Abstract
Both the genomes of the epsilonproteobacteria Wolinella succinogenes and Campylobacter jejuni contain operons (sdhABE) that encode for so far uncharacterized enzyme complexes annotated as ‘non-classical’ succinate:quinone reductases (SQRs). However, the role of such an enzyme ostensibly involved in aerobic respiration in an anaerobic organism such as W. succinogenes has hitherto been unknown. We have established the first genetic system for the manipulation and production of a member of the non-classical succinate:quinone oxidoreductase family. Biochemical characterization of the W. succinogenes enzyme reveals that the putative SQR is in fact a novel methylmenaquinol:fumarate reductase (MFR) with no detectable succinate oxidation activity, clearly indicative of its involvement in anaerobic metabolism. We demonstrate that the hydrophilic subunits of the MFR complex are, in contrast to all other previously characterized members of the superfamily, exported into the periplasm via the twin-arginine translocation (tat)-pathway. Furthermore we show that a single amino acid exchange (Ala86→His) in the flavoprotein of that enzyme complex is the only additional requirement for the covalent binding of the otherwise non-covalently bound FAD. Our results provide an explanation for the previously published puzzling observation that the C. jejuni sdhABE operon is upregulated in an oxygen-limited environment as compared with microaerophilic laboratory conditions.
Introduction
A major obstacle to the exploitation of the large volume of genome sequence data is the functional characterization of the gene products (Wild and Saqi, 2004). Annotation is normally inherited from database matches to similar sequences for which the function is known and is hence prone to error propagation. Consequently, the number of functionally characterized gene products has to be increased, especially for those which are not easily accessible, as is the case for proteins that are produced in only small or non-detectable amounts under currently known laboratory growth conditions.
Previous determination of the genome sequence of the anaerobic epsilonproteobacterium Wolinella succinogenes surprisingly revealed that it encodes for protein complexes annotated to be part of an aerobic respiratory chain (Baar et al., 2003). Among these is a protein complex ostensibly involved in aerobic respiration and annotated to be a non-classical succinate:quinone reductase (SQR) which has so far not been detected under currently known growth conditions. A similar enzyme is encoded by the Campylobacter jejuni genome (Parkhill et al., 2000). SQRs are membrane protein complexes that couple the two-electron oxidation of succinate to fumarate to the reduction of quinone to quinol (Lancaster, 2004). Together with the quinol:fumarate reductases (QFRs) of anaerobic fumarate respiration, which catalyse the reaction in the reverse direction in vivo, the SQRs form the superfamily of succinate:quinone oxidoreductases (SQORs) (Hägerhäll, 1997; Lancaster, 2002). They generally contain three or four subunits: a flavoprotein (subunit A), an iron–sulphur protein (subunit B) and one large or two small membrane anchor subunits, referred to as subunit(s) C (and D). Subunit A contains the dicarboxylate oxidation/reduction site; the quinone/quinol binding site is located in the hydrophobic subunit. Among species, the hydrophilic subunits A and B have high sequence identity, while that for the membrane anchor is much lower. Based on their hydrophobic subunit and haem content SQORs can be classified in five types established as A, B, C, D or E in the literature (see Hederstedt, 1999; Lancaster, 2002 for reviews). Both type A and B enzymes contain two haem groups, the latter possesses one large hydrophobic subunit instead of two small ones as is the case for all the other types. Type C enzymes harbour a single haem, D-type and E-type enzymes contain no haem at all. Over the past nine years, three-dimensional structures of representatives of B-type (Lancaster et al., 1999; Madej et al., 2006a), C-type (Yankovskaya et al., 2003; Sun et al., 2005; Huang et al., 2006) and D-type (Iverson et al., 1999; Iverson et al., 2002) members of the superfamily have become available. The hydrophobic subunits of the E-type SQORs are very different from the other four types. They are more similar to those of heterodisulphide reductases from methanogenic archaea. Non-classical (‘E-type’) SQORs are not well characterized; the so far best-characterized enzyme is the SQR from the crenarcheon Acidianus ambivalens (Lemos et al., 2002; Schäfer et al., 2002). To our knowledge no genetic manipulation of an operon encoding for a ‘non-classical’ SQOR has been reported.
The putative SQR from W. succinogenes consists of the two hydrophilic subunits SdhA, SdhB and a hydrophobic subunit SdhE which are encoded by the sdhABE operon (Fig. 1). The subunits SdhA, SdhB and SdhE show the highest sequence identity to the putative SQR from C. jejuni, of 63%, 71% and 69%, respectively, and of 38%, 27% and 32%, respectively, to the A. ambivalens enzyme. The similarity to the cyanobacterial enzyme subunits from Synechocystis is 37% for SdhA, 29% for SdhB1, 25% for SdhB2 and 40% for the putative ‘heterodisulphide reductase subunit b’ which could possibly be the membrane subunit for an SdhABE complex. The SQR from A. ambivalens contains a second hydrophobic subunit SdhF but no similar protein is encoded by the W. succinogenes and C. jejuni genomes.
Fig. 1.
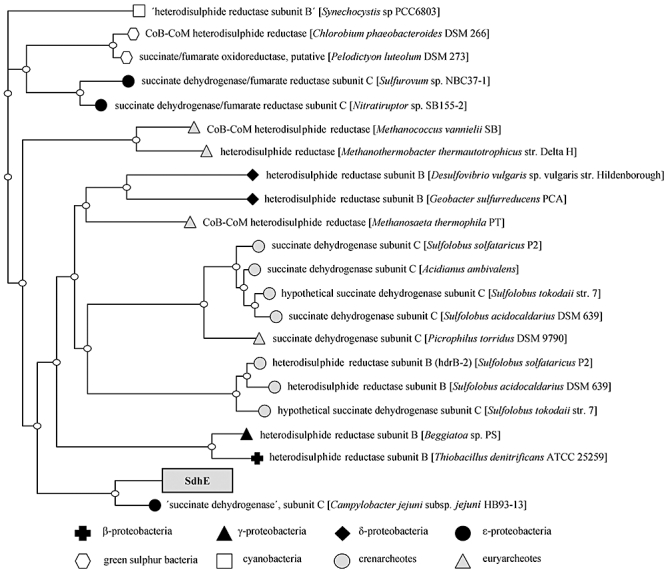
The phylogenetic relationship of the unusual membrane anchor subunit SdhE to selected organisms. A similar enzyme complex is encoded by the Campylobacter jejuni genome. The W. succinogenes sdhE gene (locus tag WS1022; see Fig. 2A) is annotated as sdhC; it was renamed to sdhE in analogy to the Acidianus ambivalens homologue (Lemos et al., 2002). The gene product was named accordingly SdhE.
In contrast to all other SQORs characterized in this respect (see Singer and McIntire, 1984; Lancaster, 2004 for reviews), the flavoproteins of the SdhABE complexes from W. succinogenes and C. jejuni are predicted to harbour non-covalently bound FAD found so far only in soluble fumarate reductases of Shewanella species (Bamford et al., 1999; Leys et al., 1999; Taylor et al., 1999). The respective histidine residue required for the covalent linkage is not conserved in these proteins. Experiments on Saccharomyces cerevisiae SQR mutants demonstrated that a covalently bound FAD is a requirement for succinate oxidation activity (Robinson et al., 1994), a first indication to doubt the assigned succinate dehydrogenase function of the W. succinogenes SdhABE complex.
The SdhA of both W. succinogenes and C. jejuni contains an approximately 40-amino-acid residue extension compared with all other SQORs and harbours a salient twin arginine motif – a signal for the protein export via the tat-pathway (Palmer et al., 2005). All other SQORs are oriented towards the cytoplasm (Lancaster, 2002). The iron–sulphur cluster composition in SdhB is characteristic for E-type SQORs; the cysteine pattern implies the presence of a second [4Fe−4S] cluster instead of a [3Fe−4S] cluster, as first discussed for A. ambivalens SQR (Gomes et al., 1999). The unusual membrane anchor SdhE contains a striking cysteine-rich region referred to as 10-cysteine motif and is phylogenetically related to succinate dehydrogenases and heterodisulphide reductases from a variety of different organisms (Fig. 1) among them not only proteobacteria and archaea but also cyanobacteria and green sulphur bacteria. Furthermore the hydrophobic subunit is predicted to contain no transmembrane domains; membrane association is most likely mediated via amphipathic helices.
Here we present, in the framework of a functional genomics project, the first genetic system for the manipulation and production of a non-classical SQOR. Biochemical characterization of this enzyme reveals that the putative succinate dehydrogenase is in fact a novel 8-methylmenaquinol:fumarate reductase (MFR) with no detectable succinate dehydrogenase activity. Studies on variant enzymes show that the hydrophilic subunits of this complex are, in contrast to all other members of the superfamily, exported into the periplasm via the tat-pathway.
