Summary
Dengue virus belongs to the family Flaviviridae and is a major emerging pathogen for which the development of vaccines and antiviral therapy has seen little success. The NS3 viral protease is a potential target for antiviral drugs since it is required for virus replication. The goal of this study was to identify novel dengue virus (type 2; DEN2V) protease inhibitors for eventual development as effective anti-flaviviral drugs. The EUDOC docking program was used to computationally screen a small-molecule library for compounds that dock into the P1 pocket and the catalytic site of the DEN2V NS3 protease domain apo-structure (Murthy et al., 1999) and the Bowman-Birk inhibitor-bound structure (Murthy et al., 2000). The top 20 computer–identified hits that demonstrated the most favorable scoring “energies” were selected for in vitro assessment of protease inhibition. Preliminary protease activity assays demonstrated that more than half of the tested compounds were soluble and exhibited in vitro inhibition of the DEN2V protease. Two of these compounds also inhibited viral replication in cell culture experiments, and thus are promising compounds for further development.
Keywords: dengue virus, NS2B-NS3, protease, small-molecule inhibitor, flavivirus
The mosquito-borne dengue virus (DENV), a member of the family Flaviviridae and genus Flavivirus, appears to be expanding its range, increasing in disease severity, and has recently been deemed by the US National Institutes of Health as a potential serious public health threat to the United States (Morens and Fauci, 2008). The U.S. Centers for Disease Control and Prevention estimate that ~2.5 billion people worldwide are at risk for dengue infection; annually 50–100 million dengue infections occur with ~500000 cases of dengue hemorrhagic fever (DHF)/dengue shock syndrome (DSS) resulting in ~25000 deaths. Primary dengue fever is caused by any of four distinct serotypes of the virus (dengue virus type 1–4). It is postulated that subsequent infection by a different serotype may promote more serious forms of the disease such as DHF/DSS (Alvarez et al., 2006; Halstead, 2003). There are no approved vaccines or antiviral therapies to combat this disease. The increasing spread and severity of DENV infections emphasize the importance of drug discovery strategies that efficiently and cost-effectively identify antiviral drug leads for development into potent drugs.
The ~ 11 kB positive-strand RNA genome of DENV is transcribed and translated as a single polyprotein that is co- and post-translationally cleaved by cellular and viral proteases (Fields, 1996). The N-terminal region of the non-structural 3 protein (NS3) is a serine protease (Bazan and Fletterick, 1989; Chambers et al., 1990) that binds a required NS2B cofactor and cleaves the polyprotein after dibasic residues in the NS2A–NS2B, NS2B-NS3, NS3-NS4A, and NS4B-NS5 cleavage sites (Chambers et al., 1990). The NS2B-NS3 protease is required for viral replication (Falgout et al., 1991) and serves as a promising target for DENV antiviral drug development (Leyson et al., 2000, Sampath et al., 2009). While other studies have identified protease inhibitors by either high throughput screening or assaying compounds that mimic the peptide substrate (Chanprapaph et al., 2005; Ganesh et al., 2005; Leung et al., 2001; Yin et al., 2006a 2006b), this study used a virtual screen to predict which chemicals in a commercially-available library could inhibit the dengue virus protease. Testing of computer-predicted hits using a rapid in vitro DEN2V protease assay confirmed the activity of several compounds that inhibited the NS2B-NS3 protease; two compounds further demonstrated antiviral activity in cell-based replication assays.
The increased availability of three-dimensional protein structures and chemical compound libraries has expanded the role of high-throughput computational screening in discovering new drug leads (e.g., Laird and Blake, 2004). The chemical library chosen for this screen was a subset of an in-house database from Mayo Clinic that contained 2.5 million three-dimensional structures of small molecules that were reportedly available from chemical vendors. Two additional filters were imposed on this dataset. First, due to concerns regarding cellular uptake and cell membrane impermeability to ions (Ghose et al., 2001, Alberts et al., 2002) only neutral non-zwitterionic compounds (at pH 7.4) were passed to the filtered chemical library Second, only compounds readily purchasable from highly reputable chemical vendors were retained in the filtered chemical library. Using the EUDOC program (Pang et al., 2001 and 2008; Wang and Pang, 2007), we computationally screened this filtered chemical library against two previously reported dengue virus type 2 (DEN2V) protease crystal structures, the NS3 protease domain alone (PDB identifier 1BEF; Murthy et al., 1999) and the NS3 protease domain complexed with the Bowman-Birk inhibitor (a soybean protein that has been shown to inhibit the dengue protease) (PDB identifier 1DF9; Murthy et al., 2000). The EUDOC program performed virtual screens by systematically and separately docking each small molecule from our chemical library into the protease active site and the P1 pocket. Docked conformations of each compound were assigned an intermolecular interaction “energy” score that included charge-charge and Van der Waals interaction energy terms (Pang et. al. 2001). From the ensemble of docked conformations for each ligand, the conformation with the lowest “energy” score was retained as representative of the bound structure. Twenty potential protease inhibitors that had some of the lowest energy scores were purchased from large chemical suppliers (e.g., Sigma-Aldrich) for further characterization.
