Abstract
Type III secretion systems (T3SSs) mediate bacterial protein translocation into eukaryotic cells, a process essential for virulence of many Gram-negative pathogens. They are composed of a cytoplasmic secretion machinery and a base bridging both bacterial membranes into which a hollow, external needle is embedded. When isolated, the latter two parts are termed ‘needle complex’ (NC). Incomplete understanding of NC structure hampers studies of T3SS function. To estimate the stoichiometry of its components, the mass f its sub-domains was measured by scanning transmission electron microscopy (STEM). Subunit symmetries were determined by analysis of top and side views within negatively stained samples in low dose transmission electron microscopy (TEM). Application of 12-fold symmetry allowed generation of a 21-25Å resolution three-dimensional (3D) reconstruction of the NC base, revealing many new features and permitting tentative docking of the crystal structure of EscJ, an inner membrane component.
T3SS are found in many Gram-negative bacteria, forming molecular injection devices to deliver bacterial effector proteins into target eukaryotic host cells during infection. Understanding their 3D structure is important for design of new broad-spectrum strategies to combat bacterial infection. These macromolecular assemblies are made of ~25 proteins. They are genetically and morphologically related to bacterial flagellar basal bodies (FBBs). Hence, functional parallels with that system have been drawn4. However, FBBs are constitutively active in sequential secretion of axial flagellar proteins while T3SS activation for effector secretion requires direct physical contact with host cells.
Shigella flexneri is the causative agent of human bacillary dysentery5. Shigella uses a T3SS for invasion of, and dissemination within, the gut epithelial lining6. In situ, its T3SS is made up of three major parts7: a cytoplasmic region or “bulb”, a region spanning both inner and outer bacterial membranes and a hollow, largely extracellular “needle”. The cytoplasmic bulb may house soluble T3SS components mediating effector export, while the transmembrane region creates a chamber containing an export channel that connects to the periplasmic end of the needle. The effectors probably travel through the needle channel9 and pass into the host cell via a translocation pore assembled from the tip of the needle into the host membrane10.
The T3SS portion corresponding to the transmembrane and extracellular parts was first purified from Salmonella typhimurium and visualized using electron microscopy (EM) by Kubori et al., who termed it ‘NC’11. Blocker et al. then produced a rotationally averaged 17Å 3D reconstruction of Shigella NCs from negatively stained molecules12. This consists of two sets of rings of differing diameters, with the smaller diameter set positioned at the outer membrane (OM) and the larger one in the inner membrane (IM). The needle protrudes from the OM rings’ centre. A 3D reconstruction of the Shigella needle alone13 showed that it is a helical polymer 70Å in diameter, containing an internal, ~30Å-diameter channel. A pseudoatomic model of the Shigella needle was produced by docking a crystal structure of its subunit, MxiH, to this 16Å EM map14. A complex of effector proteins located at the distal needle tip senses the host cell and the needle is involved in transduction of the activation signal10,14-17. However, if and how the signal travels along the needle remains unknown15,18, as is the means by which it activates the cytoplasmic portion of the apparatus for secretion. To understand this, higher resolution structural information for the NC is required. For this, a key step is determination of the stoichiometry and symmetry of the complex.
Four major components of the NC transmembrane region were identified11,12,19,20. In Shigella, MxiG and MxiJ are located in the inner membrane region (IMR21,22), where they may form a concentric ring structure19,23. The largely periplasmic MxiJ protein, carrying a single C-terminal transmembrane helix (TMH) and lipidated at its N-terminus by an unusual lipid inserted into the IM22-24, is likely located at the centre of the ring and surrounded by MxiG23. The only transmembrane region-associated NC protein for which an atomic structure is available is EscJ, the enteropathogenic E. coli (EPEC) homolog of MxiJ. It crystallized as a superhelix with 24 molecules/turn23. MxiG is predicted to have a single TMH with a small cytoplasmic domain and larger periplasmic one. MxiD is predicted to form a β-barrel “secretin”-type ring constituting most of the outer membrane region mass (OMR2,25). Minor NC components are also known: the periplasmic portion of the needle is likely built up by the sequence-related MxiI26; MxiM, a small OM lipoprotein required for efficient secretin assembly, was localized to the outside of the MxiD ring within mature NCs27,28; in affinity-purified NC preparations, Spa40 and Spa24, two of five IM proteins thought to form the T3SS core inner membrane export apparatus (CIMEA) were found28. A ‘socket/cup’ encapsulating the needle periplasmic base was identified26, equating to the ‘bulge/plate” which Blocker et al. proposed contained the CIMEA12.
NC component stoichiometry was estimated as 120 MxiH subunits, 5-10 MxiI26 subunits and 19-22 MxiG, MxiJ23 and MxiD26 subunits. By quantitative bulk amino acid analysis, Marlovits et al.26 determined a 1:1:1 molar ratio for InvG (MxiD, 62kDa), PrgH (MxiG, 43kDa) and PrgK (MxiJ, 25kDa), which would correspond to an ~1:1 OMR to IMR mass ratio. However, Yip et al.23, using radioactive labeling, found that the relative ratio of these proteins was 1:1.3:1.7. Thus, major NC components stoichiometry remains unclear. Indeed, NCs breakage during isolation, particularly within the IMR, could to be leading to inaccurate bulk measurements.
Marlovits et al.26 produced 3D reconstructions of S. typhimurium NCs from cryo-electron microscopy (cryoEM) data, applying symmetry to resolve the NC ring systems further. From side view analysis they report heterogeneity in terms of rottional symmetries, with 20-fold symmetric NCs being most abundant. Their 17Å reconstruction shows 20-fold modulations in the IMR but features of other sub-domains, including the OMR, are not resolved. Other work indicates that secretins, to which family T3SS OMR proteins belong, exist as 12-14 mers29.
Lack of a well-resolved NC 3D map containing atomic structures of sub-components hampers studies of NC structure-function relationships. Therefore, we reanalysed the stoichiometry of NC base proteins, using STEM to visually select and measure the mass of only intact NCs. We then produced a newly symmetrized reconstruction of the whole NC transmembrane region (base) from TEM images of negatively stained samples (Fig. 1).
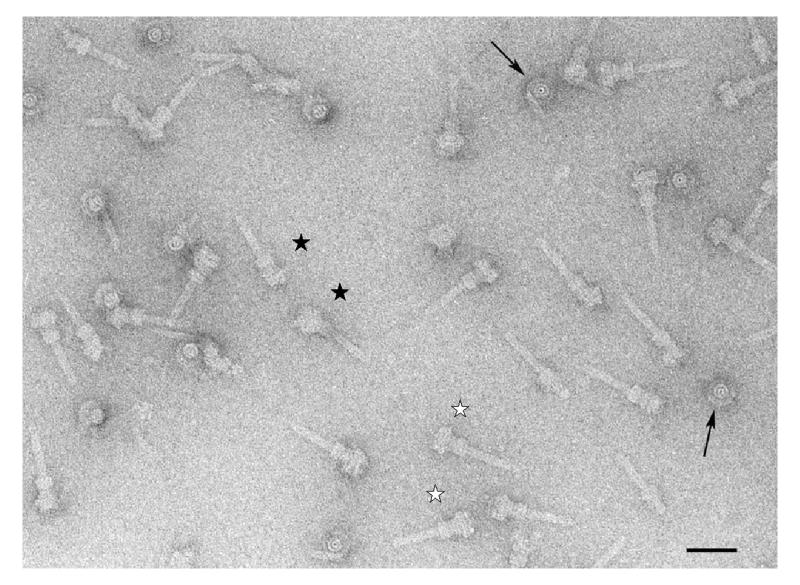
Figure 1.
