Abstract
Heart failure is a cause of significant morbidity and mortality in developed nations, and results from a complex interplay between genetic and environmental factors. To discover gene regulatory networks underlying heart failure, we analyzed DNA microarray data based on left ventricular free-wall myocardium from 59 failing (32 ischemic cardiomyopathy, 27 idiopathic dilated cardiomyopathy) and 33 non-failing explanted human hearts from the Cardiogenomics Consortium. In particular, we sought to investigate cardiac gene expression changes at the level of individual genes, as well as biological pathways which contain groups of functionally related genes. Utilizing a combination of computational techniques, including Comparative Marker Selection and Gene Set Enrichment Analysis, we identified a subset of downstream gene targets of the master mitochondrial transcriptional regulator, peroxisome proliferator-activated receptor gamma coactivator-1α (PGC-1α), whose expression is collectively decreased in failing human hearts. We also observed decreased expression of the key PGC-1α regulatory partner, estrogen-related receptor α (ERRα), as well as ERRα target genes which may participate in the downregulation of mitochondrial metabolic capacity. Gene expression of the antiapoptotic Raf-1/extracellular signal-regulated kinase (ERK) pathway was decreased in failing hearts. Alterations in PGC-1α and ERRα target gene sets were significantly correlated with an important clinical parameter of disease severity - left ventricular ejection fraction, and were predictive of failing vs. non-failing phenotypes. Overall, our results implicate PGC-1α and ERRα in the pathophysiology of human heart failure, and define dynamic target gene sets sharing known interrelated regulatory mechanisms capable of contributing to the mitochondrial dysfunction characteristic of this disease process.
Keywords: Heart failure, Mitochondria, Fatty acid oxidation, PGC-1α, ERRα, Raf-1, ERK, LCAD, Gene expression profiling
1. Introduction
Congestive heart failure is a cause of significant morbidity and mortality in developed nations. For individuals free of heart failure at age 40, data from the Framingham Heart Study indicates that the lifetime risk of developing heart failure is 20% [1]. Among the most common etiologies of end-stage human heart failure are idiopathic dilated cardiomyopathy (DCM) and ischemic cardiomyopathy (ICM). To investigate pathophysiologic mechanisms of heart failure, microarray technology has been utilized extensively in the characterization of global gene expression changes in both ischemic and idiopathic dilated (or non-ischemic) cardiomyopathy [2,3]. A number of prior microarray studies of heart failure have noted no significant gene expression changes between DCM and ICM [4] or have grouped ischemic and non-ischemic samples into a single failing category when comparing against non-failing controls [5,6]. Although end-stage heart failure may represent a common clinical endpoint with a uniform gene expression signature irrespective of etiology, additional studies have identified both shared and distinct gene expression changes between ischemic and non-ischemic heart failure [7–9].
Gene regulatory networks, or pathways, previously associated with heart failure include immune and inflammatory response [10], cell signaling and metabolism [4,6,8], sarcomere and cytoloskeletal organization [6,9,11], transcription/translation [12], and apoptosis [13,14]. Conventional methods used to discover dysregulated pathways in heart failure have relied fundamentally on single-gene approaches combined with Gene Ontology or self-defined functional annotations [7,10,11,15]. Our current study differs in that it also employs biological pathway-based analytic techniques to detect over-arching patterns of gene expression in relatively large populations of failing (DCM/ICM) vs. non-failing (NF) human hearts, as well as in individual comparisons of DCM and ICM groups.
Given that insufficient energy delivery and altered substrate utilization occur in the failing heart, gene regulatory networks modulating mitochondrial function have been implicated in the pathogenesis of heart failure [16–19]. In particular, studies investigating mechanisms of defective energy transduction in heart failure have defined a role for the transcriptional coactivator, peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α), a key regulator of post-natal cardiac mitochondrial function [16,20–22]. In the mouse heart, PGC-1α is essential for maximal and efficient cardiac mitochondrial fatty acid oxidation, ATP synthesis, and lipid homeostasis, especially in the setting of increased work [23]. Forced expression of PGC-1α in hearts of transgenic mice increases mitochondrial number and stimulates respiration via several downstream transcriptional regulatory circuits, one of which involves the nuclear respiratory factors 1 and 2 (NRF-1 and NRF-2) [24]. Additionally, the ability of PGC-1α to coactivate the peroxisome proliferator-activated receptors (PPARs) and estrogen-related receptor α (ERRα) gives it a critical role in the regulation of genes central to the mitochondrial β-oxidation of fatty acids as well as overall mitochondrial oxidative phosphorylation [16,25–30].
Reduced PGC-1α activity has been observed in the evolution of several important diseases associated with metabolic derangement, including diabetes mellitus, pathologic cardiac hypertrophy, and heart failure. Oxidative phosphorylation genes regulated by PGC-1α are coordinately depressed in human skeletal muscle with diabetes mellitus type 2 [28,31]. In pressure-overload cardiac hypertrophy in mice, PGC-1α gene expression is decreased [21], and its level of expression correlates with mitochondrial oxidative capacity in both healthy and failing rat hearts subjected to chronic pressure overload [32]. Consistent with involvement of PGC-1α in the maintenance of cardiac function following the stress of pressure overload, mice with generalized loss of PGC-1α develop accelerated heart failure two months following transverse aortic constriction [33]. The current study attempts to extend to human heart failure a role for mitochondrial transcriptional regulatory circuits characterized predominantly in normal and failing mouse hearts. This work implicates the PGC-1α/ERRα regulatory circuit in the pathophysiology of human heart failure, suggesting transcriptional regulatory mechanisms involving PGC-1α and its key partners which may contribute to the mitochondrial dysfunction characteristic of the failing heart.
2. Methods
2.1 Source of Microarray Data
Publicly available microarray data was obtained from the Cardiogenomics Consortium at Brigham and Women’s Hospital in Boston, Massachusetts (http://cardiogenomics.med.harvard.edu), an NHLBI sponsored Program in Genomic Applications. RNA was isolated from left ventricular free-wall myocardium extracted from 92 explanted human hearts and hybridized to the Affymetrix HG-U133 plus 2.0 oligonucleotide microarray (54,675 probe sets) according to the guidelines and protocols as outlined on the website. (The ‘Quality Control’ section of the Cardiogenomics Consortium website (above) offers further details.) These studies were approved by the Committee for the Protection of Human Subjects at Brigham and Women’s Hospital and Harvard Medical School. The gene expression analysis component reported in this paper was further reviewed by the Washington University Human Research Protection Office. Fifty-nine of the 92 explanted hearts were from patients with end-stage heart failure at the time of cardiac transplantation, where the etiology of failure was ischemic cardiomyopathy (ICM) in 32 patients and idiopathic dilated cardiomyopathy (DCM) in 27 patients. The remaining 33 samples were from normal, non-failing (NF) cardiac transplant donors whose cause of death was non-cardiac in origin, but whose heart could not be used for transplantation. Left ventricular tissue was harvested intraoperatively at the time of transplantation and snap frozen immediately (within 1 min) in liquid nitrogen, then stored at −80°C until RNA was isolated. Demographics and limited clinical parameters were available for the majority of patients. For the non-failing group, data on body mass index, hypertension, diabetes, and left ventricular ejection fraction were available for less than 50% of patients, while pulmonary capillary wedge pressures were not available for any of the patients in the non-failing group.
2.2 Data Scaling and Processing
The data were subjected to global scaling to correct for intensity-related biases as described on the website. Only scans with a 3′/5′ ratio<1.33 were included in our study. A single ICM sample was eliminated by this criterion, leaving a total of 91 samples - 27 DCMs, 31 ICMs, and 33 NFs.
All probe sets which were found to have greater than 93% absent calls across 91 samples were eliminated. These probe sets were considered absent, or under-expressed, in our study and were designated as ‘filtered’. Of 54,675 total probe sets on the HG-U133 plus 2.0 array, 30,608 probe sets subsequently remained after filtering. We further collapsed the data from 30,608 probe sets to 17,419 non-redundant genes with distinct HUGO gene symbols, since multiple probe sets often map to a single gene. In this instance, the probe set with the maximum absolute expression value was uniformly assigned to the gene.
In order to reduce the age variability in our population and given the imbalanced number of samples derived from female subjects, all females and marked age outliers were eliminated from our study. A simple algorithm was constructed to remove age outliers in an objective fashion, such that the oldest and youngest were systematically subtracted from each population until the mean age of the three populations was not significantly different. In effect, an age and gender ‘matched’ subset was created, consisting of 49 samples – 13 DCMs, 20 ICMs, and 16 NFs, while still preserving sample size and power. This subset is referred to as the ‘matched’ population. The remaining 42 samples were defined as the ‘unmatched’ population and used for independent validation of our class prediction model.
2.3 Single-gene Analysis
The Comparative Marker Selection algorithm from the GenePattern 2.0 software package [34,35] was employed to identify individual genes that were differentially expressed in all four pairwise comparisons (DCM/ICM vs. NF, DCM vs. NF, ICM vs. NF, DCM vs. ICM). A 2-sided t-test followed by 1000 permutations of phenotype labels was performed for this analysis. A false discovery rate q value (FDR)<0.05, accounting for multiple hypothesis testing [35], and a fold change (FC)>1.2 served as a cutoff for significant genes. A family-wise error rate (FWER)<0.05 was used as another, more stringent statistic to discover highly significant genes.
2.4 Pathway Analysis
Gene Set Enrichment Analysis (GSEA) was utilized to probe pathways, or groups of functionally related genes, dysregulated in heart failure [36]. In discovery mode, gene sets with an FDR<0.25 and a nominal p value<0.05 were considered significant [31,36]. The gene ranking metric was a signal-to-noise ratio and the number of permutations specified was 1000.
502 curated gene sets representing generic biological pathways were downloaded from the Broad Institute Molecular Signature Database (MSigDb) (http://www.broad.mit.edu/gsea). The original sources of these pathways include BioCarta, GenMapp, Kyoto Encyclopedia of Genes and Genomes (KEGG), and the Broad Institute. The ‘PGC-1α targets’ gene set was derived from the MSigDb pathway annotated as ‘PGC’, and is based on genes responsive to adenoviral-mediated PGC-1α gain-of-function in cultured mouse myoblasts (C2-C12 cells) [31]. The ‘ERRα targets 1’ and ‘ERRα targets 2’ gene sets were mined from the literature, and are based on genes influenced by adenoviral-mediated ERRα gain-of-function in primary rat neonatal cardiac myocytes [25] and ChIP-on-chip gene promoter occupancy assays [37], respectively. A total of 504 pathways were initially compiled for investigation. Gene sets with less than 15 genes or more than 500 genes were excluded from the analysis, leaving 252 pathways after application of this threshold. Gene classification using Gene Ontology (GO) Biological Process and Cellular Component terms was performed for core enrichment set genes via FatiGo online web tool [38].
