p53 interactions. In binding to p53, the M protein can efficiently induce nuclear localization of a full length NLS-deficient p53 mutant (Figure 5). Moreover, in unstressed cells expressing both M and wild-type p53, more wild-type p53 is in the nucleus compared to similar cells not expressing the M protein (Figure 4). This suggests that the interaction of the M protein with p53 facilitates its nuclear localization or inhibits its nuclear export. The M protein is missing its N-terminal p53 nuclear export signal (NES) and it is possible that the M-p53 interaction could mask the C-terminal NES in both M and p53, thus inhibiting export of full-length p53.
While p53 is unstable in unstressed cells, p53 protein levels are stabilized by the M protein. This is supported by the fact that p53+/m MEFs have greater p53 stability than p53+/− MEFs, and that expression of the M protein in U2OS cells increases p53 stability (Figure 6 and 7). That M inhibits proteasomal degradation of p53 is suggested by data in Figures 6C, 6D and 7C, where proteasome inhibitors are unable to promote further p53 accumulation in cells expressing the M protein. Despite the fact that the M protein increases p53 stability, the mechanism of stabilization remains unclear. Surprisingly, the M protein is associated with increased binding of MDM2 to p53 and subsequent increased p53 ubiquitination (Figure 8). The M protein may be uncoupling p53 ubiquitination from degradation, perhaps by interfering with proteasomal degradation in some way. One explanation for the increased p53 stability may be simply the ability of the M protein to promote p53 nuclear accumulation. The M protein could prevent the shuttling of p53 out to the cytoplasm for degradation. However, as recent evidence has determined that p53 can be degraded in the nucleus, this is unlikely to be the only explanation (Shirangi et al., 2002; von Mikecz, 2006). O'Keefe et al. also observed uncoupling of p53 ubiquitination and degradation by examining a combination of p53 NLS and NES mutants (O'Keefe et al., 2003). The results from this study suggest that p53 must acquire some modifications in addition to ubiquitination prior to proteasomal degradation and that this modification of p53 is likely to occur in the nucleus. The M protein may interfere with or mask this modification, thereby preventing degradation.
At the time the p53+/m model was originally described, the mechanism by which this mutant p53 “m” allele could be causing the observed cancer and aging phenotypes was unclear. Unfortunately, the nature of the serendipitously generated m allele mutation precluded a straightforward mechanistic interpretation. Exons 1−6 of the m allele are deleted, resulting in expression of a chimeric RNA message in which a leader sequence from a gene (Arhgef15) 600 kb upstream of p53 is fused to exons 7−11 of the p53 gene (Tyner et al., 2002; Gentry and Venkatachalam, 2005). This m message produces a truncated C-terminal p53 protein translationally initiated from an ATG codon in p53 exon 7. The m message is expressed in virtually all tissues, though at lower levels than wild-type p53 message. While we hypothesize that most of the cancer resistance and accelerated aging phenotypes can be explained primarily by effects of the M protein on wild-type p53 in the p53+/m mouse, the possibility remains that some or all of the phenotypes could be attributed to haploinsufficiency resulting from the 600 kb deletion upstream of the truncated p53 gene. In fact, there has been some controversy in the literature regarding this issue (Gentry and Venkatachalam, 2005; Vijg and Hasty, 2005). Several lines of evidence support our hypothesis that the cancer resistance and aging phenotypes are driven by chronic p53 hyperactivity in the p53+/m mouse. Initial characterization of the p53+/m mouse revealed that the effect of the m allele is largely dependent on the presence of wild-type p53; p53−/m mice have a median longevity only marginally greater than p53−/− mice (Tyner et al., 2002). Additionally, irradiated p53+/m mice show prolonged stability of wild-type p53 in their tissues in comparison with irradiated p53+/− and p53+/+ mice and expression of the M protein significantly increases wild-type p53 transactivation of the p21 promoter (a key p53 transcriptional target) (Tyner et al., 2002). Further, p53+/m MEFs are more resistant to oncogene transformation than p53+/+ MEFs (Tyner et al., 2002), and p53+/m MEFs show reduced cell cycle progression, enhanced G1 checkpoint control, and enhanced recombination suppression compared to their p53+/− counterparts also containing a single wild-type p53 allele (Figure 1). In this paper we show that the M protein can suppress growth in human osteosarcoma cells, and augments the capacity of both wild-type and mutant forms of p53 to suppress osteosarcoma cell growth (Figure 2). The data presented in this paper also demonstrate that the M protein interacts with wild-type p53 and promotes its nuclear accumulation and increased stability even in the absence of stress. The increased p53 stability in p53+/m MEFs could be recapitulated by transfecting M into U2OS cells expressing normal levels of p53. Additional work from our laboratory indicates that the biological aging phenotypes observed in the p53+/m mouse are accompanied by higher levels of molecular senescence markers, including the p53 target gene, p21 (Gatza et al., in preparation).