Results and discussion
Genetic system, homologous production and purification of the SdhABE complex
Transformation of the W. succinogenes QFR deletion mutant ΔfrdCAB with the vector pSdhA carrying the sdhA gene and the frd promoter results in the new mutant fMFR (Fig. 2). Integration of the vector at the sdh locus puts the sdhABE operon under control of the strong frd promoter (Fig. 2). The mutant fMFR does not support growth with fumarate as sole electron acceptor; fumarate was complemented with nitrate. In contrast to the deletion mutant, the cell homogenate of the nitrate-grown fMFR strain displays significant fumarate reduction activity (Table 1). Therefore the sdhABE operon encodes for a real and active protein which is produced homologously in the W. succinogenes mutant fMFR. After membrane preparation from mutant cells, a large proportion of the total activity is found in the soluble fraction, an indication that the SdhABE complex is membrane associated rather than tightly membrane bound. Compared with the A. ambivalens enzyme which is clearly a membrane bound protein (Gomes et al., 1999), a much larger part of the activity is found in the soluble fraction (Table 1). It can be speculated that the reason for the stronger membrane attachment of the A. ambivalens enzyme may be due to the presence of a second hydrophobic subunit (F) (Lemos et al., 2001) which is not encoded by the W. succinogenes and C. jejuni genome. In this work a purification procedure could be established to enrich the protein 24-fold via a combination of anion exchange and gel filtration chromatography with a yield of 36% of the initial activity in the periplasm extract (Table 2). The presence of all three subunits in the preparation could be identified by peptide mass fingerprint analysis (see Fig. S1). Subunits B and C cannot be stained well, as observed previously for the W. succinogenes QFR subunits B and C.
Fig. 2.
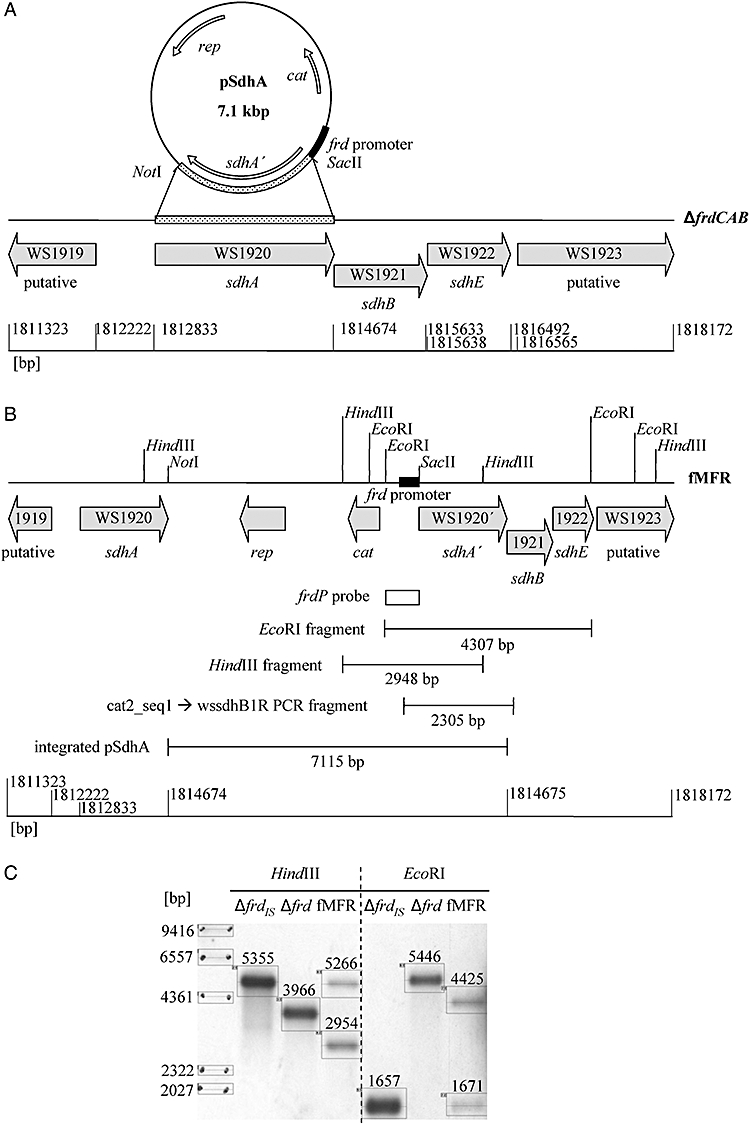
Genetic system for the production and manipulation of the SdhABE complex. A. In the vector pSdhA the structural gene sdhA is cloned next to the strong frd promoter (black box). The vector integrates via the homologous sdhA gene (dotted boxes) at the native locus in the genome of the QFR-deficient deletion strain ΔfrdCAB. B. In the resulting mutant fMFR the sdhABE operon is under control of the strong frd promoter. This system allows introduction of mutations and affinity tags in sdhA. Open reading frames are indicated by grey arrows including locus tags. The ruler gives base pairs with respect to the Wolinella succinogenes DSM 1740 complete genome sequence (NC_005090). Correct integration was verified via PCR with the primer pair Cat2_seq1 and wssdhB1R and sequencing of the resulting DNA fragment and Southern blot. C. Southern blot of cut genomic DNA from mutants fMFR and ΔfrdCAB. The frdP probe (clear box) hybridizes with the frd promoter and detects the 4307 bp EcoRI and the 2948 bp HindIII fragment, respectively, in the mutant fMFR. The probe also detects the frd promoter at its native locus (second bands in lanes fMFR). An insertion of a 1.3 kb insertion sequence is known to occur in the genome of the deletion mutant upstream of the kan gene that replaces the frdCAB genes [IS1302, see Simon and Kröger (1998) for details]. Consequently both variants with and without insertion sequence were used as positive control (lanes ΔfrdIS and Δfrd respectively). IS1302 also introduces an additional EcoRI restriction site leading to a smaller EcoRI fragment in ΔfrdIS than in Δfrd. Fragment sizes were calculated from a calibration curve with the five marker bands using the program un-scan-it™ (Silk Scientific).
Table 1.
Specific enzymatic activities of the mutant fMFR cell homogenate, soluble fraction and membranes as determined by monitoring the reduction of fumarate by benzylviologen radicals.
| Specific activities (U mg−1) | |
|---|---|
| Wild-type cell homogenate (QFR) | 1.45 |
| ΔfrdCAB cell homogenate | < 0.01 |
| Mutant fMFR cell homogenate | 0.48 |
| Mutant fMFR soluble fraction | 0.20 |
| Mutant fMFR membranes | 0.14 |
As positive and negative control cell homogenate activities of the wild type and the ΔfrdCAB mutant are shown respectively. One unit (U) of activity is defined as the consumption of 1 μmol fumarate per minute at 37°C.
Table 2.
Enrichment of the SdhABE complexes MFR, MFR–A86H, MFR–HT and MFR–AH1 and cleavage of the amino-terminal signal peptide.
| Specific activities (U mg−1) |
Enrichment fold |
Yield % |
Specific activities (U mg−1) |
|||||
|---|---|---|---|---|---|---|---|---|
| Purification step | MFR | A86H | MFR | A86H | MFR | A86H | HT | AH1 |
| Periplasm extract | 0.7 | 0.1 | 1 | 1 | 100 | 100 | 0.7 | 0.1 |
| Anion exchange | 6.5 | 0.2 | 9 | 3 | 81 | 83 | 6.5 | 0.8 |
| Gel filtration | 16.5 | 1.1 | 24 | 18 | 36 | 6 | ||
| 6xhis-tag positions | No tag | 1 | 37 | |||||
| Western blot signal | (−) | (+) | ||||||
Only the variant SdhA subunit with a 6xhis-tag on amino acid position 37 (AH1) could be detected on a Western blot treated with an anti-pentahistidine antibody.
To our knowledge, no genetic system for the manipulation and production of a non-classical SQR has been established so far. The system established here allows the study of the putative succinate degydrogenase in the non-pathogenic host W. succinogenes with relevance also to the analogous enzyme of the human pathogen C. jejuni. The genetic system established in this work allows the introduction of mutations and affinity tags in sdhA. In an experiment analogous to the heterologous production of the QFR from Helicobacter pylori and C. jejuni in W. succinogenes (Mileni et al., 2006), the initial attempt to clone the whole sdhABE operon in the vector pFrdcat2 failed. The frd promoter from W. succinogenes on the vector pFrdcat2 is also recognized by the Escherichia coli strain used for vector amplification (Körtner, 1989), so it can be speculated that the SdhABE complex is produced and toxic in E. coli.