S. flexneri needle complexes. Electron micrograph of particles negatively stained with 2% w/v uranyl acetate. Examples of well-preserved particles are indicated (white stars). Note top views of base without needles (arrowed). Clearly damaged or tilted particles (black star) were excluded from the analysis. Scale bar represents 500Å.
Results
By STEM, the OMR to IMR mass ratio is 1:2
We collected STEM data from 76 NC fields, from which we selected 340 intact side views (Fig. 2). The ratio between OMR (~1MDa) and IMR (~2MDa) mass within the base was consistently 1:2 (Table 1), with standard deviations <10% (~100kDa). We used these values to calculate a first approximation of NC component symmetries. Given the order of magnitude of standard deviations and assuming MxiD is the major OMR component, while MxiJ and MxiG are the major IMR components and share identical symmetries, these mass values are consistent with subunit numbers of the 8-14 for the OMR and of 20-26 of the IMR.
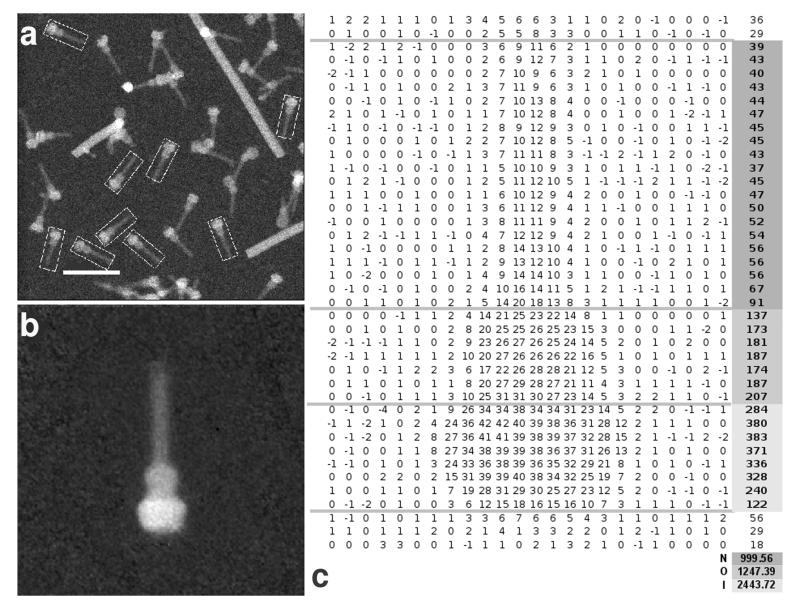
Figure 2.
STEM data collection and analysis. (a) Typical NC field used to collect data with picked particles (boxed). Long filaments are TMV used as internal calibration standard. (b) Average of boxed particles. c. Average mass map with background subtracted for picked particles in the example field shown (mass in kDa for each 2nm ]× 2nm area). Right hand column indicates mass sums (in kDa) for the horizontal slice of the mass map immediately to its left. Masses highlighted in dark grey, mid-grey, light grey, were assigned to needle, OMR and IMR respectively. The integrated mass sum of regions assigned to each component is shown at the bottom of the columns in assigned colour. Horizontal grey lines on mass map delineate range of slices summed. Scale bar is 100μm.
Table 1. Summary of STEM analysis.
| Measurement (kDa) | Base | IMR | OMR |
|---|---|---|---|
| Normalized average | 4268 | 2787 | 1472 |
| Background corrected averagea) |
3158 | 2062 | 1089 |
| Background corrected Standard deviationb) |
161 | 152 | 101 |
Pooled average masses for 2 data sets collected are shown.
Data shown are normalized between fields
corrected for background, using needle mass/length as a standard.
Top views suggest the outer IMR has 24-fold symmetry
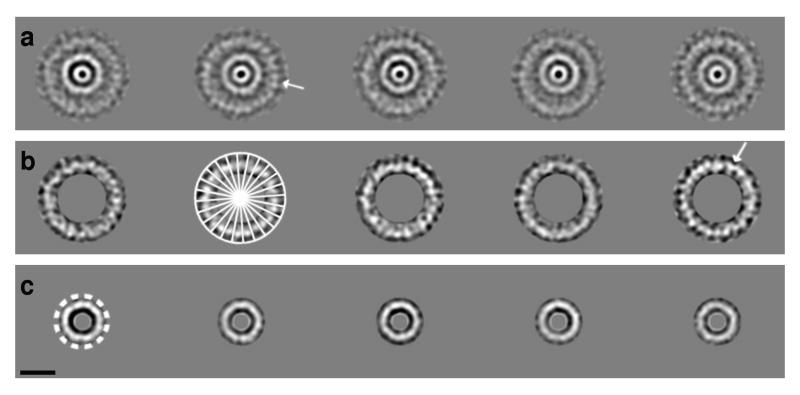
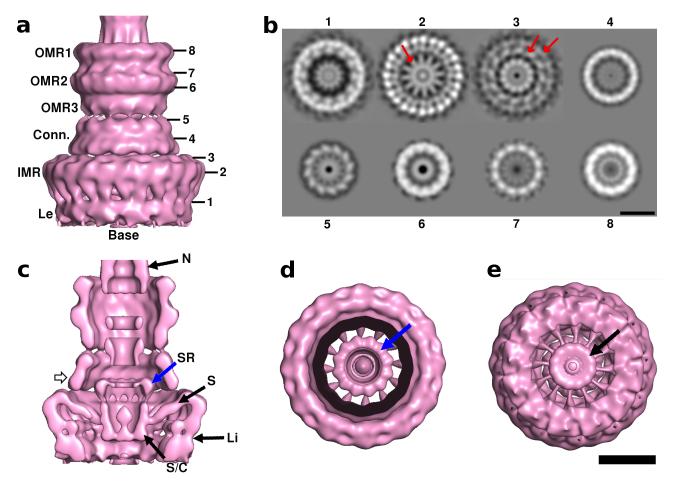
After alignment and classification according to IMR features (see Methods for details), several TEM top view classes showed a density modulation consistent with 24 apparent domains at an IMR diameter of 235Å (Fig. 3a, b). Each apparent domain measured 31Å. Further investigation of symmetry was performed using the RotaStat suite30 on the above 44 top views (Supplementary Fig. 1). This analysis revealed two broad peaks centred around 24-fold and 12-fold symmetry (Supplementary Fig. 2). Only 24-fold symmetry showed statistical significance (p<0.0001).
Figure 3.