Given that multiple probe sets may map to a single gene, the GSEA software package offers two options with respect to collapsing multiple probe sets into a single expression vector to represent a gene: the maximum probe set expression value or the median probe set expression value. In this study, the maximal probe set expression value was set as the default in the algorithm, as in other previous studies that rely on GSEA. According to the authors of the GSEA algorithm, this approach allows for more widespread and accurate signal detection [36]. Moreover, we performed GSEA employing both options and found the difference between the two analyses minimal (e.g., the PGC-1α and ERRα target gene pathways remain significantly downregulated with human heart failure in both cases).
2.5 Linear Regression Model
Linear regression models were generated using Microsoft Excel’s Regression Package. Expression levels of the PGC-1α target, ERRα target 1, and ERRα target 2 core enrichment set genes were designated as independent, explanatory variables, and left ventricular ejection fraction (LVEF) as a dependent variable. Expression values of genes in each core enrichment set were normalized to μ=0 and σ=1 across samples from the matched population. A core enrichment set mean expression level was then computed for each of the samples and regressed versus LVEF to illustrate the explanatory power of this gene set. LVEF was known for 27 of the 49 samples in the matched population.
2.6 Class Prediction Model
Using the K-nearest neighbors (KNN) algorithm of the GenePattern 2.0 software package with a Euclidean distance metric [34], prediction models were built to distinguish between failing and non-failing classes based on genes in the PGC-1α target, ERRα target 1, ERRα target 2, and ERK pathway core enrichment sets (CES). For comparison, a parallel prediction model was created employing the most significantly up and downregulated markers at FWER<0.05. All models were initially trained on samples from the matched population, and then tested on samples from the unmatched population. It is important to note that these are two mutually exclusive populations, the sum of which makes up the total population.
2.7 Source of Left Ventricular Tissue for Protein Level Quantification
For determination of left ventricular protein levels in failing versus nonfailing hearts, cardiac tissue samples were obtained from 8 hearts with no history of myocardial dysfunction and with normal left ventricular ejection fraction by echocardiography. These ‘nonfailing’ hearts (from 4 men and 4 women) were donated for orthotopic cardiac transplantation but declined for reasons related to size or ABO blood type mismatch. Consent for donation of heart tissue for research purposes was obtained from family members by the organ donor organization covering the Colorado-Wyoming-Montana region (Donor Alliance). Eight ‘failing’ heart samples were obtained from cardiac transplant candidates (4 men and 4 women) with idiopathic dilated cardiomyopathy whose age was nearly identical to the ‘nonfailing’ population. In particular, the mean ages of the ‘nonfailing’ and ‘failing’ populations were both 58±2 years, with age ranges of 52–69 years and 52–65 years, respectively. The left ventricular ejection fraction by echocardiography was 62±2% for the ‘nonfailing’ hearts and 17±5% for the ‘failing’ hearts.
2.8 Western Blotting
Generation of left ventricular whole cell protein extracts, protein immunoblotting, and immunoreactive signal quantification were performed as previously described [39]. Protein gels were transferred to a nitrocellulose membrane, prior to incubation with the following antibodies: calnexin (C4731, Sigma-Aldrich, St. Louis, MO), long-chain acyl-CoA dehydrogenase (LCAD, a gift of Arnold W. Strauss), citrate synthase (a gift of John O. Holloszy), and cytochrome c (556433, BD Pharmingen, San Jose, CA). Antibodies were diluted in 1× phosphate-buffered saline containing 3% bovine serum albumin (BSA) and 0.1% Tween, then incubated with the blot overnight at 4 C. Anti-rabbit and anti-mouse secondary antibodies were used at 1:10,000 dilution.
3. Results
We analyzed microarray data from 58 failing (31 ICMs, 27 DCMs) and 33 non-failing explanted human hearts. Since age and gender are known confounders in transcriptional profiling studies of heart failure [40,41], we created an age and gender ‘matched’ subset by eliminating all females and age outliers to minimize variability in our population (Table 1). The matched population consisted of 49 males (20 ICM, 13 DCM, 16 NF), where the difference in mean ages was not statistically significant. Consistent with the association between hypertension, diabetes, and coronary artery disease, the ICM group in the total population exhibited the highest prevalence of hypertension, along with a trend toward increased prevalence of diabetes (Table 1). In the total population, the DCM group displayed the most severe heart failure, with a mean left ventricular ejection fraction (LVEF) of 12.9±1.3% vs. ICM mean LVEF of 21.0±2.7%, although both groups demonstrated marked left ventricular systolic dysfunction. A similar trend toward greater impairment of systolic function was observed in the DCM vs. ICM groups of the matched population; this apparent difference, however, was not statistically significant.
Table 1.
Characteristics of Total and Matched Patient Populations
| Total Population |
Matched Population |
|||||
|---|---|---|---|---|---|---|
| NF | ICM | DCM | NF | ICM | DCM | |
| n | 33 | 31 | 27 | 16 | 20 | 13 |
| Gender | 19M, 14F | 26M, 5F | 17M, 10F | 16M | 20M | 13M |
| Age (y) | 39.8 ± 3.2 | 52.1 ± 1.9* | 46.9 ± 3.6 | 46.9 ± 4.1 | 49.5 ± 2.2 | 49.6 ± 4.0 |
| Age Range (y) | 1–69 | 20–69 | 10–79 | 20–69 | 20–69 | 20–79 |
| BMI (kg/m2) | 24.4 ± 1.5 | 25.7 ± 0.8 | 23.6 ± 1.1 | 24.6 ± 1.5 | 25.2 ± 1.0 | 23.1 ± 2.1 |
| HTN | 18.2% | 58.1%* | 22.2%# | 25.0% | 50.0% | 15.4%# |
| DM | 3.0% | 35.5%* | 22.2%* | 6.3% | 30.0% | 15.4% |
| LVEF (%) | 61.3 ± 4.3 | 21.0 ± 2.7* | 12.9 ± 1.3*# | 60.0 ± 5.8 | 22.3 ± 3.7* | 14.3 ± 2.2* |
| PCWP (mmHg) | N/A | 17.6 ± 1.5 | 23.5 ± 1.6# | N/A | 17.8 ± 2.0 | 24.7 ± 2.6# |
M, male; F, female. HTN, hypertension; DM, diabetes mellitus; BMI, body mass index; LVEF, left ventricular ejection fraction, PCWP, pulmonary capillary wedge pressure. Age, BMI, LVEF, and PCWP are reported as mean ± S.E.M.
Comparison with NF is significant by 2-sided Student’s -test or Chi-square test at p < 0.05.
Comparison with ICM is significant by 2-sided Student’s t -test or Chi-square test at p < 0.05. N/A denotes data not available.
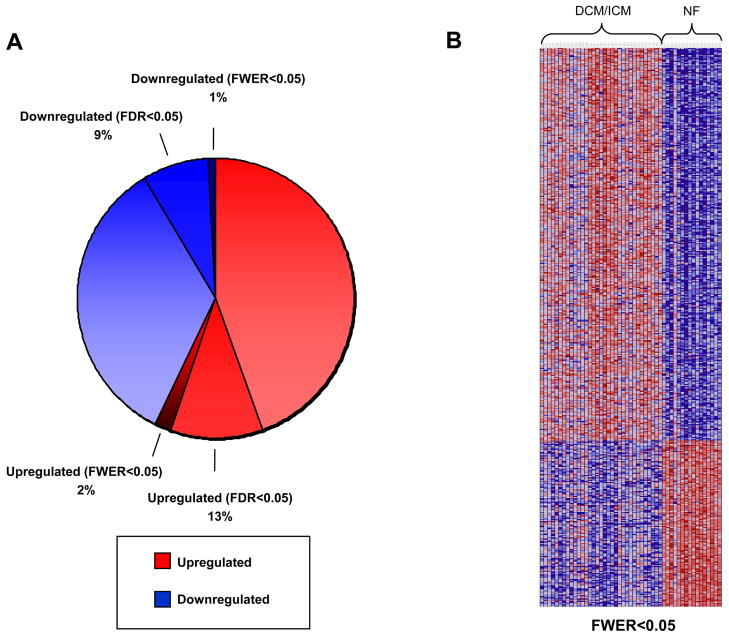
In our initial approach, the Comparative Marker Selection algorithm [34,35] was used to identify individual genes that were differentially expressed between non-failing (NF) and combined failing (DCM/ICM) classes. Out of a total of 17,419 distinct human genes in our dataset after filtering (as per Methods), 9,991 genes (57%) exhibited a fold change>0, while 7,428 genes (43%) exhibited a fold change<0. At a false discovery rate q value (FDR)<0.05 and a fold change (FC)>1.2, 2,236 genes (13% of total) were significantly upregulated and 1,509 genes (9% of total) were significantly downregulated (Fig. 1A). Using an even more stringent statistic to define up and downregulated genes, a family-wise error rate (FWER)<0.05 and FC>1.2, only 340 genes (2% of total) were upregulated and 144 genes (1% of total) were downregulated (Figs. 1A and 1B). For a complete list of significant genes in all categories, see Supplemental Table S1. This analysis was also performed for the following pair-wise comparisons: DCM vs. NF, ICM vs. NF, and DCM vs. ICM (Supplemental Tables S2, S3, and S4 respectively).
Fig. 1.
Results of comparative marker selection analysis in DCM/ICM vs. NF groups. Red indicates increased expression, while blue indicates decreased expression relative to the non-failing population mean. (A) The pie chart conveys the percentage of genes up and downregulated of 17,419 total genes. Those genes that are upregulated at FWER<0.05 are a subset of those upregulated at FDR<0.05, which are in turn a subset of all upregulated genes; the same applies to downregulated genes. (B) Heatmap of genes significantly up and downregulated at FWER<0.05.