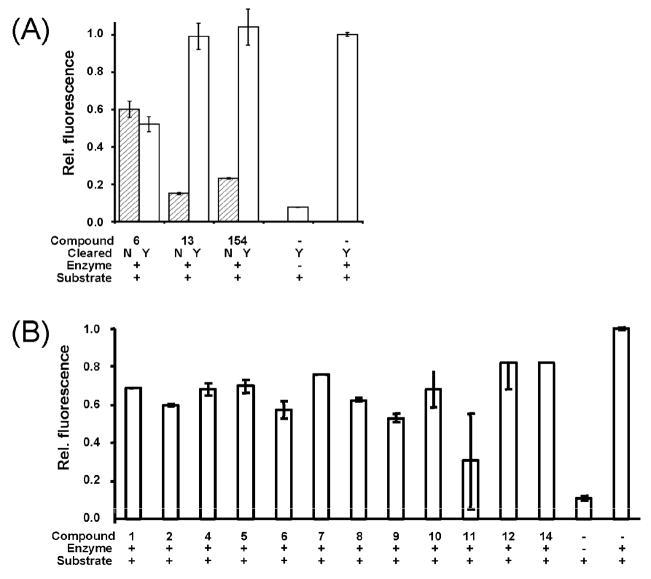
Compounds identified by the EUDOC program as potential protease inhibitors were tested for solubility and protease inhibition activity. Of the 20 potential inhibitors that were purchased, 13 were soluble at 100 μM in aqueous cleavage buffer (200 mM Tris [pH 9.0], 20% glycerol) and used in subsequent NS2B-NS3pro inhibition tests. Potential inhibitors were initially dissolved in DMSO and diluted with cleavage buffer to their desired final concentration. In addition to analysis by light scattering to determine compound solubility, protease inhibition assays were performed before and after filtering and high-speed centrifugation to remove precipitated compounds. Poorly soluble compounds generally produced false positive results, in that protease inhibition appeared to occur in pre-centrifuged solutions that contained precipitants (Fig. 1A, inhibitors 13 and 154). Soluble compounds generated similar protease inhibition results both before and after centrifugation (Fig. 1A, inhibitor 6). Compounds with poor solubility were removed from consideration to prevent false positives, and all assays were performed on solutions that were “cleared” by centrifugation to remove precipitants. In vitro DEN2V protease activity assays have been previously described (Leung et al., 2001; Yusof et al., 2000). The DEN2V NS2B cofactor linked to the protease domain of NS3 (NS2B-NS3pro; plasmid obtained from Dr. Padmanabhan, Georgetown University) was expressed and purified as previously described (Yusof et al., 2000). Protease activity was confirmed by cleavage of a dibasic fluorophore-linked peptide substrate, Boc-Gly-Lys-Arg-AMC (Fig. 1, substrate + enzyme control relative to substrate control). DEN2V NS2B-NS3pro (100 nM) was incubated with each soluble compound (100 μM, 1% DMSO) and 100 μM Boc-GKR-AMC (Bachem, USA) for 30 minutes at room temperature. Protease activity was determined by monitoring fluorescence intensity from AMC (excitation 390 nm, emission 465 nm) generated by cleavage from the peptide substrate. Protease reactions performed with 100 μM aprotinin, a known broad-spectrum serine protease inhibitor, showed fluorescence levels that were similar to that of the “substrate alone” background control (data not shown).
Figure 1.