The enhanced p53 stability and nuclear localization described here support a model in which p53 may exist in a chronic, low level, hyperactive state in the presence of the M protein. The enhanced cancer resistance of the p53+/m mice is wholly consistent with a mechanism of chronic p53 hyperactivity. Chronic p53 hyperactivation by the M protein may help explain the discrepancies between the p53+/m mouse model and the ‘super p53’ model. ‘Super p53’ mice express one additional copy of the entire p53 locus and also have a reduced tumor incidence but display a normal lifespan and no accelerated aging phenotypes (Garcia-Cao et al., 2002). One key difference between the p53+/m and ‘super’ p53 models is that the ‘super p53’ mice have no increase in basal p53 levels, only a heightened response to stress. On the other hand, we have shown that the M protein induces p53 nuclear accumulation and enhanced stability even in the absence of stress. Further support for p53-dependent aging characteristics comes from the p44 transgenic mouse model by Scrable and colleagues (Maier et al., 2004). The expression of another C-terminal truncated version of p53 (p44) as a transgene in mice recapitulates many of the early aging phenotypes seen in the p53+/m mice (Maier et al., 2004).
As the m allele was generated in a serendipitous gene targeting event, it does not represent a naturally occurring form of p53. However, several p53 isoforms have been identified including those that initiate at codon 40, similar to p44 in the mouse, as well as isoforms that contain a larger N-terminal deletion (Bourdon et al., 2005). The M protein is most similar to the Δ133p53 isoform of p53. This isoform is expressed in most normal tissue and encodes a 35 kDa protein that lacks the N-terminal transactivation domain and a small portion of the DNA binding domain (Bourdon et al., 2005). Similar to the M protein, Δ133p53 localizes predominantly to the nucleus (Bourdon et al., 2005). This isoform has also been identified in zebrafish, and its expression can lead to increased p21 activity suggesting that the M protein may behave similarly to other naturally occurring isoforms of p53 (Chen et al., 2005).
The biological manifestations of chronic low level M-induced p53 stabilization over the mouse lifespan could account for some of the aging and cancer resistance phenotypes in the p53+/m mice. We have observed an increase in the number of senescent cells in several tissues of the p53+/m mice compared to wild-type (Gatza et al., in preparation). Over time, the tissues of the p53+/m mice accumulate senescent cells and may decline in function, particularly if p53-induced senescence occurs in a significant fraction of tissue stem and progenitor cells. We have previously shown that hematopoietic stem cells from old p53+/m mice exhibit reduced proliferation and regeneration capacity compared to their age-matched p53+/+ and p53+/− counterparts (Dumble et al., 2007). Additionally, we observe a decrease in apoptosis in response to ionizing radiation in several tissues of the p53+/m mice (Gatza et al., in preparation). Thus, disruption in normal organ homeostasis is likely to be a large contributing factor to the aging phenotypes that develop in the p53+/m mice. These factors must be considered when targeting the p53 pathway for cancer therapies as aberrant activation of p53 may result in increased tumor suppression at the expense of proper organ homeostasis.