Image analysis of top views of the NC base. (a) Class averages containing 14, 21, 14, 19 and 12 images respectively. (b) Class averages of IMR (shown masked with the ring mask used for MSA for display purposes) show 24 subunits. Class 2 has a 24 fold overlay to illustrate this. These subunits often appear paired (arrow in a and b). (c) OMR only class averages (masked using MSA mask for display), although often 12 sub-units can be seen, indicated by dashed ring on image 1, these are more poorly resolved than for the IMR. Scale bar is 100Å.
OMR image analysis using the same methodology (Fig. 3c) delineated the outer OMR diameter at ~137Å. Most commonly 12 subunits were discernible in classes and images (Fig. 3c, image 1; Supplementary Fig. 1). RotaStat suggested symmetries in the range 8-14 in individual OMR images, but failed to find statistically significant symmetries (not shown). Superimposition of multiple similar sized OMR ring components in projection prevents clear results. However, a survey of the literature on related structures, supports 12-fold symmetry (Table 2).
Table 2. Summary of our and published studies of symmetry/diameter relationships in proteins belonging to the secretin family.
| Biological system | Technique used |
Outer diameter |
Inner diameter |
Subunit diameter |
Symmetry | Reference |
|---|---|---|---|---|---|---|
|
S. flexneri T3SS OMR |
Negative stain TEM |
137 | 81 | 25 | C12 | This study |
| K. oxytoca T2SS | Rotary | |||||
| PulD trypsineda) | shadowing | 130 | 86 | 25 | C12 | 1 |
| Pul-PulS complexb) | CryoEM | 137 | 80 | 20 | ||
|
Y. enterocolitica T3SS YscC |
STEM | 155 | 80 | NM | C13 | 2 |
|
E. coli filamentous phage |
Negative stain TEM |
135 | 87 | NM | C14/D14 | 3 |
| pIV multimer | CryoEM | 137 | 65 | NM | ||
|
N. meningitidis T4SS PilQ |
Negative stain TEM |
165 | 65 | NM | C12 | 8 |
All numbers are in Å.
indicates PulD without the PulS domain.
refers to native PulD-PulS complex, where the external diameter was measured without the external spokes. NM indicates where a parameter was not measurable.
Side view classification produces two NC groups
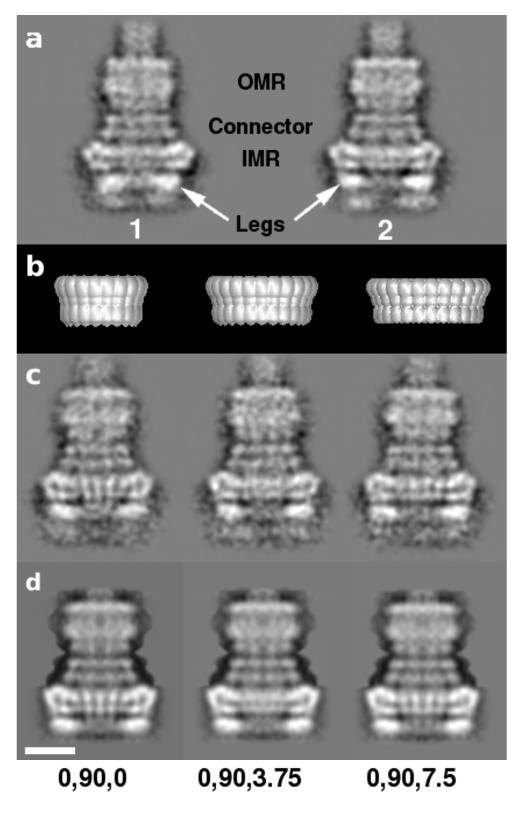
After removal of tilted or distorted particles (Fig. 1), classification of best-preserved NC side views by multivariate statistical analysis (MSA) showed 2 broad groups owing to flexibility in the lower (‘leg’) part of the IMR. We termed these states ‘closed’ and ‘open’ legs (Fig. 4a, class 1 and 2, respectively). As >2/3 of the particles belonged to the open group, it was chosen for subsequent analyses.
Figure 4.
Classification of NC side views. (a) Classification according to the IMR shows two sub-populations defined by the position of the “leg” portion (arrowed): closed legs, class 1 and open legs, class 2. (b) Surface representation of IMR models (C20, 24, 26 symmetries shown) used to check for symmetry variation in side views using composite models (see Supplementary Methods). (c) Selected class averages used for first C24 symmetry 3D reconstruction. Euler angles assigned to each class are indicated at the base of the figure. (d) Reprojections from the C24 3D reconstruction compare well with input class averages in c. Weak densities seen at extreme base end in class averages were highly disordered and did not reconstruct well and were therefore masked from final 3D reconstructions. Scale bar is 100Å.
Side view analysis does not support symmetry heterogeneity
Top view analysis suggested that a substantial proportion of the IMRs in S. flexneri have 24-fold symmetry (as supported by top view class average analysis of others27). However, studies of related systems26,31 suggest existence of sub-populations with differing symmetries. To investigate this, we checked NC side view images for symmetry variation using composite IMR models created with C20-26 symmetry (see Supplementary Methods). We used C20-26 3D models in side and slightly oblique projections as references to sort the data into groups according to any remaining out of plane tilt about the short NC axis (estimated at +/−10°) and diametric/symmetric variation. We subjected the side view images to multi-reference alignment (in SPIDER) and sorted them into groups by correlation with the IMAGIC5 projection-matching module using the IMR models (Fig. 4b) as references. We discarded any particle, which was misaligned or matched to an oblique reference tilted >3°.
Of the remaining particles, 330 (~75%) correlated best with the C24 reference. These were further partitioned by multi-reference alignment using references created by projection of the C24 model over a range of angles corresponding to rotation about its central axis (Euler angle γ). This produced classes with good internal detail in the IMR. We used three such classes associated with distinctly different orientations (57 particles, Fig. 4c) to produce a C24 3D reconstruction. This reconstruction was then projected out along the Euler angles assigned for each class. There was good correlation between the input IMR classes and the output 2D reprojections of the 3D reconstruction (Fig. 4c, d). The C24 reconstruction was then projected over the C24 asymmetric unit in γ in 1° increments and an expected range of ß angles for a negative stain dataset on a support film (70-110°) to produce a first reference set for further multi-reference alignment and angular assignment by projection matching of the 330 raw images.
3D reconstruction refinement reveals C12 symmetry
Although alignment and classification of images above was only according to the IMR (24-fold symmetric) part of the map, the structure’s OMR and connector part were also resolved in class averages, suggesting a common sub-symmetry for the whole complex. This sub-symmetry had to be even, since side view classes (Fig. 4a) are mirror symmetric. Given our STEM and top view analysis, and available information on OMR-related structures (Table 2), this symmetry was likely to be C12. This was checked by alignment with a range of OMR composite models (C8-C12; Supplementary Fig. 3) created and used as the IMR models described above. Over 50% of images from the 330 C24 image group correlated best with the C12 model (data not shown). Those that did not were obviously misaligned with respect to references and hence discarded.