Since heart failure represents a complex disease caused by alterations in multiple genes or gene sets such that single-gene strategies may be less revealing in terms of overall physiology, we next chose to pursue a pathway-based methodology for analysis. In particular, we applied Gene Set Enrichment Analysis (GSEA) [31,36] to investigate 504 curated, generic biological pathways in an unbiased fashion for the failing (DCM/ICM) vs. non-failing (NF) comparison (see Supplemental Table S5 for list of pathways). At an FDR<0.25 and p<0.05 [36], no pathways were found to be significantly upregulated, despite the fact that 57% of all genes in our dataset exhibited a fold change>0. However, 41 pathways were significantly downregulated in failing hearts by these criteria (Table 2). We also evaluated these pathways for DCM vs. NF and ICM vs. NF comparisons and found comparable patterns of downregulation (data not shown). Additionally, no pathways were significantly differentially regulated between the DCM and ICM groups (data not shown). Given that the left ventricular free-wall myocardium from which RNA was isolated for determination of gene expression contains a mixture of cardiac myocytes, fibroblasts, and vascular cells, the results of this study reflect gene expression changes integrated across several important cardiac cell types.
Table 2.
Significantly Downregulated Pathways in Heart Failure by Gene Set Enrichment Analysis
| Pathway | Description | FDR | p -value |
|---|---|---|---|
| ERK PATHWAY | Cell growth is promoted by Ras activation of the anti-apoptotic p44/42 MAP kinase pathway | 0.103 | <0.001 |
| INTEGRIN PATHWAY | Integrins are cell surface receptors that interact with the extracellular matrix and transduce signaling | 0.105 | <0.001 |
| INSULIN SIGNALING | Genes related to insulin signaling | 0.136 | <0.001 |
| B CELL ANTIGEN RECEPTOR | B cell receptors bind antigens and promote B cell activation | 0.166 | <0.001 |
| FMLP PATHWAY | The fMLP receptor in neutrophils recognizes bacterial peptides and activates NADPH oxidase | 0.159 | 0.004 |
| CCR3 PATHWAY | CCR3 is a G-protein coupled receptor that recruits eosinophils to inflammation sites via chemokines | 0.131 | 0.006 |
| MAPK PATHWAY | The MAP kinase pathway has four main sub-pathways: Erk, JNK/SAPK, p53, and ERK5 | 0.169 | 0.006 |
| UCALPAIN PATHWAY | Calpains promote formation of integrin adhesion clusters which recruit Rac | 0.182 | 0.008 |
| PYK2 PATHWAY | Pyk2 and Rac1 stimulate the JNK cascade and activate MKK3, which activates p38 | 0.161 | 0.008 |
| HEART FAILURE DOWNREGULATED (VENTRICLE) | Downregulated genes in the ventricles of failing hearts compared to healthy controls | 0.147 | 0.012 |
| GALACTOSE METABOLISM | Genes involved in galactose metabolism | 0.181 | 0.012 |
| ERRα TARGETS 1 | Genes regulated by estrogen-related receptor α (Huss et al., Mol Cell Biol 2004) | 0.213 | 0.012 |
| PURINE METABOLISM | Genes involved in purine metabolism | 0.165 | 0.013 |
| NDK/DYNAMIN PATHWAY | NDK, Phosphins and Dynamin have a role in endocytosis | 0.186 | 0.013 |
| PGC-1α TARGETS | Genes regulated by peroxisome proliferator-activated receptor gamma coactivator-1α | 0.197 | 0.014 |
| RAS PATHWAY | Ras activation stimulates many signaling cascades, including PI3K/AKT, to inhibit apoptosis | 0.166 | 0.015 |
| TRANSLATION FACTORS | Genes involved in translation | 0.143 | 0.015 |
| ERK1/ERK2/MAPK PATHWAY | The Erk1 and Erk2 MAP kinase pathways are regulated by Raf, Mos, and Tpl-2 | 0.181 | 0.015 |
| BCR SIGNALING PATHWAY | Members of the BCR signaling pathway | 0.205 | 0.016 |
| MET PATHWAY | The hepatocyte growth factor receptor c-Met stimulates proliferation and alters cell adhesion | 0.187 | 0.017 |
| IL2RB PATHWAY | The IL-2 receptor is required for IL-2 and IL-15 signal recognition and activates JAK kinase | 0.191 | 0.018 |
| NFKB PATHWAY | Active NF-kB is localized in the nucleus and regulates transcription of a variety of genes | 0.172 | 0.019 |
| INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway in cardiac myocytes | 0.164 | 0.021 |
| IGF1/MTOR PATHWAY | Growth factor IGF-1 activates AKT, Gsk3-beta, and mTOR to promote muscle hypertrophy | 0.197 | 0.024 |
| PROSTAGLANDIN AND LEUKOTRIENE METABOLISM | Genes involved in prostaglandin and leukotriene metabolism | 0.179 | 0.024 |
| SPRY PATHWAY | The Sprouty protein family bind Grb-2, preventing Ras and MAP kinase activation | 0.174 | 0.024 |
| INTEGRIN MEDIATED CELL ADHESION | Genes involved in integrin-mediated cell adhesion | 0.220 | 0.026 |
| ACTIN CYTOSKELETON REGULATION | Genes related to regulation of the actin cytoskeleton, in particular Rho GTPases | 0.220 | 0.027 |
| PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes | 0.228 | 0.027 |
| FRUCTOSE AND MANNOSE METABOLISM | Genes involved in fructose and mannose metabolism | 0.216 | 0.032 |
| ERRα TARGETS 2 | Genes regulated by estrogen-related receptor α (DuFour et al., Cell Met 2007) | 0.217 | 0.033 |
| cAMP CHEMOTAXIS PATHWAY | Genes involved in cytoskeletal organization during chemotaxis | 0.210 | 0.034 |
| TRKA RECEPTOR | TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth | 0.170 | 0.035 |
| ECM PATHWAY | Extracellular matrix induces integrin-mediated FAK phosphorylation, activating PI3 and MAP kinase | 0.170 | 0.035 |
| G13 SIGNALING PATHWAY | Genes involved in G13 signaling pathway | 0.170 | 0.037 |
| GLYCOLYSIS AND GLUCONEOGENESIS | Genes involved in glycolysis and gluconeogenesis | 0.220 | 0.040 |
| CERAMIDE PATHWAY | Ceramide is a lipid signaling molecule that can activate proliferative or apoptotic pathways | 0.217 | 0.041 |
| FAS SIGNALING PATHWAY | Fas receptor induces apoptosis and NF-kB activation when bound to Fas ligand | 0.227 | 0.042 |
| IGF1R PATHWAY | IGF-1R promotes cell growth and inhibits apoptosis via Ras activation and the AKT pathway | 0.224 | 0.042 |
| MTOR PATHWAY | Target of rapamycin, mTOR senses mitogenic factors and nutrients and induces cell proliferation | 0.213 | 0.043 |
| EGF PATHWAY | Epidermal growth factor promotes cell proliferation via the MAP kinase and Ras pathways | 0.241 | 0.048 |
Significance is defined by FDR<0.25 and p<0.05. Pathways are ordered by p-value. Descriptions are based on those provided in MSigDb.
The vast majority of downregulated pathways clustered into two broad functional categories –metabolism and cell signaling/growth, with remaining pathways related to inflammatory response, as well as cytoskeletal and membrane organization (Table 2). Cardiac remodeling in the setting of cardiac hypertrophy and heart failure progression is mediated in part by mitogen-activated protein kinase (MAPK) signaling cascades [extracellular signal-regulated kinases (ERKs), stress-activated protein kinases (SAPKs/JNK), and p38] [42,43]. Our data indicate that gene expression profiles for these signaling and growth pathways are downregulated in heart failure. This potentially represents either a later, end-stage event or negative gene regulation in response to increased activation of pathways which are regulated more at the post-translational than transcriptional level. It is possible, though, that the observed downregulated gene expression of the ERK pathway, which favors physiologic growth and protection from apoptosis, may contribute to the progression of heart failure [44]. Indeed, the ERK gene pathway was the most significantly downregulated pathway in human heart failure by the gene set enrichment analysis (Table 2), indicating that this downregulation of gene expression is a potential contributor to decreased ERK pathway activity in severe heart failure. Gene set enrichment analysis of failing human hearts in this study revealed that 14 genes in the ERK pathway form the ERK core enrichment set (ERK CES, Supplemental Table S6), driving downregulation of the ERK pathway. Members of the ERK CES include the following genes which encode key proteins of the ERK pathway three-component kinase cascade: RAF1 (Raf-1), MAP2K2 (MKK2), MAP2K1 (MKK1), and MAPK3 (ERK1). Other related pro-growth and antiapoptotic pathways demonstrated by the GSEA analysis to be downregulated with human heart failure include the insulin-like growth factor 1 receptor (IGF1R) pathway, the IGF1/MTOR pathway, the insulin signaling, and insulin receptor pathways (Table 2). Consistent with the considerable number of downregulated metabolic pathways in the failing (DCM/ICM) state, genes regulated by PGC-1α and ERRα which are central to mitochondrial energy metabolism were also significantly downregulated (p<0.035) (Table 2). Downregulation of PGC-1α and ERRα target genes may participate in the mitochondrial dysfunction previously characterized in heart failure [16]. Concomitant downregulation in heart failure of PGC-1α, ERRα, and ERK pathways would result in increased susceptibility for apoptosis in the failing heart, exacerbated by impaired overall mitochondrial oxidative capacity.