Acknowledgments
We thank David Sassoon for generous provision of plasmid vectors and Catherine Gatza for helpful discussions and provision of mouse embryo fibroblasts. We also thank Sue Hilsenbeck for statistical analyses. This work was supported by National Institute for Aging and Ellison Medical Foundation grants to L.A.D. and the Ruth L. Kirschstein National Research Service Award Viral Oncology Training Grant to L.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alarcon-Vargas D, Ronai Z. p53-Mdm2--the affair that never ends. Carcinogenesis. 2002;23(4):541–547. doi: 10.1093/carcin/23.4.541. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Malureanu L, Perez-Terzic C, Terzic A, van Deursen JMA. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J.Cell Biol. 2006;172(4):529–540. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand P, Rouillard D, Boulet A, Levalois C, Soussi T, Lopez BS. Increase of spontaneous intrachromosomal homologous recombination in mammalian cells expressing a mutant p53 protein. Oncogene. 1997;14(9):1117–1122. doi: 10.1038/sj.onc.1200931. [DOI] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19(18):2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282(5393):1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Cao L, Li W, Kim S, Brodie SG, Deng CX. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003;17(2):201–213. doi: 10.1101/gad.1050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ruan H, Ng SM, Gao C, Soo HM, Wu W, Zhang Z, Wen Z, Lane DP, Peng J. Loss of function of def selectively up-regulates {Delta}113p53 expression to arrest expansion growth of digestive organs in zebrafish. Genes Dev. 2005;19(23):2900–2911. doi: 10.1101/gad.1366405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PL, Chen YM, Bookstein R, Lee WH. Genetic mechanisms of tumor suppression by the human p53 gene. Science. 1990;250(4987):1576–1580. doi: 10.1126/science.2274789. [DOI] [PubMed] [Google Scholar]
- Craig AL, Burch L, Vojtesek B, Mikutowska J, Thompson A, Hupp TR. Novel phosphorylation sites of human tumour suppressor protein p53 at Ser20 and Thr18 that disrupt the binding of mdm2 (mouse double minute 2) protein are modified in human cancers. Biochem. J. 1999;342(1):133–141. [PMC free article] [PubMed] [Google Scholar]
- de Stanchina E, Lowe SW. Tumour suppression: something for nothing? Nat. Cell Biol. 2002;4(12):E275–E276. doi: 10.1038/ncb1202-e275. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, Mcarthur MJ, Montgomery CA, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumors. Nature. 1992;356(6366):215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Dumble M, Gatza C, Tyner S, Venkatachalam S, Donehower LA. Insights into aging obtained from p53 mutant mouse models. Ann.N.Y.Acad. Sci. 2004;1019:171–177. doi: 10.1196/annals.1297.027. [DOI] [PubMed] [Google Scholar]
- Dumble M, Moore L, Chambers SM, Geiger H, Van Zant G, Goodell MA, Donehower LA. The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood. 2007;109(4):1736–1742. doi: 10.1182/blood-2006-03-010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erster S, Mihara M, Kim RH, Petrenko O, Moll UM. In Vivo Mitochondrial p53 Translocation Triggers a Rapid First Wave of Cell Death in Response to DNA Damage That Can Precede p53 Target Gene Activation. Mol. Cell. Biol. 2004;24(15):6728–6741. doi: 10.1128/MCB.24.15.6728-6741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbro M, Henderson BR. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Experimental Cell Research. 2003;282(2):59–69. doi: 10.1016/s0014-4827(02)00019-8. [DOI] [PubMed] [Google Scholar]
- Ferrigno P, Silver PA. Regulated nuclear localization of stress-responsive factors: how the nuclear trafficking of protein kinases and transcription factors contributes to cell survival. Oncogene. 1999;18(45):6129–6134. doi: 10.1038/sj.onc.1203132. [DOI] [PubMed] [Google Scholar]
