Abstract
The THRB gene encodes the well-described thyroid hormone (T3) receptor (TR) isoforms TRβ1 and TRβ2 and two additional variants, TRβ3 and TRΔβ3, of unknown physiological significance. TRβ1, TRβ2, and TRβ3 are bona fide T3 receptors that bind DNA and T3 and regulate expression of T3-responsive target genes. TRΔβ3 retains T3 binding activity but lacks a DNA binding domain and does not activate target gene transcription. TRΔβ3 can be translated from a specific TRΔβ3 mRNA or is coexpressed with TRβ3 from a single transcript that contains an internal TRΔβ3 translation start site. In these studies, we provide evidence that the TRβ3/Δβ3 locus is present in rat but not in other vertebrates, including humans. We compared the activity of TRβ3 with other TR isoforms and investigated mechanisms of action of TRΔβ3 at specific thyroid hormone response elements (TREs) in two cell types. TRβ3 was the most potent isoform, but TR potency was TRE dependent. TRΔβ3 acted as a cell-specific and TRE-dependent modulator of TRβ3 when coexpressed at low concentrations. At higher concentrations, TRΔβ3 was a TRE-selective and cell-specific antagonist of TRα1, -β1, and -β3. Both TRβ3 and TRΔβ3 were expressed in the nucleus in the absence and presence of hormone, and their actions were determined by cell type and TRE structure, whereas TRΔβ3 actions were also dependent on the TR isoform with which it interacted. Analysis of these complex responses implicates a range of nuclear corepressors and coactivators as cell-, TR isoform-, and TRE-specific modulators of T3 action.
Thyroid hormone (T3) actions are mediated by nuclear receptors that function as ligand-inducible transcription factors. The T3 receptor (TR) genes THRA and THRB are conserved in all vertebrates and encode TRα and TRβ proteins (1-3). TRs bind to discrete thyroid hormone response elements (TREs) of varying sequence and arrangement located within the promoter regions of T3-responsive target genes (4, 5). Unliganded apoTRs bind to TREs as homodimers or in heterodimer complexes with retinoid X receptors (RXR), whereas liganded TRs bind DNA as RXR/TR heterodimers (2). ApoTRs inhibit basal transcription of T3 target genes by interacting preferentially with corepressor proteins, such as nuclear receptor corepressor (NCoR), that form additional complexes with histone deacetylases leading to repression of gene transcription. T3 binding results in a conformational change in the TR leading to release of NCoR and the recruitment of coactivator proteins, such as steroid receptor coactivator-1 (SRC-1), that possess histone acetyl transferase activity and reverse the histone deacetylation associated with basal repression (1-3, 5). Subsequent recruitment of a large transcription factor complex known as vitamin D receptor interacting protein/TR-associated protein (DRIP/TRAP) to the TR/SRC-1 coactivator leads to binding and stabilization of RNA polymerase II and hormone-dependent activation of transcription (5-7).
The well-described TRα1, -β1, and -β2 isoforms bind DNA and T3 and act as functional apo- and liganded TRs, whereas TRα2 does not bind T3 and acts as a weak antagonist in vitro (1-3, 5). The various TR isoforms are expressed in temporospatial-specific patterns during development (8, 9) and in distinct ratios in adult tissues (5), and studies of TR-knockout and mutant mice have indicated specific roles for TRα and TRβ as well as functional redundancy (10, 11). For example, TRα mediates important T3 actions during heart, bone, and intestinal development and controls basal heart rate and body temperature in adults (12-19), whereas TRβ mediates T3 action in liver (20) and is responsible for regulation of the hypothalamic-pituitary-thyroid axis (21, 22). Detailed analysis of TRβ indicates that TRβ1 is expressed in most tissues, whereas TRβ2 is restricted to the hypothalamus, pituitary, cochlea, and retina (23-25). Studies of TRβ and TRβ2 knockout mice indicate that TRβ1 is essential for development of auditory function, whereas TRβ2 is not required (22, 26, 27), but that TRβ2 alone is essential for development of M-cone photoreceptors (28). In contrast, both TRβ1 and TRβ2 are necessary for regulation of the hypothalamic-pituitary-thyroid axis (27, 29). We recently cloned two additional rat TRβ isoforms, TRβ3 and TRΔβ3 (30). TRβ3 is a functional T3 receptor, whereas TRΔβ3 lacks a DNA-binding domain but retains T3-binding activity and acts as a potent antagonist in vitro (30). Although TRβ3 and Δβ3 are expressed widely, their actions have not been characterized in detail, and their physiological importance is unknown.
Resistance to thyroid hormone (RTH) is an autosomal dominant but heterogeneous condition caused by a large number of described mutations of THRB, which result in expression of dominant-negative TRβ proteins that inhibit T3-target gene expression in a wide range of tissues by several possible mechanisms (31, 32). In vitro analyses of mutant TRs have revealed the mutant receptors fail to mediate a transcriptional response to T3 but also interfere with wild-type TRα and TRβ function. Full and potent dominant-negative activity of mutant TRs requires them to retain the ability to bind DNA and to form homodimers and RXR/TR heterodimers (32). The precise mechanism resulting in dominant-negative activity has not been determined, but mutant TRs that fail to interact with coactivators (33, 34) or are defective in T3-induced release of corepressors (35, 36) have been identified in RTH patients. These findings suggest that dominant-negative activity in RTH is mediated by transcriptionally inactive complexes that contain mutant TRs and bind to TREs (32).
The aims of these studies were to determine whether TRβ3 and -Δβ3 are conserved among vertebrate species and to characterize their functional activities in comparison with known TRs. The mechanism of TRΔβ3 action was particularly investigated because, in contrast to RTH mutant TRs, it lacks a DNA-binding domain (30). Thus, TRβ3 and -Δβ3 were studied in two cell types, and T3 responses on four TREs were characterized. Mechanisms of action were studied by generating TRβ1, -β3, and -Δβ3 mutants with impaired T3 binding, heterodimerization, NCoR release, or coactivator interaction activities.
Materials and Methods
TRβ1, -β3, and -Δβ3 mutants
ATRβ3 mutant (TRβ3mut), in which the in-frame TRΔβ3 AUG start codon at position 103 of TRβ3 is mutated to CTG, was described previously (30). Four well-characterized TRβ1 mutants (CGG3→CAG to generate R243Q, TCC3→TAC for S314Y, TTG3→GCG for L454A, and CTG3→CGG for L428R) and equivalent TRβ3 (R172Q, S243Y, L383A, and L357R) and TRΔβ3 (R70Q, S141Y, L281A, and L255R) mutants were generated by site-directed mutagenesis (QuikChange Site-Directed Mutagenesis Kit; Stratagene, Amsterdam, The Netherlands) and sequenced. TRβ1 R243Q is a naturally occurring RTH mutant that has impaired ability to release NCoR (36-39). T3-binding activity of R243Q in solution is similar to wild-type TRβ1, but when bound to DNA with NCoR, T3 binding is impaired significantly (36, 37). A 5- to 10-fold increased T3 concentration can overcome the dominant-negative activity of R243Q and restore normal transactivation function by inducing release of NCoR (36, 38). TRβ1 S314Y is a dominant-negative RTH mutant that fails to bind T3 and exhibits no response to hormone (40). TRβ1 L454A contains an artificial mutation in the activation function-2 domain. This mutant binds T3 and DNA normally and interacts with retinoid X receptors (RXR) but fails to interact with coactivators, resulting in no response to T3 (34, 41, 42). TRβ1 L428R contains an artificial mutation in the dimerization domain, which results in impaired RXR heterodimerization but preserved homodimerization activity and an inability to bind T3 and no transactivation response (43-45).