Catalytic properties of the SdhABE complex
The catalytic properties of the purified enzyme were examined using various possible electron donor and acceptor substrates (Table 3). Strikingly, for the SdhABE complex annotated as a ‘succinate dehydrogenase’ no succinate oxidation activity could be detected. This was independent of whether methylene blue, ferricenium or DCPIP was used as the electron acceptor. One might argue that this could be due to the lack of a suitable binding site for these artificial compounds. However, no succinate oxidation activity could be found using the high-potential quinone EQ-0 (Fig. 3B, Madej et al., 2006b) as electron acceptor, although the complex catalyses quinol oxidation and hence harbours a quinone binding site (Table 3). The reason for the lack of succinate oxidation activity could be explained by the absence of a covalently bound FAD which seems to be a prerequisite for succinate oxidation activity (Mewies et al., 1998). In variants of E. coli QFR (Blaut et al., 1989) and S. cerevisiae SQR (Robinson et al., 1994), the histidine residues which establish the covalent bond to the FAD were replaced by other residues. The variant enzymes were assembled correctly but contained a non-covalently bound FAD and lost the ability to oxidize succinate.
Table 3.
Catalytic properties of the enriched SdhABE (MFR) complex.
| Specific activities (U mg−1) |
||||||
|---|---|---|---|---|---|---|
| Electron donor | E (mV) | Electron acceptor | E (mV) | ΔE (mV) | MFR | QFR |
| Reduced benzylviologen | −374a | Fumarate | 25b | −399 | 16.5 | 59.0 |
| Succinate | 25b | Methylene blue | 11c | 14 | < 0.01 | 28.8 |
| Succinate | 25b | Ferricenium | 380d | −355 | < 0.01 | 14.6 |
| Succinate | 25b | DCPIP | 217e | −192 | < 0.01 | |
| Succinate | 25b | DMN | −35f | 60 | < 0.01 | |
| Succinate | 25b | EQ-0 | 56g | −31 | < 0.01 | |
| DMNH2 | −35f | Fumarate | 25b | −60 | 0.1 | 7.4 |
| TMNH2 | −124h | Fumarate | 25b | −149 | 0.4 | 1.0 |
| 5-MMKH2-6 analogue | −124h | Fumarate | 25b | −149 | 0.2 | |
| 8-MMKH2-6 analogue | −124h | Fumarate | 25b | −149 | 0.9 | |
| CoB-SH/CoM-SH | −143i | Fumarate | 25b | −168 | <0.01 | |
in situ potential in QFR, Madej et al. (2006a).
This work.
Selected corresponding values of purified QFR (Lancaster et al 2000; 2001; this work) are included for comparison.
Fig. 3.
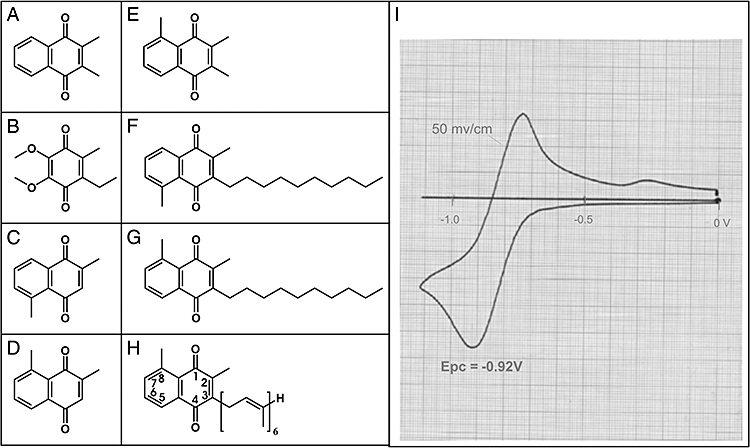
Chemical structures of quinone compounds. See also Chemical synthesis of quinones. A. 2,3-Dimethyl-1,4-naphthoquinone (DMN). B. 2,3-Dimethoxy-5-ethyl-6-methyl-1,4-benzoquinone (EQ-0). C. 2,5-Dimethyl-1,4-naphthoquinone. D. 2,8-Dimethyl-1,4-naphthoquinone. E. 2,3,5-Trimethyl-1,4-naphthoquinone (TMN). F. The 5-MMK-6 analogue: 3-decyl-2,5-dimethyl-1,4-naphthoquinone. G. The 8-MMK-6 analogue: 2-decyl-3,5-dimethyl-1,4-naphthoquinone. H. 8-Methylmenaquinone-6 (8-MMK-6). I. Cyclic voltammogram of the quinone compound 3g (8-MMK-6 analogue); c = 1 mM; x = 100 mV cm−1; i = 1 mA; v = 50 mV s−1.
In view of the phylogenetic relationship to heterodisulphide reductases which catalyse the reversible reduction of heterodisulphide (CoM-S-S-CoB) to CoM-SH and CoB-SH (Hedderich et al., 2005), the SdhABE complex was tested for CoM-SH/CoB-SH oxidation activity. However, no such activity could be detected (Table 3).
Although the complex catalyses fumarate reduction with the menaquinol-6 (MKH2-6) analogue 2,3-dimethyl-1,4-naphthoquinol [DMNH2; the quinol form of the quinone shown in Fig. 3A (Lancaster et al., 2005)] the activities are very low. In addition to menaquinol-6, membranes of C. jejuni (Carlone and Anet, 1983; Collins et al., 1984) and of W. succinogenes (Collins and Fernandez, 1984) contain a second quinol, 8-methylmenaquinol-6 (8-MMKH2-6) (Thummer et al., 2007). So far only MKH2-6 has been associated with fumarate respiration. In experiments with W. succinogenes respiratory enzymes reconstituted in proteoliposomes, it could be shown that fumarate respiration with formate or hydrogen was catalysed only at high activities when MK-6 was incorporated in the liposomes (Biel et al., 2002). However the activities of polysulphide respiration with formate or hydrogen of proteoliposomes containing 8-MMK-6 were more than an order of magnitude greater than of those prepared with MK-6 (Dietrich and Klimmek, 2002). Hence formate dehydrogenase and hydrogenase reduce both MK-6 and 8-MMK-6 whereas the respective terminal reductase preferentially oxidizes one of the two quinols.
The novel enzyme characterized in this work couples the oxidation of both the MKH2-6 analogue DMNH2 and an 8-MMKH2-6 analogue (the quinol form of the quinone shown in Fig. 3G, see also Fig. S2) with the reduction of fumarate (Table 3). However, activities are about one order of magnitude higher when the 8-MMKH2-6 analogue is used as electron donor, which correlates also with the approximately 50 mV lower redox mid-point potential of the 8-MMKH2-6 analogue (see Chemical synthesis of quinones below, and Fig. 3). However, the fumarate reductase activity of the SdhABE complex is only doubled if the methyl group is located at position 5 (5-MMKH2-6 analogue) and only four times higher when the symmetrical quinol TMNH2 (the quinol forms of the quinones shown in Fig. 3F and E respectively) is used as substrate. In analogy to previous investigations (H.R. Nasiri et al., submitted), we can safely assume that the 5-MMK-6 analogue possesses essentially the same oxidation-reduction mid-point potential as the 8-MMK-6 analogue. Consequently, the increase of activity using the 8-methylmenaquinol-6 analogue as electron donor cannot solely be attributed to its lower mid-point potential compared with that of DMNH2. Comparison with the results obtained with purified QFR from W. succinogenes (right column of Table 3) also indicated that the observed activities are not simply proportional to the driving force of the respective reaction. Thus, these results indicate that the novel enzyme displays a specificity for 8-methylmenaquinol-6 and therefore should more appropriately be referred to as an 8-methylmenaquinol:fumarate reductase (MFR) in future work.
Orientation of the SdhABE (MFR) complex: the mutants fMFR–R7/8Q, fMFR–HT and fMFR–AH1
In a strategy analogous to that performed previously to show that W. succinogenes hydrogenase is exported into the periplasm via the tat-pathway (Großet al., 1999), the genetic system presented in this work was used to create the mutant fMFR–R7/8Q to examine the orientation of the complex and the recognition of the putative amino-terminal tat-signal sequence (Fig. S3). The replacement of arginines 7 and 8 with glutamines in SdhA disrupts the putative signal peptide. The native QFR is oriented towards the cytoplasm and is used as a control in the experiment. The periplasm of wild-type, mutant fMFR and mutant fMFR–R7/8Q cells was extracted and the spheroblasts were subsequently fractionated in cytoplasm and membranes. The respective fractions were tested for fumarate reductase activity with benzylviologen radicals as electron donor. About 60% of the activity of the unmodified MFR complex was found in the periplasm in contrast to about 4% for the variant fMFR–R7/8Q and the wild-type control (Table 4). Therefore, the results demonstrate the periplasmic orientation of the unmodified MFR complex.