- Florenes VA, Maelandsmo GM, Forus A, Andreassen A, Myklebost O, Fodstad O. MDM2 Gene Amplification and Transcript Levels in Human Sarcomas: Relationship to TP53 Gene Status. J. Natl. Cancer Inst. 1994;86(17):1297–1302. doi: 10.1093/jnci/86.17.1297. [DOI] [PubMed] [Google Scholar]
- Fuchs SY, Adler V, Pincus MR, Ronai Z. MEKK1/JNK signaling stabilizes and activates p53. PNAS. 1998;95(18):10541–10546. doi: 10.1073/pnas.95.18.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao I, Garcia-Cao M, Martin-Caballero J, Criado LM, Klatt P, Flores JM, Weill JC, Blasco MA, Serrano M. ”Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002;21(22):6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry A, Venkatachalam S. Complicating the role of p53 in aging. Aging Cell. 2005;4(3):157–160. doi: 10.1111/j.1474-9726.2005.00154.x. [DOI] [PubMed] [Google Scholar]
- Giaccia AJ, Kastan MB. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12(19):2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- Grossman SR, Deato ME, Brignone C, Chan HM, Kung AL, Tagami H, Nakatani Y, Livingston DM. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 2003;300(5617):342–344. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- Harvey M, Sands AT, Weiss RS, Hegi ME, Wiseman RW, Pantazis P, Giovanella BC, Tainsky MA, Bradley A, Donehower LA. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene. 1993;8(9):2457–2467. [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Letters. 1997;420(1):25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26(9):1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- Klatt P, Serrano M. Engineering cancer resistance in mice. Carcinogenesis. 2003;24(5):817–826. doi: 10.1093/carcin/bgg057. [DOI] [PubMed] [Google Scholar]
- Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302(5652):1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- Lim DS, Vogel H, Willerford DM, Sands AT, Platt KA, Hasty P. Analysis of ku80-Mutant Mice and Cells with Deficient Levels of p53. Mol.Cell.Biol. 2000;20(11):3772–3780. doi: 10.1128/mcb.20.11.3772-3780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Lozano G, Donehower LA. Activities of wildtype and mutant p53 in suppression of homologous recombination as measured by a retroviral vector system. Mutat.Res. 2003;522(1−2):69–83. doi: 10.1016/s0027-5107(02)00261-0. [DOI] [PubMed] [Google Scholar]
- Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T, Sutherland A, Thorner M, Scrable H. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18(3):306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateu MG, Fersht AR. Nine hydrophobic side chains are key determinants of the thermodynamic stability and oligomerization status of tumour suppressor p53 tetramerization domain. EMBO J. 1998;17(10):2748–2758. doi: 10.1093/emboj/17.10.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekeel KL, Tang W, Kachnic LA, Luo CM, DeFrank JS, Powell SN. Inactivation of p53 results in high rates of homologous recombination. Oncogene. 1997;14(15):1847–1857. doi: 10.1038/sj.onc.1201143. [DOI] [PubMed] [Google Scholar]
- Mendrysa SM, O'Leary KA, McElwee MK, Michalowski J, Eisenman RN, Powell DA, Perry ME. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 2006;20(1):16–21. doi: 10.1101/gad.1378506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, Petrenko O. The MDM2-p53 Interaction. Mol. Cancer Res. 2003;1(14):1001–1008. [PubMed] [Google Scholar]
- O'Keefe K, Li HP, Zhang YP. Nucleocytoplasmic shuttling of p53 is essential for MDM2-mediated cytoplasmic degradation but not ubiquitination. Mol. Cell. Biol. 2003;23(18):6396–6405. doi: 10.1128/MCB.23.18.6396-6405.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeyer AG, Runko E, Winkfield B, Ahn B, Moll UM. Cytoplasmically sequestered wild-type p53 protein in neuroblastoma is relocated to the nucleus by a C-terminal peptide. PNAS. 1996;93(26):15190–15194. doi: 10.1073/pnas.93.26.15190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano JW, Ehrhart JC, Duthu A, Kim CM, Appella E, May P. Identification and characterization of a p53 gene mutation in a human osteosarcoma cell line. Oncogene. 1989;4(12):1483–1488. [PubMed] [Google Scholar]