In vitro DEN2V NS2B-NS3 protease inhibition assay. (A) Representative protease inhibition assays performed both before and after centrifugation and filtration to “clear” the solutions of precipitated compounds. (B) Protease inhibition assay of soluble compounds in cleared solutions; compounds were predicted by EUDOC virtual screening as potential DEN2V inhibitors. Compounds were assayed for in vitro protease inhibition along with “no inhibitor” (i.e., protease + substrate) and no protease (i.e., substrate alone) controls. Protease activities of each reaction were normalized to the “no inhibitor” control.
In vitro assay results indicated that more than half of the tested compounds showed reduced in vitro NS2B-NS3pro activity as compared to the “no inhibitor” control (Fig. 1B). The five compounds with the greatest in vitro inhibition of DEN2V protease (Fig. 1B) were further characterized for the ability to reduce DEN2V replication in cell cultures. These virus replication studies used the New Guinea C (Sabin, 1952; ATCC, Manassas, Virginia) and Thailand/16681/1964 (Halstead and Simasthien, 1970) strains of DEN2V. To test small molecules for inhibition of viral replication we optimized an ELISA-like cell-based replication assay that monitored production of DEN2V viral proteins. For this assay, confluent LLC-MK2 cells in 96-well plates were infected with DEN2V at MOI 0.2. Final concentrations of compounds ranged from 1nM to 100 μM, and were added to the cell monolayer at the time of infection. All assays were performed in quadruplicate. After 48 hr incubation at 37°C, cells were fixed, treated with polyclonal mouse DEN2V primary antibody (R. Tesh, Arbovirus Reference Collection, UTMB), washed, and incubated with HRP-conjugated anti-mouse secondary antibody. Following secondary antibody incubation, cells were washed and developed with 50 μL per well of the ELISA substrate 3,3′,5,5′-tetramethylenzidine (Sigma). Reactions were incubated at room temperature for 5 minutes, stopped with 50 μL 1N HCl, and results determined by measuring optical density at 450 nm in a microplate reader.
In parallel experiments, MTT [3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] toxicity assays were performed to determine possible cellular toxicity of the compounds. In these assays, compound was removed and MTT added to the cells. MTT reduction in the mitochondria of living cells yielded purple crystals which when solublized can be quanitified spectroscopically to determine drug toxicity as a function of living cells (Mosmann 1983; Gerlier and Thomasset, 1986). Effective concentrations (EC50) and cytotoxic concentrations (CC50) were calculated from nonlinear regression fitting (GraphPad Prism 4, Graphpad, SanDiego, CA) of signal vs concentration data points to the standard dose response equation Y=Bottom + (Top−Bottom)/(1+10^(Log(EC50)−X)). In this equation, X was the log of compound concentration, Y was the response signal, and Bottom and Top refer to plateaus of the sigmoid response curve.
Using the above cell-based virus replication assay, the small molecules ARDP0006 and ARDP0009 were observed to have the best efficacy among the tested compounds at inhibiting DEN2V replication (Table 1). ARDP0009 had no apparent toxicity at the concentrations tested thus a lower limit to its toxicity was based on the maximum tested concentration. Selectivity indices (SIs; selectivity index = CC50/EC50) calculated for ARDP0006 and ARDP0009 were comparable to the SI calculated for ribavirin (Table 1), a nucleoside analog that has demonstrated inhibition of the DENV 2′-O-methyltransferase NS5 (Benarroch et al., 2004) and hepatitis C virus replication when used in combination with interferon.
Table 1.
Inhibition of dengue virus in cell-based replication assays
| Compound | Structure | EC50a (μM) | CC50b (μM) | Slc |
|---|---|---|---|---|
| ARDP0006 |
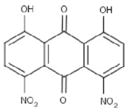
|
4.2 ± 1.9 | 69 ±4 | 16.6 |
| ARDP0009 |

|
35 ± 8 | >300 | >8.6 |
| Ribavirin (non-protease Inhibitor) |

|
37 ± 1 | >300 | >8.1 |
EC50 represents the concentration at which the ELISA signal has been reduced 50% in the cell-based replication assay.
CC50 represents the concentration at which 50% of the cells have died in the toxicity (MTT) assay.
SI (selectivity index) represents the CC50 divided by the EC50.