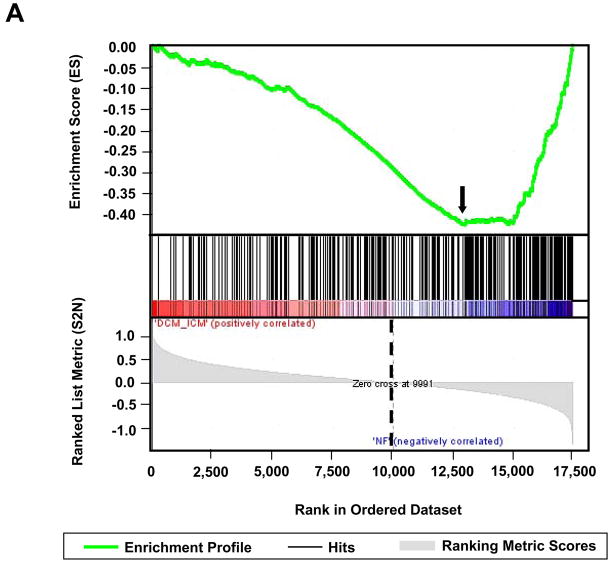
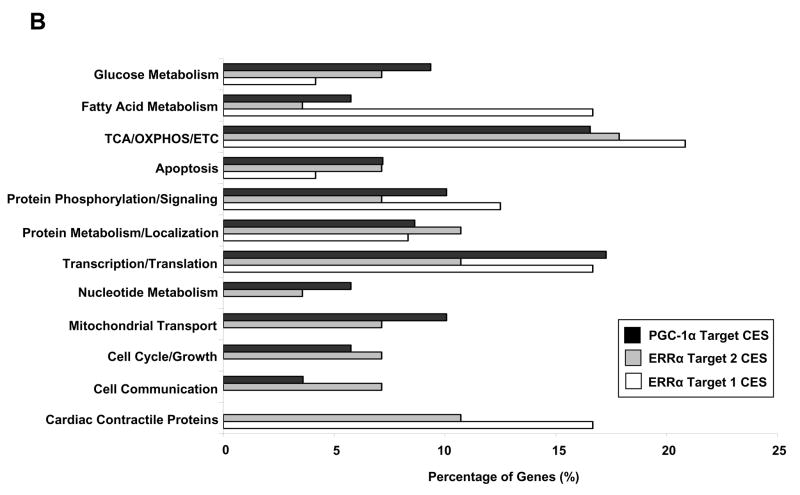
Of the 327 genes in the ‘PGC-1α targets’ gene set which were also present in our data (Supplemental Table S7), 158 genes comprised what is termed the ‘core enrichment set’ (PGC-1α target CES) based on the GSEA results (Supplemental Table S8). A core enrichment set may also be referred to as a leading edge subset [36], as it is the genes in this set that drive the statistical significance of a pathway by determining the enrichment score (Fig. 2A). Thus, the PGC-1α target CES represents PGC-1α responsive genes that are significantly downregulated in heart failure (Fig. 2A). Similarly, the ‘ERRα targets 1’ and ‘ERRα targets 2’ gene sets contained 70 and 104 genes [25,37] (Supplemental Tables S9 and S11), 24 and 38 of which formed the core enrichment sets (ERRα target 1 CES and ERRα target 2 CES), respectively (Supplemental Tables S10 and S12). Classification of genes in the PGC-1α target CES, ERRα target 1 CES, and ERRα target 2 CES using GO Biological Process terms revealed marked functional overlap (Fig. 2B). A high percentage of genes in all three core enrichment sets were involved in glucose metabolism, apoptosis, and the core mitochondrial functions of fatty acid metabolism, the citric acid cycle, electron transport, and oxidative phosphorylation – pathways known to be regulated by PGC-1α and ERRα. Additional highly represented categories in the PGC-1α target and both ERRα target core enrichment sets included transcription/translation, protein metabolism/localization, and protein phosphorylation/signaling. In contrast to the PGC-1α target CES, the ERRα target 1 and 2 CES contained a significant number of genes encoding cardiac contractile proteins. Further classification, using GO Cellular Component terms, demonstrated that mitochondria-associated genes comprised 50% of the PGC-1α target CES, 26% of the ERRα target 1 CES, and 40% of the ERRα target 2 CES.
Fig. 2.
Characterization of PGC-1α, ERRα target1, and ERRα target 2 core enrichment sets defined by the DCM/ICM vs. NF comparison. (A) Upper plot depicts the enrichment profile in green of the ‘PGC-1α targets’ pathway by Gene Set Enrichment Analysis (GSEA). The arrow denotes the point of maximal deviation of the Enrichment Score (ES) from the baseline, which is determined by genes of the ‘Core Enrichment Set’ (CES). Lower plot shows all 17,419 genes rank-ordered according to the signal-to-noise metric, with the dotted line demarcating genes that are positively (red) and negatively (blue) correlated with heart failure. (B) Comparative functional distributions using GO Biological Process terms of the 158 genes of the PGC-1α target CES, the 24 genes of the ERRα target 1 CES, and the 38 genes of the ERRα target 2 CES.
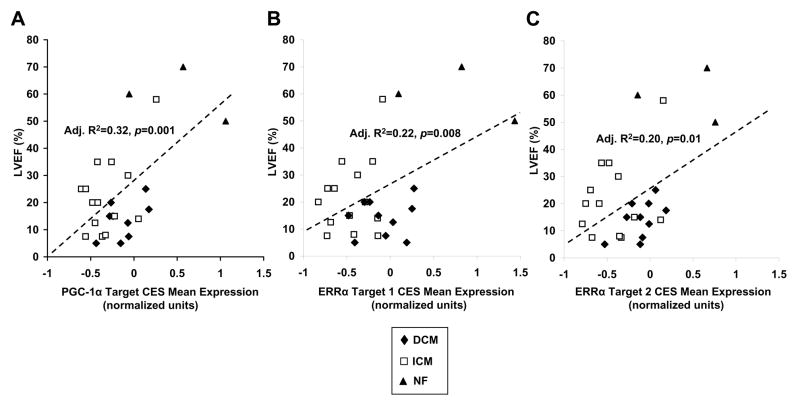
Given that diminished mitochondrial energy transduction has been linked with progression of heart failure [45,46], we investigated the relationships between PGC-1α and ERRα target CES gene expression and left ventricular ejection fraction (LVEF), a clinical parameter decreased in systolic heart failure. We utilized linear regression models to define these potential relationships (Fig. 3). As depicted (Fig. 3A), a significant positive correlation existed between LVEF and PGC-1α target CES mean expression (adjusted R2=0.32 and p= 0.001, where p refers to the F statistic of the model). The coefficients of the regression were also significant at p<0.01. Parallel analyses demonstrated positive correlation between LVEF and both ERRα target 1 and 2 CES mean expression (Figs. 3B and 3C). Thus, reduced expression of PGC-1α and ERRα target CES genes are associated with impaired left ventricular systolic function and, as such, may serve as indicators of heart failure severity.
Fig. 3.
PGC-1α target, ERRα target 1, and ERRα target 2 core enrichment set (CES) mean expression correlate with severity of heart failure. Linear regression was used to model the relationship between left ventricular ejection fraction (LVEF), a marker of disease severity, with normalized, mean expression levels of genes in the PGC-1α target CES(A), ERRα target 1 CES (B), and ERRα target 2 CES (C). The Adjusted R2 and p-value of each model is shown.
In light of the association of PGC-1α target CES expression with heart failure severity, we explored whether the relatively small number of genes comprising the PGC-1α target CES was capable of independently predicting the heart failure phenotype. Utilizing a K-nearest neighbor (KNN) class prediction model, we found that expression of PGC-1α target CES genes correctly distinguished between failing and non-failing classes perfectly (100%) in the matched population, on which the model was trained, and in 80.9% of the unmatched population, on which the model is tested (Table 3). The unmatched population consisted of the remaining members of the total population not included in the matched population (see “Methods”). The sensitivity of the PGC-1α target CES gene set in detecting heart failure was 96.0% in the unmatched population, suggesting that PGC-1α target gene downregulation serves as a signature for severe cardiac dysfunction. The related ERRα target 1 and 2 CES genes similarly predicted the heart failure phenotype in the unmatched population with a sensitivity of 96.0% and 84.0%, respectively (Table 3). The specificity of the PGC-1α target CES gene set was 58.8% in the unmatched population (Table 3), while ERRα target 1 and 2 CES genes demonstrated specificities less than 50%. Decreased specificity consistently observed in the unmatched population may reflect known increased age and gender heterogeneity of this population among the non-failing subjects (Table 1). ERK pathway CES gene set sensitivity in detecting heart failure was 88.0% in the unmatched population, indicating that the ERK pathway CES, representing the most downregulated pathway in heart failure by the GSEA analysis, is also predictive of heart failure status (Table 3). For comparison, we built a parallel model using the most significantly differentially expressed gene markers at FWER<0.05. This set contains 484 genes spanning broad functional categories in contrast to the smaller PGC-1α target CES of 158 genes which are strongly associated with mitochondrial function. Employing these significant markers at FWER<0.05 for the unmatched population, failing vs. non-failing samples were correctly assigned in 85.7% of instances, with a sensitivity and specificity of 92.0% and 84.7%, respectively (Table 3). Overall, these results demonstrated that PGC-1α and ERRα target CES downregulation is predictive of end-stage human heart failure, illustrating the power of this small, coordinately regulated, and focused gene network as an independent signature of a complex disease.
Table 3.
PGC-1α, ERRα, and ERK CES genes predict Failing vs. Non-failing Phenotypes
| Unmatched (Test) Population | Matched (Training) Population | ||
|---|---|---|---|
| PGC-1α Target CES | Correct/Total | 34/42 (80.9%) | 49/49 (100%) |
| Sensitivity | 96.0% | ||
| Specificity | 58.8% | ||
| ERRα Target 1 CES | Correct/Total | 32/42 (76.2%) | 49/49 (100%) |
| Sensitivity | 96.0% | ||
| Specificity | 47.1% | ||
| ERRα Target 2 CES | Correct/Total | 26/42 (63.4%) | 49/49 (100%) |
| Sensitivity | 84.0% | ||
| Specificity | 29.4% | ||
| ERK Pathway CES | Correct/Total | 31/42 (73.8%) | 49/49 (100%) |
| Sensitivity | 88.0% | ||
| Specificity | 52.9% | ||
| Significant Markers (FWER<0.05) | Correct/Total | 36/42 (85.7%) | 49/49 (100%) |
| Sensitivity | 92.0% | ||
| Specificity | 76.5% |
Results of KNN prediction models using the PGC-1α target, ERRα target 1, ERRα target 2, and ERK pathway CES genes vs. the most significantly differentially expressed markers at FWER < 0.05 are shown, including the ratio of samples correctly assigned by the model, as well as sensitivity and specificity. CES, ‘core enrichment set’ by Gene Set Enrichment Analysis.
To investigate potential mechanisms contributing to the downregulation of PGC-1α target CES genes, we reviewed the Comparative Marker Selection results (described earlier) for altered expression of PGC-1α and its regulatory partners in heart failure. Despite probable decreased activity of PGC-1α as evidenced by downregulation of PGC-1α target genes, expression of the PGC-1α gene itself was modestly increased by 36%, perhaps in a compensatory effort (Table 4). Gene expression of the key PGC-1α partner ERRα was significantly reduced by 35%, consistent with the above described downregulation of ERRα target genes and potentially contributing to decreased functional activity of PGC-1α. Similarly, we observed decreased expression of peroxisome proliferator-activated receptor δ (PPARδ), another important PGC-1α partner which regulates expression of genes encoding key mitochondrial proteins involved in fatty acid and glucose oxidation [47,48]. A less well characterized family member of PGC-1α, PGC-1-related coactivator (PRC) was also significantly decreased by 53% in failing (DCM/ICM) vs. non-failing hearts. These data suggest a role for ERRα, PPARδ, and PRC in the dysregulation of PGC-1α target genes in heart failure.