- Saintigny Y, Rouillard D, Chaput B, Soussi T, Lopez BS. Mutant p53 proteins stimulate spontaneous and radiation-induced intrachromosomal homologous recombination independently of the alteration of the transactivation activity and of the G1 checkpoint. Oncogene. 1999;18(24):3553–3563. doi: 10.1038/sj.onc.1202941. [DOI] [PubMed] [Google Scholar]
- Selivanova G, Iotsova V, Okan I, Fritsche M, Strom M, Groner B, Grafstrom RC, Wiman KG. Restoration of the growth suppression function of mutant p53 by a synthetic peptide derived from the p53 C-terminal domain. Nat Med. 1997;3(6):632–638. doi: 10.1038/nm0697-632. [DOI] [PubMed] [Google Scholar]
- Selivanova G, Ryabchenko L, Jansson E, Iotsova V, Wiman KG. Reactivation of mutant p53 through interaction of a C-terminal peptide with the core domain. Mol. Cell. Biol. 1999;19(5):3395–3402. doi: 10.1128/mcb.19.5.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirangi TR, Zaika A, Moll UM. Nuclear degradation of p53 occurs during down-regulation of the p53 response after DNA damage. FASEB J. 2002;16(3):420–422. doi: 10.1096/fj.01-0617fje. [DOI] [PubMed] [Google Scholar]
- Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18(6):1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturzbecher HW, Donzelmann B, Henning W, Knippschild U, Buchhop S. p53 is linked directly to homologous recombination processes via RAD51/RecA protein interaction. EMBO J. 1996;15(8):1992–2002. [PMC free article] [PubMed] [Google Scholar]
- Subramanian D, Griffith JD. Interactions between p53, hMSH2-hMSH6 and HMG I(Y) on Holliday junctions and bulged bases. Nucleic Acids Res. 2002;30(11):2427–2434. doi: 10.1093/nar/30.11.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu XB, Soron G, Cooper B, Brayton C, Park SH, Thompson T, Karsenty G, Bradley A, Donehower LA. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415(6867):45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- Varela I, Cadinanos J, Pendas AM, Gutierrez-Fernandez A, Folgueras AR, Sanchez LM, Zhou Z, Rodriguez FJ, Stewart CL, Vega JA, Tryggvason K, Freije JM, Lopez-Otin C. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signaling activation. Nature. 2005;437(7058):564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- Vijg J, Hasty P. Aging and p53: getting it straight. A commentary on a recent paper by Gentry and Venkatachalam. Aging Cell. 2005;4(6):331–333. doi: 10.1111/j.1474-9726.2005.00179.x. [DOI] [PubMed] [Google Scholar]
- von Mikecz A. The nuclear ubiquitin-proteasome system. J. Cell. Sci. 2006;119(Pt 10):1977–1984. doi: 10.1242/jcs.03008. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat. Rev. Cancer. 2002;2(8):594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Wang L, Yang L, Debidda M, Witte D, Zheng Y. Cdc42 GTPase-activating protein deficiency promotes genomic instability and premature aging-like phenotypes. PNAS. 2007;104(4):1248–1253. doi: 10.1073/pnas.0609149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, Maser RS, Bachoo RM, Menon J, Carrasco DR, Gu Y, Alt FW, DePinho RA. Telomere dysfunction and Atm deficiency compromises organ homeostasis and accelerates ageing. Nature. 2003;421(6923):643–648. doi: 10.1038/nature01385. [DOI] [PubMed] [Google Scholar]
- Yang Q, Zhang R, Wang XW, Spillare EA, Linke SP, Subramanian D, Griffith JD, Li JL, Hickson ID, Shen JC, Loeb LA, Mazur SJ, Appella E, Brosh RM, Jr., Karmakar P, Bohr VA, Harris CC. The processing of Holliday junctions by BLM and WRN helicases is regulated by p53. J. Biol. Chem. 2002;277(35):31980–31987. doi: 10.1074/jbc.M204111200. [DOI] [PubMed] [Google Scholar]