To evaluate C12 symmetry applicability to the whole base, we reconstructed the C24 group of particles with C12 symmetry. Two separate analyses of the data in C12 (Fig. 5) and C24 (Supplementary Fig. 4) symmetries were conducted, with each refined in parallel over 4 cycles of multi-reference alignment, projection matching, reconstruction and forward projection. Only with C12 symmetry did the whole NC base structure refine to produce strong density modulations throughout the entire base. The final 3D reconstruction contained 41 particles selected on image quality, agreement with reprojections and distribution of angles. The strong signal-to-noise ratio of raw images enabled use of single images rather than classes for angular assignment and 3D reconstruction. Resolution of the final reconstruction was estimated by Fourier Shell Correlation (FSC) to be 21-25Å (Supplementary Fig. 5). We thus limited the resolution of the final 3D map to 25Å by Fourier filtration.
Figure 5.
3D reconstruction of NCs with C12 symmetry. (a) Surface representation. Labels on left hand side define structural regions in the map and numbers 1-8 on right hand side define points at which slices were taken for 5b. Conn.=connector, Le= legs. The bacterial (cytoplasmic) side of the NC, is defined as the base of the complex. (b) Slices through the 3D reconstructions at 1 (linker), 2 (IMR-shoulder), 3 (Shoulder-connector transition), 4 (Connector), 5 (Connector-OMR3 transition), 6 (OMR2), 7 (OMR2-1 transition) and 8 (OMR1). Arrow in b2 shows radial density spokes extending inward from between each pair of outer IMR sub-units. Slice 3 shows the top of IMR (large radius ring indicated) and start of connector (smaller radius ring indicated). Slices 3 and 5 show azimuthal tilts of connector sub-units. 12-fold symmetry is seen throughout the OMR rings (slices 6-8). More slices are shown in Supplementary Fig. 6. (c) Longitudinal cutaway of map reveals details of internal components with a distinct “socket-ring” (SR, blue arrow) with strong density modulations and 12 fold connections to the “socket-cup” (S/C) directly below it. Twelve spokes connect the socket-cup to the IMR (S). Two slender linkers (Li) connect the legs to the underside of the IMR shoulder. The top of the connector shows 12-fold connectivity with the underside of OMR3. N=needle. Hollow arrow on left side of connector indicates point at which structure cut for view in 5d (where black ring is cut-through volume). (d) Top view surface cut back to the socket-ring shows its 12-fold symmetry and domains with azimuthal tilt (blue arrow). The spokes connecting the outer IMR to the socket/cup are seen below this ring. (e) View of the NC base from the cytoplasmic side showing 12-fold symmetry of socket cup (black arrow). The 12-fold spokes display azimuthal tilting. Scale bar for a, c, d and e is 100Å. Scale bar for b is 100Å.
The C12 3D reconstruction reveals many new features
We divided the C12 map into structural sub-domains for examination (Fig. 5). At the cytoplasmic side, leg-like densities are connected to the main IMR ring by linker densities. A skirt-like connector region connects the IMR to three OMR rings. The total height of the NC base is 300Å (OMR height is 115Å, connector height is 55Å and IMR height including legs is 112Å). The volume of each major portion of the C12 map is given in Table 3.
Table 3. Volume-mass conversion for subparts of the C12 map.
| Map subpart | Measured volume (Å3)a | Mass conversion (kDa) |
|---|---|---|
| OMR123+connector | 1550000 | 720 |
| OMR123 alone | 1050000 | 490 |
| Connector alone | 500000 | 230 |
| IMR with spokes (but without socket) | 2100000 | 980 |
| Socket ring | 110000 | 51 |
| Socket cup | 190000 | 88 |
The map was contoured at level 1.0 (see Methods for details), which gave accurate needle mass/length14.
Leg-linker
24 leg domains are seen, paired at their inner edge (Fig. 5a). Slender linker densities connect the legs to the IMR shoulder. Longitudinal cutaway sections (Fig. 5c) reveal that the legs and IMR shoulder linkage consists of inner and outer links at radii 150Å and 200Å.
IMR-Shoulder
The outer IMR consists of paired 24 stalk-like subunits arranged with slight azimuthal tilt (Fig. 5a, b). The shoulder diameter is 220Å at its widest point. 12 density spokes extend from the IMR ring shoulder toward the centre of the complex (Fig. 5b, 2). Longitudinal cutaways (Fig. 5c) show that the spokes connect the socket, defined by Marlovits26 and surrounding the needle at the complex’s centre, with the outer IMR. The spokes display azimuthal tilt (Fig 5d and e). The lower part of the socket, which we term “socket-cup” (Fig. 5c) measures 52Å at its base and 63Å at its top.
Shoulder-connector
Twelve bulges at the connector’s cytoplasmic face appose one copy within each pair of IMR-shoulder external subunits, specifically that not already interacting with one of the 12 spokes (Fig. 5a, b2 and c). Slice 3 (Fig. 5b) shows the top of the IMR subunits and the start of the larger diameter connector. There is clear azimuthal tilt to the connector with 12 densities forming a collar-like arrangement 141Å in diameter at its widest, closest to the IMR, and tapering to 115Å at the OMR junction. A 73Å-diameter 12-domain structure we term the “socket-ring” is seen inside the connector. It forms the top portion of the socket (Fig. 5c), spans the IMR-connector junction and also has 12 connections to the rest of the socket below it. Top views sliced at the level of socket ring (Fig. 5d) highlight the azimuthal tilt of the 12 subunits in the socket ring. The rest of the socket appears as a continuous structure with its cytoplasmic side also showing strong 12-fold modulation (Fig. 5e).
OMR rings
The OMR is a 3 ring-system (Fig. 5). OMR2 has the largest diameter (130Å), while OMR1 and 3 are smaller (120 and 110Å, respectively). Strong density modulations are seen throughout the whole OMR (Fig. 5a, b). The OMR3 base is a ring with 12 azimuthally tilted subunits (Fig. 5a; b5), which interact with the connector top below (Fig. 5a; 5c). OMR2 and 1 also show strong 12-fold modulation (Fig. 5b, 6 and 7) and the connections between them are tilted to ~60° (Fig. 5a).
Discussion
IMR symmetry and symmetry heterogeneity
Studies on S. typhimurium, using multi-reference alignment of side views to symmetry models, report heterogeneous symmetries within the flagellar M-ring (24-26 fold31) and NC IMR26. 19-22 fold symmetry was found in this IMR and the major 20-fold symmetry sub-population reconstructed26. In the Shigella NC IMR we counted 24 sub-domains in top view class averages produced without alignment to symmetry models. Although top view symmetry analysis (Supplementary Fig. 2) suggested some spread, only 24-fold symmetry was statistically significant. We therefore attribute this spread to using single, noisier images. Furthermore, NC side views correlated best with 24-fold symmetric models. Moreover, we saw no evidence of diametric differences between images or classes, unlike that found in Salmonella26. Crucially, given the high degree of protein conservation between systems, an IMR symmetry change from 20 to 24-fold would result in a ~20% diameter increase. Yet, the Shigella and Salmonella reconstructions IMR diameters are very similar: 220Å versus 208Å. Our IMR symmetry determination and lack of symmetry heterogeneity in Shigella NC populations are supported by a report of 24-fold symmetry in a RotaStat analysis of 95 top views27.