Cell culture and transfections
Rat osteoblastic osteosarcoma ROS 17/2.8 cells were maintained in Ham's F12 medium plus 5% fetal calf serum (FCS). Monkey kidney fibroblast COS-7 cells were cultured in DMEM plus 5% FCS. For transfections, ROS 17/2.8 or COS-7 cells were seeded in six-well plates (105 cells per well) containing Ham's F12 or DMEM plus 5% charcoal-stripped FCS (CSS) (46) and transferred to serum-free medium before transfection using Lipofectamine PLUS (Invitrogen-Life Technologies, Inc., Paisley, UK) as described (30). Cells were transfected with 500 ng luciferase reporter gene driven by a thymidine kinase promoter controlled by TREs from the rat malic enzyme (ME) or α-myosin heavy chain (MHC) genes (47), by two copies of a palindromic TRE (PAL) (48) or a synthetic TRE containing a repeat of the hexanucleotide sequence AGGTCA separated by direct repeat + 4 (DR4) (Fig. 1), 40–200 ng TR plasmid (wild-type or mutant TRα1, TRα2, TRβ1, TRβ3, or TRΔβ3) (30, 49-51), 100 ng Renilla internal control reporter (Promega, Southampton, UK), and pCDM8 empty vector carrier DNA to a total of 1.5 μg DNA per well. After 3 h, 1 ml 10% CSS medium was added and cells were incubated for 24 h. Transfected cells from each well were split into four in a 24-well plate containing 5% CSS medium without or with T3 (10−10–10−6 m) and incubated for 48 h. Reporter gene activities were determined as described (30). and luciferase activity was normalized to Renilla activity before analysis of responses to T3. Expression of transfected TRβ1, TRβ3, and TRΔβ3 proteins was analyzed by Western blotting as described (30) using a monoclonal antibody (MA1–215; Affinity Biore-agents, Cambridge BioScience, Cambridge, UK) against TRβ1 amino acids 235–414, which are conserved in TRβ3 and TRΔβ3.
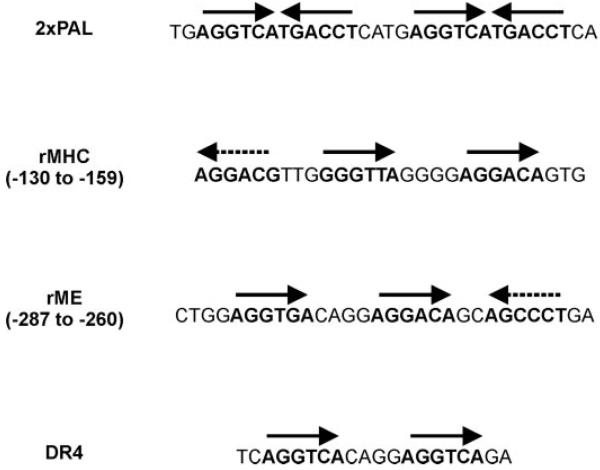
Fig. 1.
Comparison of TRE structures in reporter genes used for transfection experiments. 2xPAL shows two copies of the palindromic TRE included in the PAL reporter; rMHC shows the rat MHC gene TRE located at nucleotide position −139 to −159 upstream of the transcription start site in the native gene; rME shows the rat ME gene TRE located at position −287 to −260; DR4 shows the synthetic direct repeat TRE included in the DR4 reporter (4, 47, 48). Solid arrows indicate orientations of consensus and near-consensus hexamer sequences (in bold) that bind TR proteins in gel-shift studies and that have been shown to be required to mediate maximal T3 responses in transfections. Dashed arrows indicate sequences (in bold) required for T3 responses in transfections but that interact only weakly with TRs in gel shifts (4).
Subcellular localization of TRβ1, TRβ3, and TRΔβ3 proteins
Plasmid vectors encoding various green fluorescent protein (GFP)-TR fusion proteins were constructed as described (52). HeLa cells were maintained in DMEM with 10% FCS, antibiotics (100 U/ml penicillin and streptomycin and 0.5 ml gentamycin; Invitrogen) and 2 mm l-glutamine. For transient transfections, HeLa cells were cultured in six-well plates on coverslips at the density of 1 × 105 cells per well and transfected with various GFP-TR isoforms (100 ng/well) in serum-free medium using Lipofectamine PLUS according to the manufacturer's instructions. The cells expressing various GFP-TR fusion proteins were viewed under a Leica TCS SP laser scanning confocal microscope mounted on a DMIRBE inverted epifluorescent microscope equipped with ×63 magnification using a 1.4 numerical aperture oil immersion lens (Leica, Heidelberg, Germany). The GFP was excited at 488 nm from an air-cooled fiber-coupled argon laser at less than 10% of maximal power. GFP fluorescence was visualized using a 1.0 Airy unit pinhole and analyzed as described (53).
Results
The TRβ3/Δβ3 locus is detected only in rats but not in other species
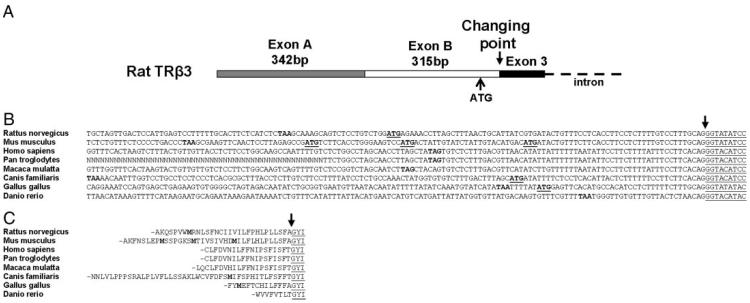
In the rat, TRβ3/Δβ3 locus exons A and B lie directly 5′ to the first common exon of TRβ (exon 3) with no intervening introns (30). To investigate the possible presence of TRβ3 and TRΔβ3 in other vertebrates, we identified genomic DNA sequences 5′ to the first common TRβ exon in eight species using Ensembl (http://www.ensembl.org/index.html) and Entrez nucleotide (http://www.ncbi.nlm.nih.gov) searches (Fig. 2). Open reading frames of between 25 and 133 bp were identified immediately upstream of an invariant splice site, termed the changing point (54). In-frame ATG codons, as previously identified in rat (GenBank accession no. AF239916.1), were also present in mouse (GenBank AC154626), dog (Ensembl no. ENSCAFG00000005741), and chicken (Ensembl ENSGALG00000011294) sequences but not in human (GenBank AC093927), chimpanzee (Ensembl ENSPTRG00000014697), macaque (Ensembl ENSMMUG00000000067), or zebra fish (GenBank BX927163). However, none of these ATG codons were positioned within a favorable Kozak translation initiation sequence context (55, 56). Blast searches (http://www.ncbi.nlm.nih.gov/BLAST) using these 5′ sequences identified the previously published rat TRβ3 and TRΔβ3 sequences but no additional TRβ transcripts or expressed sequence tags. Furthermore, amino acid sequence searches (rpsblast) using predicted sequences derived from the upstream open reading frames did not identify protein homology or conserved domain structures. Comparison of the predicted amino acid sequences upstream of the common TRβ protein revealed 50% identity between rat and mouse but no homology between rat and dog or chicken. Thus, TRβ3 may be present only in rodents and is not found in primates or other vertebrates. The lack of murine TRβ3 or TRΔβ3 expressed sequence tags, however, suggests that expression from this locus is unique to rats.
Fig. 2.
A, Schematic representation of the rat TRβ3 locus. The first TRβ common exon (exon 3) is shown as a black box, and its splice acceptor site is termed the changing point. B, The open reading frame that lies 5′ of exon 3 and in frame with the common TRβ coding sequence is shown in eight species. The common exon 3 sequence is underlined, and an arrow indicates the changing point. In-frame stop codons are shown in bold and ATG codons in bold underlined text. C, Predicted amino acid sequences of in-frame open reading frames identified 5′ to exon 3. The changing point is shown by the arrow, and the first three amino acids encoded by exon 3 are underlined. Predicted methionine amino acids are shown in bold, and the dash at the beginning of each open reading frame indicates the location of an in-frame stop codon.
TRβ3 exerts cell- and TRE-specific actions
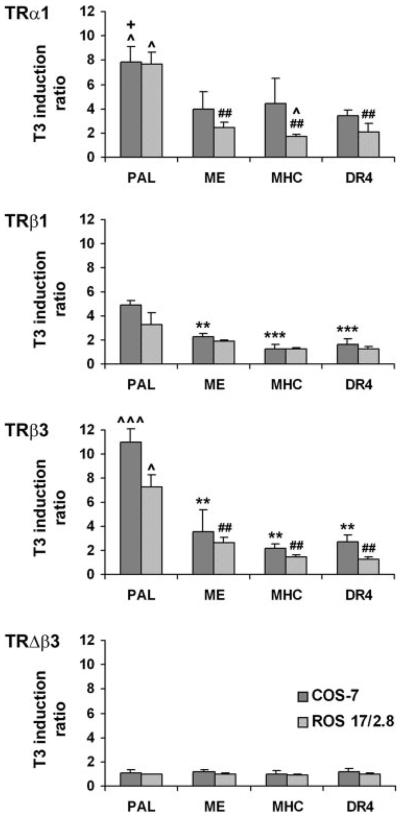
COS-7 and ROS 17/2.8 cells were transfected with PAL, ME, MHC, or DR4 reporters and increasing concentrations (0–200 ng) of TRα1, -α2, -β1, -β3, or -Δβ3 to optimize the TR concentration for additional studies. T3 responses of each TRE increased with increasing concentration of TRα1, -β1, or -β3, and the maximum response in both cell lines was seen after addition of 160 ng receptor (data not shown). Responses mediated by TRα1 (7.9 ± 1.2-fold in COS-7; 7.7 ± 1.0-fold in ROS 17/2.8), TRβ1 (4.9 ± 0.3-fold in COS-7; 3.3 ± 1.0-fold in ROS 17/2.8), or TRβ3 (11.0 ± 1.1-fold in COS-7; 7.2 ± 1.0-fold in ROS 17/2.8) were greatest on the PAL TRE in both cell types. TRα1 and -β3 mediated a greater T3 induction of PAL than TRβ1 in both cell types, whereas TRβ3 was also more potent than TRα1 in COS-7 cells (Fig. 3). In contrast, T3 responses mediated by each receptor on the ME, MHC, and DR4 TREs were similar in both cell types, although responses of all elements tended to be lower in ROS 17/2.8 compared with COS-7 cells. TRα2 and -Δβ3 did not mediate reporter gene responses on any TRE in either cell type (Fig. 3 and data not shown).