Table 4.
Degree of Differential Expression of PGC-1α and Key Transcriptional Partners
| Gene | Symbol | Fold Change | FDR |
|---|---|---|---|
| Peroxisome proliferator-activated receptor gamma coactivator 1α | PPARGC1A | 1.36 | 0.013 |
| Peroxisome proliferator-activated receptor gamma coactivator 1β | PPARGC1B | Filtered | |
| Peroxisome proliferator-activated receptor gamma coactivator-related 1 | PPRC1 | −1.53 | 0.013 |
| Nuclear respiratory factor 1 | NRF1 | Filtered | |
| Nuclear respiratory factor 2 | NRF2/GABPA | 1.29 | 0.013 |
| Mitochondrial transcription factor A | TFAM | 1.05 | 0.558 |
| Estrogen-related receptor α | ESRRA | −1.35 | 0.013 |
| Estrogen-related receptor β | ESRRB | 1.09 | 0.743 |
| Estrogen-related receptor γ | ESRRG | −1.09 | 0.480 |
| Peroxisome proliferator-activated receptor α | PPARA | 1.11 | 0.275 |
| Peroxisome proliferator-activated receptor β/δ | PPARB/D | −1.35 | 0.034 |
| Peroxisome proliferator-activated receptor γ | PPARG | 1.24 | 0.013 |
| Myocyte enhancing factor 2A | MEF2A | 1.16 | 0.028 |
| Myocyte enhancing factor 2B | MEF2B | 1.06 | 0.369 |
| Myocyte enhancing factor 2C | MEF2C | −1.01 | 0.957 |
| Myocyte enhancing factor 2D | MEF2D | 1.24 | 0.166 |
| Forkhead box C1 | FOXC1 | 1.32 | 0.263 |
| Forkhead box C2 | FOXC2 | 1.22 | 0.040 |
| Forkhead box P1 | FOXP1 | −1.07 | 0.336 |
| Forkhead box P4 | FOXP4 | 1.03 | 0.789 |
| Forkhead box O1A | FOXO1A | 1.22 | 0.021 |
| Menage-a-trois 1 | MNAT1 | Filtered |
Comparison for differential expression is DCM/ICM vs. NF. Fold change is positive if upregulated, negative if downregulated. Those genes with an FDR<0.05 by Comparative Marker Selection are highlighted in bold.
The PGC-1β and nuclear respiratory factor-1 (NRF-1) genes were filtered from our analysis due to absent signal, while gene expression of mitochondrial transcription factor A (Tfam) was unchanged. Expression of nuclear respiratory factor-2 (NRF-2) was increased by 29% (Table 4), another possible attempt at compensation in response to decreased expression of mitochondrial genes. Although the known PGC-1α partner peroxisome proliferator-activated receptor γ (PPARγ) is typically the least represented of the PPARs in the heart, its expression was increased by 24% in heart failure, while expression of the more abundant peroxisome proliferator-activated receptor α (PPARα) was unchanged. Interestingly, a murine model of ERRα null hearts also exhibited a potentially compensatory elevation in PGC-1α and PPARγ gene expression [29], paralleling the above described findings in human heart failure. Consistent with previous studies of altered gene expression in the failing heart, expression of additional PGC-1α partners, myocyte enhancing factor 2A (MEF2A) and forkhead box C2 and O1A (FOXC2 and FOXO1A), was increased [49]. Overall, these results both confirm prior heart-failure associated alterations in key transcription factors and propose important novel transcriptional regulatory patterns in the failing human heart which require further validation.
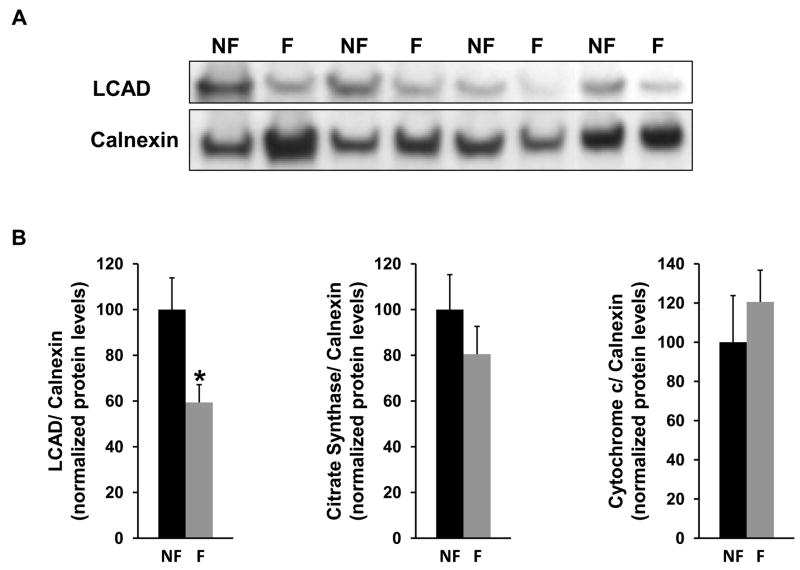
To begin to understand how downregulation of PGC-1α and ERRα heart target gene expression influences the failing heart at the level of the proteome, protein levels of three PGC-1α target genes [long-chain acyl-CoA dehydrogenase (LCAD), citrate synthase, and cytochrome c] were assessed by western blot analysis. These three mitochondrial enzymes are central to the fatty acid β-oxidation spiral, the tricarboxylic acid (TCA) cycle, and the electron transport chain, respectively. For determination of left ventricular protein levels in failing versus nonfailing hearts, cardiac tissue samples were obtained from 8 hearts with no history of myocardial dysfunction and with normal left ventricular ejection fraction by echocardiography (see “Methods”). These nonfailing hearts (from 4 men and 4 women, age range 52–69 y) were compared with failing hearts from age and gender-matched patients with idiopathic dilated cardiomyopathy. (The left ventricular ejection fraction by echocardiography was 62±2% for the nonfailing hearts and 17±5% for the failing hearts.) The protein level of the PGC-1α target gene LCAD, which catalyzes the first reaction in the mitochondrial β-oxidation of straight-chain fatty acids, was significantly reduced by 40% in failing versus non-failing hearts (Fig. 4A). LCAD protein levels were normalized to the level of calnexin, used as a loading control (Fig. 4B). (Calnexin, an integral protein of the endoplasmic reticulum, was not regulated with heart failure.) The protein level of citrate synthase, the first enzyme of the mitochondrial TCA cycle, trended toward a reduced level in the failing left ventricles, although this was not statistically significant in this limited population (Fig. 4B). The absence of reduction in cytochrome c protein levels in the failing heart (Fig. 4B) is likely consistent with complex post-transcriptional regulatory mechanisms influencing the balance of protein synthesis and degradation. Nevertheless, marked reduction in the failing heart of the PGC-1α target gene LCAD, a mitochondrial enzyme central to the heart’s ability to transduce energy from its dominant metabolic substrate – fatty acid, suggests that core components of mitochondria may be altered selectively in the failing heart.
Fig. 4.
Protein levels of PGC-1α target genes in left ventricular tissue obtained from failing and nonfailing hearts. (A) Representative western blot panel depicts levels of the PGC-1α target gene long-chain acyl-CoA dehydrogenase (LCAD) and calnexin (used as a loading control) in nonfailing (NF) versus failing (F) hearts. Failing heart tissue was obtained from the left ventricle of patients with idiopathic dilated cardiomyopathy. Patients with failing and nonfailing hearts were age and gender-matched. (B) Bar graphs depict western blot protein levels of three PGC-1α target genes (LCAD, citrate synthase, and cytochrome c) normalized to the level of calnexin in nonfailing (NF) versus failing (F) left ventricles (n=8). * significantly different (p<0.05) compared to nonfailing hearts.
4. Discussion
The ERK gene pathway, which favors physiologic growth and protection from apoptosis [44], was the most significantly downregulated pathway in human heart failure by gene set enrichment analysis in this study, with regulation at the gene expression level a consistent finding and a potential contributor to decreased ERK pathway activity in severe heart failure. Ultrasensitivity in the MAPK cascade [50] may allow changes in gene expression (and subsequent protein levels) to alter signaling of an activated MAPK pathway. Concomitant downregulation in heart failure of PGC-1α, ERRα, and ERK pathways would result in increased susceptibility for apoptosis in the failing heart, exacerbated by impaired overall mitochondrial oxidative capacity, increased reactive oxygen species, and diminished mitochondrial membrane potential. ERK pathway core enrichment set genes driving the downregulation of the ERK pathway included RAF1 (Raf-1), MAP2K2 (MKK2), MAP2K1 (MKK1), and MAPK3 (ERK1). Raf protein kinases phosphorylate and activate mitogen-activated protein kinase kinases (MKKs), which in turn phosphorylate and activate extracellular signal-regulated kinases (ERKs) [44]. ERKs regulate cardiac cellular physiology by phosphorylating numerous substrates, including transcription factors, apoptotic proteins, and translational factors. The MKK1-ERK signaling pathway stimulates physiologic cardiac hypertrophy associated with augmented cardiac function and partial resistance to apoptosis, without inducing p38 or c-Jun N-terminal kinases 1/2 (JNK 1/2) [51]. Studies in mice have demonstrated that, in response to pressure overload, Raf-1 kinase activity is essential for cardiac hypertrophy and cardiac myocyte survival through inhibition of apoptosis [52]. ERK phosphorylation of the proapoptotic caspase-9 and BIM proteins mediates its antiapoptotic function [53,54], while Raf-1 has additional antiapoptotic targets (independent of the ERK cascade) which include apoptosis signal-regulating kinase 1 (ASK1) and the mammalian sterile 20-like kinase 2 (MST2) [44]. Given a role for apoptosis in the failing human heart [55,56], the physiologic growth promoting and potent antiapoptotic functions of the Raf-1/MKK/ERK cascade suggest that reductions in the protein levels or activity of this pathway may sensitize patients to the development of heart failure [44,51]. In contrast, antagonism of the JNK and p38 activation associated with cardiomyopathy may be therapeutic[51]. Other related pro-growth and antiapoptotic pathways demonstrated by the GSEA analysis to be downregulated with human heart failure include the insulin-like growth factor 1 receptor (IGF1R) pathway, the IGF1/MTOR pathway, the insulin signaling, and insulin receptor pathways. Clinical studies suggest that growth hormone therapy may have beneficial effects in congestive heart failure in humans [57], while growth hormone therapy in failing rat hearts reduces oxidative stress and enhances energy metabolism [58].