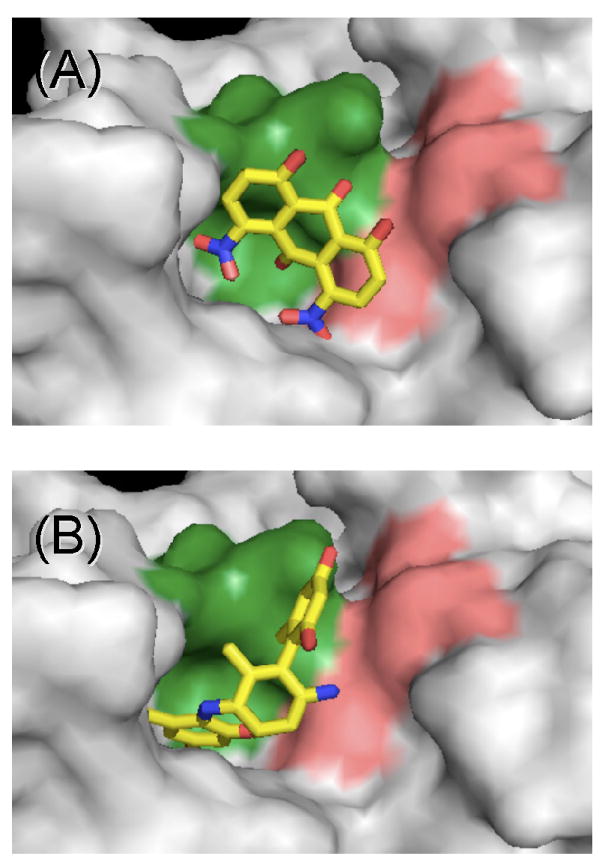
The EUDOC-predicted bound conformations of ARDP0006 and ARDP0009, the two compounds that were active against DEN2V in cell culture, were analyzed to identify intermolecular contacts that might contribute to the observed inhibition. ARDP0006 was predicted to interact with active site and P1 pocket residues, including Gln 35 (adjacent to the active site), His 51 and Ser 135 of the active site, and Gly 151 and Gly 153 of the P1 pocket (Fig. 3A). Additional contacts (< 4Å) were made between ARDP0006 and protease residues Ser 131, Pro 132, Gly 133, Thr 134, Asn 152, and Val 155. ARDP0009 was also predicted to interact with the protease active site and P1 pocket residues, including Gln 35, His 51, Ser 135, Tyr 150, Gly 151, and Gly 153 (Fig. 3B). Additional contacts (< 4Å) were made between ARDP0009 and protease residues Leu 115, Asn 152, Val 155, and Ala 160. Both small-molecule inhibitors were also docked into the recently solved structure of active DEN2V NS2B-NS3 protease (PDB identifier 2FOM; Erbel et al., 2006). Although the NS2B-NS3 protease complex and the NS3 protease had several significant structural differences, docking predictions identified similar intermolecular contacts between the inhibitors and the DEN2V protease structures. The intermolecular contact analysis supported the inference that protease inhibition observed in vitro was responsible for inhibition of virus replication observed in the cell-based assays. However, further investigations will be required to unequivocally delineate the molecular mechanism of virus replication inhibition of our protease inhibitors.
Figure 3.
EUDOC-predicted models of bound conformations of compounds that demonstrated dengue antiviral activity in cell culture. (A) ARDP0006 and (B) ARDP0009 were predicted to interact with active site (pink) and P1 pocket (green) residues of the DEN2V protease. Images were generated with SWISS PDB (Guex and Peitsch, 1997).
In summary, we report a strategy that correctly predicted the protease inhibitory activity of several small molecules and assays to rapidly validate the computer predictions. This strategy incorporated virtual screening of a chemical library to identify small molecules that fit into defined target sites (active site and P1 pocket) on the DEN2V NS3 protease crystal structure. These computer “hits” were tested for solubility and in vitro protease inhibition to establish activity and mechanism of inhibition. Several EUDOC “hits” demonstrated in vitro inhibition of the DEN2V NS3 protease. Significantly, these compounds were subsequently analyzed for antiviral activity in cell-based replication assays, in which two protease inhibitors showed dengue antiviral activity in cell culture. While others have reported limited in vitro studies to identify DEN2V protease inhibitors (Ganesh et al., 2005), we extended these studies to analyze protease inhibitors for cellular toxicity and inhibition of viral replication in cell culture. This strategy reflects a logical progression for early-stage drug discovery that can be used to successfully identify candidate antiviral drugs.
Figure 2.
Dose response curves for compounds that demonstrated antiviral activity in cell-based ELISA replication assays. Toxicity was measured using a standard MTT assay. Curves were fit using Graphpad Prism 4. (A) Efficacy and toxicity dose response curves for ARDP0006. (B) Efficacy dose response curve for ARDP0009. Since this compound had no observed toxicity at the concentrations tested, its toxicity curve was not included.