Table 4.
Comparison of enzyme activities in the cytoplasm, periplasm and membranes of unmodified SdhABE (MFR) complex with the MFR–R7/8Q variant.
| % of total activity |
|||
|---|---|---|---|
| Strain | Periplasm | Cytoplasm | Membranes |
| Wild type (QFR) | 4.4 | 0.7 | 94.9 |
| Mutant fMFR | 59.5 | 0.2 | 40.3 |
| Mutant fMFR–R7/8Q | 3.8 | 12.5 | 83.8 |
Activities were determined by monitoring the oxidation of reduced benzylviologen by fumarate. Wild-type QFR is oriented towards the cytoplasm and is used for comparison.
According to previous studies (Berks et al., 2005), the tat signal peptide is cleaved during export at a peptidase recognition sequence corresponding to amino acids 31–33 in SdhA (Fig. 4). The genetic system established in this work allowed us to create variant SdhABE (MFR) complexes containing a 6xhis-tag at amino acid positions 1 and 37 in the mutants fMFR–HT and fMFR–AH1 respectively. The active variant proteins were enriched by anion exchange chromatography and analysed on a Western blot treated with anti-polyhistidine antibodies (Fig. S4). Only the SdhA variant with the his-tag on amino acid position 37 is detectable (Table 2); the amino-terminal signal peptide is most likely cleaved during export.
Fig. 4.
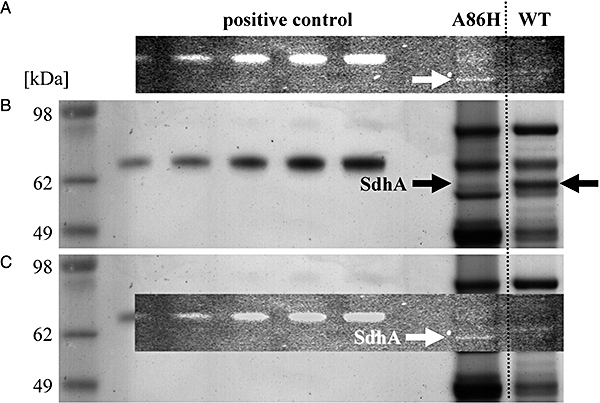
SDS-PAGE of the variant enzyme A86H and the unmodified protein (WT). A. The unstained gel was exposed to UV light; in contrast to the unmodified enzyme a fluorescent band can be detected in the variant A86H. B. The coomassie-stained gel shows the SdhA band for both the variant and the unmodified enzyme. C. Superimposition of the pictures identifies the fluorescent band as SdhA. In the variant enzyme A86H the FAD is covalently bound to the protein.
Attachment of the FAD: the mutant fMFR–A86H
The amino acid sequence alignment of SdhA with other SQORs (Fig. S5) indicates a non-covalent attachment of the FAD to the protein, the respective histidine is not conserved in the MFR and corresponds to alanine 86. To elucidate the requirements for the covalent attachment of the FAD to a protein the mutant fMFR–A86H was created which encodes for a histidine on position 86 instead of an alanine in SdhA. The active variant enzyme could be enriched (Table 2) and analysed via SDS-PAGE (Fig. 4). In contrast to the unmodified enzyme a fluorescent band corresponding to SdhA could be detected in the variant. The exchange of alanine 86 to histidine is the single additional requirement for the covalent attachment of the FAD to the protein.
In the W. succinogenes QFR the FAD is covalently bound to histidine 43 of subunit A (Kenney and Kröger, 1977; Lancaster et al., 1999). Amino acids close to the FAD binding site are highly conserved (Fig. S5), mutations in this area in the SQR from Bacillus subtilis resulted in the loss of the flavin and enzyme activity (Maguire et al., 1986). Both in the SQR from S. cerevisiae (Robinson and Lemire, 1996) and in the E. coli QFR (Blaut et al., 1989; Ackrell et al., 1992) the exchange of the FAD-binding histidine resulted in a non-covalently bound FAD, and the enzyme complex remained intact and active. Interestingly these complexes lost their ability to oxidize succinate; the covalent FAD is most likely a prerequisite for succinate oxidation. The observation that the MFR is also unable to catalyse succinate oxidation in detectable amounts supports this hypothesis. Interestingly the variant complex MFR–A86H is still not able to catalyse succinate oxidation with either methylene blue, ferricenium, DCPIP, DMN or EQ-0 as electron acceptors so the covalent linkage alone is not sufficient to support succinate oxidation activity.
Apart from the SdhABE complexes from C. jejuni and W. succinogenes the FAD-binding histidine is conserved among all SQORs but not in the periplasmic fumarate reductase of Shewanella species (Pealing et al., 1992). However these enzymes harbour neither iron–sulphur clusters nor a 10-cystein motif but haem groups which distinguishes them from the MFR (Bamford et al., 1999; Leys et al., 1999; Taylor et al., 1999).
Physiological role
Although no growth conditions have been identified so far under which the W. succinogenes MFR complex is produced, it is known that the expression of the sdhABE genes encoding for the highly similar putative ‘succinate dehydrogenase’ from the human pathogen C. jejuni are regulated in response to growth conditions. The genes are upregulated in vivo in the chicken caecum relative to the expression of laboratory-grown bacteria (Woodall et al., 2005). It can be reasoned that in the caecum, C. jejuni grows under oxygen-limited conditions in contrast to the microaerophilic environment in the laboratory-grown strain. In contrast to findings for E. coli, where succinate dehydrogenase is upregulated under aerobic conditions and fumarate reductase in anaerobiosis (Park et al., 1995), both the sdhABE genes and the fumarate reductase genes are upregulated in C. jejuni in an oxygen-limited environment (Woodall et al., 2005). Our results reconcile this initially puzzling observation, if we conclude that the C. jejuni putative succinate dehydrogenase is in fact a methylmenaquinol:fumarate reductase with properties similar to those of the W. succinogenes MFR.
The novel methylmenaquinol:fumarate reductase could be part of an alternative metabolic pathway using the low-potential methylmenaquinol-6 reduced by hydrogenase or formate dehydrogenase. Due to the periplasmic orientation the enzyme could use fumarate directly without the need of importing it into the cytoplasm via the dcu dicarboxylate transporters (Janausch et al., 2002).
An unsolved question is why the Wolinella fMFR mutant is unable to grow by fumarate respiration, although the overproduced MFR enzyme has substantial 8-methylmenaquinol:fumarate reductase activity. Further work is required to examine the role of this enzyme at a physiological level – its properties can provide hints for the screening of new growth conditions in the laboratory. Quantitative RT-PCR of W. succinogenes grown under different conditions could clarify some points discussed here. In conclusion, this work is an important step towards the mechanistic understanding of this type of SQORs which are so far not well characterized.
Experimental procedures
Bacterial growth
Wolinella succinogenes was grown with formate as electron donor and fumarate or nitrate as electron acceptor (Bronder et al., 1982; Lorenzen et al., 1993). The medium was supplemented with Brain-Heart-Infusion (0.5% w/v; Gibco BRL)
Plasmid construction
Due to its central significance for the W. succinogenes expression system, the sequence of the plasmid pFrdcat2 (Simon et al., 1998) was determined in this work and deposited in the EMBL nucleotide sequence database with the Accession No. AM909725.
pSdhA
DNA fragments corresponding to the region, which encodes for the SdhA subunit (1850 bp) and the pFrdcat2 vector fragment without frdCAB genes (5283 bp), were synthesized by PCR using primers carrying suitable restriction sites for cloning at their 5′ ends. Genomic DNA from W. succinogenes with primer pair Sac_Ws-Sdh_Fw and Not_Ws-SdhA_Rv and plasmid pFrdcat2 with primer pair Not_cat2_Fw and Sac_cat2_Rv were used respectively (Table S1). The sdhA gene was cloned in the plasmid fragment using SacII and NotI restriction (Fig. 2A). The entire sdhA gene of the resulting plasmid was sequenced.
pSdhAHT
The oligonucleotides SacIIHis6TEV1 and SacIIHis6TEV2 (Table S1) were hybridized by heating them for 10 min at 95°C and slowly cooled to 4°C at a rate of 0.1°C min−1. The double-stranded oligonucleotide was inserted at the start of the sdhA gene in pSdhA using SacII restriction. The orientation of the oligonucleotide was verified by sequencing.
pSdhAH1
To insert a 6xhis-tag at amino acid position 37 in SdhA a PCR fragment was synthesized using suitable primers carrying an ApaI restriction site and one of them the bases encoding for the 6 histidines as overhang on their 5′ ends. The vector pSdhA was used as template with the primer pair SdhH1_Fw and SdhH1_Rv (Table S1). The resulting linear PCR fragment was circularized using ApaI restriction.