The entire NC base is resolved by C12 symmetry application
Our reconstruction shows an overall size, shape and major sub-component arrangement consistent with previous studies12,26. However, we now see 12-fold symmetric features and details of connections between sub-domains both internally and externally throughout the NC base. Details of individual subunits and azimuthal tilt, which changes direction over the structure’s length, are also revealed. Furthermore, outer IMR symmetry is seen to be not 24-fold27 but pseudo-24-fold.
Novel OMR-connector features
The main OMR protein is MxiD, a secretin. Our OMR structure is similar to that of other secretins1-3,8,32,33: all show a similar sized 3-ring structure and the majority also 12-fold symmetry. Secretins carry a conserved C-terminal domain, predicted as a β-barrel, probably embedded in the OM along with a bound lipoprotein (located on the outside of some secretin OMRs, including MxiD28). Reconstructions of the Klebsiella oxytoca T2SS secretin PulD and of PulD+PulS (lipoprotein) complexes suggest that the larger two rings form the OM-integral portion. This would make it equivalent to our OMR1+2, to which it compares well dimensionally. A less conserved extended N-terminal domain, predicted as predominantly α-helical, may form the secretin periplasmic portion. Its sequence-divergence outside closely-related secretin subfamilies29 may tailor it to interact with components of particular secretion machineries. This could correspond to our OMR3-connector. The Yersinia enterocolitica T3SS YscC structure2 is shorter (140Å versus our 170Å). Yet, it possesses extra mass at the periplasmic end, which may equate to our connector domain but become disordered in the absence of interaction with IMR proteins. This region may also fold to form the central ‘plug’ seen in maps of isolated secretins and needleless NCs26,29.
Our map resolves both predicted regions of secretins, making it the first complete secretin EM map reported. Previous reconstructions imposing D143 or C121 symmetry appeared featureless or showed weak 12-fold modulations. Our more detailed structure probably results from better preservation of the structure in complex with other NC proteins and the strategy used to refine NC alignment and reconstruction.
Despite lack of sequence conservation, our OMR1/2 structure and dimension resembles that of the FBB’s L- and P-rings. The L-ring sits in the OM and the P-ring is peptidoglycan-anchored. A third, possibly OMR3-equivalent, but smaller ring sits below34. The peptidoglycan layer’s position relative to our NC structure is unknown but may lie between OMR3 and connector, in view of the density reduction at this junction.
New IMR features and attempted EscJ docking
The IMR’s pseudo-24-fold paired subunits merge into the 12-spoked, internal IMR. MxiJ is proposed to lie on the IMR’s inside, surrounded by MxiG because during surface biotinylation of intact NCs, PrgK (Salmonella MxiJ homolog) lysines are inaccessible while those of PrgH (MxiG homolog) are modified23.
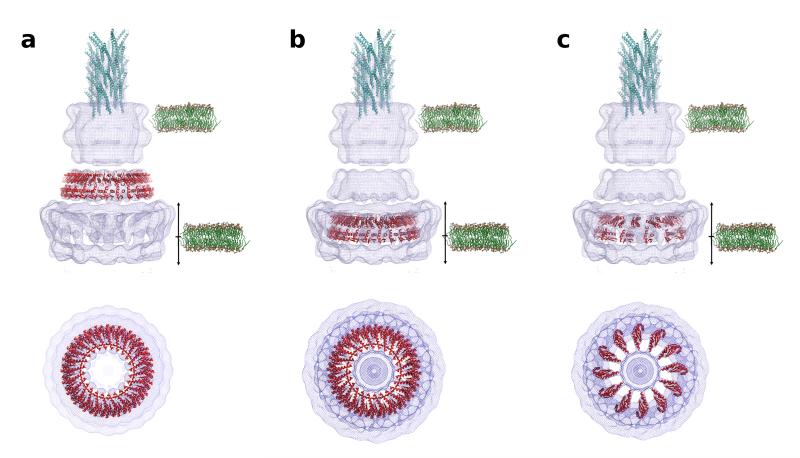
Crystal packing and molecular modelling suggests that EscJ (EPEC MxiJ homolog) forms a 24-subunit ring23,35. The ring’s orientation relative to the cytoplasmic membrane is dictated by EscJ’s IM N-terminal lipid modification23,24. However, automated fitting methods persistently placed the EscJ 24mer in the connector (Fig. 6a). When we fitted the EscJ model manually to the IMR, its radius matched that of the 12 inner spokes (Fig. 6b). Yet, there was more mass in the model than provided by the spokes (Fig 6b, lower panel). If every other EscJ subunit was removed and some orientational alterations made to the remaining EscJs, these fitted approximately into the spoke densities (Fig 6c). The correlation coefficients between these different fits and their surrounding EM densities (cut using a mask contoured at 12Å around the 24mer model) are 63.3, 9.5 and 32.4%36, respectively. This suggests that there are 12, not 24, copies of MxiJ in the Shigella NC. However, T3SSs from other bacterial species, such as EPEC, which support a robust and extended filament structure37, may need more EscJ subunits. In the FBB, which supports rotation of a μm-long filament, the IMR is made of only one protein, FliF38. This aligns at secondary-structure level with both MxiJ and MxiG homologs. Therefore, this ring is unlikely to display internal symmetry differences. Alternatively, MxiJ may not form the spokes. Site-directed mutagenesis of PscJ39 (Pseudomonas EscJ homolog) identified surface-located amino acids important for function. Unfortunately, our map’s resolution and therefore our docking’s imprecision makes it impossible to judge whether their location support our fit or not. Finally, the IM’s location relative to the IMR remains speculative (Fig. 6). An IMR-shoulder position, required for MxiJ and its lipid anchor to sit in the connector, places the socket-cup and most of MxiG in the cytoplasm. A location at IMR leg height also seems unlikely. The two thin “linkers” in the IMR area showing flexibility and leading to “open/closed” leg conformations are level with the socket-cup bottom (Fig. 5c). Whether these represent the single TMHs of MxiG and MxiJ, indicating the IM’s location, needs experimental testing.
Figure 6.
Possible fits of crystal structure of EscJ (MxiJ homolog) in the IMR/connector portions of the map. (a) Position of EscJ 24-mer (35; red) fitted by automated methods (Molrep55) to the outline of the C12 NC reconstruction (blue). (b) EscJ 24-mer placed manually in a position equivalent to Moraes et al.56 fit to 3D reconstruction of Marlovits et al.26. In our map this placement was only possible via molecular replacement if that specific region of density was cut out of the map. (c) An EscJ 12-mer created by removing every other copy of the EscJ 24-mer, followed by a manual rotation of the subunits to best fit the spoke density. This 12-mer does position itself with Molrep in the full C12 NC reconstruction. The IM and OM (green) are shown for perspective. The membrane model used is POPC (palmitoyloleoylphosphatidylcholine). The IM is positioned at a mid-point of possible placements suggested in the literature. The up arrow indicates where the top of the top leaflet could reach and the down arrow indicates where the bottom of the bottom leaflet could reach. The needle model (70Å diameter, cyan; PDB code 2V6L14) is shown for scaling and was docked manually.