Fig. 3.
COS-7 or ROS 17/2.8 cells transfected with TRα1, -β1, -β3, or -Δβ3 (160 ng) and a luciferase reporter controlled by either an ME, MHC, PAL, or DR4 TRE. T3 induction of each element mediated by each receptor in both cell lines is shown. Luciferase activity was normalized to activity of a Renilla internal control vector, and results are expressed as mean T3 induction ratio (± sem), calculated by dividing normalized luciferase activities after T3 treatment by basal values (n = 3–5 experiments, three to six replicates per experiment; ANOVA followed by Tukey's multiple comparison post hoc tests: **, P < 0.01, ***, P < 0.001 vs. T3 induction of PAL by TRβ1 or TRβ3 in COS-7 cells; ##, P < 0.01 vs. T3 induction of PAL by TRα1 or TRβ3 in ROS 17/2.8 cells; ^, P < 0.05; ^^^, P < 0.001 vs. T3 induction mediated by TRβ1; +, P < 0.05 vs. T3 induction mediated by TRβ3).
In the absence of T3, unliganded apoTRs bind corepressors and inhibit basal target gene expression. An exchange of cofactors occurs after addition of T3, and ligand-bound TRs interact with coactivators to stimulate target gene expression. Thus, the response to T3 is a two-stage process; T3 activation of gene transcription follows relief of apoTR-mediated repression (1-3, 5), and the T3 induction ratio mediated by TRs is calculated by dividing reporter gene activity in the presence of T3 by reporter gene activity in the absence of T3. To investigate whether the differing activities of TRα1, -β1, or -β3 result from differences in apoTR-mediated repression, TRE responses to each receptor were determined in the absence of T3. ApoTRα1 repressed ME by 40 ± 13% in COS-7 cells (P < 0.05) but increased its expression by 40 ± 23% in ROS 17/2.8 cells (P < 0.05), indicating cell-specific activity of apoTRα1 on the ME element. ApoTRβ1 repressed PAL by 37 ± 16% in COS-7 cells (P < 0.05) and 36 ± 20% in ROS 17/2.8 cells (P < 0.05) and repressed ME by 36 ± 8% in ROS 17/2.8 cells (P < 0.01). ApoTRβ3 repressed DR4 expression by 36 ± 11% in ROS 17/2.8 cells (P < 0.05) only. ApoTRΔβ3 repressed PAL by 18 ± 4% in ROS 17/2.8 cells (P < 0.01) and MHC by 32 ± 12% in COS-7 cells (P < 0.05). Thus, effects of unliganded apoTRs on basal gene expression were dependent on the cell type and TRE and contribute to the complexity of cell- and gene-specific responses to T3. ApoTRβ3, however, had the weakest effect on basal gene transcription, indicating that TRβ3 activity results predominantly from T3-mediated effects rather than actions of the unliganded aporeceptor.
TRΔβ3 is coexpressed at low concentrations along with TRβ3 from a single transcript and acts as a TRE-selective modulator of TRβ3 action
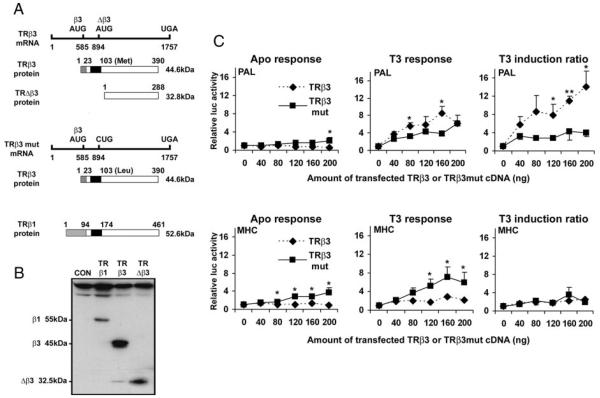
In previous studies, in vitro transcription-translation of TRβ3 cDNA resulted in expression of a 45-kDa TRβ3 protein together with a 32.5-kDa TRΔβ3 protein originating from an in-frame AUG codon, whereas transcription-translation of TRβ1 resulted in a single 55-kDa TRβ1 protein (30). In the current studies, transfection of COS-7 cells with TRβ3 similarly resulted in coexpression of a low relative concentration of TRΔβ3, whereas transfection with TRβ1 resulted only in expression of TRβ1 (Fig. 4, A and B).
Fig. 4.
A, Diagram of TRβ3 and TRβ3mut mRNAs and translated TRβ3 and TRΔβ3 proteins in comparison with TRβ1 protein. TRβ3 mRNA contains a 584-nucleotide 5′-untranslated region, an open reading frame at nucleotide 585 that encodes TRβ3 (390-amino-acid protein, predicted molecular mass, 44.6 kDa), an in-frame open reading frame at nucleotide 894 that encodes TRΔβ3 (288-amino-acid protein, predicted molecular mass, 32.8 kDa), and a stop codon at position 1757. In TRβ3mut mRNA, the AUG codon at nucleotide 894 is mutated to CUG so that only TRβ3 protein (in which methionine at position 103 is replaced by leucine) is translated. TRβ1 is a 461-amino-acid protein of predicted molecular mass 52.6 kDa that shares 100% identity with TRβ3 apart from the first 94 amino acids of TRβ1, which are replaced by 23 different amino acids in TRβ3 (30). B, Extracts prepared from COS-7 cells transfected with 20 ng empty vector (CON) or TRβ1, -β3, or -Δβ3 were analyzed by Western blotting using a TRβ-specific MA1–215 antibody that recognizes all TRβ isoforms. Cells transfected with TRβ1 express a single 55-kDa protein. Transfection of TRβ3 results in coexpression of 45- and 32.5-kDa β3 and Δβ3 proteins. Transfection with TRΔβ3 results in expression of a single 32.5-kDa protein. C, COS-7 cells transfected with either TRβ3 or TRβ3mut cDNA (0–200 ng), in which the AUG at position 894 that initiates translation of TRΔβ3 was mutated to CTG. Responses of PAL or MHC luciferase reporters to TRβ3 (◆) or TRβ3 mut (■) are plotted. Reporter gene activity was normalized to Renilla, and results are shown as luciferase activity in the absence or presence of T3 relative to activity under each condition in the absence of cotransfected receptor, which was normalized to a value of 1. Values less than 1 in the absence of T3 indicate repression by unliganded aporeceptor; values greater than 1 after addition of hormone indicate T3 response. Results are also expressed as mean T3 induction ratios (± SEM), calculated by dividing relative luciferase activities after T3 treatment by basal values (n = 3–5 experiments, three to six replicates per experiment; ANOVA followed by Tukey's multiple comparison post hoc tests: *, P < 0.05; **, P < 0.01 response to TRβ3 vs. TRβ3mut).
To investigate the effect of TRΔβ3 on activity of β3, COS-7 cells were transfected with either a wild-type TRβ3 cDNA (from which both TRβ3 and TRΔβ3 are coexpressed) or a TRβ3mut cDNA (in which the in-frame AUG codon at position 103 is mutated to prevent coexpression of TRΔβ3) (30).