Site-directed mutagenesis
Site-directed mutagenesis was performed using the QuikChange® site-directed mutagenesis kit from Stratagene and a modified protocol as described in Wang and Malcolm (1999). The vectors pR7/8Q and pA86H were synthesized with the plasmid pSdhA as template and specifically synthesized oligonucleotides carrying the desired nucleotide mismatches (Table S1). The sequence of the respective modified region of the resulting plasmid was determined.
Transformation of W. succinogenes
The transformation of the fumarate reductase activity-deficient deletion mutant ΔfrdCAB with the constructed plasmids was performed as described in Simon et al. (1998). Integration of the plasmid occurs at the native sdh locus in the W. succinogenes genome and was verified via PCR, Southern blot and sequencing (Fig. 2 and Fig. S4).
Computer analysis
Database searches and phylogenetical tree representation of resulting data made use of the program blast (Altschul et al., 1997). Plasmid/primer design and multiple sequence alignments were performed with Clone Manager 9 (SciEd).
Chemical synthesis of quinones
General remarks
NMR spectra were recorded on a Bruker spectrometer AM250 operating at a 1H frequency of 250 MHz and 13C frequency of 62.9 MHz. Mass spectra (MALDI) were recorded on a Fisons VG TofSpec spectrometer. All reactions were monitored by thin-layer chromatography (TLC), performed on silica gel POLYgram® (Macherey-Nagel). Chromatographic purifications were done with Merck silica gel 60.
Chemistry
2,3-Dimethyl-1,4-naphthoquinone (DMN; Fig. 3A; Lancaster et al., 2005) and 2,3-dimethoxy-5-ethyl-6-methyl-1,4-benzoquinone (EQ-0; Fig. 3B; Madej et al., 2006b) were synthesized previously. 2,5-Dimethyl-1,4-naphthoquinone (Fig. 3C), 2,8-dimethyl-1,4-naphthoquinone (Fig. 3D) and 2,3,5-trimethyl-1,4-naphthoquinone (TMN; Fig. 3E) were prepared starting from commercial naphthalene 1,6-dimethylnaphthalene, 1,7-dimethylnaphthalene and 1,6,7-trimethylnaphthalene via chromium trioxide oxidation (Schmid and Labahn, 1998). The 5-MMK-6 analogue 3-decyl-2,5-dimethyl-1,4-naphthoquinone (Fig. 3F) and the 8-MMK-6 analogue 2-decyl-3,5-dimethyl-1,4-naphthoquinone (Fig. 3G) were synthesized starting from 2,5-dimethyl-1,4-naphthoquinone (Fig. 3C) and 2,8-dimethyl-1,4-naphthoquinone (Fig. 3D) respectively. The formal introduction of the alkyl side-chain was achieved by radical alkylation via a Hunsdiecker oxidative decarboxylation of undecanoic acid, promoted by silver nitrate as a catalyst in the presence of ammonium peroxydisulphate in a acetonitrile/water mixture as previously described (Madej et al., 2006a). The purification of the crude products by silica chromatography (hexane : ethylacetate 6:1) yielded 8-MMK-6 analogue (3g) and 5-MMK-6 analogue (3c) at 44.2% and 48% yields respectively.
Electrochemistry
Cyclovoltammetry experiment (Fig. 3I) was measured using a conventional three-electrode cell in dimethoxyethane with tetrabutylammoniumperchlorate as supporting electrolyte on platinum working and counter electrodes. The concentration of 2-decyl-3,5-dimethyl-1,4-naphthoquinone (Fig. 3G) was 1 mM. The potential is given versus Ag/AgCl reference electrode, with a voltage sweep rate of 50 mV s−1. 1,2-Dimethoxyethane (DME) was purchased from Fluka, puriss; dried over molecular sieves (H2O < 0.005%). Solution of 2-decyl-3,5-dimethyl-1,4-naphthoquinone was deoxygenated with dry nitrogen and maintained under an inert nitrogen atmosphere at 25°C (H.R. Nasiri et al., submitted).
2,5-Dimethyl-1,4-naphthoquinone (Fig. 3C)
1H-NMR (250.13 MHz, CDCl3) as previously reported (Schmid and Labahn, 1998); 13C-NMR (62.9 MHz, CDCl3): δ (p.p.m.) = 187.0; 185.9 (C=O), 146.2; 140.9; 133.6; 129.7 (C), 137.8; 137.4; 132.6; 125.4 (CH), 22.5; 15.9 (CH3).
2,8-Dimethyl-1,4-naphthoquinone (Fig. 3D)
1H-NMR (250.13 MHz, CDCl3) as previously reported (Schmid and Labahn, 1998); 13C-NMR (62.9 MHz, CDCl3): δ (p.p.m.) = 187.4; 185.3 (C=O), 149.4; 141.2; 133.7; 129.7 (C), 137.8; 134.2; 132.7; 124.9 (CH), 22.8; 16.8 (CH3).
2,3,5-Trimethyl-1,4-naphthoquinone (Fig. 3E)
1H-NMR (250.13 MHz, CDCl3) as previously reported (Schmid and Labahn, 1998); 13C-NMR (62.9 MHz, CDCl3): δ (p.p.m.) = 186.9; 185.2 (C=O), 144.1; 141.7; 140.7; 133.6; 129.9 (C), 137.2; 132.3; 125.1 (CH), 22.7; 13.0; 12.5 (CH3).
The 5-MMK-6 analogue 3-decyl-2,5-dimethyl-1,4-naphthoquinone (Fig. 3F)
1H-NMR (250.13 MHz, CDCl3): δ (p.p.m.) = 8.00 (m, 1H, aromatic proton), 7.60 (m, 2H, aromatic protons), 2.98 (s, 3H, CH3), 2.70 (m, 2H, CH2CH2), 2.1 (s, 3H, CH3) 1.1–1.7 (m, 16H, 8xCH2), 0.89 (s, 3H, CH3). 13C-NMR (62.9 MHz, CDCl3): δ (p.p.m.) = 186.7; 185.8 (C=O), 148.8; 141.3; 140.7; 133.6; 129.9 (C), 137.3, 132.3, 125.0 (CH), 31.9; 30.1; 29.5; 29.4; 29.3; 28.9; 27.3; 22.9; 22.8 (CH2), 22.6; 14.1; 12.4 (CH3).
The 8-MMK-6 analogue 2-decyl-3,5-dimethyl-1,4-naphthoquinone (Fig. 3G)
1H-NMR (250.13 MHz, CDCl3): δ (p.p.m.) = 7.90 (d, J = 7.5 Hz, 1H, aromatic proton), 7.45 (m, 2H, aromatic protons), 2.67 (s, 3H, CH3), 2.39 (t, J = 7.3 Hz 2H, CH2CH2), 2.14 (s, 3H, CH3) 1.1–1.6 (m, 16H, 8xCH2), 0.83 (s, 3H, CH3). 13C-NMR (62.9 MHz, CDCl3): δ (p.p.m.) = 187.4; 185.1 (C=O), 145.8; 144.8; 140.6; 133.6; 125.1 (C), 137.2, 133.8, 125.1 (CH), 31.8; 29.9; 29.5; 29.5; 29.4; 29.2; 28.6; 26.8; 22.8 (CH2), 22.6; 14.1; 12.8 (CH3). MS (MALDI): 326.48 (M).
E0 = −920 mV, 50 mV lower than that for DMN/DMNH2 (Madej et al., 2006a).
Preparation of the SdhABE (MFR) complex
Frozen W. succinogenes cells were three times thawed and frozen in liquid nitrogen and suspended in 2 ml g−1 wet cells anoxic buffer containing 50 mM TRIS·Cl (pH 8.25), 1 mM malonate and dithiothreitol (DTT) respectively. The suspension was centrifuged for 15 min at 20 000 g and 0.1% w/v laurylmaltoside (LM) was added to the supernatant and all buffers. After stirring for 30 min at room temperature the mixture was centrifuged in an ultracentrifuge for 45 min at 190 000 g. The supernatant was applied on a HiPrep™ 16/10 DEAE Sepharose™ column using an ÄKTApurifier™ system (GE Healthcare) and eluted with 0.15 M NaCl. The eluated fractions were tested for fumarate reductase activity using benzylviologen radicals as electron donor. Fractions containing at least 10% of the maximum activity were pooled and concentrated in a pressure dialysis cell (50 kDa cut-off) and further purified via gel filtration using a HiLoad™ 16/60 Superdex™ 200 column on the ÄKTApurifier™ system with anoxic buffer containing 50 mM TRIS·Cl (pH 8.25), 0.2 M NaCl, 1 mM malonate, 1 mM DTT and 0.1% LM.