This study implicates the transcriptional energy metabolic regulator PGC-1α, along with its target genes, in the pathogenesis of human heart failure. Furthermore, regulatory mechanisms involving PGC-1α and its key transcriptional partners, including ERRα and PPARδ, may contribute to the mitochondrial dysfunction characteristic of the failing heart. Impaired mitochondrial function leads to insufficient energy delivery in heart failure by reducing overall mitochondrial oxidative efficiency and by altering the heart’s use of its preferred energy substrate – fatty acid [16,17,19,59]. Studies in humans with pathologic left ventricular hypertrophy demonstrate not only a decrease in myocardial fatty acid uptake and oxidation but also compromised myocardial efficiency (i.e., the ratio of myocardial work to myocardial oxygen consumption) [60–62]. Moreover, the phosphocreatine-to-ATP ratio (PCr/ATP), a gauge of cellular high energy stores, is reduced in heart failure. Magnetic resonance spectroscopy has demonstrated in animals and humans that the reduction in high-energy phosphate stores in compensated pressure overload-induced cardiac hypertrophy further declines with the transition to heart failure [18,63–65]. Indeed, PCr/ATP ratios are correlated with heart failure severity as defined by New York Heart Association (NYHA) classification [66] and are strongly predictive of total and cardiovascular mortality [45,46,67].
Potential mechanisms underlying the imbalance in energy metabolism associated with pathologic cardiac hypertrophy and heart failure include reduced activity both of PGC-1α, a critical transcriptional regulator of mitochondrial function [21,22], and of ERRα, which recruits PGC-1α to cardiac metabolic target genes. Interestingly, mice lacking PGC-1α exhibit decreased levels of ATP [22], as do failing canine hearts [68]. In the mouse heart, PGC-1α is essential for maximal and efficient cardiac mitochondrial fatty acid oxidation, ATP synthesis, and lipid homeostasis, especially in the setting of increased work [23]. Loss of PGC-1α in the mouse heart reduces mitochondrial coupling as evidenced by reduction in the maximal ATP/O ratio (ratio of ATP produced to oxygen consumed with State 3 respiration) [23]. Furthermore, hearts of mice deficient in ERRα demonstrate abnormal phosphocreatine depletion and functional decline when subjected to hemodynamic stress [30]. Our current finding of a positive correlation between PGC-1α and ERRα target CES gene expression and left ventricular systolic function parallels the previously described relationship between the phosphocreatine-to-ATP ratio and heart failure severity, and is consistent with a role for PGC-1α and ERRα as important regulators of myocardial energy homeostasis.
In this study, the ability of PGC-1α and ERRα target gene expression to predict the heart failure phenotype with high sensitivity suggests that this transcriptional profile represents a signature of human heart failure. Despite the multiple biologic pathways dysregulated in the failing heart, it is notable that the PGC-1α regulatory network, with its relatively confined metabolic focus, is so uniformly downregulated with severe cardiac dysfunction. As with any changes observed in end-stage heart failure, these alterations may reflect primary mechanisms of pathology or compensatory changes, potentially adaptive initially but maladaptive chronically. Gene expression patterns with heart failure may also be influenced by treatment of patients with extensive pharmacologic regimens, for which limited information was available in our study population. Although logistically difficult, obtaining human cardiac tissue at different stages in the progression of heart failure may clarify a causal link between these gene expression changes, impaired cardiac energy transduction, and the evolution of heart failure. Interestingly, recent work demonstrates that polymorphisms in the PGC-1α gene are associated with hypertrophic cardiomyopathy [69].
Despite the downregulation of PGC-1α target genes, expression of PGC-1α itself was minimally induced, potentially consistent with multiple complex mechanisms influencing PGC-1α target gene expression. Given that the PGC-1α family members, PGC-1β and PRC, may have overlapping gene expression profiles [16,70], one such mechanism may involve reduced transcriptional activity of one or both of these family members. The PGC-1β gene was filtered in our study due to absent signal. PRC, which also coactivates transcription factors involved in mitochondrial biogenesis including NRF-1[70–72], was found to be significantly decreased, suggestive of a novel role for PRC in heart failure in reducing expression of nuclear respiratory factor gene targets. Further detailed knowledge of PRC regulation and its target gene set in the cardiac myocyte is necessary. The observed minimal increased expression of the PGC-1α gene in the setting of energetic insufficiency in heart failure may be compensatory and reflective of altered signaling or energy sensing pathways involving AMP-activated protein kinase [73].
Mechanisms by which PGC-1α target gene expression may be decreased independent of the expression of the PGC-1α gene itself include reduced activity of PGC-1α as a transcriptional coactivator secondary to its post-translational modification or diminished activity of its key transcriptional partners. PGC-1α integrates input from developmental cues, fasting, and exercise, as well as calcium and stress-activated pathways, such as the p38 MAPK pathway [16]. Alterations in p38 MAPK with associated post-translational modification of PGC-1α have been demonstrated to impact the transcriptional capacity of PGC-1α by increasing PGC-1α protein stability [74], promoting dissociation of a repressor [75,76], and enhancing selective activation of PPARα [77]. Deacetylation of PGC-1α by the NAD+-dependent histone deacetylase SIRT1 has also been shown to modify PGC-1α activity [78]. Decreased gene expression and possibly decreased activity of the key PGC-1α transcriptional partners ERRα and PPARδ likely represent an additional mechanism negatively influencing expression of PGC-1α target genes, including those encoding mitochondrial proteins central to fat and glucose oxidation. Indeed, both gene expression and activity of ERRα are reduced, as indirectly evidenced by ERRα target gene downregulation. Given that ERRα target genes encoding key mitochondrial proteins are enriched in nuclear respiratory factor promoters [37], decreased ERRα gene expression and activity observed in heart failure may contribute to a decrease in overlapping NRF target gene induction, despite the modest 29% increase in NRF-2 gene expression observed in this study with heart failure.
Notably, the gene encoding pyruvate dehydrogenase kinase 4 (PDK4) contributes to the PGC-1α target core enrichment set of genes which demonstrate decreased expression in heart failure (Supplemental Table S8). This finding is consistent with prior single-gene analysis of failing hearts [4]. A negative regulator of glucose oxidation, PDK4 has previously been described as a PGC-1α/ERRα target in skeletal muscle and provides one mechanism whereby PGC-1α exerts reciprocal inhibition of glucose catabolism while increasing fatty acid oxidation pathways [79]. A model by which altered PGC-1α and ERRα target gene expression contributes to energy metabolic substrate switches in the developing and failing heart is depicted in Fig. 5. Further confirmation of this model in human heart failure will require tissue based validation in the form of RT-PCR, gene promoter occupancy assays, and proteomics studies which explore regulation at the level of protein abundance and activity rather than gene expression [80]. A previous survey of the mitochondrial proteome in various tissue types, including cardiac tissue, however, does suggest overall concordance between mRNA abundance and protein expression [81]. Indeed, the marked 40% protein level reduction in the failing heart of the PGC-1α target gene LCAD, a mitochondrial enzyme which catalyzes the first step of the mitochondrial fatty acid β-oxidation (FAO) spiral, suggests that core components of mitochondria are altered selectively in the failing heart leading to the observed metabolic remodeling. Downregulation of LCAD protein in heart failure is consistent with the previously described downregulation in human heart failure of the mitochondrial FAO enzyme medium-chain acyl-CoA dehydrogenase (MCAD) [59]. Gene expression of both LCAD and MCAD is under cooperative control by PGC-1α and PPARα [82,83], and LCAD gene expression is reduced in the hearts of mice deficient in PGC-1α [23]. Through their ability to restore appropriate energy metabolic balance in the pathologic hypertrophied and failing human heart, PGC-1α and its key transcriptional partner ERRα may represent novel therapeutic targets in heart failure.
Fig. 5.
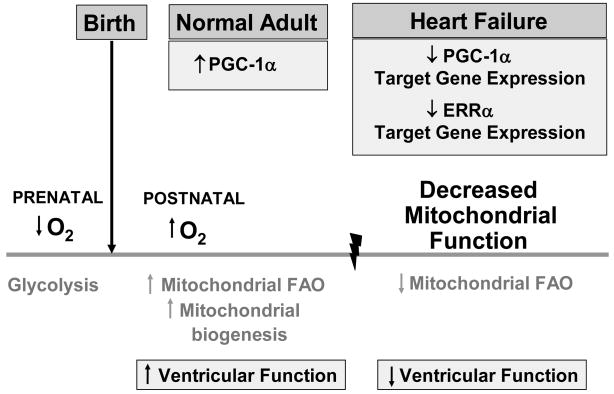
A role for altered PGC-1α and ERRα target gene expression in energy metabolic substrate switches in the developing and failing heart. In the setting of dietary and physiologic cues, developmentally programmed gene regulatory events facilitate the postnatal “switch” in myocardial fuel preference from glucose as the predominant source for ATP production to fatty acids metabolized via the mitochondrial fatty acid oxidation (FAO) pathway in the adult heart. Coordinate with this increased mitochondrial FAO is mitochondrial biogenesis promoted by PGC-1α, and associated enhanced left ventricular function. With heart failure, cardiac mitochondrial function declines with changes reminiscent of fetal metabolic re-programming, concomitant with decline in PGC-1α and ERRα target gene expression.