Coexpression of TRβ3 and TRΔβ3 proteins in cells transfected with increasing concentrations of wild-type TRβ3 cDNA in the absence of T3 resulted in reduced activity of the PAL element only after transfection of the highest concentration of plasmid (200 ng). By contrast, there was a concentration-dependent reduction in activity of MHC in the absence of T3 (Fig. 4C), and similar effects were seen with ME (not shown). These findings indicate that coexpressed TRΔβ3 protein cooperates with TRβ3 protein to repress basal gene expression in the absence of T3 and that the MHC and ME TREs were more sensitive to this effect than PAL (Fig. 4C and data not shown).
T3 responses in the absence of TRΔβ3 (cells transfected with TRβ3mut cDNA) on the MHC and ME TREs were greater than responses in the presence of TRΔβ3 (cells transfected with TRβ3 cDNA) (Fig. 4C and data not shown), indicating that low concentrations of TRΔβ3 protein also inhibit TRβ3-mediated T3 responses on these elements. By contrast, the T3 response of PAL in the absence of TRΔβ3 (cells transfected with TRβ3mut cDNA) was much lower than the response in the presence of TRΔβ3 (cells transfected with TRβ3 cDNA). Thus, transcriptional repression and activation were both impaired in the absence of TRΔβ3 protein, whereas in the presence of TRΔβ3, basal transcription was repressed but T3 responsiveness was enhanced. These data indicate that low levels of coexpressed TRΔβ3 protein potentiate T3 activation of the PAL element by the TRβ3 receptor (Fig. 4C).
TRΔβ3 exerts dominant-negative cell-,TRE-, and TR-specific actions
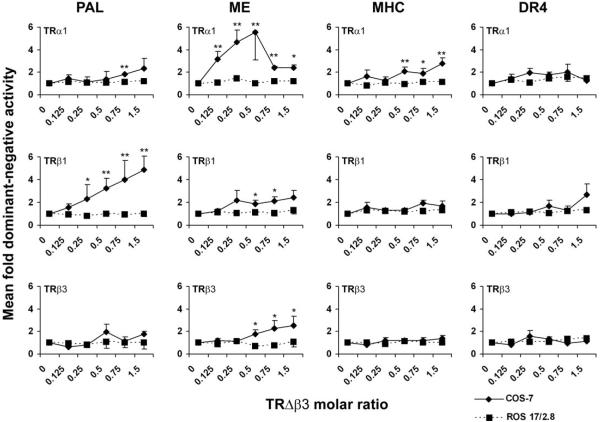
To determine whether TRΔβ3 actions were concentration dependent or whether they differed between cell types, on distinct TREs, or in the presence of different TR isoforms, COS-7 or ROS 17/2.8 cells were transfected with a PAL, ME, MHC, or DR4 reporter and an optimized concentration of TR together with increasing concentrations of TRΔβ3 (Fig. 5). In ROS 17/2.8 cells, TRΔβ3 did not influence T3 responses mediated by TRα1, -β1, or -β3 on any TRE. In COS-7 cells, TRΔβ3 repressed TRα1 activity on PAL [up to 2.3-fold (57%) repression], ME [up to 5.6-fold (82%) repression], and MHC [up to 2.8-fold (64%) repression] but not on DR4. TRΔβ3 also repressed TRβ1 on PAL [up to 5.6-fold (82%) repression] and ME [up to 2.4-fold (59%) repression] but not on MHC or DR4. TRΔβ3 also repressed TRβ3 on ME [up to 2.5-fold (61%) repression] but not on PAL, MHC, or DR4. Thus, TRΔβ3 acted as a TR isoform-specific and TRE-selective dominant-negative antagonist in COS-7 cells.
Fig. 5.
COS-7 or ROS 17/2.8 cells transfected with TRα1, -β1, or -β3 (160 ng) and a PAL, ME, MHC, or DR4 reporter together with an increasing concentration of TRΔβ3. TRΔβ3-mediated dominant-negative activity at each element in the presence of each TR isoform in both cell lines is shown. Luciferase activity was normalized to activity of a Renilla internal control vector. T3 induction ratios mediated by each TR in the absence of cotransfected TRΔβ3 were normalized to a value of 1. The mean fold dominant-negative activity was obtained by calculating the reciprocal of the T3 induction ratio in the presence of increasing concentrations of TRΔβ3 (± SEM). Values greater than 1 show the degree of repression mediated by TRΔβ3 and indicate its dominant-negative activity (n = 3–5 experiments, three to eight replicates per experiment; two-tailed paired Student's t tests: *, P < 0.05; **, P < 0.01-fold dominant-negative activity mediated by TRΔβ3 in COS-7 vs. ROS 17/2.8 cells).
The mechanism of action of TRβ3 is similar to TRβ1 in COS-7 cells
To investigate the mechanism of TRβ3 and -Δβ3 action in comparison with the known activity of TRβ1, four well-characterized TRβ1 (R243Q, S314Y, L454A, and L428R) and equivalent TRβ3 (R172Q, S243Y, L383A, and L357R) and TRΔβ3 (R70Q, S141Y, L281A, and L255R) mutants were generated. Mutant M1 (R243Q) has impaired release of NCoR, and its dominant-negative activity can be overcome by increased concentrations of T3 (36-39). M2 (S314Y) does not bind or respond to T3 and acts as a dominant-negative antagonist (40). M3 (L454A) binds T3 but fails to respond because it does not interact with coactivators (34, 41, 42). M4 (L428R) interacts poorly with RXR, fails to bind T3, and does not respond to hormone (43-45).
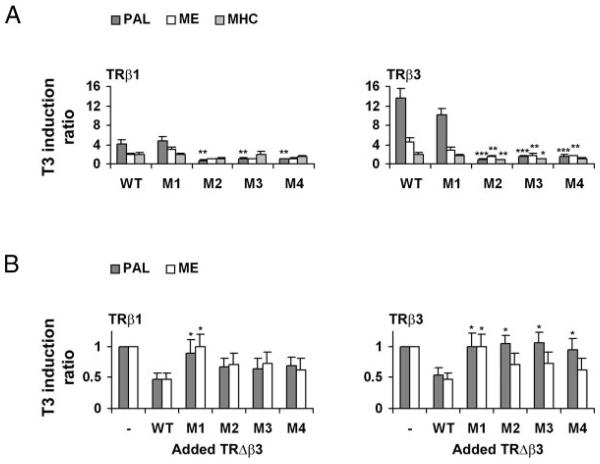
Activities of each of the TRβ1 and TRβ3 mutants were determined on PAL, ME, and MHC in COS-7 cells to compare their functional properties. There were no significant differences in the activities of TRβ1 and TRβ3 M1 mutants in response to saturating concentrations of T3 (100 nm) compared with the responses of wild-type TRβ1 and TRβ3 on each TRE (Fig. 6A). Treatment of TRβ1 or TRβ3 M1 with a lower concentration of T3 revealed impaired activities in response to 1 nm T3 on the PAL TRE (TRβ1, 2.69 ± 0.60 vs. 1.21 ± 0.16, P < 0.05; TRβ3, 2.36 ± 0.48 vs. 1.08 ± 0.18, P < 0.05; T3 induction ratio mediated by wild-type TR vs. M1 mutant in response to 1 nm T3, two-tailed unpaired Student's t tests; n = 4–7), whereas activities of wild-type β1 and β3 and M1 mutants were not different in the presence of saturating concentrations of ligand. Similar findings were obtained using the ME and MHC TREs (data not shown). The TRβ1 M1 mutant releases NCoR only in the presence of higher concentrations of T3 compared with wild-type receptor (36-39), and these data indicate the TRβ3 M1 mutant acts similarly. The TRβ1 and -β3 M2, M3, and M4 mutants failed to respond to T3 on each of the three TREs studied. All effects of the four mutations on TR activity resulted from blockade of the T3 response rather than by an effect on apoTR activity (data not shown).
Fig. 6.
A, COS-7 cells transfected with TRβ1 or TRβ3 (160 ng) or mutant TRs (β1 M1 = R243Q, M2 = S314Y, M3 = L454A, M4 = L428R; β3 M1 = R172Q, M2 = S243Y, M3 = L383A, M4 = L357R) and a ME, MHC, or PAL reporter. T3 induction of each element mediated by each receptor or mutant is shown. Luciferase activity was normalized to Renilla and results expressed as mean T3 induction ratio (± SEM), calculated by dividing normalized luciferase activities after T3 treatment by basal values (n = 5–10 experiments, three replicates per experiment; ANOVA followed by Tukey's multiple comparison post hoc tests: *, P < 0.05; **, P < 0.01; ***, P < 0.001 induction mediated by mutant TR vs. wild type). B, COS-7 cells transfected with TRβ1 or TRβ3 (160 ng) and a PAL or ME reporter in the absence or presence of 240 ng wild-type TRΔβ3 or mutant TRΔβ3 (Δβ3 M1 = R70Q, M2 = S141Y, M3 = L281A, M4 = L255R). Luciferase was normalized to Renilla, and T3 induction ratio mediated by TR in the absence of cotransfected Δβ3 was normalized to a value of 1. Responses are shown as the mean T3 induction ratio mediated by TRβ1 or TRβ3 in the absence (−) or presence of wild-type or mutant TRΔβ3 (± SEM)(n = 3–5 experiments, six to eight replicates per experiment; two-tailed paired Student's t tests: *, P < 0.05 repression mediated by TRΔβ3 mutant vs. wild-type TRΔβ3). WT, Wild type.