Enzymatic activities
All enzymatic assays were performed at 37°C in 0.4 cm path-length anoxic quartz cuvettes.
Fumarate reduction activity was measured in anoxic buffer containing 50 mM TRIS·Cl (pH 8.0) by monitoring photometrically the oxidation of dithionite-reduced benzylviologen (BV, ε546 19.5 mM−1 cm−1) by fumarate as described (Unden and Kröger, 1986). The 1 ml assay mixture contained 1 mM benzylviologen titrated with sodium dithionite (20 mg ml−1) to an optical density at 546 nm of 2 and 10 μg of enriched MFR solution; the reaction was started by the addition of 10 mM fumarate. The SdhABE (MFR) complex was tested for succinate oxidation activity using either methylene blue (MB, ε578 17.1 mM−1 cm−1) (Kröger et al., 1980), dichlorophenolindophenol (DCPIP, ε578 15.3 mM−1 cm−1) (Lemma et al., 1990), ferricenium hexafluorophosphate (Lehman et al., 1990), dimethylnaphthoquinone (DMN) (Unden and Kröger, 1981) or EQ-0 (Madej et al., 2006b) as electron acceptor. The 1 ml assay mixture contained 10–80 μg of enriched MFR solution as well as 0.2 mM methylene blue, DCPIP, ferricenium, DMN or EQ-0 respectively. The reaction was started by the addition of 10 mM succinate. The reduction of fumarate with CoM-S-H and Co-B-S-H was tested as described (Heim et al., 1998). The reaction mixture contained 10 μg of enriched MFR solution and 4 mM of both CoM-S-H and Co-B-S-H. The reaction was started by the addition of 10 mM fumarate. The oxidation of the menaquinol analogue DMNH2 (ε270−290 15.2 mM−1 cm−1) with fumarate was determined as described (Unden and Kröger, 1981). The 1 ml assay mixture contained 0.2 mM DMNH2 reduced with about 10 μl of NaBH4 (5 mg ml−1) and 20 μg of enriched MFR solution. The reaction was started by the addition of 1 mM fumarate.
The synthesized quinones (TMN, the 5-MMK-6 analogue and the 8-MMK-6 analogue) were reduced with dithionite according to Rich (1978). The quinones were put in an anoxic vial and dissolved in diethylether (5 mM). An equal volume of aqueous sodium dithionite solution (20 mg ml−1) was added and the whole was vigorously shaken. The quinol layer was decanted and transferred to a fresh anoxic vial using a gas-tight syringe. The ether was evaporated under vacuum and the vial was flushed with argon. The quinol was then dissolved in anoxic ethanol. The quinol oxidation activity was measured in 50 mM sodium phosphate buffer (pH 7.4) by monitoring the change of the absorbance difference between 270 and 290 nm. The 1 ml reaction mixture contained 20 μg of enriched MFR solution and 0.2 mM TMN, 0.06 mM 5-MMK-6 analogue or 0.06 mM 8-MMK-6 analogue respectively. The reaction was started by the addition of 1 mM fumarate. The molar extinction coefficient of the 8-MMK-6 analogue was calculated from air-oxidized and NaBH4-reduced spectra: ε270−290 10.5 ± 1.5 mM−1 cm−1 (Fig. S2). The same value was used to calculate the activity with the TMN and the 5-MMK-6 analogue.
Membrane preparation
Frozen cells were suspended with anoxic buffer containing 50 mM TRIS·Cl (pH 7.8), 2 mM malonate, 1 mM DTT and disrupted in a French Pressure Cell (130 MPa; 10 ml min−1 flow). Membranes were prepared from the cell homogenate via ultracentrifugation (200 000 g, 60 min).
Separation of the periplasm, cytoplasm and membranes
Wolinella succinogenes mutant fMFR cells were harvested in the exponential growth phase, suspended in cold anoxic buffer containing 50 mM TRIS·Cl (pH 8.0), 0.5 M mannitol, 10 mM EDTA, 0.5 mg ml−1 lysozyme and gently mixed with a vortexer for 1 min. The mixture was subsequently centrifuged (14 000 g, 1.5 min; 4°C) to separate the periplasm extract (supernatant) from the spheroblasts which were suspended in cold anoxic buffer containing 10 mM TRIS·Cl (pH 8.0), 15 mM magnesium chloride and 1.5 g l−1 DNase I. To lyse the spheroblasts and separate the membranes from the cytoplasm the mixture was gently mixed with a vortexer for 5 min and centrifugated (20 000 g, 15 min, 4°C) (Krafft et al., 1995).
Protein characterization
SDS-PAGE was performed according to Laemmli (1970) using 4–12% polyacrylamide gradient gels (Invitrogen) and Coomassie Blue for staining. Detection of covalently bound FAD was carried out on a SDS-PAGE gel washed for 10 min in 10% acetic acid and irradiated with UV light (λ = 302 nm) before staining. The total protein concentration was measured with the BCA (bicinchoninic acid) assay (Pierce Biotechnology). For Western blot analysis, protein was transferred from the gel to nitrocellulose by electroblotting in a discontinuous buffer system (Kyhse-Andersen, 1984). The anti-pentahistidine-alkaline phosphatase antibody (Sigma) was applied at a dilution of 1:2000.
Peptide mass fingerprint analysis
The mass-spectrometric protein identification was performed as described (Rais, 2005). The respective protein bands were cut from an coomassie-stained SDS-PAGE gel and destained with a 100 mM ammoniumhydrogencarbonate (AHC)/50% acetonitrile (ACN) mixture before shrinking the gel pieces with pure ACN. The dried pieces were soaked with trypsin solution (20 μg ml−1, Promega), covered with 40 mM AHC and incubated overnight at 37°C. Extraction of the cut peptides was performed via ultrasonification and addition of 2 μl of 10% trifluor acetic acid (TFA) to the supernatant. The solution was evaporated in a vacuum centrifuge and the pellet dissolved in 70% ACN/0.1% TFA. A saturated solution of α-cyano-4-hydroxy-cinnamic acid in 70% ACN/0.1% TFA was prepared. Equal parts of the peptide solution and a suitable matrix dilution were applied to the target, dried and washed with ice-cold formic acid. The peptide fragments were examined using in a ‘Omni Flex®’ MALDI-TOF spectrometer (Bruker). Resulting peaklists (Fig. S1) were compared with the MSDB database using the program ‘Mascot’ (Matrix Science).
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft [Cluster of Excellence ‘Macromolecular Complexes’ (DFG Project EXC 115); SFB 472 ‘Molecular Bioenergetics’], a grant from the German Federal Ministry of Education, Science, Research and Technology framework programme (‘BMBF-Verbundprojekt’) on the ‘Proteome-wide Analysis of Membrane Proteins’ (ProAMP, 0312890C), the VolkswagenStiftung (I/77 595), and the Max Planck Society. We thank Jörg Simon for providing the pFrdcat2 vector and for helpful discussions, Monica Sänger for introducing H.D.J. to W. succinogenes genetics, and Rainer Hedderich for a generous gift of CoB.