Supplementary Material
Acknowledgments
The authors acknowledge the following public source for the microarray data: Genomics of Cardiovascular Development, Adaptation, and Remodeling, NHLBI Program for Genomic Applications, Harvard Medical School, URL: http://www.cardiogenomics.org [(November, 2006) accessed]. This work is supported in part by an N.I.H. KO8 AG024844 grant (J.J.L.) and an Alpha Omega Alpha Medical Student Research Fellowship (S.S.). The authors thank Paul D. Allen for help in obtaining microarray data from the Cardiogenomics Consortium, Vamsi K. Mootha for advice on data processing and analysis, Daniel P. Kelly, Anthony J. Muslin, and Rakesh Nagarajan for comments on the manuscript, and Mary Wingate for assistance in manuscript preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lloyd-Jones DM, Larson MG, Leip EP, Beiser A, D’Agostino RB, Kannel WB, et al. Lifetime risk for developing congestive heart failure: The Framingham heart study. Circulation. 2002;106:3068–72. doi: 10.1161/01.cir.0000039105.49749.6f. [DOI] [PubMed] [Google Scholar]
- 2.Nanni L, Romualdi C, Maseri A, Lanfranchi G. Differential gene expresion profiling in genetic and multifactorial cardiovascular diseases. J Mol Cell Cardiol. 2006;41:934–48. doi: 10.1016/j.yjmcc.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Sharma UC, Pokharel S, Evelo CTA, Maessen JG. A systematic review of large scale and heterogeneous gene array data in heart failure. J Mol Cell Cardiol. 2005;38:425. doi: 10.1016/j.yjmcc.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 4.Steenman M, Chen Y-W, Le Cunff M, Lamirault G, Varró A, Hoffman E, et al. Transcriptomal analysis of failing and nonfailing human hearts. Physiol Genomics. 2003;12:97–112. doi: 10.1152/physiolgenomics.00148.2002. [DOI] [PubMed] [Google Scholar]
- 5.Kääb S, Barth AS, Margerie D, Dugas M, Gebauer M, Zwermann L, et al. Global gene expression in human myocardium-oligonucleotide microarray analysis of regional diversity and transcriptional regulation in heart failure. Journal of Molecular Medicine. 2004;82:308–16. doi: 10.1007/s00109-004-0527-2. [DOI] [PubMed] [Google Scholar]
- 6.Yang J, Moravec CS, Sussman MA, DiPaola NR, Fu D, Hawthorn L, et al. Decreased SLIM1 expression and increased gelsolin expression in failing human hearts measured by high-density oligonucleotide arrays. Circulation. 2000;102:3046–52. doi: 10.1161/01.cir.102.25.3046. [DOI] [PubMed] [Google Scholar]
- 7.Beisvag V, Lehre PK, Midelfart H, Aass H, Geiran O, Sandvik AK, et al. Aetiology-specific patterns in end-stage heart failure patients identified by functional annotation and classification of microarray data. The European Journal of Heart Failure. 2006;8:381–9. doi: 10.1016/j.ejheart.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Kittleson MM, Minhas KM, Irizarry RA, Ye SQ, Edness G, Breton E, et al. Gene expression analysis of ischemic and nonischemic cardiomyopathy: Shared and distinct fenes in the development of heart failure. Physiol Genomics. 2005;21:299–307. doi: 10.1152/physiolgenomics.00255.2004. [DOI] [PubMed] [Google Scholar]
- 9.Tan F-L, Moravec CS, Li J, Apperson-Hansen C, McCarthy PM, Young JB, et al. The gene expression fingerprint of human heart failure. Proc Natl Acad Sci USA. 2002;99:11387–92. doi: 10.1073/pnas.162370099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barth AS, Kuner R, Buness A, Ruschhaupt M, Merk S, Zwermann L, et al. Identification of a common gene expression signature in dilated cardiomyopathy across independent microarray studies. Journal of American College of Cardiology. 2006;48:1618–20. doi: 10.1016/j.jacc.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 11.Barrans JD, Allen PD, Stamatiou D, Dzau VJ, Liew C-C. Global gene expression profiling of end-stage dilated cardiomyopathy using a human cardiovascular-based cDNA microarray. American Journal of Pathology. 2002;160:2035–43. doi: 10.1016/S0002-9440(10)61153-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang J-J, Allen PD, Tseng GC, Lam C-W, Fananapazir L, Dzau VJ, et al. Microarray gene expression profiles in dilated and hypertrophic cardiomyopathic end-stage heart failure. Physiol Genomics. 2002;10:31–44. doi: 10.1152/physiolgenomics.00122.2001. [DOI] [PubMed] [Google Scholar]
- 13.Steenbergen C, Afshari CA, Petranka JG, Collins J, Martin K, Bennett L, et al. Alterations in apoptotic signaling in human idiopathic cardiomyopathic hearts in failure. Am J Physiol Heart Circ Physiol. 2002;284:H268–H276. doi: 10.1152/ajpheart.00707.2002. [DOI] [PubMed] [Google Scholar]
- 14.Yung CK, Halperin VL, Tomaselli GF, Winslow RL. Gene expression profiles in end-stage human idiopathic dilated cardiomyopathy: altered expression of apoptotic and cytoskeletal genes. Genomics. 2004;83:281–97. doi: 10.1016/j.ygeno.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Hwang DM, Dempsey AA, Wang R-X, Rezvani M, Barrans JD, Dai K-S, et al. A genome-based resource for moleuclar cardiovascular medicine. Circulation. 1997;96:4146–203. doi: 10.1161/01.cir.96.12.4146. [DOI] [PubMed] [Google Scholar]
- 16.Huss JM, Kelly DP. Mitochondrial energy metabolism in heart failure: A question of balance. J Clin Invest. 2005;115:547–55. doi: 10.1172/JCI200524405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abozguia K, Clarke K, Lee L, Frenneaux M. Modification of myocardial substrate use as a therapy for heart failure. Nature Clinical Practice Cardiovascular Medicine. 2006;3:490–8. doi: 10.1038/ncpcardio0583. [DOI] [PubMed] [Google Scholar]
- 18.Ingwall JS, Weiss RG. Is the failing heart energy starved? On using chemical energy to support cardiac function. Circ Res. 2004;95:135–45. doi: 10.1161/01.RES.0000137170.41939.d9. [DOI] [PubMed] [Google Scholar]
- 19.Neubauer S. The failing heart-an engine out of fuel. N Engl J Med. 2007;356:1140–51. doi: 10.1056/NEJMra063052. [DOI] [PubMed] [Google Scholar]
- 20.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros D, Kelly DP. PPARγ coactivator-1 (PGC-1) promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–56. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehman JJ, Kelly DP. Transcriptional activation of energy metabolic switches in the developing and hypertrophied heart. Clin Exp Pharmacol Physiol. 2002;29:339–45. doi: 10.1046/j.1440-1681.2002.03655.x. [DOI] [PubMed] [Google Scholar]
- 22.Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, et al. Transcriptional coactivator PGC-1α controls the energy state and contractile function of cardiac muscle. Cell Metabolism. 2005;1:259–71. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Lehman JJ, Boudina S, Banke NH, Sambandam N, Han X, Young DM, et al. The Transcriptional Coactivator PGC-1α is Essential for Maximal and Efficient Cardiac Mitochondrial Fatty Acid Oxidation and Lipid Homeostasis. Am J Physiol Heart Circ Physiol. 2008 May 23; doi: 10.1152/ajpheart.00081.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–24. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 25.Huss JM, Pinéda Torra I, Staels B, Giguère V, Kelly DP. ERRα directs PPARα signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol Cell Biol. 2004;24:9079–91. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huss JM, Kopp RP, Kelly DP. PGC-1α coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-α and -γ. J Biol Chem. 2002;277:40265–74. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 27.Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor α (ERRα) J Biol Chem. 2003;278:9013–8. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 28.Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, et al. ERRα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci USA. 2004;101:6570–5. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, et al. Genome-wide orchestration of heart functions by orphan nuclear receptors ERRα andγ. Cell Metabolism. 2007;5:345–56. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Huss JM, Imahashi K-I, Dufour CR, Weinheimer CJ, Courtois M, Kovacs A, et al. The nuclear receptor ERRα is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metabolism. 2007;6:25–37. doi: 10.1016/j.cmet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 32.Garnier A, Fortin D, Deloménie C, Momken I, Veksler V, Ventura-Clapier R. Depressed mitochondrial transcription factors and oxidative capacity in rat failing cardiac and skeletal muscles. J Physiol. 2003;551:491–501. doi: 10.1113/jphysiol.2003.045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arany Z, Novikov M, Chin S, Ma Y, Rosenzweig A, Spiegelman BM. Transverse aortic constriction leads to accelerated heart failure in mice lacking PPARγ coactivator 1α. Proc Natl Acad Sci USA. 2006;103:10086–91. doi: 10.1073/pnas.0603615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 35.Gould J, Getz G, Monti S, Reich M, Mesirov JP. Comparative gene marker selection suite. Bioinformatics. 2006;22:1924–5. doi: 10.1093/bioinformatics/btl196. [DOI] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, et al. Genome-wide orchestration of cardiac functions by orphan nuclear receptors ERRα and γ. Cell Metabolism. 2007;5:345–56. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Al-Shahrour F, Díaz-Uriarte R, Dopazo J. FatiGO: A web tool for finding significant associatons of gene ontology terms with groups of genes. Bioinformatics. 2004;20:578–80. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 39.Sucharov CC, Mariner P, Long C, Bristow M, Leinwand L. Yin Yang 1 is increased in human heart failure and represses the activity of the human alpha-myosin heavy chain promoter. J Biol Chem. 2003 Aug 15;278(33):31233–9. doi: 10.1074/jbc.M301917200. [DOI] [PubMed] [Google Scholar]
- 40.Boheler KR, Volkova M, Morrell C, Garg R, Zhu Y, Margulies K, et al. Sex- and age-dependent human transcriptome variability: Implicatoins for chronic heart failure. Proc Natl Acad Sci USA. 2003;100:2754–9. doi: 10.1073/pnas.0436564100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volkova M, Garg R, Dick S, Boheler KR. Aging-associated changes in cardiac gene expression. Cardiovasc Res. 2005;66:194–204. doi: 10.1016/j.cardiores.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 42.Bueno OF, Molkentin JD. Involvement of extracellular signal-regulated kinases 1/2 in cardiac hypertrophy and cell death. Circ Res. 2002;91:776–81. doi: 10.1161/01.res.0000038488.38975.1a. [DOI] [PubMed] [Google Scholar]