Differential dominant-negative inhibition of TRβ1 and TRβ3 by wild-type and mutant TRΔβ3
TRΔβ3 mutants were employed to investigate the mechanism of dominant negative activity of this TR isoform (Fig. 6B). In contrast to wild-type TRΔβ3, TRΔβ3 mutant M1 did not inhibit the activities of wild-type TRβ1 or TRβ3 on either the PAL or ME TRE. The TRΔβ3 mutants M2, M3, and M4, however, inhibited wild-type TRβ1 activity on both TREs to a similar degree as the level of inhibition mediated by wild-type TRΔβ3. In contrast, TRΔβ3 mutants M2, M3, and M4 failed to inhibit wild-type TRβ3 on PAL but inhibited its activity on ME. None of the TRΔβ3 mutants affected apoTRβ1 or apoTRβ3 activities (data not shown).
TRα1, TRβ1, TRβ3, or TRΔβ3 proteins are predominantly localized to the nucleus in the absence and presence of T3
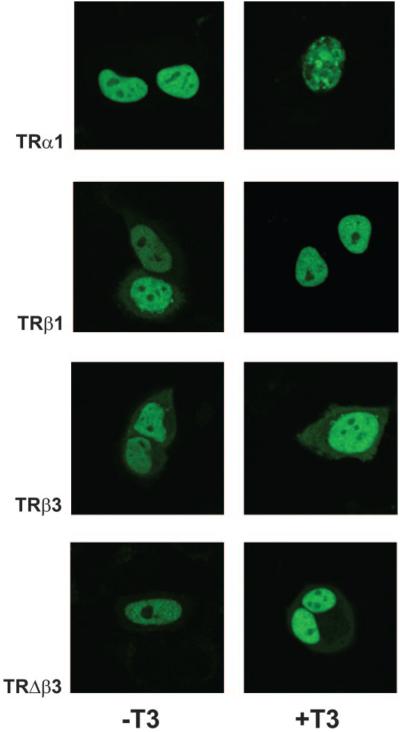
To determine whether T3 affected the subcellular localization of TRs, HeLa cells were transfected with GFP-tagged TRα1, TRβ1, TRβ3, or TRΔβ3 cDNAs in the absence or presence of T3, and localization of expressed proteins were determined by fluorescence confocal microscopy (Fig. 7). These studies revealed that TRα1 was exclusively localized in the nucleus in the absence and presence of T3. Treatment with T3, however, resulted in a change from a homogeneous nuclear distribution of TRα1 to a punctate pattern after addition of T3. Similar findings have been observed previously for TRs and other nuclear receptors, suggesting that ligand-induced intranuclear reorganization of nuclear receptors may be an important process required for hormone responsiveness (52, 53, 57-59). TRβ1, -β3, and -Δβ3 proteins were also localized predominantly in the nucleus in the absence of T3, although a small fraction of receptor was also present in the cytoplasm.
Fig. 7.
HeLa cells transfected with GFP-tagged TRα1, TRβ1, TRβ3, or TRΔβ3 constructs (100 ng/well) in the absence or presence of T3 (10 nM). Transfected cells were examined using a confocal fluorescence 3 microscope, and GFP staining reveals the effect of T3 on the cellular localization of TR isoforms.
Discussion
TRα1, TRβ1, and TRβ3 activities were highest on the PAL TRE and similar on the ME, MHC, and DR4 elements in both COS-7 and ROS 17/2.8 cells. The increased responsiveness of PAL is likely to be due to inclusion of two copies of the TRE in the reporter construct. TRβ3 mediated the greatest T3 response on PAL and was more potent than both TRα1 and TRβ1 in both cell types. The TRβ3>TRα1>TRβ1 hierarchy of potencies, however, was not evident for activation of ME, MHC, and DR4 elements. Consequently, the reduced T3 sensitivities of ME, MHC, and DR4 resulted in a failure to discriminate between the relative potencies of TRα1, TRβ1, and TRβ3 on these elements. This suggests that differences in activities of TR isoforms may only be apparent on certain genes that contain more highly responsive TREs. In this model, the sequence and arrangement of the TRE would determine T3 sensitivity of a particular target gene and provide a mechanism to account for TR isoform selectivity, whereas less responsive genes would lack such selectivity.
Analysis of apoTR- and ligand-induced TR actions also revealed differences between TR isoforms. ApoTRα1 repressed basal activity of ME in COS-7 cells but activated the same element in ROS 17/2.8 cells, whereas no differences were observed on other TREs. These divergent effects on ME expression in COS-7 and ROS 17/2.8 cells suggests the two cell types express a different repertoire of cofactors that interact with TRα1 in the absence of T3. Alternatively, the same cofactor could be modified differently in the two cell types, for example by methylation, to alter its functional properties from that of a coactivator to a corepressor (60). By contrast, the similar actions of apoTRβ1 and apoTRβ3 in both cell types suggest that cofactors interacting with TRβ in COS-7 and ROS 17/2.8 cells are not functionally distinct. Thus, apoTRα1 mediated responses that discriminate between TREs and cell types, whereas apoTRβ1 and apoTRβ3 did not. Studies of TRα/SRC-1 and TRβ/SRC-1 double-knockout mice have revealed that SRC-1 interacts differently with TRα and TRβ in the pituitary (61), with data supporting the hypothesis that a functional interaction occurs between TRα and SRC-1 in the absence of ligand, whereas in the presence of T3, SRC-1 interacts functionally with TRβ. This study indicates that TR-interacting cofactors can also discriminate between TR isoforms.
TRβ3 is coexpressed with TRΔβ3 from a single TRβ3 mRNA transcript. Deletion of the TRΔβ3 initiation codon in TRβ3mut results in expression of TRβ3 alone and allows investigation of TRΔβ3 action. Basal repression of ME and MHC was absent after transfection of TRβ3mut and the response to T3 was increased. These findings indicate that coexpressed TRΔβ3 modulates basal activity of ME and MHC in the absence of ligand and inhibits TRβ3-mediated T3 activation in the presence of ligand. TRΔβ3 lacks a DNA-binding domain but binds T3 with equal affinity to TRβ3 (30), indicating TRΔβ3 is functional in the absence and presence of T3 and its actions are independent of DNA binding. Inhibition of the activities of TRβ3 on the ME and MHC elements by TRΔβ3 is likely, therefore, to result from sequestration of cofactors that are necessary for the actions of apoand liganded TRβ3 (1-3, 5, 7). The actions of TRΔβ3 at the PAL TRE, however, were different. Absence of basal repression after transfection of TRβ3mut and a reduced response of PAL to liganded TRβ3 indicates that coexpressed TRΔβ3 also inhibits basal expression of PAL but surprisingly increases TRβ3-mediated T3 activation. This finding suggests the sequence or arrangement of PAL influences the activity of TRΔβ3 even though TRΔβ3 lacks a DNA-binding domain. Previous studies indicate that RXR/TR heterodimers adopt different conformations when bound to different TREs, and RXR/TR interactions with TREs are also modulated by cofactors (43, 62-64). Thus, RXR/TRβ3, when bound to PAL in the presence of T3, may interact with a different coactivator complex compared with when it is bound to ME or MHC. Such subtle TRE- or tissue-specific alteration of TR action (65-67) could influence the activity of TRΔβ3. In this model, interaction of TRΔβ3 with distinct coactivators would result in specific modifications that alter how coactivators interact with RXR/TRβ3/TRE complexes, thereby accounting for divergent effects of TRΔβ3 on TRβ3 function that are determined by TRE structure. Although our experiments suggest a functional role for coexpressed TRΔβ3 in modulating TRβ3 action, it is important to consider that some of the observed effects after transfection of TRβ3mut could result from increased TRβ3 expression rather than a lack of TRΔβ3. Nevertheless, in Fig. 4B, it is clear that the ratio of coexpressed TRβ3 to TRΔβ3 is very high after transfection of intact TRβ3. Thus, as in previous studies with in vitro translated TRβ3 and TRβ3mut (30), transfection of an equivalent concentration of TRβ3mut would have a negligible effect on the total amount of expressed TRβ3, supporting the view that coexpressed TRΔβ3 regulates TRβ3 activity.