Acknowledgments
We thank Drs. A. Barrett, S. Gilbertson, J. Halpert, J. Lee, and J. Stevens for helpful discussions. We thank Drs. S. Weaver and N. Vasilakis for providing dengue 2 strain 16681, Dr. R. Tesh for the dengue 2 antibodies, and Dr. Padmanabhan for the plasmid expressing dengue NS2B-NS3pro.
Abbreviations
- DENV
dengue virus
- DEN2V
dengue 2 virus
- NS
nonstructural
- NS2B-NS3pro
NS3 protease with NS2B cofactor
- DHF
dengue hemorrhagic fever
- DSS
dengue shock syndrome
- GKR
glycine-lysine-arginine
- AMC
7-amino-4-methylcoumarin
Footnotes
This research was supported in part by a subcontract to SJW from NIH/NIAID NGA 5 U54 AI057156-03, The Welch Foundation H-1642 (SJW), a training fellowship to SMT from the Computational and Structural Biology in Biodefense Training Program of the W. M. Keck Center for Interdisciplinary Bioscience Training of the Gulf Coast Consortia (NIAID Grant No. 1 T32 AI065396-01), and the Defense Advanced Research Projects Agency DAAD19-01-1-0322 (YPP).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4. Garland Science; New York, New York: 2002. Principles of membrane transport; pp. 616–619. [Google Scholar]
- Alvarez M, Rodriguez-Roche R, Bernardo L, Vasquez S, Morier L, Gonzalez D, Castro O, Kouri G, Halstead S, Guzman M. Dengue hemorrhagic fever caused by sequential dengue 1–3 virus infections over a long time interval: Havana epidemic, 2001–2002. Am J Trop Med Hyg. 2006;75:113–117. [PubMed] [Google Scholar]
- Bazan J, Fletterick R. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology. 1989;171:637–639. doi: 10.1016/0042-6822(89)90639-9. [DOI] [PubMed] [Google Scholar]
- Benarroch D, Egolff M, Mulard L, Guerreiro C, Romette J, Canard B. A structural basis for the inhibition of the NS5 dengue virus mRNA 2′-O-methyltransferase domain by ribavirin 5′-triphosphae. J Biol Chem. 2004;279:35638–35643. doi: 10.1074/jbc.M400460200. [DOI] [PubMed] [Google Scholar]
- Chambers T, Weir R, Grakoui A, McCourt D, Bazan J, Fletterick R, Rice C. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci USA. 1990;87:8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanprapaph S, Saparpakorn P, Sangma C, Niyomrattanakit P, Hannongbua S, Angsuthanasombat C, Katzenmeier G. Competitive inhibition of the dengue virus NS3 serine protease by synthetic peptides representing polyprotein cleavage sites. Biochem Biophys Res Commun. 2005;330:1237–1246. doi: 10.1016/j.bbrc.2005.03.107. [DOI] [PubMed] [Google Scholar]
- Erbel P, Schiering N, D’Arcy A, Renatus M, Kroemer M, Lim S, Yin Z, Keller T, Vasudevan S, Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol. 2006;13:372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- Falgout B, Pethel M, Zhang Y, Lai C. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991;65:2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler D, Juno G, Markoff L. Flaviviruses. In: Fields B, Knipe D, Howley P, Chanock R, Melnick J, Monath T, Roizman B, Strauss S, editors. Field’s Virology. 3. Vol. 1. Lippincott William & Wilkins; Philadelphia, PA: 1996. pp. 961–1034. [Google Scholar]
- Ganesh VK, Muller N, Judge K, Luan CH, Padmanabhan R, Murthy KHM. Identification and characterization of nonsubstrate based inhibitors of the essential dengue and West Nile virus proteases. Bioorg Med Chem. 2005;13:257–264. doi: 10.1016/j.bmc.2004.09.036. [DOI] [PubMed] [Google Scholar]
- Gerlier D, Thomasset N. Use of MTT colorimetric assay to measure cell activation. J Immunol Methods. 1986;94:57–63. doi: 10.1016/0022-1759(86)90215-2. [DOI] [PubMed] [Google Scholar]