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ackrell BAC, Johnson MK, Gunsalus RP, Cecchini G. Structure and Function of Succinate Dehydrogenase and Fumarate Reductase. Boca Raton, FL: CRC Press; 1992. [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baar C, Eppinger M, Raddatz G, Simon J, Lanz C, Klimmek O, et al. Complete genome sequence and analysis of Wolinella succinogenes. Proc Natl Acad Sci USA. 2003;100:11690–11695. doi: 10.1073/pnas.1932838100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford V, Dobbin PS, Richardson DJ, Hemmings AM. Open conformation of a flavocytochrome c3 fumarate reductase. Nat Struct Biol. 1999;6:1104–1107. doi: 10.1038/70039. [DOI] [PubMed] [Google Scholar]
- Berks BC, Palmer T, Sargent F. Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr Opin Microbiol. 2005;8:174–181. doi: 10.1016/j.mib.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Biel S, Simon J, Groß R, Ruiz T, Ruitenberg M, Kröger A. Reconstitution of coupled fumarate respiration in liposomes by incorporating the electron transport enzymes isolated from Wolinella succinogenes. Eur J Biochem. 2002;269:1974–1983. doi: 10.1046/j.1432-1033.2002.02842.x. [DOI] [PubMed] [Google Scholar]
- Blaut M, Whittaker K, Valdovinos A, Ackrell BAC, Gunsalus RP, Cecchini G. Fumarate reductase mutants of Escherichia coli that lack covalently bound flavin. J Biol Chem. 1989;264:13599–13604. [PubMed] [Google Scholar]
- Bronder M, Mell H, Stupperich E, Kröger A. Biosynthetic pathways of Vibrio succinogenes growing with fumarate as terminal electron acceptor and sole carbon source. Arch Microbiol. 1982;131:216–223. doi: 10.1007/BF00405882. [DOI] [PubMed] [Google Scholar]
- Carlone GM, Anet FA. Detection of menaquinone-6 and a novel methyl-substituted menaquinone-6 in Campylobacter jejuni and Campylobacter fetus subsp. fetus. J Gen Microbiol. 1983;129:3385–3393. doi: 10.1099/00221287-129-11-3385. [DOI] [PubMed] [Google Scholar]
- Clark WM. Oxidation Reduction Potentials of Organic Systems. Baltimore, MD: Williams and Wilklns; 1960. [Google Scholar]
- Collins MD, Fernandez F. Menaquinone-6 and Thermoplasmaquinone-6 in Wolinella succinogenes. FEMS Microbiol Lett. 1984;22:273–276. [Google Scholar]
- Collins MD, Costas M, Owen RJ. Isoprenoid quinone composition of representatives of the genus Campylobacter. Arch Microbiol. 1984;137:168–170. doi: 10.1007/BF00414461. [DOI] [PubMed] [Google Scholar]
- Dietrich V, Klimmek O. The function of methyl-menaquinone-6 and polysulfide reductase membrane anchor (PsrC) in polysulfide respiration of Wolinella succinogenes. Eur J Biochem. 2002;269:1086–1095. doi: 10.1046/j.0014-2956.2001.02662.x. [DOI] [PubMed] [Google Scholar]
- Gomes CM, Lemos RS, Teixeira M, Kletzin A, Huber H, Stetter KO, et al. The unusual iron–sulfur composition of the Acidianus ambivalens succinate dehydrogenase complex. Biochim Biophys Acta. 1999;1411:134–141. doi: 10.1016/s0005-2728(99)00046-8. [DOI] [PubMed] [Google Scholar]
- Groß R, Simon J, Kröger A. The role of the twin-arginine motif in the signal peptide encoded by the hydA gene of the hydrogenase from Wolinella succinogenes. Arch Microbiol. 1999;172:227–232. doi: 10.1007/s002030050764. [DOI] [PubMed] [Google Scholar]
- Hägerhäll C. Succinate: quinone oxidoreductases – variations on a conserved theme. Biochim Biophys Acta. 1997;1320:107–141. doi: 10.1016/s0005-2728(97)00019-4. [DOI] [PubMed] [Google Scholar]
- Hedderich R, Hamann N, Bennati M. Heterodisulfide reductase from methanogenic archaea: a new catalytic role for an iron–sulfur cluster. Biol Chem. 2005;386:961–970. doi: 10.1515/BC.2005.112. [DOI] [PubMed] [Google Scholar]
- Hederstedt L. Bioenergetics – respiration without O-2. Science. 1999;284:1941–1942. doi: 10.1126/science.284.5422.1941. [DOI] [PubMed] [Google Scholar]
- Heim S, Kunkel A, Thauer RK, Hedderich R. Thiol: fumarate reductase (Tfr) from Methanobacterium thermoautotrophicum– identification of the catalytic sites for fumarate reduction and thiol oxidation. Eur J Biochem. 1998;253:292–299. doi: 10.1046/j.1432-1327.1998.2530292.x. [DOI] [PubMed] [Google Scholar]
- Huang LS, Sun G, Cobessi D, Wang AC, Shen JT, Tung EY, et al. 3-Nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by Complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. J Biol Chem. 2006;281:5965–5972. doi: 10.1074/jbc.M511270200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson TM, Luna-Chavez C, Cecchini G, Rees DC. Structure of the Escherichia coli fumarate reductase respiratory complex. Science. 1999;284:1961–1966. doi: 10.1126/science.284.5422.1961. [DOI] [PubMed] [Google Scholar]
- Iverson TM, Luna-Chavez C, Croal LR, Cecchini G, Rees DC. Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the quinol-binding site. J Biol Chem. 2002;277:16124–16130. doi: 10.1074/jbc.M200815200. [DOI] [PubMed] [Google Scholar]
- Janausch IG, Zientz E, Tran QH, Kröger A, Unden G. C-4-dicarboxylate carriers and sensors in bacteria. Biochim Biophys Acta. 2002;1553:39–56. doi: 10.1016/s0005-2728(01)00233-x. [DOI] [PubMed] [Google Scholar]
- Kenney WC, Kröger A. Covalently bound flavin of Vibrio succinogenes succinate dehydrogenase. FEBS Lett. 1977;73:239–243. doi: 10.1016/0014-5793(77)80989-7. [DOI] [PubMed] [Google Scholar]
- Körtner C. Fachbereich Biologie. Frankfurt am Main: Johann Wolfgang Goethe-Universität; 1989. Das Operon des Fumaratreduktase-Komplexes aus Wolinella succinogenes. Doctoral thesis. [Google Scholar]
- Krafft T, Groß R, Kröger A. The function of Wolinella succinogenes psr genes in electron transport with polysulphide as the terminal electron acceptor. Eur J Biochem. 1995;230:601–606. doi: 10.1111/j.1432-1033.1995.0601h.x. [DOI] [PubMed] [Google Scholar]
- Kröger A, Dorrer E, Winkler E. Orientation of the substrate sites of formate dehydrogenase and fumarate reductase in the membrane of Vibrio succinogenes. Biochim Biophys Acta. 1980;589:118–136. doi: 10.1016/0005-2728(80)90136-x. [DOI] [PubMed] [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels – a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lancaster CRD. Succinate: quinone oxidoreductases: an overview. Biochim Biophys Acta. 2002;1553:1–6. doi: 10.1016/s0005-2728(01)00240-7. [DOI] [PubMed] [Google Scholar]
- Lancaster CRD. Structure and function of succinate:quinone oxidoreductases and the role of quinol: fumarate reductases in fumarate respiration. In: Zannoni D, editor. Respiration in Archaea and Bacteria Volume 1: Diversity of Prokaryotic Electron Transport Carriers. Dordrecht: Kluwer Scientific; 2004. pp. 57–85. [Google Scholar]
- Lancaster CRD, Kröger A, Auer M, Michel H. Structure of fumarate reductase from Wolinella succinogenes at 2.2 Å resolution. Nature. 1999;402:377–385. doi: 10.1038/46483. [DOI] [PubMed] [Google Scholar]
- Lancaster CRD, Groß R, Haas A, Ritter M, Mäntele W, Simon J, Kröger A. Essential role of Glu-C66 for menaquinol oxidation indicates transmembrane electrochemical potential generation by Wolinella succinogenes fumarate reductase. Proc Natl Acad Sci USA. 2000;97:13051–13056. doi: 10.1073/pnas.220425797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster CRD, Groß R, Simon J. A third crystal form of Wolinella succinogenes quinol:fumarate reductase reveals domain closure at the site of fumarate reduction. Eur J Biochem. 2001;268:1820–1827. [PubMed] [Google Scholar]
- Lancaster CRD, Sauer US, Groß R, Haas AH, Graf J, Schwalbe H, et al. Experimental support for the ‘E pathway hypothesis’ of coupled transmembrane e– and H+ transfer in dihemic quinol:fumarate reductase. Proc Natl Acad Sci USA. 2005;102:18860–18865. doi: 10.1073/pnas.0509711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster CRD, Herzog E, Juhnke HD, Madej MG, Müller FG, Paul R, Schleidt PG. Electroneutral and electrogenic catalysis by dihaem-containing succinate:quinone oxidoreductases. Biochem Soc Trans. 2008;5:996–1000. doi: 10.1042/BST0360996. [DOI] [PubMed] [Google Scholar]
- Lehman TC, Hale DE, Bhala A, Thorpe C. An acyl-coenzyme A dehydrogenase assay utilizing the ferricenium ion. Anal Biochem. 1990;186:280–284. doi: 10.1016/0003-2697(90)90080-s. [DOI] [PubMed] [Google Scholar]
- Lemma E, Unden G, Kröger A. Menaquinone is an obligatory component of the chain catalyzing succinate respiration in Bacillus subtilis. Arch Microbiol. 1990;155:62–67. doi: 10.1007/BF00291276. [DOI] [PubMed] [Google Scholar]