Activity of TRΔβ3 was examined further by cotransfecting increasing concentrations of TRΔβ3 with TRα1, TRβ1, or TRβ3 in either COS-7 or ROS 17/2.8 cells. TRΔβ3 displayed cell type-, TRE-, and TR isoform-selective antagonist actions. TRΔβ3 was inactive in ROS 17/2.8 cells, suggesting TRΔβ3-interacting cofactors are not expressed or that ROS 17/2.8 cells express an inhibitor that prevents TRΔβ3 interaction with cofactors. Similarly, TRE- and TR isoform-selective actions of TRΔβ3 are likely due to sequestration of discrete cofactor complexes that differentially interact with RXR/TRα1, RXR/TRβ1, or RXR/TRβ3 on specific TREs. To address mechanisms underlying the complex actions of TRΔβ3, well-described TRβ1 mutants with defective NCoR interaction (36-39), T3 binding affinity (40), coactivator interaction (34, 41, 42), or RXR heterodimerization activity (43-45) were synthesized in TRΔβ3. Comparison of the activities of the TRΔβ3 mutants with the activity of wild-type TRΔβ3 on PAL and ME TREs suggests that TRΔβ3 inhibition of TRβ1 on both elements and TRβ3 on ME requires TRΔβ3 to interact fully with NCoR but does not require interactions between TRΔβ3 and T3, SRC-1, or RXR. In contrast, TRΔβ3 inhibition of TRβ3 on PAL requires TRΔβ3 to interact with T3 and all these cofactors. These results suggest further that TRE structure can determine TRΔβ3 activity indirectly via modification of cofactor interactions.
It is noteworthy that TRβ3 and TRΔβ3 have restricted patterns of expression in vivo (30). In the current studies, TRβ3 acted mainly as an activator in response to T3, whereas TRΔβ3 was mainly a repressor. Thus, tissues such as kidney and liver that express high levels of TRβ3 would be expected to be more sensitive to T3 than spleen and lung, which express mainly TRΔβ3 (30). In contrast, cerebral cortex and heart express similar levels of TRβ3 and TRΔβ3. The adult cerebral cortex is regarded as being largely unresponsive to T3, whereas heart is a classical T3 target tissue. Interestingly, TRΔβ3 expression was markedly reduced in heart from thyroidectomized rats, and TRβ3 expression was increased, whereas no effect of thyroid status on the TRβ3:Δβ3 ratio was found in cerebral cortex (30). These findings suggest that TRΔβ3 may inhibit T3 responses in cerebral cortex and heart in euthyroid animals but that down-regulation of TRΔβ3 and up-regulation of TRβ3 in cardiac muscle after thyroidectomy may account in part for increased T3 sensitivity of the hypothyroid heart (68). Changes in the relative levels of TRβ3 and TRΔβ3 in other tissues in response to alterations of thyroid status (30) may also contribute to the diverse effects of hypothyroidism and thyrotoxicosis by modulating tissue-specific interactions among TR isoforms and cofactors.
In summary, these studies demonstrate that TR actions are highly specific. Combinatorial interactions between TRα1, TRβ1, or TRβ3 and TRΔβ3 isoforms, individual TREs, and cell-specific cofactors result in enormous potential for modification of T3 responses and a high level of complexity. All the TR isoforms studied, including TRΔβ3, were mainly localized in the nucleus in the absence and presence of T3, indicating that fine tuning of T3 action is predominantly a nuclear event. Although the ratios of expressed TR isoforms and available target gene TREs in individual cell types are important determinants of T3 action, these studies also implicate a range of TR-interacting nuclear cofactors as key cell-and TRE-specific modulators of T3 action. A major challenge for the future will be to identify and characterize them.
Acknowledgments
We are grateful to Liza Jinadu for valuable technical assistance.
This work was supported by: Medical Research Council (MRC) Career Establishment Grant (G9803002) and Wellcome Trust Project Grant (50570) to G.R.W. and an MRC Clinician Scientist Fellowship to J.H.D.B.
Abbreviations
- CSS
Charcoal-stripped FCS
- DR4
direct repeat + 4
- FCS
fetal calf serum
- GFP
green fluorescent protein
- ME
malic enzyme
- MHC
α-myosin heavy chain
- NCoR
nuclear receptor corepressor
- PAL
palindromic TRE
- RTH
resistance to thyroid hormone
- RXR
retinoid X receptor
- SRC-1
steroid receptor coactivator-1
- TR
T3 receptor
- TRE
thyroid hormone response element
Footnotes
Disclosure Statement: The authors have nothing to disclose.
References
- 1.Zhang J, Lazar MA. The mechanism of action of thyroid hormones. Annu Rev Physiol. 2000;62:439–466. doi: 10.1146/annurev.physiol.62.1.439. [DOI] [PubMed] [Google Scholar]
- 2.Yen PM. Physiological and molecular basis of thyroid hormone action. Physiol Rev. 2001;81:1097–1142. doi: 10.1152/physrev.2001.81.3.1097. [DOI] [PubMed] [Google Scholar]
- 3.Harvey CB, Williams GR. Mechanism of thyroid hormone action. Thyroid. 2002;12:441–446. doi: 10.1089/105072502760143791. [DOI] [PubMed] [Google Scholar]
- 4.Williams GR, Brent GA. Thyroid hormone response elements. In: Weintraub B, editor. Molecular endocrinology: basic concepts and clinical correlations. New York: Raven Press; 1995. pp. 217–239. [Google Scholar]
- 5.Cheng SY. Multiple mechanisms for regulation of the transcriptional activity of thyroid hormone receptors. Rev Endocr Metab Disord. 2000;1:9–18. doi: 10.1023/a:1010052101214. [DOI] [PubMed] [Google Scholar]
- 6.Ito M, Roeder RG. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol Metab. 2001;12:127–134. doi: 10.1016/s1043-2760(00)00355-6. [DOI] [PubMed] [Google Scholar]
- 7.McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 8.Forrest D, Hallbook F, Persson H, Vennstrom B. Distinct functions for thyroid hormone receptors α and β in brain development indicated by differential expression of receptor genes. EMBO J. 1991;10:269–275. doi: 10.1002/j.1460-2075.1991.tb07947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forrest D, Sjoberg M, Vennstrom B. Contrasting developmental and tissue-specific expression of α and β thyroid hormone receptor genes. EMBO J. 1990;9:1519–1528. doi: 10.1002/j.1460-2075.1990.tb08270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flamant F, Samarut J. Thyroid hormone receptors: lessons from knockout and knock-in mutant mice. Trends Endocrinol Metab. 2003;14:85–90. doi: 10.1016/s1043-2760(02)00043-7. [DOI] [PubMed] [Google Scholar]
- 11.O'Shea PJ, Williams GR. Insight into the physiological actions of thyroid hormone receptors from genetically modified mice. J Endocrinol. 2002;175:553–570. doi: 10.1677/joe.0.1750553. [DOI] [PubMed] [Google Scholar]
- 12.Gauthier K, Plateroti M, Harvey CB, Williams GR, Weiss RE, Refetoff S, Willott JF, Sundin V, Roux JP, Malaval L, Hara M, Samarut J, Chassande O. Genetic analysis reveals different functions for the products of the thyroid hormone receptor α locus. Mol Cell Biol. 2001;21:4748–4760. doi: 10.1128/MCB.21.14.4748-4760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraichard A, Chassande O, Plateroti M, Roux JP, Trouillas J, Dehay C, Legrand C, Gauthier K, Kedinger M, Malaval L, Rousset B, Samarut J. The T3Rα gene encoding a thyroid hormone receptor is essential for post-natal development and thyroid hormone production. EMBO J. 1997;16:4412–4420. doi: 10.1093/emboj/16.14.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mai W, Janier MF, Allioli N, Quignodon L, Chuzel T, Flamant F, Samarut J. Thyroid hormone receptor α is a molecular switch of cardiac function between fetal and postnatal life. Proc Natl Acad Sci USA. 2004;101:10332–10337. doi: 10.1073/pnas.0401843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wikstrom L, Johansson C, Salto C, Barlow C, Campos Barros A, Baas F, Forrest D, Thoren P, Vennstrom B. Abnormal heart rate and body temperature in mice lacking thyroid hormone receptor α1. EMBO J. 1998;17:455–461. doi: 10.1093/emboj/17.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johansson C, Gothe S, Forrest D, Vennstrom B, Thoren P. Cardiovascular phenotype and temperature control in mice lacking thyroid hormone receptor-β or both α1 and β. Am J Physiol. 1999;276:H2006–H2012. doi: 10.1152/ajpheart.1999.276.6.H2006. [DOI] [PubMed] [Google Scholar]