- Ghose A, Viswanadhan V. Combinatorial Library Design and Evaluation: Principles, Software, Tools, and Applications in Drug Discovery. Vol. 223. CRC Press; New York, New York: 2001. Fast continuum electrostatics methods for structure-based ligand design. [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Halstead S. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Vir Res. 2003;60:421–467. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- Halstead SB, Simasthien P. Observations related to the pathogenesis of dengue hemorrhagic fever. II Antigenic and biological properties of dengue viruses and their association with disease response in the host. Yale J Biol Med. 1970;42:276–292. [PMC free article] [PubMed] [Google Scholar]
- Laird E, Blake J. Structure-based generation of viable leads from small combinatorial libraries. Curr Opin Drug Discov Devel. 2004;7:354–359. [PubMed] [Google Scholar]
- Leung D, Schroder K, White H, Fang N, Stoermer M, Abbenante G, Martin J, Young P, Fairlie D. Activity of recombinant dengue 2 virus NS3 protease in the presence of a truncated NS2B co-factor, small peptide substrates, and inhibitors. J Biol Chem. 2001;276:45762–45771. doi: 10.1074/jbc.M107360200. [DOI] [PubMed] [Google Scholar]
- Leyssen P, De Clercq E, Neyts J. Perspectives for the treatment of infections with Flaviviridae. Clin Microbiol Rev. 2000;13:67–82. doi: 10.1128/cmr.13.1.67-82.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D, Fauci A. Dengue and hemorrhagic fever: a potential threat to public health in the United States. JAMA. 2008;299:214–216. doi: 10.1001/jama.2007.31-a. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunological Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Murthy K, Clum S, Padmanabhan R. Crystal structure and insights into interaction of the active site with substrates by molecular modeling and structural analysis of mutational effects. J Biol Chem. 1999;274:5573–5580. doi: 10.1074/jbc.274.9.5573. [DOI] [PubMed] [Google Scholar]
- Murthy K, Judge K, DeLucas L, Padmanabhan R. Crystal structure of dengue virus NS3 protease in complex with a Bowman-Birk inhibitor: implications for flaviviral polyprotein processing and drug design. J Mol Biol. 2000;301:759–767. doi: 10.1006/jmbi.2000.3924. [DOI] [PubMed] [Google Scholar]
- Pang YP, Perola E, Xu K, Prendergast F. EUDOC: a computer program for identification of drug interaction sites in macromolecules and drug leads from chemical databases. J Comput Chem. 2001;22:1750–1771. doi: 10.1002/jcc.1129. [DOI] [PubMed] [Google Scholar]
- Pang YP, et al. EUDOC on Blue Gene: accelerating the transfer of drug discoveries from laboratory to patient. IBM J Res Dev. 2008;52:69–81. [Google Scholar]
- Sabin A. Research on dengue during World War II. Am J Trop Med Hyg. 1952;1:30–50. doi: 10.4269/ajtmh.1952.1.30. [DOI] [PubMed] [Google Scholar]
- Sampath A, Padmanabhan R. Molecular targets for flavivirus drug discovery. Antiviral Research. 2009;81:6–15. doi: 10.1016/j.antiviral.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Pang YP. Accurate reproduction of 161 small-molecule complex crystal structures using the EUDOC program: expanding the use of EUDOC to supramolecular chemistry. PLoS ONE. 2007;2:e531. doi: 10.1371/journal.pone.0000531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Patel SJ, Wang WL, Wang G, Chan WL, Rao KRR, Alam J, Jeyaraj DA, Ngew X, Patel V, Beer D, Lim SP, Vasudevan SG, Keller TH. Peptide inhibitors of dengue virus NS3 protease. Part 1: Warhead. Bioorg Med Chem Lett. 2006a;16:36–39. doi: 10.1016/j.bmcl.2005.09.062. [DOI] [PubMed] [Google Scholar]
- Yin Z, Patel SJ, Wang WL, Chan WL, Rao KRR, Wang G, Ngew X, Patel V, Beer D, Knox JE, Ma NL, Ehrhardt C, Lim SP, Vasudevan SG, Keller TH. Peptide inhibitors of dengue virus NS3 protease. Part 2: SAR study of tetrapeptide aldehyde inhibitors. Bioorg Med Chem Lett. 2006b;16:40–43. doi: 10.1016/j.bmcl.2005.09.049. [DOI] [PubMed] [Google Scholar]
- Yusof R, Clum S, Wetzel M, Murthy K, Padmanabhan R. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J Biol Chem. 2000;275:9963–9969. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]