Remaining NC stoichiometry uncertainties
By STEM we find the mass ratio between OMR and IMR is 1:2, significantly different to that reported using bulk stoichiometric measurements of only major base components23,26. STEM allows pre-selection of intact NCs. However, these measurements include the difficult-to-estimate mass of lipids and detergents.
Our NC base mass is ~3200 +/−160kDa. The FBB mass was estimated by STEM as ~4400+/−500kDa by Sosinski et al.40, who also generated FBB sub-complexes using biochemical dissociation methods. Subtracting the flagellar OMR and internal rod mass from that of the FBB leads to ~1800kDa mass for a region equivalent to the ~2000+/−150kDa NC IMR. This seems appropriate given that it is in these organelles’ IM portion that their components are most similar41. Thus, NC tilting on STEM grids does not lead to errors larger than those associated with dissociation procedures. Similarity of both data sets with measurements of known FBB component stoichiometries using radioactive labeling42 confirms that, when isolated complexes are stable, both methods give equivalent accuracy40.
The NC versus FBB mass difference thus stems from their OMRs, where components differ. Yersinia T3SS YscC and Klebsiella T2SS PulD secretin rings masses are ~1000+/−150kDa2,43, equal to our OMR mass estimates. However, subtraction of the needle/rod mass from the OMR portion gives ~700kDa, close to the figure obtained using our determined OMR symmetry (12MxiDs), assuming 40% occupancy by the 15kDa MxiM28, which belongs to a protein family binding secretins 1:1. As these other secretins are not traversed by a needle, using Tobacco Mosaic Virus (TMV) for calibration may not be as accurate as using a subcomponent of known mass40.
The mass of the major IMR components summed using our most probable symmetries of 24MxiGs+12MxiJs is 1332kDa which, given our IMR estimate of ~2000+/−150kDa, leaves 400-800kDa possibly contributed by the needle/rod26 and a portion of CIMEA28. Both FBB studies found a similar size discrepancy40,42, suggesting CIMEA components stoichiometry and/or degree of detergent extraction is similar between FBBs and NCs.
When volume measurements of the map’s different parts are converted to masses (Table 3), the OMR+connector is 720kDa, which agrees with the theoretical mass of 12MxiDs (730kDa; substoichiometric binding of MxiM would make it invisible in our map) and our STEM data. However, the IMR (without socket/rod) mass is only 980kDa. This is significantly smaller than that calculated for 24MxiGs+12MxiJs and that determined by STEM and most consistent with 12MxiJs+12MxiGs (816KDa). Possible explanations for this discrepancy are: 1) IMR mass STEM measurements are artefactually high, because more lipid/detergent is bound there; 2) IMR protein portions are disordered and hence invisible in the map, leading to volume underestimation; 3) in our side-view TEM images, the larger IMR may not be as completely coated with stain as other NC parts, leading to non-uniform volume representation across the map and/or 4) TEM image analysis symmetry determinations for this portion of the map are inaccurate.
Novel socket features
Socket shape and dimensions are as described previously12,26,44 and similar also to a structure at the same relative position in FBBs31. However, in our map, its upper and lower parts are additionally characterized by strong 12-fold modulation and azimuthal tilt and are seen to interact with the 12 IMR spokes. Full socket composition/stoichiometry is unknown for any T3SS-related system, but its volumetric mass of <200KDa (Table 3) is similar to the summed masses of one 1 copy of each CIMEA component, suggesting it is of far lower than 12-fold symmetry.
Implications for T3SS function
The socket-cup shows 12-fold symmetry where it may interact with the cytoplasmic export ATPase, known to form hexamers resembling the F1-mitochondrial ATPase45-48. Whether and how this “symmetry jump” is bridged requires further investigation.
12-fold symmetry is also seen at the socket-ring’s periplasmic face, which is optimally located to form a nucleating base for the rod/needle. The socket also links to the IMR via 12 spokes and 12-fold symmetric connections exist between the rod/needle and the connector’s lower portion, OMR3 and OMR2. How the 11 helical rod/needle protofilaments integrate with these connections must now be addressed. These “symmetry mismatches” could allow control of rod/needle assembly and host-cell contact signal transduction via a “gear system”. We will test this using our map to obtain higher resolution reconstructions of wild-type and mutant10,17 NCs from cryoEM images without imposition of symmetry, where future NC component structures can be precisely fitted.
Methods
Needle complex purification
We purified NCs from a mxiG- strain expressing His6-tagged MxiG at native levels28. After release from bacterial membranes via peptidoglycan dissolution and addition of non-ionic detergent, NCs were enriched by rounds of low- and high-speed ultracentrifugation. After the last pelleting step, Ni-agarose beads were added to resuspended NCs. Following overnight binding, the beads were washed and NCs eluted using imidazole. For STEM, we added a gel filtration and concentration step28.
Scanning transmission electron microscopy
NCs (250 μg ml-1) were diluted 4- or 10- fold in 25mM Tris pH 8, 2mM EDTA. Grids were prepared for STEM by the wet film method49 washed with several drops of buffer then TMV solution, buffer, sample solution, then more buffer and blotted to ~1μm thickness, fast frozen in liquid nitrogen slush and freeze dried overnight in an ion-pumped chamber. Samples were transferred under vacuum to the STEM, operating at 40keV with a 512×512 element scan, a 3Å probe and a typical dose of 10el/Å2. The specimen was maintained at -160°C during low-dose imaging. We collected data in two independent sets from different NC preparations. For image analysis, using the PCMASS29 software49 (Fig. 2) all NCs selected were masked by an automatic algorithm and the background computed in the unmasked area. An NC side view model was fitted automatically to each particle, then subtracted from the image data to produce a difference map to highlight imperfections in particles as an aid in quality control of particle selection. TMV served as an internal control for specimen preservation and mass normalization. The software was used to produce an average mass map for all selected particles in a given field (Fig. 2). We measured the masses of the different parts of the complex e.g. needle, OMR and IMR by summing, in a spreadsheet, a set number of 2nm sections within the mass map which clearly contributed to that feature within the majority of mass maps (Fig. 2c). We used the known needle mass/length14 as a secondary standard for background correction between fields.
Transmission electron microscopy
We used 400 mesh copper grids (Athene) covered with holey carbon film over which thin plain carbon was laid. Grids were glow discharged in argon (Edwards 306) and fresh (non-frozen) samples were applied at 50μg ml-1 in 10mM Tris pH 8, 0.1% v/v Triton X-100 and 1mM EDTA buffer. Samples were negatively stained with 2% w/v uranyl acetate for ~30 seconds. EM was carried out on a Philips CM200 FEG transmission electron microscope at a calibrated magnification of 48600. We used an underfocus value of ~800nm, placing the first zero in the CTF at around 14Å, i.e. well beyond the expected resolution of the negative stain dataset hence obviating the need for CTF correction. We took micrographs under low-dose conditions on Kodak SO163 film and digitized them on a Nikon Coolscan 9000 at 6.35μm step size resulting in a resolution of 1.31Å per pixel. We selected particles using Ximdisp software, cutting them out to an initial box size of 400 by 400 pixel and coarsening them to 2.62Å per pixel using Label50 prior to further processing.