- 17.Plateroti M, Gauthier K, Domon-Dell C, Freund JN, Samarut J, Chassande O. Functional interference between thyroid hormone receptor α (TRα) and natural truncated TRΔα isoforms in the control of intestine development. Mol Cell Biol. 2001;21:4761–4772. doi: 10.1128/MCB.21.14.4761-4772.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Shea PJ, Harvey CB, Suzuki H, Kaneshige M, Kaneshige K, Cheng SY, Williams GR. A thyrotoxic skeletal phenotype of advanced bone formation in mice with resistance to thyroid hormone. Mol Endocrinol. 2003;17:1410–1424. doi: 10.1210/me.2002-0296. [DOI] [PubMed] [Google Scholar]
- 19.O'Shea PJ, Bassett JH, Sriskantharajah S, Ying H, Cheng SY, Williams GR. Contrasting skeletal phenotypes in mice with an identical mutation targeted to thyroid hormone receptor α1 or β. Mol Endocrinol. 2005;19:3045–3059. doi: 10.1210/me.2005-0224. [DOI] [PubMed] [Google Scholar]
- 20.Gullberg H, Rudling M, Forrest D, Angelin B, Vennstrom B. Thyroid hormone receptor β-deficient mice show complete loss of the normal cholesterol 7α-hydroxylase (CYP7A) response to thyroid hormone but display enhanced resistance to dietary cholesterol. Mol Endocrinol. 2000;14:1739–1749. doi: 10.1210/mend.14.11.0548. [DOI] [PubMed] [Google Scholar]
- 21.Gauthier K, Chassande O, Plateroti M, Roux JP, Legrand C, Pain B, Rousset B, Weiss R, Trouillas J, Samarut J. Different functions for the thyroid hormone receptors TRα and TRβ in the control of thyroid hormone production and post-natal development. EMBO J. 1999;18:623–631. doi: 10.1093/emboj/18.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forrest D, Hanebuth E, Smeyne RJ, Everds N, Stewart CL, Wehner JM, Curran T. Recessive resistance to thyroid hormone in mice lacking thyroid hormone receptor β: evidence for tissue-specific modulation of receptor function. EMBO J. 1996;15:3006–3015. [PMC free article] [PubMed] [Google Scholar]
- 23.Sjoberg M, Vennstrom B, Forrest D. Thyroid hormone receptors in chick retinal development: differential expression of mRNAs for α and N-terminal variant β receptors. Development. 1992;114:39–47. doi: 10.1242/dev.114.1.39. [DOI] [PubMed] [Google Scholar]
- 24.Hodin RA, Lazar MA, Chin WW. Differential and tissue-specific regulation of the multiple rat c-erbA messenger RNA species by thyroid hormone. J Clin Invest. 1990;85:101–105. doi: 10.1172/JCI114398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradley DJ, Towle HC, Young WS., 3rd α and β thyroid hormone receptor (TR) gene expression during auditory neurogenesis: evidence for TR isoform-specific transcriptional regulation in vivo. Proc Natl Acad Sci USA. 1994;91:439–443. doi: 10.1073/pnas.91.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rusch A, Erway LC, Oliver D, Vennstrom B, Forrest D. Thyroid hormone receptor β-dependent expression of a potassium conductance in inner hair cells at the onset of hearing. Proc Natl Acad Sci USA. 1998;95:15758–15762. doi: 10.1073/pnas.95.26.15758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abel ED, Boers ME, Pazos-Moura C, Moura E, Kaulbach H, Zakaria M, Lowell B, Radovick S, Liberman MC, Wondisford F. Divergent roles for thyroid hormone receptor β isoforms in the endocrine axis and auditory system. J Clin Invest. 1999;104:291–300. doi: 10.1172/JCI6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng L, Hurley JB, Dierks B, Srinivas M, Salto C, Vennstrom B, Reh TA, Forrest D. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet. 2001;27:94–98. doi: 10.1038/83829. [DOI] [PubMed] [Google Scholar]
- 29.Ng L, Rusch A, Amma LL, Nordstrom K, Erway LC, Vennstrom B, Forrest D. Suppression of the deafness and thyroid dysfunction in Thrb-null mice by an independent mutation in the Thra thyroid hormone receptor α gene. Hum Mol Genet. 2001;10:2701–2708. doi: 10.1093/hmg/10.23.2701. [DOI] [PubMed] [Google Scholar]
- 30.Williams GR. Cloning and characterization of two novel thyroid hormone receptor β isoforms. Mol Cell Biol. 2000;20:8329–8342. doi: 10.1128/mcb.20.22.8329-8342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss RE, Refetoff S. Resistance to thyroid hormone. Rev Endocr Metab Disord. 2000;1:97–108. doi: 10.1023/a:1010072605757. [DOI] [PubMed] [Google Scholar]
- 32.Yen PM. Molecular basis of resistance to thyroid hormone. Trends Endocrinol Metab. 2003;14:327–333. doi: 10.1016/s1043-2760(03)00114-0. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Takeshita A, Misiti S, Chin WW, Yen PM. Lack of coactivator interaction can be a mechanism for dominant negative activity by mutant thyroid hormone receptors. Endocrinology. 1998;139:4197–4204. doi: 10.1210/endo.139.10.6218. [DOI] [PubMed] [Google Scholar]
- 34.Collingwood TN, Rajanayagam O, Adams M, Wagner R, Cavailles V, Kalkhoven E, Matthews C, Nystrom E, Stenlof K, Lindstedt G, Tisell L, Fletterick RJ, Parker MG, Chatterjee VK. A natural transactivation mutation in the thyroid hormone β receptor: impaired interaction with putative transcriptional mediators. Proc Natl Acad Sci USA. 1997;94:248–253. doi: 10.1073/pnas.94.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoh SM, Chatterjee VK, Privalsky ML. Thyroid hormone resistance syndrome manifests as an aberrant interaction between mutant T3 receptors and transcriptional corepressors. Mol Endocrinol. 1997;11:470–480. doi: 10.1210/mend.11.4.9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Safer JD, Cohen RN, Hollenberg AN, Wondisford FE. Defective release of corepressor by hinge mutants of the thyroid hormone receptor found in patients with resistance to thyroid hormone. J Biol Chem. 1998;273:30175–30182. doi: 10.1074/jbc.273.46.30175. [DOI] [PubMed] [Google Scholar]
- 37.Yagi H, Pohlenz J, Hayashi Y, Sakurai A, Refetoff S. Resistance to thyroid hormone caused by two mutant thyroid hormone receptors β, R243Q and R243W, with marked impairment of function that cannot be explained by altered in vitro 3,5,3′-triiodothyroinine binding affinity. J Clin Endocrinol Metab. 1997;82:1608–1614. doi: 10.1210/jcem.82.5.3945. [DOI] [PubMed] [Google Scholar]
- 38.Huber BR, Desclozeaux M, West BL, Cunha-Lima ST, Nguyen HT, Baxter JD, Ingraham HA, Fletterick RJ. Thyroid hormone receptor-β mutations conferring hormone resistance and reduced corepressor release exhibit decreased stability in the N-terminal ligand-binding domain. Mol Endocrinol. 2003;17:107–116. doi: 10.1210/me.2002-0097. [DOI] [PubMed] [Google Scholar]
- 39.Collingwood TN, Wagner R, Matthews CH, Clifton-Bligh RJ, Gurnell M, Rajanayagam O, Agostini M, Fletterick RJ, Beck-Peccoz P, Reinhardt W, Binder G, Ranke MB, Hermus A, Hesch RD, Lazarus J, Newrick P, Parfitt V, Raggatt P, de Zegher F, Chatterjee VK. A role for helix 3 of the TRβ ligand-binding domain in coactivator recruitment identified by characterization of a third cluster of mutations in resistance to thyroid hormone. EMBO J. 1998;17:4760–4770. doi: 10.1093/emboj/17.16.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gurnell M, Rajanayagam O, Agostini M, Clifton-Bligh RJ, Wang T, Zelissen PM, van der Horst F, van de Wiel A, Macchia E, Pinchera A, Schwabe JW, Chatterjee VK. Three novel mutations at serine 314 in the thyroid hormone β receptor differentially impair ligand binding in the syndrome of resistance to thyroid hormone. Endocrinology. 1999;140:5901–5906. doi: 10.1210/endo.140.12.7203. [DOI] [PubMed] [Google Scholar]