- Lemos RS, Gomes CM, Teixeira M. Acidianus ambivalens complex II typifies a novel family of succinate dehydrogenases. Biochem Biophys Res Commun. 2001;281:141–150. doi: 10.1006/bbrc.2001.4317. [DOI] [PubMed] [Google Scholar]
- Lemos RS, Fernandes AS, Pereira MM, Gomes CM, Teixeira M. Quinol:fumarate oxidoreductases and succinate:quinone oxidoreductases: phylogenetic relationships, metal centres and membrane attachment. Biochim Biophys Acta. 2002;1553:158–170. doi: 10.1016/s0005-2728(01)00239-0. [DOI] [PubMed] [Google Scholar]
- Leys D, Tsapin AS, Nealson KH, Meyer TE, Cusanovich MA, Beeumen JJV. Structure and mechanism of the flavocytochrome c fumarate reductase of Shewanella putrefaciens MR-1. Nat Struct Biol. 1999;6:1113–1117. doi: 10.1038/70051. [DOI] [PubMed] [Google Scholar]
- Lorenzen JP, Kröger A, Unden G. Regulation of anaerobic respiratory pathways in Wolinella succinogenes by the presence of electron-acceptors. Arch Microbiol. 1993;159:477–483. [Google Scholar]
- Madej MG, Nasiri HR, Hilgendorff NS, Schwalbe H, Lancaster CRD. Evidence for transmembrane proton transfer in a dihaem-containing membrane protein complex. EMBO J. 2006a;25:4963–4970. doi: 10.1038/sj.emboj.7601361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madej MG, Nasiri HR, Hilgendorff NS, Schwalbe H, Unden G, Lancaster CRD. Experimental evidence for proton motive force-dependent catalysis by the diheme-containing succinate: Menaquinone oxidoreductase from the gram-positive bacterium Bacillus licheniformis. Biochemistry. 2006b;45:15049–15055. doi: 10.1021/bi0618161. [DOI] [PubMed] [Google Scholar]
- Maguire JJ, Magnusson K, Hederstedt L. Bacillus subtilis mutant succinate dehydrogenase lacking covalently bound flavin – identification of the primary defect and studies on the iron sulfur clusters in mutated and wild-type enzyme. Biochemistry. 1986;25:5202–5208. doi: 10.1021/bi00366a033. [DOI] [PubMed] [Google Scholar]
- Mewies M, McIntire WS, Scrutton NS. Covalent attachment of flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) to enzymes: the current state of affairs. Protein Sci. 1998;7:7–20. doi: 10.1002/pro.5560070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileni M, MacMillan F, Tziatzios C, Zwicker K, Haas AH, Mäntele W, et al. Heterologous production in Wolinella succinogenes and characterization of the quinol: fumarate reductase enzymes from Helicobacter pylori and Campylobacter jejuni. Biochem J. 2006;395:191–201. doi: 10.1042/BJ20051675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Moser CC, Page CC, Dutton PL, Yano T. Simple redox-linked proton-transfer design: new insights from structures of quinol:fumarate reductase. Structure. 2000;8:R23–R32. doi: 10.1016/s0969-2126(00)00098-8. [DOI] [PubMed] [Google Scholar]
- Palmer T, Sargent F, Berks BC. Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol. 2005;13:175–180. doi: 10.1016/j.tim.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Park SJ, Tseng CP, Gunsalus RP. Regulation of succinate dehydrogenase (sdhCDAB) operon expression in Escherichia coli in response to carbon supply and anaerobiosis – role of ArcA and Fnr. Mol Microbiol. 1995;15:473–482. doi: 10.1111/j.1365-2958.1995.tb02261.x. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Pealing SL, Black AC, Manson FDC, Ward FB, Chapman SK, Reid GA. Sequence of the gene encoding flavocytochrome c from Shewanella putrefaciens– a tetraheme flavoenzyme that is a soluble fumarate reductase related to the membrane-bound enzymes from other bacteria. Biochemistry. 1992;31:12132–12140. doi: 10.1021/bi00163a023. [DOI] [PubMed] [Google Scholar]
- Rais I. Fachbereich Chemische und Pharmazeutische Wissenschaften. Frankfurt am Main: Johann Wolfgang Goethe-Universität; 2005. Neue Strategien zur massenspektrometrischen und gelelektrophoretischen Identifizierung von Membranproteinkomplexen. Doctoral thesis. [Google Scholar]
- Rich PR. Quinol oxidation in Arum maculatum mitochondria and its application to assay, solubilization and partial purification of alternative oxidase. FEBS Lett. 1978;96:252–256. [Google Scholar]
- Robinson KM, Lemire BD. Covalent attachment of FAD to the yeast succinate dehydrogenase flavoprotein requires import into mitochondria, presequence removal, and folding. J Biol Chem. 1996;271:4055–4060. doi: 10.1074/jbc.271.8.4055. [DOI] [PubMed] [Google Scholar]
- Robinson KM, Rothery RA, Weiner JH, Lemire BD. The covalent attachment of FAD to the flavoprotein of Saccharomyces cerevisiae succinate dehydrogenase is not necessary for import and assembly into mitochondria. Eur J Biochem. 1994;222:983–990. doi: 10.1111/j.1432-1033.1994.tb18949.x. [DOI] [PubMed] [Google Scholar]
- Schäfer G, Anemüller S, Moll R. Archaeal complex II: ‘classical’ and ‘non-classical’ succinate:quinone reductases with unusual features. Biochim Biophys Acta. 2002;1553:57–73. doi: 10.1016/s0005-2728(01)00232-8. [DOI] [PubMed] [Google Scholar]
- Schmid R, Labahn A. The influence of quinone structure on quinone binding to the QA site in bacterial reaction centers from Rhodobacter sphaeroides. In: Garab G, editor. Photosynthesis: Mechanisms and Effects. Vol. 2. Dordrecht, NL: Kluwer Academic Publishers; 1998. pp. 877–880. Proceedings of the 11th International Congress on Photosynthesis in Budapest. [Google Scholar]
- Simon J, Kröger A. Identification and characterization of IS1302, a novel insertion element from Wolinella succinogenes belonging to the IS3 family. Arch Microbiol. 1998;170:43–49. doi: 10.1007/s002030050613. [DOI] [PubMed] [Google Scholar]
- Simon J, Groß R, Ringel M, Schmidt E, Kröger A. Deletion and site-directed mutagenesis of the Wolinella succinogenes fumarate reductase operon. Eur J Biochem. 1998;251:418–426. doi: 10.1046/j.1432-1327.1998.2510418.x. [DOI] [PubMed] [Google Scholar]
- Singer TP, McIntire WS. Covalent attachment of flavin to flavoproteins – occurrence, assay, and synthesis. Methods Enzymol. 1984;106:369–378. doi: 10.1016/0076-6879(84)06039-0. [DOI] [PubMed] [Google Scholar]
- Sun F, Huo X, Zhai YJ, Wang AJ, Xu JX, Su D, et al. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121:1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- Taylor P, Pealing SL, Reid GA, Chapman SK, Walkinshaw MD. Structural and mechanistic mapping of a unique fumarate reductase. Nat Struct Biol. 1999;6:1108–1112. doi: 10.1038/70045. [DOI] [PubMed] [Google Scholar]
- Thummer R, Klimmek O, Schmitz RA. Biochemical studies of Klebsiella pneumoniae NifL reduction using reconstituted partial anaerobic respiratory chains of Wolinella succinogenes. J Biol Chem. 2007;282:12517–12526. doi: 10.1074/jbc.M609826200. [DOI] [PubMed] [Google Scholar]
- Tietze M, Beuchle A, Lamla I, Orth N, Dehler M, Greiner G, Beifuss U. Redox potentials of methanophenazine and CoB-S-S-CoM, factors involved in electron transport in methanogenic archaea. Chem Bio Chem. 2003;4:333–335. doi: 10.1002/cbic.200390053. [DOI] [PubMed] [Google Scholar]
- Unden G, Kröger A. The function of the subunits of the fumarate reductase complex of Vibrio succinogenes. Eur J Biochem. 1981;120:577–584. doi: 10.1111/j.1432-1033.1981.tb05739.x. [DOI] [PubMed] [Google Scholar]
- Unden G, Kröger A. Reconstitution of a functional electron-transfer chain from purified formate dehydrogenase and fumarate reductase complexes. Methods Enzymol. 1986;126:387–399. doi: 10.1016/s0076-6879(86)26039-5. [DOI] [PubMed] [Google Scholar]
- Wang WY, Malcolm BA. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange(TM) site-directed mutagenesis. Biotechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- Wardman P. The reduction potential of benzyl viologen – an important reference compound for oxidant radical redox couples. Free Radic Res Commun. 1991;14:57–67. doi: 10.3109/10715769109088942. [DOI] [PubMed] [Google Scholar]
- Wild DL, Saqi MAS. Structural proteomics: inferring function from protein structure. Curr Proteomics. 2004;1:59–65. [Google Scholar]
- Woodall CA, Jones MA, Barrow PA, Hinds J, Marsden GL, Kelly DJ, et al. Campylobacter jejuni gene expression in the chick cecum: evidence for adaptation to a low-oxygen environment. Infect Immun. 2005;73:5278–5285. doi: 10.1128/IAI.73.8.5278-5285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, et al. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