- 43.Petrich BG, Wang Y. Stress-activated MAP kinases in cardiac remodeling and heart failure. Trends in Cardiovascular Medicine. 2004;14:50–5. doi: 10.1016/j.tcm.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 44.Muslin AJ. Role of raf proteins in cardiac hypertrophy and cardiomyocyte survival. Trends Cardiovasc Med. 2005 Aug;15(6):225–9. doi: 10.1016/j.tcm.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 45.Neubauer S, Horn M, Cramer M, Harre K, Newell JB, Peters W, et al. Myocardial phosphocreatine-to-ATP ratio is a predictor of mortality in patients with dilated cardiomyopathy. Circulation. 1997;96:2190–6. doi: 10.1161/01.cir.96.7.2190. [DOI] [PubMed] [Google Scholar]
- 46.Conway MA, Allis J, Ouwerkerk R, Niioka T, Rajagopalan B, Radda GK. Detection of low phosphocreatine to ATP ration in failing hypertrophied human myocardium by 31P magnetic resonance spectroscopy. Lancet. 1991;338:973–6. doi: 10.1016/0140-6736(91)91838-l. [DOI] [PubMed] [Google Scholar]
- 47.Burkart EM, Sambandam N, Han X, Gross RW, Courtois M, Gierasch CM, et al. Nuclear receptors PPARbeta/delta and PPARalpha direct distinct metabolic regulatory programs in the mouse heart. J Clin Invest. 2007 Dec;117(12):3930–9. doi: 10.1172/JCI32578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hancock CR, Han DH, Chen M, Terada S, Yasuda T, Wright DC, et al. High-fat diets cause insulin resistance despite an increase in muscle mitochondria. Proc Natl Acad Sci U S A. 2008 Jun 3;105(22):7815–20. doi: 10.1073/pnas.0802057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, et al. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006;114:1269–76. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- 50.Huang CY, Ferrell JE., Jr Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci U S A. 1996 Sep 17;93(19):10078–83. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bueno OF, De Windt LJ, Tymitz KM, Witt SA, Kimball TR, Klevitsky R, et al. The MEK1-ERK1/2 signaling pathway promotes compensated cardiac hypertrophy in transgenic mice. EMBO J. 2000 Dec 1;19(23):6341–50. doi: 10.1093/emboj/19.23.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris IS, Zhang S, Treskov I, Kovacs A, Weinheimer C, Muslin AJ. Raf-1 kinase is required for cardiac hypertrophy and cardiomyocyte survival in response to pressure overload. Circulation. 2004 Aug 10;110(6):718–23. doi: 10.1161/01.CIR.0000138190.50127.6A. [DOI] [PubMed] [Google Scholar]
- 53.Allan LA, Morrice N, Brady S, Magee G, Pathak S, Clarke PR. Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat Cell Biol. 2003 Jul;5(7):647–54. doi: 10.1038/ncb1005. [DOI] [PubMed] [Google Scholar]
- 54.Harada H, Quearry B, Ruiz-Vela A, Korsmeyer SJ. Survival factor-induced extracellular signal-regulated kinase phosphorylates BIM, inhibiting its association with BAX and proapoptotic activity. Proc Natl Acad Sci U S A. 2004 Oct 26;101(43):15313–7. doi: 10.1073/pnas.0406837101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang PM, Yue P, Izumo S. New insights into the role of apoptosis in cardiovascular disease. Circ J. 2002 Jan;66(1):1–9. doi: 10.1253/circj.66.1. [DOI] [PubMed] [Google Scholar]
- 56.Olivetti G, Abbi R, Quaini F, Kajstura J, Cheng W, Nitahara JA, et al. Apoptosis in the failing human heart. N Engl J Med. 1997;336:1131–41. doi: 10.1056/NEJM199704173361603. [DOI] [PubMed] [Google Scholar]
- 57.Tritos NA, Danias PG. Growth hormone therapy in congestive heart failure due to left ventricular systolic dysfunction: a meta-analysis. Endocr Pract. 2008 Jan;14(1):40–9. doi: 10.4158/EP.14.1.40. [DOI] [PubMed] [Google Scholar]
- 58.Seiva FR, Ebaid GM, Castro AV, Okoshi K, Nascimento A, Rocha KK, et al. Growth hormone and heart failure: Oxidative stress and energetic metabolism in rats. Growth Horm IGF Res. 2008 Aug;18(4):275–83. doi: 10.1016/j.ghir.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 59.Sack MN, Rader TA, Park S, Bastin J, McCune SA, Kelly DP. Fatty acid oxidation enzyme gene expression is downregulated in the failing heart. Circulation. 1996 Dec 1;94(11):2837–42. doi: 10.1161/01.cir.94.11.2837. [DOI] [PubMed] [Google Scholar]
- 60.Laine H, Katoh C, Luotolahti M, Yki-Järvinen H, Kantola I, Jula A, et al. Myocardial oxygen consumption is unchanged but efficiency is reduced in patients with essential hypertension and left ventricular hypertrophy. Circulation. 1999;100:2425–30. doi: 10.1161/01.cir.100.24.2425. [DOI] [PubMed] [Google Scholar]
- 61.de las Fuentes L, Soto PF, Cupps BP, Pasque MK, Herrero P, Gropler RJ, et al. Hypertensive left ventricular hypertrophy is associated with abnormal myocardial fatty acid metabolism and myocardial efficiency. J Nucl Cardiol. 2006;13:369–77. doi: 10.1016/j.nuclcard.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 62.Sorokina N, O’Donnell JM, McKinney RD, Pound KM, Woldegiorgis G, LaNoue KF, et al. Recruitment of compensatory pathways to sustain oxidative flux with reduced carnitine palmitoyltransferase I activity characterizes inefficiency in energy metabolism in hypertrophied hearts. Circulation. 2007;115:2033–41. doi: 10.1161/CIRCULATIONAHA.106.668665. [DOI] [PubMed] [Google Scholar]
- 63.de Roos A, Doornbos J, Luyten P, Oosterwaal L, van der Wall E, den Hollander J. Cardiac metabolism in patients with dilated and hypertrophic cardiomyopathy: Assessment with proton-decoupled P-31 MR spectroscopy. J Magn Reson Imaging. 1992;2:711–9. doi: 10.1002/jmri.1880020616. [DOI] [PubMed] [Google Scholar]
- 64.Ingwall JS, Kramer MF, Fifer MA, Lorell BH, Shemin R, Grossman W, et al. The creatine kinase system in normal and diseased human myocardium. N Engl J Med. 1985;313:1050–4. doi: 10.1056/NEJM198510243131704. [DOI] [PubMed] [Google Scholar]
- 65.Tian R, Nascimben L, Kaddurah-Daouk R, Ingwall JS. Depletion of energy reserve via the creatine kinase reaction during the evolution of heart failure in cardiomyopathic hamsters. J Mol Cell Cardiol. 1996;28:755–65. doi: 10.1006/jmcc.1996.0070. [DOI] [PubMed] [Google Scholar]
- 66.Neubauer S, Krahe T, Schindler R, Horn M, Hillenbrand H, Entzeroth C, et al. 31P magnetic resonance spectroscopy in dilated cardiomyopathy and coronary artery disease. Altered cardiac high-energy phosphate metabolism in heart failure. Circulation. 1992;86:1810–8. doi: 10.1161/01.cir.86.6.1810. [DOI] [PubMed] [Google Scholar]
- 67.Hardy CJ, Weiss RG, Bottomley PA, Gerstenblith G. Altered myocardial high-energy phosphate metabolites in patients with dilated cardiomyopathy. Am Heart J. 1991;122:795–801. doi: 10.1016/0002-8703(91)90527-o. [DOI] [PubMed] [Google Scholar]
- 68.Shen W, Asai K, Uechi M, Mathier MA, Shannon RP, Vatner SF, et al. Progressive loss of myocardial ATP due to a loss of total purines during the development of heart failure in dogs: a compensatory role for the parallel loss of creatine. Circulation. 1999;100:2113–8. doi: 10.1161/01.cir.100.20.2113. [DOI] [PubMed] [Google Scholar]
- 69.Wang S, Fu C, Wang H, Shi Y, Xu X, Chen J, et al. Polymorphisms of the peroxisome proliferator-activated receptor-γ coactivator-1α gene are associated with hypertrophic cardiomyopathy and not with hypertension hypertrophy. Clin Chem Lab Med. 2007;45:962–7. doi: 10.1515/CCLM.2007.189. [DOI] [PubMed] [Google Scholar]
- 70.Finck BN, Kelly DP. PGC-1 coactivators: Inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–22. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andersson U, Scarpulla RC. PGC-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor-1-dependent transcription in mammalian cells. Mol Cell Biol. 2001;21:3738–49. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Savagner F, Mirebeau D, Jacques C, Guyetant S, Morgan C, Franc B, et al. PGC-1-related coactivator and targets are upregulated in thyroid oncocytoma. Biochem Biophys Res Commun. 2003;310:779–84. doi: 10.1016/j.bbrc.2003.09.076. [DOI] [PubMed] [Google Scholar]
- 73.Zong H, Ren JM, Young LH, Pypaert M, Mu J, Birnbaum MJ, et al. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci USA. 2002;99:15983–7. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Puigserver P, Rhee J, Lin J, Wu Z, Yoon JC, Zhang C-Y, et al. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Mol Cell. 2001;8:971–82. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 75.Knutti D, Kressler D, Kralli A. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a corepressor. Proc Natl Acad Sci USA. 2001;98:9713–8. doi: 10.1073/pnas.171184698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fan M, Rhee J, St-Pierre J, Handschin C, Puigserver P, Lin J, et al. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1alpha: modulation by p38 MAPK. Genes Dev. 2004;18(3):278–89. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barger PM, Browning AC, Garner AN, Kelly DP. p38 MAP kinase activates PPARα: A potential role in the cardiac metabolic stress response. J Biol Chem. 2001;276:44495–501. doi: 10.1074/jbc.M105945200. [DOI] [PubMed] [Google Scholar]
- 78.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 79.Wende AR, Huss JM, Schaeffer PJ, Giguère V, Kelly DP. PGC-1α coactivates PDK4 gene expression via the orphan nuclear receptor ERRα: A mechanism for transcriptional control of muscle glucose metabolism. Mol Cell Biol. 2005;25:10684–94. doi: 10.1128/MCB.25.24.10684-10694.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arab S, Gramolini AO, Ping P, Kislinger T, Stanley B, vanEyk J, et al. Cardiovascular proteomics: Tools to develop novel biomarkers and potential applications. J Am Coll Cardiol. 2006;48:1733–41. doi: 10.1016/j.jacc.2006.06.063. [DOI] [PubMed] [Google Scholar]
- 81.Mootha VK, Bunkenborg J, Olsen JV, Hjerrid M, Wisniewski JR, Stahl E, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–40. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 82.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor a in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20(5):1868–76. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Z, Zhou Y, Mendelsohn NJ, Bauer GS, Strauss AW. Regulation of the human long chain acyl-CoA dehydrogenase gene by nuclear hormone receptor transcription factors. Biochim Biophys Acta. 1997 Jan 3;1350(1):53–64. doi: 10.1016/s0167-4781(96)00141-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.