Symmetry analysis of top views
For purposes of symmetry analysis all top view images were high pass filtered to 50Å and low pass filtered to 35Å. Initial alignment was to a centred, rotationally averaged, single top view followed by several rounds of alignment to rotationally averaged total sum of all aligned particles. As the IMR extends to a higher radius than the rest of the NC base, its symmetry can be analysed by selecting the high radius region of top views. Accordingly, after alignment and classification of the 44 best preserved and aligned individual, unmasked top view images a ring mask was generated for MSA in IMAGIC at a radius excluding obvious OMR contribution. OMR symmetry and classification of the unmasked aligned images was also carried out by MSA using a ring mask. Refinement of alignment of unmasked images to masked references was carried out in SPIDER51, MSA using a ring mask and classification in IMAGIC552. Analysis of rotational symmetry of individual images of top views was carried out using RotaStat software30. Symmetries taken as significant had to pass a t-test.
Image analysis of side views
3000 images were initially high pass filtered to 100Å, low pass filtered to 15Å and aligned vertically to a single manually aligned NC side view. Images were aligned using SPIDER and classification carried out by MSA in IMAGIC5. We removed particles tilted significantly about the short axis, distorted, or lacking needles. After initial alignment, refinement was carried out with MSA and classification of whole NCs to the IMR portion of the structure only. Particles with detectable out of plane tilt were eliminated to simplify further analysis. This enabled sorting of the remaining images for symmetry/diameter variation of the IMR part of the map by alignment to model reference sets31 and hence identification of a large symmetry group for further analysis. Three-dimensional reconstruction was carried out in IMAGIC5 using assigned symmetry. OMR symmetry analysis from side views was carried out using model OMR reference sets. Resolution testing of the final 3D-reconstruction was carried out using Fourier Shell Correlation in IMAGIC5 using the 3σ and 0.5 correlation coefficient criteria. Surface representations of 3D reconstructions are thresholded at the expected volume of the NC complex base. Measurements of distances in the reconstructions were always mass centre-to-centre. Volumes measurements were made in UCSF Chimera53 (using combinations of the “Select Subregions”, “Volume Erase” and “Measure and Color Blobs” tools) and corresponding molecular masses derived using an established protein density value (2.15Å3/Da)54.
Fitting of the EscJ crystal structure within 3D-reconstructions
Fitting of crystallographic data into electron density map was carried out either manually or by using the automated molecular replacement package Molrep55. Graphical display was done using Pymol (http://www.pymol.org). The C12 and C24 3D maps have been deposited in the EMDEP database under accession number 6391.
Supplementary Material
Acknowledgments
Beth Lin (Brookhaven) is thanked for assistance with STEM sample preparation and Pietro Roversi (Oxford) for the script to run the program for statistical assessment of docking of atomic structures to EM maps. We are indebted to Natalie Strynadka and co-workers (British Columbia) for the EscJ ring coordinates and to David DeRosier (Brandeis) and Keiichi Namba (GSFBS, Osaka) for advice and encouragements at all key stages of this work. They, along with Frank Booy (Bristol), Sarah Daniell (Bristol) and Andreas Veenendaal (Utrecht), Walter Steffen (MHH, Hanover) are also thanked for critical comments on the manuscript. JLH was funded by UK Medical Research Council project grant G0401595 to AJB and DFG grant (BR 849/29-1) to Bernhard Brenner. AJB was supported by the Guy G. F. Newton Senior Research Fellowship. SJ was funded by UK Medical Research Council project grant G0400389 to SML. JSW’s laboratory is supported by the US National Institutes of Health and Department of Energy. PCdF and EPM received funding from Cancer Research UK.
References
- 1.Chami M, et al. Structural insights into the secretin PulD and its trypsin-resistant core. J Biol Chem. 2005;280:37732–41. doi: 10.1074/jbc.M504463200. [DOI] [PubMed] [Google Scholar]
- 2.Burghout P, et al. Structure and electrophysiological properties of the YscC secretin from the type III secretion system of Yersinia enterocolitica. J. Bacteriol. 2004;186:4645–54. doi: 10.1128/JB.186.14.4645-4654.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Opalka N, et al. Structure of the filamentous phage pIV multimer by cryo-electron microscopy. J Mol Biol. 2003;325:461–70. doi: 10.1016/s0022-2836(02)01246-9. [DOI] [PubMed] [Google Scholar]
- 4.Blocker A, Komoriya K, Aizawa S. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc Natl Acad Sci U S A. 2003;100:3027–30. doi: 10.1073/pnas.0535335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hale TL. Bacilliary dysentery. In: Hansler W.J.a.S., M., editor. Topley and Wilson’s Microbiology and Microbial Infections. Vol. 3. Arnold; London: 1998. pp. 479–493. [Google Scholar]
- 6.Cossart P, Sansonetti PJ. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science. 2004;304:242–8. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- 7.Blocker A, et al. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J Cell Biol. 1999;147:683–93. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins RF, Davidsen L, Derrick JP, Ford RC, Tonjum T. Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J Bacteriol. 2001;183:3825–32. doi: 10.1128/JB.183.13.3825-3832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh P. Process of protein transport by the type III secretion system. Microbiol Mol Biol Rev. 2004;68:771–95. doi: 10.1128/MMBR.68.4.771-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veenendaal AK, et al. The type III secretion system needle tip complex mediates host cell sensing and translocon insertion. Mol Microbiol. 2007;63:1719–30. doi: 10.1111/j.1365-2958.2007.05620.x. [DOI] [PubMed] [Google Scholar]
- 11.Kubori T, et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–5. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 12.Blocker A, et al. Structure and composition of the Shigella flexneri “needle complex”, a part of its type III secreton. Mol Microbiol. 2001;39:652–63. doi: 10.1046/j.1365-2958.2001.02200.x. [DOI] [PubMed] [Google Scholar]
- 13.Cordes FS, et al. Helical structure of the needle of the type III secretion system of Shigella flexneri. J Biol Chem. 2003;278:17103–7. doi: 10.1074/jbc.M300091200. [DOI] [PubMed] [Google Scholar]
- 14.Deane JE, et al. Molecular model of a type III secretion system needle: Implications for host-cell sensing. Proc Natl Acad Sci U S A. 2006;103:12529–33. doi: 10.1073/pnas.0602689103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordes FS, et al. Helical packing of needles from functionally altered Shigella type III secretion systems. J Mol Biol. 2005;354:206–11. doi: 10.1016/j.jmb.2005.09.062. [DOI] [PubMed] [Google Scholar]
- 16.Johnson S, et al. Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J Biol Chem. 2007;282:4035–44. doi: 10.1074/jbc.M607945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenjale R, et al. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J Biol Chem. 2005;280:42929–37. doi: 10.1074/jbc.M508377200. [DOI] [PubMed] [Google Scholar]