- 41.Collingwood TN, Butler A, Tone Y, Clifton-Bligh RJ, Parker MG, Chatterjee VK. Thyroid hormone-mediated enhancement of heterodimer formation between thyroid hormone receptor β and retinoid X receptor. J Biol Chem. 1997;272:13060–13065. doi: 10.1074/jbc.272.20.13060. [DOI] [PubMed] [Google Scholar]
- 42.Tone Y, Collingwood TN, Adams M, Chatterjee VK. Functional analysis of a transactivation domain in the thyroid hormone β receptor. J Biol Chem. 1994;269:31157–31161. [PubMed] [Google Scholar]
- 43.Zavacki AM, Harney JW, Brent GA, Larsen PR. Structural features of thyroid hormone response elements that increase susceptibility to inhibition by an RTH mutant thyroid hormone receptor. Endocrinology. 1996;137:2833–2841. doi: 10.1210/endo.137.7.8770904. [DOI] [PubMed] [Google Scholar]
- 44.Nagaya T, Jameson JL. Thyroid hormone receptor dimerization is required for dominant negative inhibition by mutations that cause thyroid hormone resistance. J Biol Chem. 1993;268:15766–15771. [PubMed] [Google Scholar]
- 45.Monden T, Yamada M, Ishii S, Hosoya T, Satoh T, Wondisford FE, Hollenberg AN, Mori M. Leucine at codon 428 in the ninth heptad of thyroid hormone receptor β1 is necessary for interactions with the transcriptional cofactors and functions regardless of dimer formations. Thyroid. 2003;13:427–435. doi: 10.1089/105072503322021089. [DOI] [PubMed] [Google Scholar]
- 46.Samuels HH, Stanley F, Casanova J. Depletion of l-3,5,3′-triiodothyronine and l-thyroxine in euthyroid calf serum for use in cell culture studies of the action of thyroid hormone. Endocrinology. 1979;105:80–85. doi: 10.1210/endo-105-1-80. [DOI] [PubMed] [Google Scholar]
- 47.Williams GR, Bland R, Sheppard MC. Characterization of thyroid hormone (T3) receptors in three osteosarcoma cell lines of distinct osteoblast phenotype: interactions among T3, vitamin D3, and retinoid signaling. Endocrinology. 1994;135:2375–2385. doi: 10.1210/endo.135.6.7988420. [DOI] [PubMed] [Google Scholar]
- 48.Tagami T, Kopp P, Johnson W, Arseven OK, Jameson JL. The thyroid hormone receptor variant α2 is a weak antagonist because it is deficient in interactions with nuclear receptor corepressors. Endocrinology. 1998;139:2535–2544. doi: 10.1210/endo.139.5.6011. [DOI] [PubMed] [Google Scholar]
- 49.Koenig RJ, Warne RL, Brent GA, Harney JW, Larsen PR, Moore DD. Isolation of a cDNA clone encoding a biologically active thyroid hormone receptor. Proc Natl Acad Sci USA. 1988;85:5031–5035. doi: 10.1073/pnas.85.14.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prost E, Koenig RJ, Moore DD, Larsen PR, Whalen RG. Multiple sequences encoding potential thyroid hormone receptors isolated from mouse skeletal muscle cDNA libraries. Nucleic Acids Res. 1988;16:6248. doi: 10.1093/nar/16.13.6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rentoumis A, Chatterjee VK, Madison LD, Datta S, Gallagher GD, Degroot LJ, Jameson JL. Negative and positive transcriptional regulation by thyroid hormone receptor isoforms. Mol Endocrinol. 1990;4:1522–1531. doi: 10.1210/mend-4-10-1522. [DOI] [PubMed] [Google Scholar]
- 52.Maruvada P, Baumann CT, Hager GL, Yen PM. Dynamic shuttling and intranuclear mobility of nuclear hormone receptors. J Biol Chem. 2003;278:12425–12432. doi: 10.1074/jbc.M202752200. [DOI] [PubMed] [Google Scholar]
- 53.Baumann CT, Maruvada P, Hager GL, Yen PM. Nuclear cytoplasmic shuttling by thyroid hormone receptors. Multiple protein interactions are required for nuclear retention. J Biol Chem. 2001;276:11237–11245. doi: 10.1074/jbc.M011112200. [DOI] [PubMed] [Google Scholar]
- 54.Yaoita Y, Shi YB, Brown DD. Xenopus laevis α and β thyroid hormone receptors. Proc Natl Acad Sci USA. 1990;87:7090–7094. doi: 10.1073/pnas.87.18.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stenoien DL, Nye AC, Mancini MG, Patel K, Dutertre M, O'Malley BW, Smith CL, Belmont AS, Mancini MA. Ligand-mediated assembly and real-time cellular dynamics of estrogen receptor α-coactivator complexes in living cells. Mol Cell Biol. 2001;21:4404–4412. doi: 10.1128/MCB.21.13.4404-4412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Htun H, Holth LT, Walker D, Davie JR, Hager GL. Direct visualization of the human estrogen receptor α reveals a role for ligand in the nuclear distribution of the receptor. Mol Biol Cell. 1999;10:471–486. doi: 10.1091/mbc.10.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baumann CT, Ma H, Wolford R, Reyes JC, Maruvada P, Lim C, Yen PM, Stallcup MR, Hager GL. The glucocorticoid receptor interacting protein 1 (GRIP1) localizes in discrete nuclear foci that associate with ND10 bodies and are enriched in components of the 26S proteasome. Mol Endocrinol. 2001;15:485–500. doi: 10.1210/mend.15.4.0618. [DOI] [PubMed] [Google Scholar]
- 60.Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM. A transcriptional switch mediated by cofactor methylation. Science. 2001;294:2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- 61.Sadow PM, Koo E, Chassande O, Gauthier K, Samarut J, Xu J, O'Malley BW, Seo H, Murata Y, Weiss RE. Thyroid hormone receptor-specific interactions with steroid receptor coactivator-1 in the pituitary. Mol Endocrinol. 2003;17:882–894. doi: 10.1210/me.2002-0174. [DOI] [PubMed] [Google Scholar]
- 62.Wu Y, Xu B, Koenig RJ. Thyroid hormone response element sequence and the recruitment of retinoid X receptors for thyroid hormone responsiveness. J Biol Chem. 2001;276:3929–3936. doi: 10.1074/jbc.M006743200. [DOI] [PubMed] [Google Scholar]
- 63.Ikeda M, Wilcox EC, Chin WW. Different DNA elements can modulate the conformation of thyroid hormone receptor heterodimer and its transcriptional activity. J Biol Chem. 1996;271:23096–23104. doi: 10.1074/jbc.271.38.23096. [DOI] [PubMed] [Google Scholar]
- 64.Gloss B, Giannocco G, Swanson EA, Moriscot AS, Chiellini G, Scanlan T, Baxter JD, Dillmann WH. Different configurations of specific thyroid hormone response elements mediate opposite effects of thyroid hormone and GC-1 on gene expression. Endocrinology. 2005;146:4926–4933. doi: 10.1210/en.2005-0631. [DOI] [PubMed] [Google Scholar]
- 65.Takeshita A, Yen PM, Ikeda M, Cardona GR, Liu Y, Koibuchi N, Norwitz ER, Chin WW. Thyroid hormone response elements differentially modulate the interactions of thyroid hormone receptors with two receptor binding domains in the steroid receptor coactivator-1. J Biol Chem. 1998;273:21554–21562. doi: 10.1074/jbc.273.34.21554. [DOI] [PubMed] [Google Scholar]
- 66.Liu Y, Xia X, Fondell JD, Yen PM. Thyroid hormone-regulated target genes have distinct patterns of coactivator recruitment and histone acetylation. Mol Endocrinol. 2006;20:483–490. doi: 10.1210/me.2005-0101. [DOI] [PubMed] [Google Scholar]
- 67.Paul BD, Buchholz DR, Fu L, Shi YB. Tissue- and gene-specific recruitment of steroid receptor coactivator-3 by thyroid hormone receptor during development. J Biol Chem. 2005;280:27165–27172. doi: 10.1074/jbc.M503999200. [DOI] [PubMed] [Google Scholar]
- 68.Dillmann WH. Cellular action of thyroid hormone on the heart. Thyroid. 2002;12:447–452. doi: 10.1089/105072502760143809. [DOI] [PubMed] [Google Scholar]