- 18.Blocker AJ, et al. What’s the point of the type III secretion system needle? Proc Natl Acad Sci U S A. 2008;105:6507–13. doi: 10.1073/pnas.0708344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimbrough TG, Miller SI. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc Natl Acad Sci U S A. 2000;97:11008–13. doi: 10.1073/pnas.200209497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamano K, et al. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. Embo J. 2000;19:3876–87. doi: 10.1093/emboj/19.15.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allaoui A, et al. MxiG, a membrane protein required for secretion of Shigella spp. Ipa invasins: involvement in entry into epithelial cells and in intercellular dissemination. Mol Microbiol. 1995;17:461–70. doi: 10.1111/j.1365-2958.1995.mmi_17030461.x. [DOI] [PubMed] [Google Scholar]
- 22.Allaoui A, Sansonetti PJ, Parsot C. MxiJ, a lipoprotein involved in secretion of Shigella Ipa invasins, is homologous to YscJ, a secretion factor of the Yersinia Yop proteins. J Bacteriol. 1992;174:7661–9. doi: 10.1128/jb.174.23.7661-7669.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yip CK, et al. Structural characterization of the molecular platform for type III secretion system assembly. Nature. 2005;435:702–7. doi: 10.1038/nature03554. [DOI] [PubMed] [Google Scholar]
- 24.Silva-Herzog E, Ferracci F, Jackson MW, Joseph SS, Plano GV. Membrane localization and topology of the Yersinia pestis YscJ lipoprotein. Microbiology. 2008;154:593–607. doi: 10.1099/mic.0.2007/013045-0. [DOI] [PubMed] [Google Scholar]
- 25.Allaoui A, Sansonetti PJ, Parsot C. MxiD, an outer membrane protein necessary for the secretion of the Shigella flexneri lpa invasins. Mol Microbiol. 1993;7:59–68. doi: 10.1111/j.1365-2958.1993.tb01097.x. [DOI] [PubMed] [Google Scholar]
- 26.Marlovits TC, et al. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306:1040–2. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sani M, et al. Structural organization of the needle complex of the type III secretion apparatus of Shigella flexneri. Micron. 2007;38:291–301. doi: 10.1016/j.micron.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Zenk SF, Stabat D, Veenendaal AKJ, Johnson S, Blocker A. Identification of minor inner membrane components of the Shigella T3SS “needle complex”. . Microbiology. 2007;153:2405–2415. doi: 10.1099/mic.0.2007/007781-0. [DOI] [PubMed] [Google Scholar]
- 29.Bayan N, Guilvout I, Pugsley AP. Secretins take shape. Mol Microbiol. 2006;60:1–4. doi: 10.1111/j.1365-2958.2006.05084.x. [DOI] [PubMed] [Google Scholar]
- 30.Kocsis E, Cerritelli ME, Trus BL, Cheng N, Steven AC. Improved methods for determination of rotational symmetries in macromolecules. Ultramicroscopy. 1995;60:219–28. doi: 10.1016/0304-3991(95)00070-2. [DOI] [PubMed] [Google Scholar]
- 31.Thomas DR, Francis NR, Xu C, DeRosier DJ. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:7039–48. doi: 10.1128/JB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collins RF, et al. Three-dimensional structure of the Neisseria meningitidis secretin PilQ determined from negative-stain transmission electron microscopy. J Bacteriol. 2003;185:2611–7. doi: 10.1128/JB.185.8.2611-2617.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nouwen N, Stahlberg H, Pugsley AP, Engel A. Domain structure of secretin PulD revealed by limited proteolysis and electron microscopy. EMBO J. 2000;19:2229–36. doi: 10.1093/emboj/19.10.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 35.Andre I, Bradley P, Wang C, Baker D. Prediction of the structure of symmetrical protein assemblies. Proc Natl Acad Sci U S A. 2007;104:17656–61. doi: 10.1073/pnas.0702626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleywegt GJ, Jones TA. xdlMAPMAN and xdlDATAMAN - programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr D Biol Crystallogr. 1996;52:826–8. doi: 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
- 37.Knutton S, et al. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 1998;17:2166–76. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macnab RM. How bacteria assemble flagella. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 39.Burns RE, McDaniel-Craig A, Sukhan A. Site-directed mutagenesis of the Pseudomonas aeruginosa type III secretion system protein PscJ reveals an essential role for surface-localized residues in needle complex function. Microb Pathog. 2008;45:225–30. doi: 10.1016/j.micpath.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 40.Sosinsky GE, et al. Mass determination and estimation of subunit stoichiometry of the bacterial hook-basal body flagellar complex of Salmonella typhimurium by scanning transmission electron microscopy. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:4801–5. doi: 10.1073/pnas.89.11.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aizawa SI. Bacterial flagella and type III secretion systems. FEMS Microbiol Lett. 2001;202:157–64. doi: 10.1111/j.1574-6968.2001.tb10797.x. [DOI] [PubMed] [Google Scholar]
- 42.Jones CJ, Macnab RM, Okino H, Aizawa S. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J Mol Biol. 1990;212:377–87. doi: 10.1016/0022-2836(90)90132-6. [DOI] [PubMed] [Google Scholar]
- 43.Nouwen N, et al. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc Natl Acad Sci U S A. 1999;96:8173–7. doi: 10.1073/pnas.96.14.8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marlovits TC, et al. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature. 2006;441:637–40. doi: 10.1038/nature04822. [DOI] [PubMed] [Google Scholar]
- 45.Claret L, Calder SR, Higgins M, Hughes C. Oligomerization and activation of the FliI ATPase central to bacterial flagellum assembly. Mol Microbiol. 2003;48:1349–55. doi: 10.1046/j.1365-2958.2003.03506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imada K, Minamino T, Tahara A, Namba K. Structural similarity between the flagellar type III ATPase FliI and F1-ATPase subunits. Proc Natl Acad Sci U S A. 2007;104:485–90. doi: 10.1073/pnas.0608090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minamino T, et al. Oligomerization of the bacterial flagellar ATPase FliI is controlled by its extreme N-terminal region. J Mol Biol. 2006;360:510–9. doi: 10.1016/j.jmb.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 48.Zarivach R, Vuckovic M, Deng W, Finlay BB, Strynadka NC. Structural analysis of a prototypical ATPase from the type III secretion system. Nat Struct Mol Biol. 2007;14:131–7. doi: 10.1038/nsmb1196. [DOI] [PubMed] [Google Scholar]
- 49.Wall JS, Simon MN, Lin BY, Vinogradov SN. Mass mapping of large globin complexes by scanning transmission electron microscopy. Methods Enzymol. 2008;436:487–501. doi: 10.1016/S0076-6879(08)36027-3. [DOI] [PubMed] [Google Scholar]
- 50.Crowther RA, Henderson R, Smith JM. MRC image processing programs. J Struct Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 51.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 52.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 53.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 54.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–7. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 55.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56:1622–4. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 56.Moraes TF, Spreter T, Strynadka NC. Piecing together the type III injectisome of bacterial pathogens. Curr Opin Struct Biol. 2008;18:258–66. doi: 10.1016/j.sbi.2007.12.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.